Abstract
It is well established that major depressive disorder (MDD) is partly heritable. We present a genome-wide linkage study aiming to find regions on the genome that influence the vulnerability for MDD. Our sample consists of 110 Australian and 23 Dutch pedigrees with two or more siblings affected with MDD (total N = 278). Linkage analysis was carried out in MERLIN. Three regions showed suggestive linkage signals. The highest LOD-score of 2.1 was found on chromosome 17 at 52.6 cM along with LOD scores of 1.9 and 1.7 on chromosome 8 at 2.7 cM and chromosome 2 at 90.6 cM, respectively. The result on chromosome 8 seems most promising as two previous studies also found linkage in this region, once suggestive and once significant. The linkage peak on chromosome 17 harbors the serotonin transporter gene. In the Australian and Dutch sample, the serotonin transporter length polymorphism did not show evidence for association, thus other genes in this region or other polymorphisms in the serotonin transporter gene might be associated with MDD. Further replication is needed to establish the relevance of our linkage finding on chromosome 2.
Keywords: genome-wide linkage, depression, genetics, family study, serotonin transporter gene
INTRODUCTION
It is well established that major depressive disorder (MDD) is partly heritable. A meta-analysis estimated the heritability of MDD around 37% [Sullivan et al., 2000]. If measurement error due to unreliability is taken into account by analyzing MDD assessed at two occasions, the heritability estimate might be as high as 66% [Foley et al., 1998]. These findings have led to attempts to identify genes influencing the vulnerability for MDD. As part of these efforts, seven genome-wide linkage analyses have been performed, aiming to locate genes for MDD on the genome [Nurnberger et al., 2001; Zubenko et al., 2003;Holmans et al., 2004, 2007; Camp et al., 2005; McGuffin et al., 2005]. Other studies have focused on quantitative traits associated with a diagnosis of MDD, such as neuroticism [Cloninger et al., 1998; Fullerton et al., 2003; Nash et al., 2004; Neale et al., 2005; Kuo et al., 2007; Wray et al., 2008]. Table I summarizes the most promising results of these studies, excluding the study of Holmans et al. [2004] as this is based on the same sample as used by Holmans et al. [2007]. Several regions have shown a linkage signal with an LOD-score >3 in at least one study. The following five regions have reached an LOD-score >3 in one study and an LOD-score >1.5 in a second study: chromosome 1 between 126 and 137 cM [Fullerton et al., 2003; Neale et al., 2005], chromosome 8 between 8 and 38 cM [Cloninger et al., 1998; Fullerton et al., 2003], chromosome 11 between 2 and 35 cM [Zubenko et al., 2003; Camp et al., 2005], chromosome 11 between 85 and 99 cM [Fullerton et al., 2003; Zubenko et al., 2003] and chromosome 12 between 105 and 124 cM [Fullerton et al., 2003; McGuffin et al., 2005].
TABLE I.
Most Promising Linkage Results in the Order of the Chromosomes for Neuroticism, Harm Avoidance, MDD or the Subtypes Recurrent MDD (R-MDD) and Recurrent Early-Onset MDD (RE-MDD)
| Location (chromosome, cM) | LOD | N subjectsa/families | Phenotype |
|---|---|---|---|
| 1, 42, and 90 cM [Camp et al., 2005; Nash et al., 2004] | 1.7 | 426/90 | MDD-RE |
| 1.6 | 711/283 | Neuroticism | |
| 1, 126–137 cM [Fullerton et al., 2003; Neale et al., 2005] | 4.0b | 1,122/561 | Neuroticism |
| 2.5b | 293/129 | Neuroticism | |
| 2, 237–248 cM [Nurnberger et al., 2001; Zubenko et al., 2003] | 2.2 | 224 possible pairs | MDD comorbid with alcoholism |
| 2.5b | ?/81 | MDD | |
| 4, 176 cM Fullerton et al. [2003] | 3.8b | 1,122/561 | Neuroticism |
| 7, 42 cM Fullerton et al. [2003] | 3.9b | 1,122/561 | Neuroticism |
| 8, 8–38 cM [Cloninger et al., 1998; Fullerton et al., 2003] | 3.2 | 987/105 | Harm avoidance |
| 2.9b | 1,122/561 | Neuroticism | |
| 10, 5–9 cM [Camp et al., 2005; Wray et al., 2008] | 1.6 | 426/90 | RE-MDD |
| 2.0 | 2,030/564 | Neuroticism | |
| 10, 76 cM Zubenko et al. [2003] | 3.0 | ?/81 | MDD |
| 11, 2–35 cM [Camp et al., 2005; Zubenko et al., 2003] | 1.6 | 426/90 | RE-MDD and anxiety |
| 4.2 | ?/81 | R-MDD | |
| 11, 85–99 cM [Fullerton et al., 2003; Zubenko et al., 2003] | 3.7b | 1,122/561 | Neuroticism |
| 2.5 | ?/81 | RE-MDD | |
| 12, 105–124 cM [Fullerton et al., 2003; McGuffin et al., 2005] | 4.7b | 1,122/561 | Neuroticism |
| 1.6 | 994/497 | R-MDD | |
| 13, 64 cM Fullerton et al. [2003] | 3.8b | 1,122/561 | Neuroticism |
| 18, 73 cM Camp et al. [2005] | 3.8 | 96/21 | RE-MDD and anxiety |
| 18, 109–117 cM [Cloninger et al., 1998; Wray et al., 2008] | 1.6 | 987/105 | Harm avoidance |
| 1.9 | 8,552/2,509 | Neuroticism |
Regions with an LOD ≥3 or with an LOD ≥1.5 found at least twice are shown. Studies including subjects with bipolar disorder are excluded. Sex specific effects are not included.
For the MDD studies the number of affected individuals is given.
This is the −log P, not the LOD score.
Five of these studies focused on phenotypes considered to be most influenced by genetic factors, that is recurrent MDD [McGuffin et al., 2005] or recurrent, early-onset MDD [Zubenko et al., 2003; Holmans et al., 2004, 2007; Camp et al., 2005]. This seemed a useful strategy to increase the chance to find genes involved in MDD. However, comparing their results with the linkage analysis of MDD [Nurnberger et al., 2001] or with the additional linkage analysis of major mood disorders [Zubenko et al., 2003] shows that this approach does not seem to result in higher LOD scores than using a diagnosis of lifetime MDD. Although Zubenko et al. [2003] have found the highest LOD score for recurrent, early-onset and recurrent MDD, additional linkage peaks were identified for major mood disorders.
We present a new linkage study of MDD performed on affected sibling pairs selected from the Australian and Dutch Twin registers. Although these samples are geographically disparate, we have shown previously that the genetic distance between these Dutch and Australian subjects (Fst = 0.30%) is comparable to that between most Northern European populations [Sullivan et al., 2006]. Also, the genetic architecture of MDD is similar in the two countries with a heritability estimate for MDD of 36%and without evidence for sex differences in genetic architecture, allowing us to pool data from men and women in the linkage scan [Middeldorp et al., 2005].
MATERIALS AND METHODS
Subjects
Data were collected in the Australian and Netherlands Twin Registers as part of a project aiming to find the genes underlying the susceptibility to anxiety and MDD. Detailed descriptions of the data collection have been published previously [Boomsma et al., 2000; Kirk et al., 2000; Middeldorp et al., 2006]. In short, in 1998, the most informative families for a linkage study on anxiety and/or MDD were selected, based on survey data. Families were selected with extreme scoring discordant (low–high) or concordant (high–high and low–low) sib pairs on a quantitative scale that correlates with a diagnosis of MDD [Eaves and Meyer, 1994; Risch and Zhang, 1995; Dolan and Boomsma, 1998]. In Australia, the selection variable was a normalized neuroticism score, adjusted for age and sex, obtained in 1989. This score was available for 18,578 twins, siblings and adult offspring of twins [Kirk et al., 2000]. In The Netherlands, the selection variable was a genetic factor score expressing a subject’s genetic susceptibility to “anxious depression.” Factor scores were calculated as a weighted sum of the scores on four measures of neuroticism, anxiety, and depression assessed on three different occasions (1991, 1993, and 1997). The weights were derived from a multivariate genetic analysis and were different for men and women [Boomsma et al., 2000]. A factor score was available for 7,836 subjects [Middeldorp et al., 2006]. Extreme discordant and concordant scoring sibling pairs were asked to participate in a diagnostic psychiatric interview. In addition, other offspring in these families, regardless of their value on the selection variable, were invited to take part in the study.
In the Australian and Netherlands Twin Register, 2,918 and 1,517 subjects, respectively were approached to participate in the interview. In Australia, 256 subjects declined and 192 could not be contacted [Kirk et al., 2000]. In The Netherlands, 154 subjects refused and 107 could not be contacted [Middeldorp et al., 2006]. In total, 2,470 Australian and 1,256 Dutch individuals were interviewed. Thus, a participation rate of around 90% was achieved in both samples.
Genotyping was carried out in 1,943 subjects, including 737 men and 1,206 women from Australia and 904 subjects, including 370 men and 534 women, from The Netherlands.
Genotyping
The genotypic data available for the Australian study resulted from submission of DNA samples to one or more of six genotyping centers, namely Gemini P/L (G), Sequana Therapeutics Inc. (S), Leiden University Medical Center (L), the Center for Mammalian Genetics at the Marshfield Clinic Research Foundation (M), the Australian genotyping research facility (AGRF) (A), and the Finnish Genome Center, University of Helsinki (H). A description of the G, L, M, S genotyping and the subsequent merging and cleaning of the marker data sets is described in detail elsewhere [Cornes et al., 2005; Nyholt et al., 2005]. Since then, additional M, A, and H genotypes have been merged using the same protocol. Family members were submitted to the same genotyping facility. Data cleaning based on Mendelian errors, unlikely genotypes and consistency of pedigree and marker relationships was undertaken as described by Cornes et al. [2005]. In brief, pedigree structures for each scan were examined using graphic representation of relationships (GRR) [Abecasis et al., 2001] and RELPAIR [Epstein et al., 2000; Duren et al., 2003], to identify inconsistencies between the genotypic data and pedigree relationships. Once any discrepancies were resolved, data for the scans were merged and then checked again for pedigree errors by GRR and for Mendelian inconsistencies by SIB-PAIR [Duffy, 2002]. Duplicate markers in the combined genome scan data were included separately on the genetic map, separated by a very small distance (0.001 cM). The consistency of genotype information between these duplicate markers was checked via cross-tabulations of allele calls between different scans. Markers with genotypic data inconsistent between different genome scans were removed from further analysis. The Mendelian error rate, counted as the number of genotypes removed divided by total number of genotypes using all markers genotyped in more than one lab was 0.57%. Unlikely genotypes were identified and wiped using MERLIN [Abecasis et al., 2002]. Error rates were similar for the different labs.
For the Dutch samples, DNA was extracted from either whole blood or buccal swabs following standard protocols [Miller et al., 1988; Meulenbelt et al., 1995]. Samples were genotyped by M and L. The genotype data from these screens were combined. Allele calling and binning were equalized between markers that were present in multiple scans, using ~30 control samples. In case there were inconsistencies, the data were set to unknown for tested markers (binning and allele calling inconsistencies) and persons (genotyping errors). Sex and prior measured zygosity were checked with the marker data. Pedigree relations in the entire samples were checked with the GRR program [Abecasis et al., 2001]. Errors of Mendelian inheritance were detected with Pedstats [Wigginton and Abecasis, 2005]. Markers and samples were removed if their total error rate was more than 1%; in all other cases the specific erroneous genotypes were set as unknown. The mendelian error rate was 0.58%, Unlikely recombinants were detected using Merlin and erroneous genotypes were removed with Pedwipe [Abecasis et al., 2002]. Error rates were similar for the different labs.
Map positions of all genotyped markers were estimated in Kosambi cM by locally weighted linear regression http://www.qimr.edu.au/davidd from the NCBI Build 35.1 physical map positions and published Decode and Marshfield genetic map positions [Duffy, 2006]. Joint analysis of common markers genotyped in the Australian and Dutch samples estimated Fst between the populations to be only 0.30% implying that these samples can be combined for joint genetic analysis [Sullivan et al., 2006]. Individuals were required to have genotypes on more than 280 markers resulting in an average inter-marker distance of 8.2 cM (AU) and 9.6 cM (NL). The average number of markers genotyped in siblings was 719 in the Australian sample (range 334–1,242) and 400 in the Dutch sample (range 291–761).
Instruments
A telephone interview, during which the computerized version of the composite international diagnostic interview (CIDI) [World Health Organization, 1992] was administered to obtain lifetime DSM-IV diagnoses of mood and anxiety disorders [American Psychiatric Association, 1994]. The CIDI is a fully standardized diagnostic interview. Good reliability and validity have been reported for the CIDI [Andrews and Peters, 1998]. All interviewers were trained by the Dutch and Australian World Health Organization training centers. In order to minimize observer bias, interviewers were unaware of interviewees’ status on the initial selection variables throughout the study. The dependent variable used for linkage analysis was a lifetime diagnosis of MDD (DSM-IV codes: 296.21, 296.22, 296.23, 296.31, 296.32, and 296.33). Subjects who might have experienced a (hypo)manic episode as measured with the CIDI screening question (Australia) or the total “bipolar disorder” section (The Netherlands) were excluded.
Statistical Analyses
Linkage analysis was carried out using MERLIN [Abecasis et al., 2002]. Estimation of identity by descent (IBD) was based on the allele frequencies in the total genotyped sample and additional genotype data of family members of the affected siblings. Using the Whittemore and Halpern NPL all statistic [Whittemore and Halpern, 1994], MERLIN calculates at which markers affected siblings share more alleles IBD than expected by chance. LOD scores are calculated using the Kong and Cox linear model [Kong and Cox, 1997].
The 1-LOD-drop support interval was used as an estimate for the 90% confidence interval of any QTL locations [Abecasis et al., 2001] and was used to examine linkage replication compared to previously reported locations (using Sullivan Lab Evidence Project http://slep.unc.edu) and candidate genes (using Ensembl: www.ensembl.org and OMIM: www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=omim).
Empirically derived P values were calculated from 1,000 simulated data sets using the gene-dropping method implemented in MERLIN to calculate the threshold LOD-scores for suggestive and significant linkage.
RESULTS
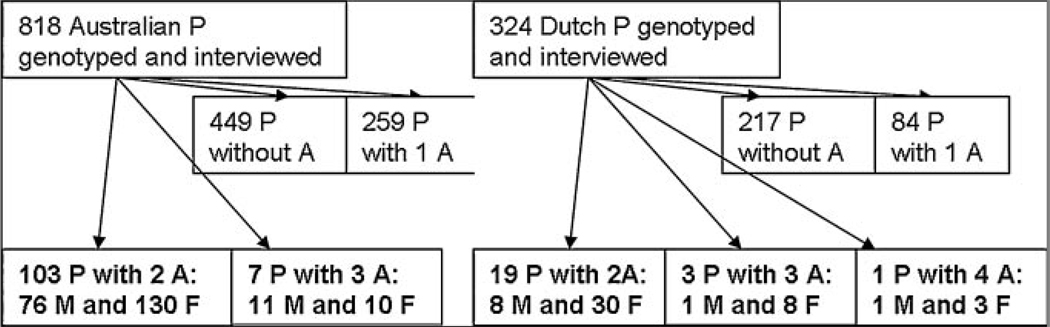
Mean age at the time of the interview was 43.1 years (SD 11.2) and 29.0 years (SD 11.2) for the 1,943 Australian subjects and the 904 Dutch subjects, respectively. Figure 1 shows that 110 Australian and 23 Dutch pedigrees included more than one affected sibling (total N = 278 affected siblings), and thus were eligible for the linkage analysis. From the families with two or more affected offspring, genotypes were available on 35 fathers and 44 mothers and an additional 130 siblings in the Australian sample and on 14 fathers and 14 mothers and an additional 43 siblings in the Dutch sample.
FIG. 1.
Number of Pedigrees (P) with and without affected (A) subjects for the Australian (left) and Dutch (right) samples. Printed in bold are the number of pedigrees with the corresponding number of affected males (M) and females (F) which are included in the linkage analysis.
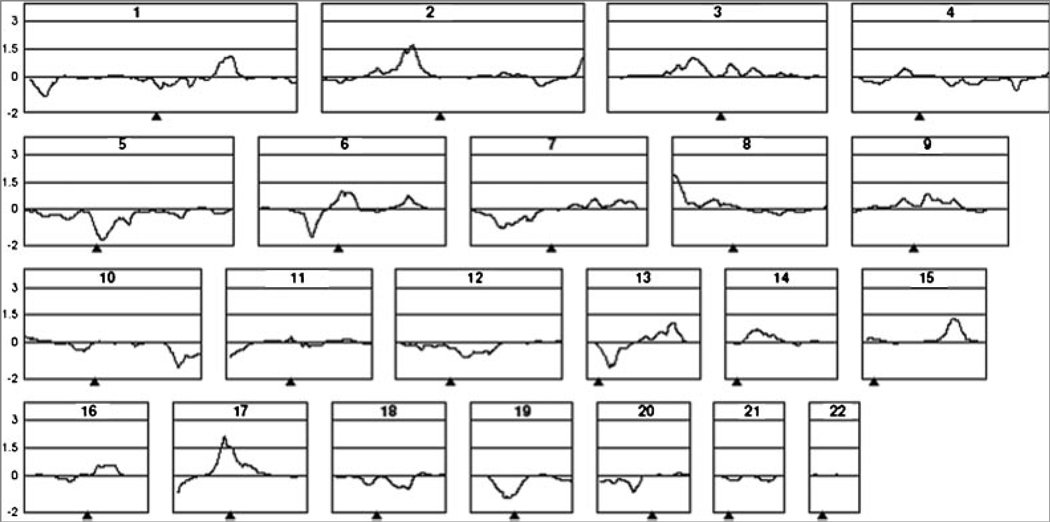
Figure 2 shows the linkage results for the analysis of a lifetime diagnosis of MDD and Table II shows the chromosomal regions with an LOD score exceeding 1.0. Simulations yielded an empirically derived suggestive and significant LOD threshold of 1.66 and 2.95. Three regions showed suggestive linkage signals. The highest LOD-score of 2.1 was found on chromosome 17 at 52.6 cm, marker ATA58E08 (90% confidence interval: 46.6–62.5 cM bounded by markers D17S921 and GGAA19G04). In addition, LOD scores of 1.9 and 1.7 were found on chromosome 8 at 2.7 cm, marker D8S504 (90% confidence interval: 0–12.4 cM, bounded by 8ptel and marker D8S277) and chromosome 2 at 90.6 cM, marker GATA66D01 (90% confidence interval: 78.1–96.9 cM, bounded by markers ATA27D04 and GATA181G08).
FIG. 2.
MDD linkage results in a combined Australian and Dutch sample.
TABLE II.
Chromosomal Regions With an LOD Scores >1.0
| Chromosome | cM | LOD |
|---|---|---|
| 17 | 52.585 | 2.11 |
| 8 | 2.652 | 1.87 |
| 2 | 90.646 | 1.7 |
| 15 | 95.943 | 1.27 |
| 13 | 97.493 | 1.08 |
| 1 | 221.663 | 1.04 |
DISCUSSION
A linkage analysis on a combined Australian and Dutch sample aiming to identify regions that might harbor genes influencing the vulnerability for MDD found suggestive linkage for two new regions on chromosome 2 and chromosome 17. In addition, we found suggestive linkage on chromosome 8 in a region that showed evidence for linkage in two previous studies, once significant and once suggestive [Cloninger et al., 1998; Fullerton et al., 2003].
The region on chromosome 17 harbors the serotonin transporter gene. In Dutch and Australian samples, partly overlapping with the current samples, no evidence was found for an association with the serotonin transporter gene length s/l polymorphism (5-HTTLPR) and MDD (Dutch and Australian sample) or neuroticism and symptoms of anxiety or depression (in the Dutch sample only) [Gillespie et al., 2005; Middeldorp et al., 2007]. These findings imply that another gene in this region is associated with MDD or another polymorphism in the serotonin transporter gene, for example, a recently identified third variant [Hu et al., 2006]. None of the other usual candidate genes for major depression serotonin receptor 1A (5HT1A), tryptophan hydroxylase (TPH) 1 and 2, catechol-o-methyltransferase (COMT), or brain derived neurotrophic factor (BDNF) were found to lie under one of the linkage peaks [Levinson, 2006].
None of the regions clearly overlapped with syntenic regions found in linkage analyses onemotionality in mice, but the region on chromosome 2 was very close, with 5 cM between the borders of the confidence intervals [Smoller et al., 2001].
An important limitation of our study, as with many other linkage studies of MDD (Table II), is the small sample size. A power analysis performed in ASP [Krawczak, 2002] showed that we had 10% power to find a P-value <0.001(LOD score~2) assuming a lifetime prevalence of MDD of 15%. Therefore, it is likely that other regions that are involved in the development of MDD were not identified. Individual risk variants underlying complex genetic disease are likely to explain only a fraction of the genetic variance [Welcome Trust Case Control Consortium, 2007] but multiple risk variants within a gene or chromosomal region may be more easily detected by linkage than association, and consistently identified regions from linkage analyses an be used to inform prioritization of results from genome-wide association studies. In this study, we identified three regions that exceeded the suggestive threshold for linkage, defined as the threshold expected to be exceeded only once per genome scan.
The current study was carried out in a sub-sample of a larger set of families used for a linkage analysis of the mean neuroticism scores over 4 (Australia) or 5 (The Netherlands) time points [Wray et al., 2008]. Neuroticism is a personality trait measuring emotional instability and those scoring highest on the neuroticism scale express feelings of both anxiety and depression. The motivation for the current study was greater phenotypic homogeneity despite the loss of power in sample size. The three regions of suggestive linkage identified in the neuroticism analysis (10p, 14q, and 18q) showed no evidence for linkage in the current analysis of MDD, although the linkage to 14q is most likely driven by anxiety phenotypes in the Dutch sample [Middeldorp et al., 2008]. Of the three regions identified in the current MDD linkage analysis, chromosome 2 at 90 cM achieved LOD of 1 in the analysis of neuroticism in the Dutch sample, but the regions on chromosomes 8 and 17 showed no evidence for linkage with mean neuroticism score. However, the measure of neuroticism collected at the same interview as the CIDI interview in the Australian sample (AU99) did show evidence for linkage (LOD > 1) across the wide region of 0–85 cM on chromosome 17 [results for individual measures of neuroticism were not presented in Wray et al., 2008], but not at other time points.
To conclude, the region on chromosome 8 around 5 cM has shown most evidence for harboring genes that are involved in the development of MDD. Further research is warranted aiming to identify the specific genes. Future studies are necessary to clarify the significance of our finding on chromosome 2 and 17.
ACKNOWLEDGMENTS
The Dutch study was supported by The Netherlands Organization for Scientific Research NWO/ZonMW (400-05-717, 911-03-016, 904-61-193, 985-10-002, 575-25-006), NIH R01 HL55976 and NHBLI Mammalian Genotyping Service (Marshfield). Statistical analyses were carried out on the Genetic Cluster Computer (http://www.geneticcluster.org) which is financially supported by The Netherlands Scientific Organization (NWO 480-05-003). CM was supported by the Hersenstichting Nederland (13F05(2).47) and NWO-ZonMw (91676125). Analytic support was provided by NIMH R01 MH059160 (to PFS). The Australian study was funded by grants to NGM Gavin Andrews from the Australian National Health and Medical Research Council (971232, 339450) and by Gemini Genomics plc (now defunct). We acknowledge the role of Dr. Andrew Heath and NIH grants (AA07535 and AA07728) in earlier projects in which selection variables were collected. For the genome scans we acknowledge and thank: the Mammalian Genotyping Service, Marshfield WI (Director: Dr. James Weber) for genotyping conducted under grants awarded to Dr. Daniel T. O’Connor, Dr. David Duffy, Dr. Dale Nyholt, and Dr. Patrick Sullivan; Dr. Eline Slagboom (Leiden scan); Dr. Peter Reed (Gemini scan); Dr. Jeff Hall (Sequana); Dr. Aarno Palotie (Finnish Genome Center, Helsinki); and Dr. Sue Forrest (Australian Genome Research Facility). We thank our interviewers and clerical and administrative support staff supervised by DJS. We thank David Smyth, Scott Gordon, Harry Beeby, and Olivia Zheng for data management and computer support and our laboratory staff, especially Megan Campbell, Anjali Henders, and Leanne McNeil, for sample processing and preparation. Most of all, we thank the twins and their relatives for their willing participation in the study.
Grant sponsor: Netherlands Organization for Scientific Research NWO; Grant numbers: 400-05-717, 911-03-016, 904-61-193, 985-10-002, 575-25-006, 480-05-003; Grant sponsor: NIH; Grant numbers: R01 HL55976; Grant sponsor: NHBLI Mammalian Genotyping Service (Marshfield); Grant sponsor: Hersenstichting Nederland; Grant numbers: 13F05(2).47; Grant sponsor: NWO-ZonMw; Grant numbers: 91676125; Grant sponsor: NIMH; Grant numbers: R01 MH059160; Grant sponsor: Australian National Health and Medical Research Council; Grant numbers: 971232, 339450; Grant sponsor: NIH; Grant numbers: AA07535, AA07728.
REFERENCES
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. GRR: Graphical representation of relationship errors. Bioinformatics. 2001;17:742–743. doi: 10.1093/bioinformatics/17.8.742. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin–rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 4th ed. Washington, DC: APA; 1994. [Google Scholar]
- Andrews G, Peters L. The psychometric properties of the Composite International Diagnostic Interview. Soc Psychiatry Psychiatr Epidemiol. 1998;33:80–88. doi: 10.1007/s001270050026. [DOI] [PubMed] [Google Scholar]
- Boomsma DI, Beem AL, van den Berg M, Dolan CV, Koopmans JR, Vink JM, de Geus EJ, Slagboom PE. Netherlands twin family study of anxious depression (NETSAD) Twin Res. 2000;3:323–334. doi: 10.1375/136905200320565300. [DOI] [PubMed] [Google Scholar]
- Camp NJ, Lowry MR, Richards RL, Plenk AM, Carter C, Hensel CH, Abkevich V, Skolnick MH, Shattuck D, Rowe KG, Hughes DC, Cannon-Albright LA. Genome-wide linkage analyses of extended Utah pedigrees identifies loci that influence recurrent, early-onset major depression and anxiety disorders. Am J Med Genet Part B. 2005;135B:85–93. doi: 10.1002/ajmg.b.30177. [DOI] [PubMed] [Google Scholar]
- Cloninger CR, Van Eerdewegh P, Goate A, Edenberg HJ, Blangero J, Hesselbrock V, Reich T, Nurnberger J, Jr, Schuckit M, Porjesz B, Crowe R, Rice JP, Foroud T, Przybeck TR, Almasy L, Bucholz K, Wu W, Shears S, Carr K, Crose C, Willig C, Zhao J, Tischfield JA, Li TK, Conneally PM. Anxiety proneness linked to epistatic loci in genome scan of human personality traits. Am J Med Genet. 1998;81:313–317. doi: 10.1002/(sici)1096-8628(19980710)81:4<313::aid-ajmg7>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Cornes BK, Medland SE, Ferreira MA, Morley KI, Duffy DL, Heijmans BT, Montgomery GW, Martin NG. Sex-limited genome-wide linkage scan for body mass index in an unselected sample of 933 Australian twin families. Twin Res Hum Genet. 2005;8:616–632. [PubMed] [Google Scholar]
- Dolan CV, Boomsma DI. Optimal selection of sib pairs from random samples for linkage analysis of a QTL using the EDAC test. Behav Genet. 1998;28:197–206. doi: 10.1023/a:1021423214032. [DOI] [PubMed] [Google Scholar]
- Duffy DL. Sib-Pair Version 0.99.9, Brisbane: Queensland Institute of Medical Research. 2002 [Google Scholar]
- Duffy DL. An integrated genetic map for linkage analysis. Behav Genet. 2006;36:4–6. doi: 10.1007/s10519-005-9015-x. [DOI] [PubMed] [Google Scholar]
- Duren WL, Epstein MP, Li M, Boehnke M. Ann Arbor, MI: University of Michigan; 2003. RELPAIR: A program that infers the relationships of pairs of individuals based on marker data (Version 2.0) [Google Scholar]
- Eaves L, Meyer J. Locating human quantitative trait loci: Guidelines for the selection of sibling pairs for genotyping. Behav Genet. 1994;24:443–455. doi: 10.1007/BF01076180. [DOI] [PubMed] [Google Scholar]
- Epstein MP, Duren WL, Boehnke M. Improved inference of relationship for pairs of individuals. Am J Hum Genet. 2000;67:1219–1231. doi: 10.1016/s0002-9297(07)62952-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley DL, Neale MC, Kendler KS. Reliability of a lifetime history of major depression: Implications for heritability and co-morbidity. Psychol Med. 1998;28:857–870. doi: 10.1017/s0033291798006977. [DOI] [PubMed] [Google Scholar]
- Fullerton J, Cubin M, Tiwari H, Wang C, Bomhra A, Davidson S, Miller S, Fairburn C, Goodwin G, Neale MC, Fiddy S, Mott R, Allison DB, Flint J. Linkage analysis of extremely discordant and concordant sibling pairs identifies quantitative-trait loci that influence variation in the human personality trait neuroticism. Am J Hum Genet. 2003;72:879–890. doi: 10.1086/374178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie NA, Whitfield JB, Williams B, Heath AC, Martin NG. The relationship between stressful life events, the serotonin transporter (5-HTTLPR) genotype and major depression. Psychol Med. 2005;35:101–111. doi: 10.1017/s0033291704002727. [DOI] [PubMed] [Google Scholar]
- Holmans P, Zubenko GS, Crowe RR, DePaulo JR, Jr, Scheftner WA, Weissman MM, Zubenko WN, Boutelle S, Murphy-Eberenz K, MacKinnon D, McInnis MG, Marta DH, Adams P, Knowles JA, Gladis M, Thomas J, Chellis J, Miller E, Levinson DF. Genomewide significant linkage to recurrent, early-onset major depressive disorder on chromosome15q. Am J Hum Genet. 2004;74:1154–1167. doi: 10.1086/421333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmans P, Weissman MM, Zubenko GS, Scheftner WA, Crowe RR, DePaulo JR, Jr, Knowles JA, Zubenko WN, Murphy-Eberenz K, Marta DH, Boutelle S, McInnis MG, Adams P, Gladis M, Steele J, Miller EB, Potash JB, Mackinnon DF, Levinson DF. Genetics of recurrent early-onset major depression (GenRED): Final genome scan report. AmJ Psychiatry. 2007;164:248–258. doi: 10.1176/ajp.2007.164.2.248. [DOI] [PubMed] [Google Scholar]
- Hu XZ, Lipsky RH, Zhu G, Akhtar LA, Taubman J, Greenberg BD, Xu K, Arnold PD, Richter MA, Kennedy JL, Murphy DL, Goldman D. Serotonin transporter promoter gain-of-function genotypes are linked to obsessive-compulsive disorder. Am J Hum Genet. 2006;78:815–826. doi: 10.1086/503850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk KM, Birley AJ, Statham DJ, Haddon B, Lake RI, Andrews JG, Martin NG. Anxiety and depression in twin and sib pairs extremely discordant and concordant for neuroticism: Prodromus to a linkage study. Twin Res. 2000;3:299–309. doi: 10.1375/136905200320565274. [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ. Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet. 1997;61:1179–1188. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawczak M. ASP (Version 1.2), Kiel, Germany: Institut für Medizinische Informatik und Statistik. 2002 [Google Scholar]
- Kuo PH, Neale MC, Riley BP, Patterson DG, Walsh D, Prescott CA, Kendler KS. A genome-wide linkage analysis for the personality trait neuroticism in the Irish affected sib-pair study of alcohol dependence. Am J Med Genet Part B. 2007;144B:463–468. doi: 10.1002/ajmg.b.30478. [DOI] [PubMed] [Google Scholar]
- Levinson DF. The genetics of depression: A review. Biol Psychiatry. 2006;60:84–92. doi: 10.1016/j.biopsych.2005.08.024. [DOI] [PubMed] [Google Scholar]
- McGuffin P, Knight J, Breen G, Brewster S, Boyd PR, Craddock N, Gill M, Korszun A, Maier W, Middleton L, Mors O, Owen MJ, Perry J, Preisig M, Reich T, Rice J, Rietschel M, Jones L, Sham P, Farmer AE. Whole genome linkage scan of recurrent depressive disorder from the depression network study. Hum Mol Genet. 2005;14:3337–3345. doi: 10.1093/hmg/ddi363. [DOI] [PubMed] [Google Scholar]
- Meulenbelt I, Droog S, Trommelen GJ, Boomsma DI, Slagboom PE. High-yield noninvasive human genomic DNA isolation method for genetic studies in geographically dispersed families and populations. Am J Hum Genet. 1995;57:1252–1254. [PMC free article] [PubMed] [Google Scholar]
- Middeldorp CM, Birley AJ, Cath DC, Gillespie NA, Willemsen G, Statham DJ, de Geus EJ, Andrews JG, Van Dyck R, Beem AL, Sullivan PF, Martin NG, Boomsma DI. Familial clustering of major depression and anxiety disorders in Australian and Dutch twins and siblings. Twin Res Hum Genet. 2005;8:609–615. doi: 10.1375/183242705774860123. [DOI] [PubMed] [Google Scholar]
- Middeldorp CM, Cath DC, van den Berg M, Beem AL, van Dyck R, Boomsma DI. The association of personality with anxious and depressive psychopathology. In: Canli T, editor. The biological basis of personality and individual differences. New York: Guilford Press; 2006. pp. 251–272. [Google Scholar]
- Middeldorp CM, de Geus EJ, Beem AL, Lakenberg N, Hottenga JJ, Slagboom PE, Boomsma DI. Family based association analyses between the serotonin transporter gene polymorphism (5-HTTLPR) and neuroticism, anxiety and depression. Behav Genet. 2007;37:294–301. doi: 10.1007/s10519-006-9139-7. [DOI] [PubMed] [Google Scholar]
- Middeldorp CM, Hottenga JJ, Slagboom PE, Sullivan PF, de Geus EJ, Posthuma D, Willemsen G, Boomsma DI. Linkage on chromosome 14 in a genomewide linkage study of a broad anxiety phenotype. Mol Psychiatry. 2008;13:84–89. doi: 10.1038/sj.mp.4002061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash MW, Huezo-Diaz P, Williamson RJ, Sterne A, Purcell S, Hoda F, Cherny SS, Abecasis GR, Prince M, Gray JA, Ball D, Asherson P, Mann A, Goldberg D, McGuffin P, Farmer A, Plomin R, Craig IW, Sham PC. Genome-wide linkage analysis of a composite index of neuroticism and mood-related scales in extreme selected sibships. Hum Mol Genet. 2004;13:2173–2182. doi: 10.1093/hmg/ddh239. [DOI] [PubMed] [Google Scholar]
- Neale BM, Sullivan PF, Kendler KS. A genome scan of neuroticism in nicotine dependent smokers. Am J Med Genet Part B. 2005;132B:65–69. doi: 10.1002/ajmg.b.30095. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Foroud T, Flury L, Su J, Meyer ET, Hu K, Crowe R, Edenberg H, Goate A, Bierut L, Reich T, Schuckit M, Reich W. Evidence for a locus on chromosome 1 that influences vulnerability to alcoholism and affective disorder. Am J Psychiatry. 2001;158:718–724. doi: 10.1176/appi.ajp.158.5.718. [DOI] [PubMed] [Google Scholar]
- Nyholt DR, Morley KI, Ferreira MA, Medland SE, Boomsma DI, Heath AC, Merikangas KR, Montgomery GW, Martin NG. Genomewide significant linkage to migrainous headache on chromosome 5q21. Am J Hum Genet. 2005;77:500–512. doi: 10.1086/444510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Zhang H. Extreme discordant sib pairs for mapping quantitative trait loci in humans. Science. 1995;268:1584–1589. doi: 10.1126/science.7777857. [DOI] [PubMed] [Google Scholar]
- Smoller JW, Acierno JS, Jr, Rosenbaum JF, Biederman J, Pollack MH, Meminger S, Pava JA, Chadwick LH, White C, Bulzacchelli M, Slaugenhaupt SA. Targeted genome screen of panic disorder and anxiety disorder proneness using homology to murine QTL regions. Am J Med Genet. 2001;105:195–206. doi: 10.1002/ajmg.1209. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: Review and meta-analysis. Am J Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- Sullivan PF, Montgomery GW, Hottenga JJ, Wray NR, Boomsma DI, Martin NG. Empirical evaluation of the genetic similarity of samples from twin registries in Australia and the Netherlands using 359 STRP markers. Twin Res Hum Genet. 2006;9:600–602. doi: 10.1375/183242706778025026. [DOI] [PubMed] [Google Scholar]
- Welcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittemore AS, Halpern J. A class of tests for linkage using affected pedigree members. Biometrics. 1994;50:118–127. [PubMed] [Google Scholar]
- Wigginton JE, Abecasis GR. PEDSTATS: Descriptive statistics, graphics and quality assessment for gene mapping data. Bioinformatics. 2005;21:3445–3447. doi: 10.1093/bioinformatics/bti529. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Composite International Diagnostic Interview (Version 2.1) Geneva: WHO; 1992. [Google Scholar]
- Wray NR, Middeldorp CM, Birley AJ, Gordon SD, Sullivan PF, Visscher PM, Willemsen G, De Geus EJC, Slagboom EP, Montgomery GW, Martin NG, Boomsma DI. Genome wide linkage analysis of multiple measures of neuroticism from two large cohorts from Australia and the Netherlands. Arch Gen Psychiatry. 2008;65:649–658. doi: 10.1001/archpsyc.65.6.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubenko GS, Maher B, Hughes HB, III, Zubenko WN, Stiffler JS, Kaplan BB, Marazita ML. Genome-wide linkage survey for genetic loci that influence the development of depressive disorders in families with recurrent, early-onset, major depression. Am J Med Genet Part B. 2003;123B:1–18. doi: 10.1002/ajmg.b.20073. [DOI] [PubMed] [Google Scholar]