Abstract
Background
A proteinase with a disintegrin and a metalloproteinase domain-8 (ADAM8) has been linked to asthma.
Objective
To explore whether ADAM8 is a therapeutic target for asthma.
Methods
We reviewed literature on ADAM8’s function and expression and activities in lungs of humans and mice with allergic airway inflammation (AAI). We used these data to generate hypotheses about the contributions of ADAM8 to asthma pathogenesis.
Conclusions
ADAM8 levels are increased in airway epithelium and airway inflammatory cells in mice with AAI and human asthma patients. Data from murine models of AAI indicate that ADAM8 dampens airway inflammation. It is not clear whether ADAM8 contributes directly to structural remodeling in asthmatic airways. Additional studies are required to validate ADAM8 as a therapeutic target for asthma.
Keywords: ADAM, airway hyper-responsiveness, allergy, asthma, disintegrin, eosinophil, epithelium, inflammation, leukocyte, macrophage, metalloproteinase, proteinase, remodeling, signaling
1. Introduction: asthma
1.1. Healthcare and economic burden
Asthma is a chronic inflammatory disease of the airways characterized by airway hyperresponsivness (AHR) and intermittent airflow obstruction. Asthma is associated with high morbidity and mortality and its incidence is increasing in most countries. Asthma affects approximately 300 million people worldwide and causes an estimated 255,000 deaths per year [1]. In 2006, 7.3% of the US population had asthma and asthma was responsible for 17 million physician consultations [2]. The total annual financial cost for all forms of asthma in the US is estimated to be ~$14 billion [3]. Thus, asthma represents a large economic and healthcare burden. Although patients with mild disease respond well to inhaled bronchodilators and/or corticosteroids, other patients need oral corticosteroids, which are associated with undesirable side effects, and some patients are refractory to these therapies. Thus, there is a pressing need to develop new and improved therapies for asthma.
1.2. Asthma pathogenesis
Asthma is thought to be caused by complex interactions between genetic and environmental factors, but its pathogenesis is still incompletely understood. Patients with asthma have increased numbers of CD4+ T cells in the airways, which are predominantly of the T helper type 2 (TH2) subtype. These TH2 lymphocytes produce TH2 cytokines including IL-4, −5, −9, and −13 which orchestrate the inflammatory response in asthmatic airways by recruiting and activating additional TH2 cells and also mast cells and eosinophils [4]. IL-4 and −13 also promote isotype switching of B cells to IgE production. Inhalation of allergens in sensitized individuals increases B cell production of IgE, which binds to IgE receptors on mast cells inducing mast cell degranulation. Degranulating mast cells release cytokines, histamine, cysteinyl-leukotrienes, prostanoids and proteinases, which induce bronchoconstriction, edema, and additional recruitment of eosinophils and TH2 lymphocytes to the airways thereby amplifying airway inflammation. However, this is a gross oversimplification of the complex inflammatory events occurring in asthma. Natural killer T cells [5], TH17 lymphocytes [6], regulatory T-cells [7], dendritic cells (DCs [8]), macrophages [9], and polymorphonuclear neutrophils (PMNs [10]) also play important roles in asthma pathogenesis.
Structural changes also occur in asthmatic airways including mucus metaplasia and hyperplasia, activation of fibroblasts, interstitial collagen deposition and airway smooth muscle hyperplasia, which contribute fundamentally to asthma chronicity and severity [11-13]. Although these airway remodeling processes were initially thought to be secondary to chronic airway inflammation, extensive airway remodeling occurs early in this disease in children and often before the onset of symptoms [14,15]. It has been suggested that in asthma the airway epithelium is abnormally sensitive to the damaging effects of oxidants [16,17]. The injured epithelium communicates in a dysregulated and bi-directional fashion with the underlying mesenchyme to form the epithelial-mesenchymal trophic unit, which initiates and amplifies airway remodeling. Poorly controlled repair processes in the asthmatic epithelium lead to excessive release of growth factors that activate subepithelial fibroblasts to form myofibroblasts that deposit extracellular matrix (ECM) proteins along the airway basement membrane and secrete mitogens for airway smooth muscle cells (SMCs) and cytokines that amplify remodeling and inflammation [18].
1.3. Asthma and proteinases
Serine and matrix metalloproteinases (MMPs) have been shown to regulate mucus hyper-secretion [19], airway inflammation [20-22], subepithelial fibrosis [23], and airway SMC proliferation [24,25] in asthma. In 2002, the a disintegrin and a metalloproteinase (ADAM) subfamily of MPs was implicated in asthma pathogenesis when genetic polymorphisms in the ADAM33 gene were found to be associated with asthma and AHR [26,27]. ADAM33, a product of mesenchymal cells, probably regulates airway remodeling rather than airway inflammation in asthma [17,28]. More recently, ADAM8 has been strongly associated with allergic airway inflammation (AAI) in humans and mice and additional studies of ADAM8 are beginning to shed light on its roles in asthma pathogenesis. Below we outline what is known about the biology of ADAM8 and its expression in AAI in humans and mice. We will also speculate about its potential contributions to pathologies occurring in the airways of asthmatic subjects and its potential as a new therapeutic target for asthma.
2. ADAM8
2.1 ADAM8 structure and chromosomal localization
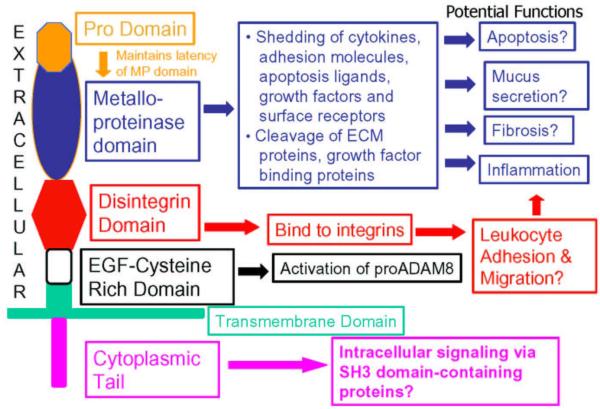
ADAM proteinases are a subfamily of zinc-dependent MPs and are type I transmembrane proteins with a multi-domain structure [29]. ADAM8 is also known as membrane-spanning 2 (MS2) or cluster of differentiation antigen 156a (CD156a) and was originally cloned in 1990 from murine macrophages and macrophage cell lines [30]. The human ADAM8 gene maps to chromosome 10q26.3 and the mouse ADAM8 gene to region F3–F4 on chromosome 7 [31,32]. There is 65.6% and 61.7% homology between human and murine ADAM8 at the nucleotide and protein levels, respectively [31]. The functions of ADAM proteins are related to their multiple domain structure which includes a pro-domain, a metalloproteinase (MP) domain, a disintegrin domain, a cysteine-rich (CR) domain, an EGF-like domain, a transmembrane domain, and a cytoplasmic tail (Figure 1). ADAM8 has all of these domains and the human protein contains 808 amino acids, including 637 residues in the ectodomain, 25 residues in the transmembrane domain, and 146 amino acids in the cytoplasmic tail [31].
Figure 1. The domain structure of ADAM8 and known or potential functions of each domain.
Structure & Potential Function of ADAM8
Most is known about the metalloproteinase (MP) and disintegrin domains of ADAM8. ADAM8 is an active MP and may cleave several cell proteins including adhesion molecules, cytokines, cytokine receptors, growth factors and leukocyte immunoglobulin receptors from cell surfaces. The disintegrin domain of ADAM8 binds to α9β1 integrin on osteoclasts but it is not clear whether it binds to other integrins expressed by leukocytes to regulate leukocyte adhesion or migration. The cytoplasmic tail of ADAM8 has SH3 binding domains but its role in binding to SH3-domain-containing intracellular proteins to regulate intracellular signaling has not been examined. ECM: extracellular matrix.
2.1.1 The pro-domain
Like other ADAMs, ADAM8 is initially synthesized as a latent pro-enzyme. The pro-domain maintains the MP domain in an inactive form through an interaction between a conserved cysteine residue in the pro-domain and the active site zinc atom. Although many proADAMs are activated by furin-mediated cleavage of the pro-domain in the trans-Golgi, proADAM8 is activated in the trans-Golgi by autocatalytic cleavage of the prodomain [33,34].
2.1.2 The MP domain
ADAM8 contains the catalytic site zinc-binding consensus sequence (HEXXHXXGXXHD) and is an active proteinase [33-35]. After proADAM8 is activated in the trans-Golgi it translocates to the cell surface. In some cells, the MP domain can further proteolytically cleave active ADAM8 with loss of the MP domain itself leaving a truncated form of the enzyme with the disintegrin domain at the NH2 terminus [34,36]. The main function of the MP domain of ADAMs is thought to be in proteolytically cleaving and releasing (or shedding) signaling molecules and their receptors from cell surfaces. The best-known example of an ADAM ‘sheddase’ is ADAM17 which cleaves latent, membrane-bound, 26 kDa pro-TNF-α thereby releasing soluble, active 17 kDa TNF-α [37]. Recombinant active ADAM8 sheds adhesion molecules and surface receptors from cell surfaces (Table 1) and also cleaves short peptide substrates containing sequences in cytokines, cytokine receptors, and growth factors that are susceptible to cleavage by other proteinases (Table 1). Some of these potential substrates for ADAM8 have been implicated in asthma pathogenesis as outlined below. The role of ADAM8 in degrading ECM proteins has not been examined, but ADAM proteinases are generally weak ECM protein-degrading enzymes [29]. The MP domain of ADAM8 is not inhibited by any of the four members of tissue inhibitor of metalloproteinase family [33,34]. Although physiological inhibitor(s) of the ADAM8 MP domain are likely to exist, they have not yet been identified.
Table 1. Known and potential substrates for ADAM8 metalloproteinase (MP) domain.
| Substrate | Effect of ADAM8-mediated cleavage of the substrate | References |
|---|---|---|
| Cytokines: | ||
| Pro-TNF-α* | Sheds active, soluble TNF-α Increase AAI |
[33,35] |
| Fractalkine (CX3CL1)* | Decrease leukocyte accumulation in the airways | [35] |
| Stem cell factor (SCF)* | Regulate mast cell activation and survival | [33,35] |
| Receptors: | ||
| TNF receptor 1* | Decrease AAI | [35] |
| IL-1 receptor II (decoy receptor)* | Decrease AAI | [33,35] |
| CD16* | Decreased leukocyte activation | [35] |
| CD23* | Increase IgE release by B cells Increase cytokine release by leukocytes |
[35,96] |
| Adhesion molecules: | ||
| L-selectin† | Limit accumulation of PMN and monocytes in the airways | [35,48] |
| VCAM-1† | Limit accumulation of eosinophils and B cells in the airways | [35,56,70] |
| P-selectin glycoprotein ligand-1* | Limit accumulation of PMN and monocytes in the airways | [35] |
| Neural cell adhesion molecule (NCAM or CHL1)† |
Suppress neuronal cell death and regulate neuronal development | [141,142] |
| Growth factors: | ||
| TGF-α* | EGF receptor activation to increase mucin production and airway remodeling |
[35] |
| Other proteins: | ||
| Amyloid precursor protein* | α-secretase activity generating anti-amyloidogenic peptides | [33,35] |
| Basic myelin protein* | *Promotes demyelination | [33-35] |
Indicates that a short peptide containing sequences in the protein known to be cleaved by proteinases was tested as a substrate for ADAM8 MP domain.
Indicates that the full length protein was tested as a substrate for ADAM8 MP domain.
AAI: allergic airway inflammation; CHL1: close homolog of L1; PMN: polymorphonuclear neutrophil; VCAM: vascular cell adhesion molecule
2.1.3 The disintegrin, cysteine-rich, and EGF-like domains
ADAMs have a disintegrin domain that is structurally similar to the disintegrin domain of hemorrhagic snake venom proteins, which contain RGD sequences that bind to platelet integrins to inhibit platelet activation and thrombosis. The disintegrin domain of ADAM8 contains an RX6DLPEF sequence rather than an RGD integrin-binding sequence. On osteoclast surfaces, ADAM8 disintegrin binds to α9β1 integrins on other osteoclasts in an RGD-independent manner to promote osteoclast fusion, activation, and differentiation [38,39]. However, it is not clear whether ADAM8 disintegrin domain binds to other integrins. ADAM8 overexpression in cell lines increases their capacity to adhere to ECM proteins and migrate [34,40], but the role of ADAM8 disintegrin in regulating the adhesion or migration of primary cells is not known. The disintegrin domain, along with the CR and EGF-like domains of ADAM8, is also required for optimal activation and processing of proADAM8 by its own MP domain [34,41]. However, it is not clear whether the CR and EGF-like domains of ADAM8 have other functions.
2.1.4. The cytoplasmic tail
This proline-rich domain contains a sequence similar to the consensus Src kinase homology 3 (SH3) binding sequence. SH3 domains are small, non-catalytic domains found in various intracellular signaling proteins including Src and Abl family protein tyrosine kinases. Although ADAM-9, −12, and −15 bind to Src family kinases [42-44], there have been no studies examining whether ADAM8 binds to SH3 domains in intracellular proteins.
2.2. ADAM8 expression and regulation
In healthy adult mice, ADAM8 is expressed at low levels in the brain, bone marrow, ovary, and the respiratory tract epithelium ([45-47]; Table 2). However, in mice, ADAM8 is not required for embryogenesis, reproduction, or survival since ADAM8−/− mice have normal development, fertility, and lifespan [47]. In humans, ADAM8 is expressed by most leukocytes [36,45,48], lung epithelial cells [49], and osteoclasts ([38,39]; Table 2). In contrast, ADAM33, the other ADAM family member that has been linked to asthma, is expressed mainly by mesenchymal cells in the airways including SMCs and fibroblasts [50,51]. However, there is conflicting literature on whether lung epithelial cells express ADAM33 [52,53].
Table 2. Expression and regulation of ADAM8 in humans and mice.
| Tissue/cells | Stimuli increasing ADAM8 | References |
|---|---|---|
| Murine lung | OVA-induced AAI, IL-4, and −13 | [57] |
| Mouse bronchial epithelial cells | Low level basal expression | [47] |
| OVA-induced AAI, IL-4 and IL-13 | [57] | |
| Human bronchial epithelial cells | Asthma | [49] |
| Leukocytes: | ||
| Eosinophils | OVA-induced AAI, IL-4, IL-13 Adhesion to endothelial cells |
[31,56,57] |
| PMN | Phorbol ester and adhesion to endothelium | [48] |
| Monocytes | Macrophage colony stimulating factor | [71] |
| Murine monocytic cell lines | LPS and interferon-γ | [32] |
| Murine macrophages | OVA-induced AAI | [30,57] |
| Myeloid DC | LPS, TNF-α | |
| IL-4, IL-4 and GM-CSF | [36] | |
| B cells | Low level basal expression | [36] |
| ADAM8 inducers not known | ||
| T cells | Do not express ADAM8 | [31,36] |
| Basophils | ADAM8 expression has not been assessed | |
| Mast cells | ADAM8 expression has not been assessed | |
| Murine central nervous system: | ||
| Oligodendrocytes and neurons | Low-level basal expression | [47] |
| Neurons and glial cells | TNF-α | |
| ADAM8 levels are increased in neurodegenerative diseases | [45] | |
| Murine ovarian follicles | Phorbol ester, human chorionic gonadotrophin and forskolin | [46] |
| Osteoclasts, bone, cartilage | Low-level basal expression | [47] |
| Macrophage colony stimulating factor | [38] | |
| Murine salivary gland epithelium | Low-level basal expression | [47] |
| Murine kidney epithelium | Low-level basal expression | [47] |
AAI: allergic airway inflammation; LPS: lipopolysaccharide OVA: ovalbumin;
Pharmacological agonists, cytokines, bacterial products and hormones upregulate ADAM8 levels in various cells (Table 2). The murine ADAM8 promoter contains a nuclear factor-1 (NF-1) binding site which may regulate phorbol ester responsiveness [46], two purine rich (PU) boxes, two IL-6 response elements, three NF-IL-6 binding sites which may regulate bacterial lipopolysaccharide (LPS) responsiveness [32], and an interferon regulatory factor-1 (IRF-1) binding sequence which may regulate TNF-α responsiveness [45]. ADAM8 levels are increased in inflammatory disorders of the lung, bone and joints, and central nervous system (Table 3). ADAM8 is also expressed in several tumors where its expression correlates positively with tumor invasiveness, metastasis, and poor prognosis (Table 3).
Table 3. ADAM8 expression and activities in diseases other than asthma.
| Disease (species) | Expression and activities of ADAM8 | References |
|---|---|---|
| Eosinophilic pneumonia (humans) | Soluble ADAM8 is increased in the BAL of patients with non-drug- induced eosinophilic pneumonias. Soluble ADAM8 levels correlate positively with soluble levels of CD23 and VCAM1 in BALF. |
[59] |
| Osteoporosis (humans) | ADAM8 is upregulated in osteoclasts and promotes osteoclast fusion and differentiation. |
[38,39] |
| Rheumatoid arthritis (humans) | Increased ADAM8 levels on PMNs and in joint pannus and synovial fluid. ADAM8 levels correlate positively with joint destruction. |
[38,48] |
| Reperfusion injury in the kidney (mice) | ADAM8 expression is increased in hypoxia-induced renal injury. | [143] |
| Neurodegeneration (mice) | ADAM8 shed NCAM1 which promotes neurite outgrowth and branching. |
[141,142] |
| Preterm delivery during pregnancy | Elevated ADAM8 levels in mid-trimester amniotic fluid are associated with an increased risk of preterm delivery. |
[144] |
| Spinal cord injury (mice) | ADAM8 is expressed in the endothelial cells of mice with spinal cord injuries and its expression correlates with angiogenesis. |
[145] |
| Tumors (all human): | ||
| Brain | ADAM8 is highly expressed especially in glioblastoma multiforme. ADAM8 expression is associated with increased tumor invasiveness. |
[146] |
| Lung | ADAM8 is increased in lung cancer tissue, especially in higher stage adenocarcinomas. sADAM8 is increased in serum of lung cancer patients. |
[40] |
| Pancreatic | ADAM8 levels are increased in pancreatic ductal adenocarcinoma and associated with decreased patient survival. ADAM8 increases the invasiveness of pancreatic cancer cell lines. |
[147] |
| Prostate | ADAM8 expression is increased and associated with unfavorable prognosis. |
[148] |
BAL: bronchoalveolar lavage; BALF: bronchoalveolar lavage fluid; PMN: polymorphonuclear neutrophil; NCAM: Neural cell adhesion molecule; VCAM: vascular cell adhesion molecule.
2.3. Biology of ADAM8 in cells relevant to asthma pathogenesis
ADAM8 is expressed by all leukocytes expect T lymphocytes [36,45,48] and epithelial cells [49]. ADAM8 is expressed at low levels in the airway epithelia of healthy mice and humans where its expression increases during AAI ([47,49]; Table 2). However, ADAM8 is not known to be expressed by airway SMCs or fibroblasts. ADAM8 is expressed at low levels on the surface of unstimulated leukocytes but pro-inflammatory stimuli including TH2 cytokines strikingly upregulate ADAM8 levels on leukocyte surfaces (Table 2). In most leukocytes, agonists increase ADAM8 at the steady state mRNA level. However, in PMNs, ADAM8 is stored as a preformed enzyme in their gelatinase and specific granules from where it translocates to their surface when PMNs are activated to degranulate [48]. The ectodomain of ADAM8 is also shed by unidentified MP(s) from the surface of activated PMNs [48] but the function of ADAM8 that is shed from PMN surfaces is not known.
ADAM8 may regulate the recruitment of eosinophils and PMNs into asthmatic airways. For example, ADAM8 may inhibit eosinophil recruitment into the airways during AAI by shedding vascular adhesion molecule-1 (VCAM-1). During AAI, activated eosinophils express α4 integrin, which binds to VCAM-1 that is induced on endothelial cells activated by TH2 cytokines. The binding of α4 integrin to VCAM-1 promotes the initial rolling and tethering of eosinophils on endothelial cells prior to their trans-endothelial migration [54]. When eosinophils adhere to endothelial cells they form podosomes which are punctuate and highly dynamic adhesive structures [55]. ADAM8 localizes to the podosomes of adherent eosinophils [56] where it may contribute to shedding of VCAM-1 from subjacent endothelial cells which may reduce eosinophil extravasation during TH2 airway inflammation. ADAM8 also sheds L-selectin from PMN surfaces in vitro [48]. Since L-selectin promotes PMN rolling on endothelial cells by binding to its ligands on endothelial cells during PMN transendothelial migration, ADAM8-mediated L-selectin shedding may also limit excessive or inappropriate efflux of PMNs and other leukocytes expressing both ADAM8 and L-selectin into sites of inflammation.
3. ADAM8 associations with asthma
3.1. ADAM8 levels are increased in the lung during AAI
ADAM8 was first linked to asthma in 2004 when King et al showed that ADAM8 (but not ADAM33) mRNA transcripts are increased in lungs of mice with Aspergillus- and ovalbumin (OVA)-induced AAI [57]. Initial robust increases in ADAM8 levels were linked to eosinophils, PMNs and macrophages infiltrating the airways. Less impressive increases in ADAM8 that persisted after inflammation had resolved were attributed to increases in epithelial-cell-derived ADAM8 [57]. Delivering or overexpressing TH2 cytokines (IL-4 and 13) in murine lungs also increased ADAM8 mRNA levels in lung homogenates by an IL4Rα and signal transducer and activator of transcription-6 (STAT-6)-dependent pathway [57]. More recently, bronchial tissue from healthy human subjects was shown to have little or no ADAM8 staining in airway epithelia or submucosa. However, intense staining for ADAM8 protein was present in bronchial inflammatory cell infiltrates and airway epithelia from patients with moderate and severe asthma which was greater than the ADAM8 staining in bronchial biopsies from subjects with mild asthma [49]. ADAM8 mRNA levels were also increased in sputum cells from patients with mild asthma compared with healthy control subjects and modest positive correlations were observed between sputum ADAM8 mRNA levels and sputum PMN and eosinophil counts and sCD23 levels [58]. Although these authors reported a negative correlation between sputum ADAM8 levels and forced expiratory volume in 1 second (FEV1) measurements, the correlation was weak, only a small number of patients was studied, and most patients had mild disease (mean FEV1 measurement were ~90% of predicted values). Thus, this reported negative correlation between FEV1 and ADAM8 is not likely to be physiologically relevant.
Soluble ADAM8 levels were also increased in bronchoalveolar lavage (BAL) samples from patients with cigarette-smoke-induced acute eosinophilic pneumonia (AEP) and chronic idiopathic eosinophilic pneumonia (CEP [59]). However, for reasons that are not clear, ADAM8 was not increased in BAL samples from patients with drug-induced pulmonary eosinophilia [59]. This literature indicates that in most types of eosinophilic lung inflammation in humans and mice, ADAM8 levels are increased in the lung, and increases are mainly due to increased expression of ADAM8 in lung inflammatory cells and to a lesser extent in airway epithelial cells [60].
3.2. ADAM8 polymorphisms, asthma, and atopy
Interest in ADAMs in lung biology was sparked in 2002 when van Eerdewegh and coworkers used positional cloning to identify ADAM33 as a putative asthma susceptibility gene in an outbred Caucasian population [26]. Since then, strong associations between single-nucleotide polymorphisms (SNPs) in the ADAM33 gene and asthma have been reported in several other Caucasian populations [61-64]. However, for reasons that are not clear, other studies have shown only weak associations [65,66], or no association between ADAM33 SNPs and asthma [67]. In 2008, Tremblay et al [68] subjected a genome-wide association scan performed on 609 subjects in a Northeastern Quebec population with familial asthma and atopy to a ‘Genes to Disease’ computational analysis. This was done to identify candidate genes in genetic loci known to be associated with asthma (6q26) and atopy (10q26.3). This study identified a single SNP in the ADAM8 gene that was initially suggested to confer protection against allergic asthma and atopy but this polymorphism was not associated with non-allergic asthma. However, the associations between the SNP in the ADAM8 gene and asthma and atopy were modest in magnitude and not significant after multiple comparisons testing. No other studies assessing whether SNPs in the ADAM8 gene are associated with asthma or atopy in other populations have yet been published.
3.3. Studies of ADAM8 transgenic and ADAM8 deficient mice in murine models of AAI
In 2002, Higuchi at al generated a conditional C57BL/6 transgenic mouse line expressing a soluble form of the entire ADAM8 ectodomain (sADAM8) under the control of the α1-antitrypsin promoter, which is mainly expressed by hepatocytes in the mice [69]. These transgenic mice had detectable levels of sADAM8 in their plasma and plasma sADAM8 levels increased when the mice were given LPS, which is a potent inducer of the acute phase response in hepatocytes [69]. When sensitized and challenged with OVA, sADAM8 transgenic mice had decreased inflammatory cell infiltrates in bronchial submucosal and perivascular areas in lung sections compared with non-transgenic control mice [70] but the leukocyte subsets that were increased were not identified. While the ADAM8 transgenic and non-transgenic control mice did not differ in BAL fluid (BALF) levels of IL-4 or −5, OVA-treated sADAM8 transgenic mice had reduced serum IL-5 levels, but the mechanism underlying this difference was not investigated. Serum from the sADAM8-transgenic mice had greater capacity than serum from non-transgenic control mice to cleave VCAM-1 from activated endothelial cells in vitro [70]. The authors concluded that sADAM8 limits AAI by shedding VCAM-1 from endothelial cells. However, this sADAM8-induced increased VCAM-1 shedding was very modest in magnitude and the authors did not measured levels of other potential ADAM8 substrates (see Table 1) in the transgenic versus non-transgenic mice. Thus, ADAM8 may limit the influx of leukocytes into the airways of the transgenic mice by shedding substrates other than, or in addition to VCAM-1, and/or by reducing levels of IL-5 which is critical for inducing eosinophil production and survival [4].
Recently, mice genetically deficient in ADAM8 (ADAM8−/− mice) versus wild type (WT) littermate control mice in a TH1 skewed mixed SvEV129 × C57BL/6 strain were compared in an acute OVA-induced AAI model system. ADAM8−/− mice had increased BAL total leukocytes counts (due mainly to increased BAL macrophage counts), increased peribronchial infiltrates in lung sections, and increased AHR to methacholine challenge during OVA-induced AAI [71]. Although the levels of candidate substrates for the ADAM8 MP domain have not yet been measured in the mice, these data provide additional evidence that ADAM8 has potent anti-inflammatory activities during AAI in mice. In contrast, ADAM33, the other ADAM family member that has been linked to asthma, is not upregulated in the lungs of mice with AAI [57]. In addition, studies of ADAM33−/− mice in a murine model of acute allergen-induced AAI showed that ADAM33 plays no role in regulating AAI and AHR in mice. Since ADAM33 is predominantly a mesenchymal cell product and its catalytic domain promotes angiogenesis in vitro [72], it has been proposed that ADAM33 contributes mainly to airway remodeling in asthma [17,27]. However, this possibility has not yet been tested in vivo.
4. Potential activities of ADAM8 in asthma
Based upon experimental data from murine models systems and the known activities of ADAM8 MP domain in vitro (Table 1) we suggest that ADAM8 plays significant roles in limiting airway inflammation and mucus hyper-secretion but has more limited activities in structural airway remodeling in asthmatic airways. We predict that ADAM8 expressed by activated leukocytes and/or bronchial epithelial cells has critical activities in limiting airway inflammation and mucus hyper-secretion but has limited activities in other structural airway remodeling processes occurring in asthmatic airways as outlined below.
4.1. Potential activities for ADAM8 in reducing AAI
Experimental data from ADAM8 transgenic and ADAM8−/− mice indicate that ADAM8 reduces AAI and AHR in acute allergen challenge models [70,71] but there are some caveats when generating hypotheses from the results of these studies. First, it is not clear how relevant the activities of circulating soluble ADAM8 ectodomain present in the ADAM8 transgenic mice are to physiological and pathological processes occurring in asthmatic airways given that ADAM8 is thought to function as a transmembrane proteinase and signaling through its cytoplasmic tail may contribute in critical ways to ADAM8’s function in airway leukocytes and bronchial epithelial cells. While ADAM8 is shed from the surface of human PMNs [48] and sADAM8 has been detected in human plasma [40], the levels are low (267 pg/ml) and plasma levels of sADAM8 were not measured in the ADAM8-transgenic mice. Thus, it is not clear whether plasma sADAM8 levels in the transgenic mice are similar to those in human plasma especially since ADAM8 is mainly expressed by leukocytes and epithelial cells, which probably produce less sADAM8 than hepatocytes in the ADAM8 transgenic mice. Nevertheless, a recent study of ADAM8−/− mice provided additional evidence that ADAM8 limits OVA-induced AAI [71]. Second, the ADAM8 transgenic and ADAM8−/− mice studied thus far were both in TH1-skewed strains that have reduced recruitment of eosinophils and TH2 lymphocytes during AAI compared with TH2-skewed strains of mice. In order to determine whether ADAM8 regulates eosinophil and TH2 lymphocyte recruitment, studies of ADAM8−/− mice in a pure TH2-skewed strain (such as the BALB/c stain) are needed.
Although studies of ADAM8 transgenic and ADAM8−/− mice have not yet dissected the mechanisms by which ADAM8 reduces AAI in mice, based upon ADAM8’s known activities in vitro, we speculate that ADAM8’s anti-inflammatory activities in AAI are due to ADAM8 MP domain shedding adhesion molecules, cytokines and/or receptors from the surface of leukocytes, bronchial epithelial cells, and/or endothelial cell as outlined below.
4.1.1. Shedding of adhesion molecules
Most probably, ADAM8 limits AAI by shedding adhesion molecules (Figure 2A) since ADAM8 expressed on activated leukocytes cleaves L-selectin and possibly P-selectin glycoprotein ligand-1 (PSGL-1) from leukocyte surfaces [48] and VCAM-1 from endothelial cells during leukocyte-endothelial cell interactions in vitro [70]. Since L-selectin promotes PMN rolling on endothelial cells by binding to its ligands on endothelial cells during PMN transendothelial migration, ADAM8-mediated L-selectin shedding may also limit excessive or inappropriate recruitment of PMNs and other leukocytes expressing both ADAM8 and L-selectin into asthmatic airways. L-selectin may be an important substrate for leukocyte-derived ADAM8 in vivo since L-selectin levels were elevated and correlated positively with ADAM8 levels in synovial fluid from patients with rheumatoid arthritis and in lung samples from patients with eosinophilic pneumonias and mild asthma [48,58,59]. In addition, when compared with non-transgenic mice with peritonitis, ADAM8 transgenic mice with peritonitis had reduced surface L-selectin levels on peripheral blood PMNs and peritoneal exudate PMNs, which were associated with reduced peritoneal PMN counts [69]. ADAM8 also cleaves a PSGL-1-like peptide in vitro [35] and PSGL-1 binds to CD34 on endothelial cells to promote leukocyte transendothelial migration. Thus, ADAM8 expressed by leukocytes may shed L-selectin and/or PSGL-1 from leukocyte surfaces to limit the influx of leukocyte subsets that express both ADAM8 and these adhesion molecules into the airways of subjects with asthma (Figure 2A).
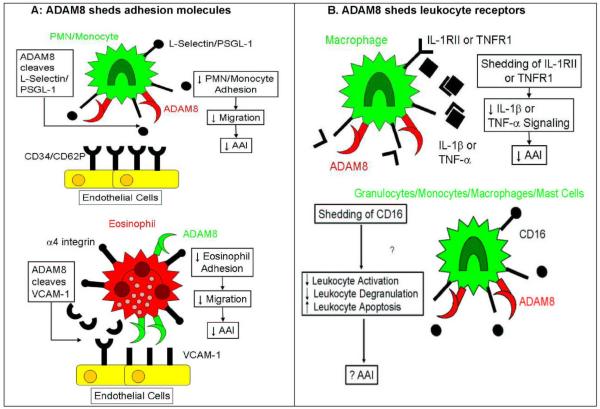
Figure 2. Schematic representation of the potential mechanism(s) by which ADAM8 may limit allergic airway inflammation (AAI).
Potential Mechanisms by which ADAM8 Reduces Allergic Airway Inflammation
In A (top panel), L-selectin and P-selectin glycoprotein ligand-1 (PSGL-1) on polymorphonuclear neutrophil (PMNs) and monocytes bind to their ligands (CD62P and CD34, respectively) on endothelial cells to promote PMN and monocyte rolling on endothelial cells which is the initial step in the transendothelial migration of these leukocytes. ADAM8 expressed on the surface of activated leukocytes in the vasculature of asthmatic subjects may shed L-selectin and possibly PSGL-1 from leukocyte surfaces to reduce the egress of PMNs and monocytes from the vasculature into the airways. In A (bottom panel), activated eosinophils express α4 integrin which binds to vascualar cell adhesion molecule-1 (VCAM-1) expressed on activated endothelial cells leading to the formation of podosome-like adhesive structures in which ADAM8 is localized on the surface of eosinophils. Eosinophil-derived ADAM8 may shed VCAM-1 from endothelial surfaces to reduce α4 integrin-VCAM-1-mediated eosinophil-endothelial-cell adhesion which may limit the migration of eosinophils into asthmatic airways. In B (top panel), ADAM8 expressed on activated leukocytes or bronchial epithelial cells in asthmatic subjects may cleave IL-1 RII and TNFR1 from leukocyte or epithelial surfaces which may decrease the pro-inflammatory signaling of IL-1β or TNF-α through their receptors in asthmatic airways. In B (bottom panel), ADAM8 may shed CD16 from the surfaces of various leukocytes. CD16 is a low-affinity IgG receptor and binding of IgG to this receptor promotes leukocyte activation, degranulation and survival. ADAM8 expressed on the surface of activated leukocytes in asthmatic airways may shed CD16 to reduce leukocyte activation and decrease leukocyte survival, which may reduce inflammation in asthmatic airways.
ADAM8 that is expressed on the surface of activated eosinophils may inhibit eosinophil recruitment into the airways during AAI by shedding VCAM-1. ADAM8 cleaves VCAM-1-like peptides in vitro [35] and serum from sADAM8 transgenic mice has increased capacity to shed VCAM-1 from activated endothelial cells in vitro [70]. Although the latter activity was modest in magnitude, more efficient VCAM-1 shedding may occur when high levels of membrane-associated ADAM8 are concentrated in the sequestered microenvironments of the podosomes of activated eosinophils attached to lung capillaries [56]. Since binding of α4 integrin expressed by eosinophils to VCAM-1 expressed by endothelial cells is a critical event during eosinophil transendothelial migration [54,73], shedding of VCAM-1 by ADAM8 expressed on the surface of eosinophils bound to endothelial cells may limit airway TH2 inflammation during AAI (Figure 2A).
4.1.2 Shedding of cytokines and their receptors
ADAM8 cleaves a number of peptides having sequences in cytokines and cytokine receptors that are cleaved by other proteinases (Table 1). ADAM8 cleaves a proTNF-α-like peptide in vitro [33,35] suggesting that ADAM8 expressed on activated macrophages or bronchial epithelial cells in asthmatic airways may shed and activate pro-TNF-α from the surfaces of these cells. However, ADAM17 is thought to be the main pro-TNF-α-activating enzyme in vivo [37]. If ADAM8 is confirmed to play important roles in the shedding of TNF receptor 1 (TNFR1 [35]), this might limit AAI by reducing the pro-inflammatory signaling of TNF-α in leukocytes and/or bronchial epithelial cells human subjects with asthma ([74] and Figure 2B). For example, if ADAM8 expressed by activated bronchial epithelial cells sheds TNFR1 from epithelial cells, this may reduce TNF-α-induced upregulation of epithelial ICAM-1 expression [75]. This, in turn, may reduce leukocyte adhesion to bronchial epithelial cells to limit leukocyte-mediated epithelial cell injury and loss of bronchial epithelial cells in asthmatic airways. IL-1β has pro-inflammatory activities during AAI in mice [76] and ADAM8 cleaves a peptide sequence occurring in one of the two IL1 receptors (IL-1 receptor II, IL-1 RII) in vitro [35]. IL-1 RII is a non-signaling decoy receptor which has a short cytoplasmic tail lacking the signaling Toll-IL-1R domain. IL-1RII is proteolytically shed from the surface of leukocytes and epithelial cells by other MPs and sIL-1RII acts as a decoy receptor that sequesters soluble IL-1β and reduces its binding to and signaling through the functional type I IL-1R [77]. If this receptor is confirmed to be an important substrate for ADAM8 in vivo, ADAM8 expressed by activated leukocytes and/or epithelial cells in asthmatic airways may shed IL-1RII from the surfaces of these cells to limit AAI and AHR (Figure 2B).
ADAM8 also cleaves a fractalkine-like peptide in vitro [35]. Fractalkine (CX3CL) is produced as a membrane-bound protein on endothelial, epithelial, and SMCs which promotes adhesion of leukocytes to these cells [78]. ADAM-10 and −17 shed CX3CL from these cells generating sCX3CL which binds to CX3CR1 on monocytes, T cells, and DCs to increase their migration [78,79]. Both forms of CX3CL are upregulated in asthmatic subjects [80]. Thus, if ADAM8 expressed on the apical surface of activated bronchial epithelial cells in asthmatic airways sheds CX3CL from the epithelial cell surface, this may increase the trafficking of leukocytes into asthmatic airways. Alternatively, ADAM8-mediated shedding of CX3CL from epithelial surfaces may reduce adhesion of leukocytes to bronchial epithelial cells and limit leukocyte-mediated epithelial cell injury and loss in asthmatic airways.
Stem cell factor (SCF, kit ligand) is produced by bronchial epithelial cells [81], eosinophils [82], alveolar macrophages [83], mast cells [83], and PMNs [84]. SCF is initially produced as a membrane-associated protein and its expression is increased in epithelial cells and macrophages in the airways of asthmatic subjects [83,85]. Mast cell chymase and unidentified MPs cleave membrane-associated SCF generating soluble SCF [86,87]. Both forms of SCF are active cytokines that bind to c-kit receptor on mast cells to promote mast cell chemotaxis, activation, degranulation and survival, and mast-cell-induced AHR [88,89]. Although ADAM8 cleaves a SCF-like peptide in vitro [35], it is not clear whether SCF is cleaved from the surface of ADAM8 expressed by activated leukocytes or bronchial epithelial cells in the airways of asthmatic subjects. It is also not clear whether ADAM8-mediated cleavage of SCF alters the biological activity of SCF.
4.1.3. Shedding of leukocyte immunoglobulin receptors
ADAM8 cleaves a CD16-like peptide in vitro [35] but it is not clear whether ADAM8 sheds CD16 from leukocyte surfaces. CD16 is a low-affinity receptor for IgG which is expressed by various leukocytes including granulocytes, mononuclear phagocytes, and mast cells [90]. IgG binding to CD16 promotes leukocyte degranulation and release of cytokines, enzymes, and other mediators. Neutrophil elastase [91] and unidentified ADAM proteinase(s) shed CD16 from leukocyte surfaces, and CD16 shedding is associated with increased PMN apoptosis [92]. While CD16 expression is increased on circulating and tissue leukocytes of asthmatic patients compared with healthy subjects [93,94], surface CD16 levels are reduced on alveolar macrophages after allergen challenge [95]. This suggests that significant CD16 shedding occurs from macrophage surfaces during AAI. If ADAM8 that is expressed on activated leukocytes in asthmatic airways induces significant shedding of CD16 from leukocyte surfaces, this may limit airway inflammation in subjects with asthma by inhibiting leukocyte degranulation and release of pro-inflammatory mediators and/or by reducing the survival of airway granulocytes (Figure 2B).
CD23 is a low affinity receptor for IgE which is expressed on B cells, monocytes, and macrophages. Soluble forms of CD23 (sCD23) are shed from cells by several ADAMs including ADAM8 in vitro [96]. Patients with asthma and atopy have increased leukocyte surface CD23 levels and increased sCD23 levels in plasma [97-99]. In addition, ADAM8 levels in lung samples correlate positively with lung sCD23 levels in patients with mild asthma and eosinophilic pneumonias [58,59]. However, it is not clear whether leukocyte-derived ADAM8 is a significant CD23 sheddase since CD23 shedding has not been measured in leukocytes isolated from WT versus ADAM8−/− mice. The effect of ADAM8-mediated shedding of CD23 during AAI is unclear. Genetic deletion or transgenic overexpression of CD23 increases or decreases serum IgE levels and AAI and AHR in mice, respectively [100,101]. However, at the cellular level, the function(s) of CD23 may depend upon the form that is expressed. For example, although membrane-bound CD23 on B cells increases B cell IgE-dependent antigen presentation to T cells [102], IgE binding to membrane-bound CD23 on B cells downregulates further IgE production. In addition, the binding of sCD23 to CD21 on B cells increases their IgE production [103,104] and sCD23 binding to CD11b/CD18, CD11c/CD18, and αv integrins on monocytes increase their production of pro-inflammatory cytokines [105]. Thus, shedding of leukocyte CD23 by ADAM8 expressed on the surface of activated leukocytes in asthmatic airways may increase rather than decrease airway inflammation.
4.2 Potential of ADAM8 to regulate airway remodeling in asthma
The activities of ADAM8 in airway remodeling are not clear since ADAM8 transgenic or deficient mice have not been studied in murine models of allergen-induced airway remodeling. Based upon ADAM8’s expression profile (it is expressed by leukocytes and bronchial epithelial cells but not by mesenchymal cells) and its activities in vitro, we suggest that, unlike ADAM33, ADAM8 does not play a major role in directly regulating structural remodeling in asthmatic airways. However, by reducing airway inflammation ADAM8 may reduce mucus hyper-secretion indirectly in the airways of subjects with asthma (Figure 3).
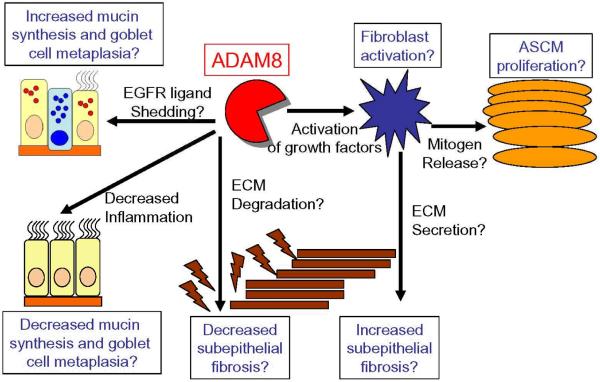
Figure 3. Schematic representation of the potential mechanisms by which ADAM8 may regulate airway remodeling in asthma.
Potential Activities of ADAM8 in Remodeling of Asthmatic Airways
We currently have no data linking ADAM8 to airway remodeling in asthma. Based upon its anti-inflammatory activities in murine models of allergic airway inflammation (AAI), we speculate that ADAM8 may indirectly reduce mucin gene expression and goblet cell metaplasia by decreasing the airway burden of inflammatory cells and their mucin-inducing proteinases. The growth factor-activating activities of ADAM8 have not yet been tested. However, if the metalloproteinase domain of ADAM8 that is expressed on the surface of activated leukocytes or bronchial epithelial cells in asthmatic airways plays a significant role in activating TGF-β family members, IGFs, or ligands for the EGFR, this would promote fibroblast activation and deposition of extracellular matrix (ECM) proteins to increase subepithelial fibrosis and fibroblast secretion of mitogens that induce airway smooth muscle cells (ASMC) proliferation. If ADAM8 that is expressed by activated bronchial epithelial cells sheds EGFR ligands from epithelial cell surfaces, this may also promote mucin secretion and goblet cell metaplasia in the airway epithelium of subjects with asthma. Alternatively, ADAM8 that is expressed by activated airway leukocytes or activated bronchial epithelial cells in asthmatic airways may reduce subepithelial fibrosis by degrading ECM proteins deposited by airway myofibroblasts.
4.2.1. Mucus hyper-secretion
Patients with asthma have goblet cell metaplasia and increased mucus secretion, and airway plugging by mucus contributes to asthma mortality [106]. TH2 cytokines and other stimuli induce mucus hyper-secretion not only by increasing EGFR expression on airway epithelial cells but also by inducing shedding of membrane-associated ligands for this receptor, generating the mature active forms of these ligands which activate the EGFR [107]. Neutrophil elastase, MMP-9, ADAM10, and ADAM17 directly or indirectly shed membrane-associated ligands for the EGFR from lung epithelial cells to increase mucin gene transcription and promote goblet cell metaplasia [108-111]. Although the EGFR-ligand-shedding activities of epithelial-derived ADAM8 have not been studied, ADAM-10 and −17 are thought to be the main EGFR-ligand sheddases contributing to increased mucus production in airway epithelium [112]. Instead, ADAM8 may reduce airway mucus hyper-secretion by limiting AAI and reducing the airway burden of other leukocyte proteinases that increase mucus production (Figure 3).
4.2.2. Myofibroblast activation, subepithelial fibrosis, and airway SMC hyperplasia
In patients with chronic asthma, there is increased production of several growth factors that drive differentiation of fibroblasts into myofibroblasts. Activated myofibroblasts secrete ECM proteins such as interstitial collagens and fibronectin and also release factors such as vascular endothelial growth factor and endothelin I which are mitogens for airway SMCs [113,114]. The MP domain of other ADAMs and metalloproteinases cleave and thereby activate latent growth factors [115,116], alter biological activity of cytokines that control fibroblast survival [117-119], or degrade ECM proteins produced by myofibroblasts [120-122]. Since ADAM8 has an active MP domain, epithelial-derived ADAM8 may also promote these processes as outlined below and as illustrated in Figure 3.
The activity of growth factors is regulated by proteinases. TGF-β1-3, which are important growth factors in asthmatic airways, are produced as large latent complexes in which a latency-associated peptide (LAP) is bound to a latent TGF-β-binding protein. These large latent complexes bind to ECM proteins. Neutrophil elastase and plasmin release complexes containing latent TGF-β from these ECM stores [123,124] and MMPs cleave LAP to activate TGF-β [115,116]. While the activities of ADAMs in activation of TGF-β have not been studied, ADAM proteinases activate IGFs, which also contribute to airway remodeling in asthma [125]. IGFs circulate in an inactive form bound to circulating IGF binding proteins (IGFPBs) which prevent IGFs from binding to their receptors on fibroblasts and airway SMCs [126]. ADAM9, ADAM12, ADAM28, and several MMPs degrade IGFBP thereby releasing active IGFs that can signal through their receptors on myofibroblasts to increase collagen deposition in tissues [117-119] and degraded IGFBP have been detected in asthmatic airways [125]. However, ADAM8’s activities as an IGFBP-degrading enzyme have not been assessed. The MP domain of ADAM8 expressed by activated bronchial epithelial cells or leukocytes in asthmatic airways could also increase remodeling of asthmatic airways by shedding IL-1RII from the surface of epithelial cells or leukocytes [35] which may bind and sequester soluble IL-1β and thereby reduce IL-1β-mediated induction of fibroblast apoptosis [127]. Thus, if ADAM8 is confirmed to be a key IL1 RII sheddase, it could increase the survival of fibroblasts in asthmatic airways and promote collagen deposition in the basement membrane. Alternatively, ADAM8 expressed by activated leukocytes or epithelial cells may limit subepithelial fibrosis in asthmatic airways by the MP domain of the proteinase degrading ECM proteins deposited by activated (myo)fibroblasts (Figure 3). By altering the ECM composition of asthmatic airways, ADAM8 MP domain may also reduce the survival and differentiation of airway SMCs which require specific ECM components for these processes [128]. However, it is noteworthy that while the ECM-degrading activity of ADAM8 has not been assessed, most ADAMs studied thus far have limited capacity to degrade ECM components, especially interstitial collagens in vitro [29].
5. Summary and conclusions
Surprisingly little is known about the contributions of ADAM proteinases to pathologies occurring in the airways of asthmatic patients. ADAM33 was the first member of the ADAM family of MPs to be linked to asthma and AHR [26]. While ADAM33 plays no role in regulating allergen-induced AAI or AHR in mice [28], we and other have proposed that ADAM33 regulates airway remodeling processes in asthmatic airways [17,27,72]. More recently, ADAM8 has been linked to asthma since it is impressively upregulated by TH2 cytokines in the lungs of mice and its expression is increased in the inflammatory cell infiltrates and airway epithelium of humans [49,129] and mice [57] with AAI and in BALF from patients with some eosinophilic pneumonias [59]. Although ADAM8 levels are increased during AAI in mice and humans, the role of ADAM8 in this disease is to dampen airway inflammation, since studies of OVA-sensitized and -challenged ADAM8 transgenic and ADAM8−/− mice show that this proteinase limits accumulation of leukocytes in the airways [70,71]. Although the mechanism(s) by which ADAM8 exerts its anti-inflammatory activities have not yet been dissected, we speculate that the MP domain of ADAM8 sheds adhesion molecules such as VCAM-1 and L-selectin and/or leukocyte surface cytokine receptors (TNFR1 or IL-1RII) and/or CD16, all of which play important roles in leukocyte recruitment and activation in asthmatic airways. However, this needs to be confirmed by measuring the shedding of these molecules in transgenic mice over-expressing ADAM8 or in ADAM8−/− mice versus control mice with allergen-induced AAI.
There have been no studies of transgenic mice overexpressing ADAM8 or ADAM8−/− mice in murine models of allergen-induced airway remodeling. Thus, it is not clear whether ADAM8 plays a significant role in regulating airway remodeling processes in asthma. However, based upon its expression profile and the activities of ADAM8 MP domain in vitro, we predict that overall, ADAM8 would play a minimal role or no role in directly regulating subepithelial fibrosis or airway SMC hyperplasia. However, by dampening airway inflammation, ADAM8 may reduce mucus hyper-secretion indirectly by limiting the airway burden of other inflammatory cell proteinases known to promote mucus hyper-secretion. Additional studies of transgenic mice overexpressing ADAM8 or ADAM8−/− mice in models of acute and chronic allergen-induced airway inflammation and remodeling are needed to determine the activities of ADAM8 in these pathologies, to identify the critical cellular sources of ADAM8 (inflammatory cells versus bronchial epithelial cells), and to dissect the mechanism involved.
6. Expert opinion
There is an urgent need to develop new treatment strategies that more effectively reduce inflammation in asthmatic airways since there are many asthmatics with chronic symptoms that are refractory to current anti-inflammatory therapies and account for up to 50% of the health care costs of this disease [130]. In addition, current asthma treatments are ineffective at preventing and reversing the chronic remodeling processes in asthmatic airways, which are linked to AHR and a steeper decline in lung function in asthmatic patients.
Most previous studies of asthma pathogenesis have focused on pro-inflammatory molecules and pathways. There have been relatively few studies examining counter-regulatory anti-inflammatory molecules and pathways in asthma. Lipid mediators that inhibit inflammation or promote resolution of inflammation have recently been found to be reduced in patients with asthma and can limit AAI when administered to mice with AAI [131-134]. Thus, anti-inflammatory pathways are an emerging area for designing novel treatment strategies for asthma. Based upon experimental data in mice, ADAM8 is an important anti-inflammatory protein in AAI [70,71] and acute peritonitis [69] but the contributions of ADAM8 to airway remodeling processes in asthma have not been assessed.
Herein, we have speculated that ADAM8 limits AAI by its MP domain shedding adhesion molecules and/or cytokine receptors and IgG receptors to limit AAI. This hypothesis is based upon the results of studies examining the activities of ADAM8 MP domain in vitro and the results of correlative studies of ADAM8 levels and the levels of shed candidate substrates in lung samples from humans and mice with AAI. There are several important caveats when drawing conclusions from the results of these studies. First, other ADAMs and MMPs are increased in asthmatic airways, which can also shed these surface molecules [29,58]. Second, our conclusions about important substrates for the ADAM8 MP domain in vivo have mainly been based upon the activities of recombinant ADAM8 against peptide sequences containing sites known to be cleaved in intact surface proteins by other sheddases (see Table 1). Thus, it is not clear whether intact proteins that have secondary and/or tertiary structure are cleaved by the transmembrane form of ADAM8 expressed on cell surface and, if so, whether this increases or decreases the biological activity of the products generated. Third, as we have learned from studying MMPs, substrates cleaved efficiently by MMPs in vitro were often found not to be important substrates for MMPs in vivo, as assessed by subsequent studies of MMP-gene-targeted mice in murine models of disease. MMPs were initially thought to function as ECM-degrading enzymes, but are now know to play more prominent roles in regulating inflammation and immunity in vivo [135]. Thus, it is not clear whether substrates for ADAM8 identified in vitro will later be confirmed to be important substrates for ADAM8 MP domain in vivo. Fourth, it is possible that ADAM8 domains other than the MP domain are involved in ADAM8’s anti-inflammatory activities. The disintegrin domain could bind to leukocyte integrins to inhibit leukocyte-endothelial cell adhesion since ADAM8 disintegrin binds to α9β1 integrin on osteoclasts [38,39] and this integrin is also expressed on leukocytes and promotes leukocyte transendothelial migration [136]. Future studies should also investigate whether the SH3 domain binding sequences in the cytoplasmic tail of ADAM8 bind to and regulate the activity of SH3-domain-containing proteins such as Src family kinases, which play critical roles in regulating leukocyte function [137]. Before ADAM8 can be validated as a new target for asthma, we need more information about the mechanisms by which ADAM8 limits AAI, the critical cellular sources of ADAM8 in the lung, the target cells upon which ADAM8 acts, the ADAM8 domains(s) that contribute to its anti-inflammatory activities, and its activities (if any) in structural remodeling in asthmatic airways. This might be best achieved by additional studies of ADAM8−/− and transgenic mice overexpressing ADAM8 in lung epithelium or leukocytes in strains that develop robust TH2 inflammation (such as the BALB/c strain) in acute and chronic allergen exposure models. We also need to address whether, as demonstrated for ADAM33 [26,138,139], there are SNPs in the ADAM8 gene that are associated with asthma. If so, it will also be critically important to assess whether SNPs in the ADAM8 gene limit its expression or reduce its functioning and thereby predispose individuals to develop asthma.
Although ADAM8 has not yet been tested as a new therapeutic target in mice with AAI, Schluesener used ADAM8 disintegrin domain as an adjuvant by fusing it to a polyvalent autoantigen vaccine and this modified vaccine reduced disease severity in rats with experimental autoimmune encephalitis [140]. These results suggest that ADAM8 disintegrin domain can inhibit immune reactions to antigens in vivo. If future studies confirm that ADAM8 is a promising new therapeutic target for asthma, approaches to increase its expression, translocation to cell surfaces, and/or retention in an active form on cell surfaces might be effective strategies in limiting airway inflammation in subjects with asthma. However, we need more information about the factors that control ADAM8 transcription, trafficking in leukocytes and epithelial cells, and its half life on cell surfaces. We also need more information on the physiological activities of ADAM8 in leukocytes, including whether ADAM8 plays roles in host defense against infection. We also need to determine whether ADAM8 has important functions in other organ systems, especially bone, since: (1) ADAM8 is expressed by osteoclasts and promotes osteoclast differentiation in vitro [38,39]; and (2) ADAM8 levels are increased in the joints of patients with rheumatoid arthritis and levels correlate positively with joint destruction [38,48]. ADAM8 is also expressed by several tumors and its expression correlates with invasiveness, metastasis, and poor prognosis (Table 3). Thus, the roles of ADAM8 in leukocyte, osteoclast, and tumor cell biology need to be clarified before strategies to increase the ADAM8 levels of activity in vivo can be validated as a new and safe target for asthma.
Acknowledgements
The authors were supported by grants RO1 HL063137, RO1 HL086814, T32 HL007633, and F32 HL095276 from the National Heart, Lung and Blood Institute of the National Institutes of Health and grant #052349 from the Flight Attendants Medical Research Institute.
Abbreviations
- AAI
allergic airway inflammation
- ADAM
a disintegrin and a metalloproteinase
- AEP
acute eosinophilic pneumonia
- AHR
airway hyper-responsiveness
- BAL
bronchoalveolar lavage
- CD
cluster of differentiation
- CEP
chronic eosinophilic pneumonia
- CR
cysteine rich
- DCs
dendritic cells
- ECM
extracellular matrix
- EMTU
epithelial mesenchymal trophic unit
- IGFBP
insulin-like growth factor binding protein
- IRF
interferon-response element
- LAP
latency associated peptide
- LPS
lipopolysaccharide
- MP
metalloproteinase
- MMP
matrix metalloproteinase
- NF-1
nuclear factor-1
- OVA
ovalbumin
- PMN
polymorphonuclear neutrophil
- PSGL-1
P-selectin glycoprotein ligand-1
- sADAM8
soluble ADAM8
- sCD23
soluble CD23
- SCF
stem cell factor
- SH3
Src homology 3
- SMC
smooth muscle cell
- SNP
single-nucleotide polymorphisms
- STAT-6
signal transducer and activator of transcription-6
- TNFR1
TNF receptor-1
- VCAM-1
vascular adhesion molecule-1
- WT
wild-type
Bibliography
- 1.Masoli M, Fabian D, Holt S, Beasley R. The global burden of asthma: executive summary of the GINA Dissemination Committee report. Allergy. 2004;59:469–78. doi: 10.1111/j.1398-9995.2004.00526.x. [DOI] [PubMed] [Google Scholar]
- 2.Cherry DK, Hing E, Woodwell DA, Rechtsteiner EA. National ambulatory medical care survey summary: 2006. National health statistics reports. 2008;6:1–39. [PubMed] [Google Scholar]
- 3.National Heart Lung, and Blood Institute . Morbidity and Mortality: Chart Book on Cardiovascular, Lung, and Blood Diseases. US Department of Health and Human Services; Bethesda, MD: 2002. [Google Scholar]
- 4.Barnes PJ. The cytokine network in asthma and chronic obstructive pulmonary disease. J Clin Invest. 2008;118:3546–56. doi: 10.1172/JCI36130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akbari O, Stock P, Meyer E, et al. Essential role of NKT cells producing IL-4 and IL-13 in the development of allergen-induced airway hyperreactivity. Nat Med. 2003;9:582–88. doi: 10.1038/nm851. [DOI] [PubMed] [Google Scholar]
- 6.McKinley L, Alcorn JF, Peterson A, et al. TH17 cells mediate steroid-resistant airway inflammation and airway hyperresponsiveness in mice. J Immunol. 2008;181:4089–97. doi: 10.4049/jimmunol.181.6.4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akbari O, Stock P, DeKruyff RH, Umetsu DT. Role of regulatory T cells in allergy and asthma. Curr Opin Immunol. 2003;15:627–33. doi: 10.1016/j.coi.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Lambrecht BN, Hammad H. Lung dendritic cells: targets for therapy in allergic disease. Chem Immunol Allergy. 2008;94:189–200. doi: 10.1159/000155087. [DOI] [PubMed] [Google Scholar]
- 9.Peters-Golden M. The alveolar macrophage: the forgotten cell in asthma. Am J Respir Cell Mol Biol. 2004;31:3–7. doi: 10.1165/rcmb.f279. [DOI] [PubMed] [Google Scholar]
- 10.Linden A, Hoshino H, Laan M. Airway neutrophils and interleukin-17. Eur Respir J. 2000;15:973–77. doi: 10.1034/j.1399-3003.2000.15e28.x. [DOI] [PubMed] [Google Scholar]
- 11.Ordonez CL, Khashayar R, Wong HH, et al. Mild and moderate asthma is associated with airway goblet cell hyperplasia and abnormalities in mucin gene expression. Am J Respir Crit Care Med. 2001;163:517–23. doi: 10.1164/ajrccm.163.2.2004039. [DOI] [PubMed] [Google Scholar]
- 12.Johnson PR, Roth M, Tamm M, et al. Airway smooth muscle cell proliferation is increased in asthma. Am J Respir Crit Care Med. 2001;164:474–77. doi: 10.1164/ajrccm.164.3.2010109. [DOI] [PubMed] [Google Scholar]
- 13.Brewster CE, Howarth PH, Djukanovic R, et al. Myofibroblasts and subepithelial fibrosis in bronchial asthma. Am J Respir Cell Mol Biol. 1990;3:507–11. doi: 10.1165/ajrcmb/3.5.507. [DOI] [PubMed] [Google Scholar]
- 14.Payne DN, Rogers AV, Adelroth E, et al. Early thickening of the reticular basement membrane in children with difficult asthma. Am J Respir Crit Care Med. 2003;167:78–82. doi: 10.1164/rccm.200205-414OC. [DOI] [PubMed] [Google Scholar]
- 15.Pohunek P, Warner JO, Turzikova J, et al. Markers of eosinophilic inflammation and tissue re-modelling in children before clinically diagnosed bronchial asthma. Pediatr Allergy Immunol. 2005;16:43–51. doi: 10.1111/j.1399-3038.2005.00239.x. [DOI] [PubMed] [Google Scholar]
- 16.Bucchieri F, Puddicombe SM, Lordan JL, et al. Asthmatic bronchial epithelium is more susceptible to oxidant-induced apoptosis. Am J Respir Cell Mol Biol. 2002;27:179–85. doi: 10.1165/ajrcmb.27.2.4699. [DOI] [PubMed] [Google Scholar]
- 17.Holgate ST, Davies DE, Powell RM, Holloway JW. ADAM33: a newly identified protease involved in airway remodelling. Pulm Pharmacol Ther. 2006;19:3–11. doi: 10.1016/j.pupt.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 18.Puddicombe SM, Polosa R, Richter A, et al. Involvement of the epidermal growth factor receptor in epithelial repair in asthma. FASEB J. 2000;14:1362–74. doi: 10.1096/fj.14.10.1362. [DOI] [PubMed] [Google Scholar]
- 19.Nadel JA. Role of mast cell and neutrophil proteases in airway secretion. Am Rev Respir Dis. 1991;144:S48–S51. doi: 10.1164/ajrccm/144.3_pt_2.S48. [DOI] [PubMed] [Google Scholar]
- 20.Corry DB, Rishi K, Kanellis J, et al. Decreased allergic lung inflammatory cell egression and increased susceptibility to asphyxiation in MMP2-deficiency. Nat Immunol. 2002;3:347–53. doi: 10.1038/ni773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMillan SJ, Kearley J, Campbell JD, et al. Matrix metalloproteinase-9 deficiency results in enhanced allergen-induced airway inflammation. J Immunol. 2004;172:2586–94. doi: 10.4049/jimmunol.172.4.2586. [DOI] [PubMed] [Google Scholar]
- 22.Gueders MM, Balbin M, Rocks N, et al. Matrix metalloproteinase-8 deficiency promotes granulocytic allergen-induced airway inflammation. J Immunol. 2005;175:2589–97. doi: 10.4049/jimmunol.175.4.2589. [DOI] [PubMed] [Google Scholar]
- 23.Lim DH, Cho JY, Miller M, et al. Reduced peribronchial fibrosis in allergen-challenged MMP-9-deficient mice. Am J Physiol Lung Cell Mol Physiol. 2006;291:L265–L271. doi: 10.1152/ajplung.00305.2005. [DOI] [PubMed] [Google Scholar]
- 24.Johnson S, Knox A. Autocrine production of matrix metalloproteinase-2 is required for human airway smooth muscle proliferation. Am J Physiol. 1999;277:L1109–L1117. doi: 10.1152/ajplung.1999.277.6.L1109. [DOI] [PubMed] [Google Scholar]
- 25.Vlahos R, Lee KS, Guida E, et al. Differential inhibition of thrombin- and EGF-stimulated human cultured airway smooth muscle proliferation by glucocorticoids. Pulm Pharmacol Ther. 2003;16:171–80. doi: 10.1016/S1094-5539(02)00183-9. [DOI] [PubMed] [Google Scholar]
- 26.Van Eerdewegh P, Little RD, Dupuis J, et al. Association of the ADAM33 gene with asthma and bronchial hyperresponsiveness. Nature. 2002;418:426–30. doi: 10.1038/nature00878. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro SD, Owen CA. ADAM-33 surfaces as an asthma gene. N Engl J Med. 2002;347:936–38. doi: 10.1056/NEJMcibr022144. [DOI] [PubMed] [Google Scholar]
- 28.Chen C, Huang X, Sheppard D. ADAM33 is not essential for growth and development and does not modulate allergic asthma in mice. Mol Cell Biol. 2006;26:6950–56. doi: 10.1128/MCB.00646-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owen CA. Leukocyte cell surface proteinases: regulation of expression, functions, and mechanisms of surface localization. Int J Biochem Cell Biol. 2008;40:1246–72. doi: 10.1016/j.biocel.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida S, Setoguchi M, Higuchi Y, et al. Molecular cloning of cDNA encoding MS2 antigen, a novel cell surface antigen strongly expressed in murine monocytic lineage. Int Immunol. 1990;2:585–91. doi: 10.1093/intimm/2.6.585. [DOI] [PubMed] [Google Scholar]
- 31.Yoshiyama K, Higuchi Y, Kataoka M, et al. CD156 (human ADAM8): expression, primary amino acid sequence, and gene location. Genomics. 1997;41:56–62. doi: 10.1006/geno.1997.4607. [DOI] [PubMed] [Google Scholar]
- 32.Kataoka M, Yoshiyama K, Matsuura K, et al. Structure of the murine CD156 gene, characterization of its promoter, and chromosomal location. J Biol Chem. 1997;272:18209–15. doi: 10.1074/jbc.272.29.18209. [DOI] [PubMed] [Google Scholar]
- 33.Amour A, Knight CG, English WR, et al. The enzymatic activity of ADAM8 and ADAM9 is not regulated by TIMPs. FEBS Lett. 2002;524:154–58. doi: 10.1016/s0014-5793(02)03047-8. [DOI] [PubMed] [Google Scholar]
- 34.Schlomann U, Wildeboer D, Webster A, et al. The metalloprotease disintegrin ADAM8. Processing by autocatalysis is required for proteolytic activity and cell adhesion. J Biol Chem. 2002;277:48210–19. doi: 10.1074/jbc.M203355200. [DOI] [PubMed] [Google Scholar]
- 35.Naus S, Reipschlager S, Wildeboer D, et al. Identification of candidate substrates for ectodomain shedding by the metalloprotease-disintegrin ADAM8. Biol Chem. 2006;387:337–46. doi: 10.1515/BC.2006.045. [DOI] [PubMed] [Google Scholar]
- 36.Richens J, Fairclough L, Ghaemmaghami AM, et al. The detection of ADAM8 protein on cells of the human immune system and the demonstration of its expression on peripheral blood B cells, dendritic cells and monocyte subsets. Immunobiology. 2007;212:29–38. doi: 10.1016/j.imbio.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 37.Black RA, Rauch CT, Kozlosky CJ, et al. A metalloproteinase disintegrin that releases tumour-necrosis factor-α from cells. Nature. 1997;385:729–33. doi: 10.1038/385729a0. [DOI] [PubMed] [Google Scholar]
- 38.Ainola M, Li TF, Mandelin J, Hukkanen M, et al. Involvement of ADAM8 in osteoclastogenesis and pathological bone destruction. Ann Rheum Dis. 2008:427–34. doi: 10.1136/ard.2008.088260. [DOI] [PubMed] [Google Scholar]
- 39.Choi SJ, Han JH, Roodman GD. ADAM8: a novel osteoclast stimulating factor. J Bone Miner Res. 2001;16:814–22. doi: 10.1359/jbmr.2001.16.5.814. [DOI] [PubMed] [Google Scholar]
- 40.Ishikawa N, Daigo Y, Yasui W, et al. ADAM8 as a novel serological and histochemical marker for lung cancer. Clin Cancer Res. 2004;10:8363–70. doi: 10.1158/1078-0432.CCR-04-1436. [DOI] [PubMed] [Google Scholar]
- 41.Hall T, Leone JW, Wiese JF, et al. Autoactivation of human ADAM8: A novel pre-processing step is required for catalytic activity. Biosci Rep. 2008 doi: 10.1042/BSR20080145. published online 23 September 2008, doi:10.1042/BSR20080145. [DOI] [PubMed] [Google Scholar]
- 42.Howard L, Nelson KK, Maciewicz RA, Blobel CP. Interaction of the metalloprotease disintegrins MDC9 and MDC15 with two SH3 domain-containing proteins, endophilin I and SH3PX1. J Biol Chem. 1999;274:31693–99. doi: 10.1074/jbc.274.44.31693. [DOI] [PubMed] [Google Scholar]
- 43.Kang Q, Cao Y, Zolkiewska A. Metalloprotease-disintegrin ADAM 12 binds to the SH3 domain of Src and activates Src tyrosine kinase in C2C12 cells. Biochem J. 2000;352(Pt 3):883–892. [PMC free article] [PubMed] [Google Scholar]
- 44.Poghosyan Z, Robbins SM, Houslay MD, et al. Phosphorylation-dependent interactions between ADAM15 cytoplasmic domain and Src family protein-tyrosine kinases. J Biol Chem. 2002;277:4999–5007. doi: 10.1074/jbc.M107430200. [DOI] [PubMed] [Google Scholar]
- 45.Schlomann U, Rathke-Hartlieb S, Yamamoto S, et al. Tumor necrosis factor α induces a metalloprotease-disintegrin, ADAM8 (CD 156): implications for neuron-glia interactions during neurodegeneration. J Neurosci. 2000;20:7964–71. doi: 10.1523/JNEUROSCI.20-21-07964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sriraman V, Eichenlaub-Ritter U, Bartsch JW, et al. Regulated expression of ADAM8 (a disintegrin and metalloprotease domain 8) in the mouse ovary: evidence for a regulatory role of luteinizing hormone, progesterone receptor, and epidermal growth factor-like growth factors. Biol Reprod. 2008;78:1038–48. doi: 10.1095/biolreprod.107.066340. [DOI] [PubMed] [Google Scholar]
- 47.Kelly K, Hutchinson G, Nebenius-Oosthuizen D, et al. Metalloprotease-disintegrin ADAM8: expression analysis and targeted deletion in mice. Dev Dyn. 2005;232:221–31. doi: 10.1002/dvdy.20221. [DOI] [PubMed] [Google Scholar]
- 48.Gomez-Gaviro M, Dominguez-Luis M, Canchado J, et al. Expression and regulation of the metalloproteinase ADAM-8 during human neutrophil pathophysiological activation and its catalytic activity on L-selectin shedding. J Immunol. 2007;178:8053–63. doi: 10.4049/jimmunol.178.12.8053. [DOI] [PubMed] [Google Scholar]
- 49.Foley SC, Mogas AK, Olivenstein R, et al. Increased expression of ADAM33 and ADAM8 with disease progression in asthma. J Allergy Clin Immunol. 2007;119:863–71. doi: 10.1016/j.jaci.2006.12.665. [DOI] [PubMed] [Google Scholar]
- 50.Haitchi HM, Powell RM, Shaw TJ, et al. ADAM33 expression in asthmatic airways and human embryonic lungs. Am J Respir Crit Care Med. 2005;171:958–965. doi: 10.1164/rccm.200409-1251OC. [DOI] [PubMed] [Google Scholar]
- 51.Powell RM, Wicks J, Holloway JW, et al. The splicing and fate of ADAM33 transcripts in primary human airways fibroblasts. Am J Respir Cell Mol Biol. 2004;31:13–21. doi: 10.1165/rcmb.2003-0330OC. [DOI] [PubMed] [Google Scholar]
- 52.Yang Y, Haitchi HM, Cakebread J, et al. Epigenetic mechanisms silence a disintegrin and metalloprotease 33 expression in bronchial epithelial cells. J Allergy Clin Immunol. 2008;121:1393–99. doi: 10.1016/j.jaci.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 53.Dijkstra A, Postma DS, Noordhoek JA, et al. Expression of ADAMs (“a disintegrin and metalloprotease”) in the human lung. Virchows Arch. 2009 doi: 10.1007/s00428-009-0748-4. published online 3 March 2009, doi: 10.1007/s00428-009-0748-4. [DOI] [PubMed] [Google Scholar]
- 54.Rose DM, Han J, Ginsberg MH. α4 integrins and the immune response. Immunol Rev. 2002;186:118–24. doi: 10.1034/j.1600-065x.2002.18611.x. [DOI] [PubMed] [Google Scholar]
- 55.Adams JC. Regulation of protrusive and contractile cell-matrix contacts. J Cell Sci. 2002;115:257–65. doi: 10.1242/jcs.115.2.257. [DOI] [PubMed] [Google Scholar]
- 56.Johansson MW, Lye MH, Barthel SR, et al. Eosinophils adhere to vascular cell adhesion molecule-1 via podosomes. Am J Respir Cell Mol Biol. 2004;31:413–22. doi: 10.1165/rcmb.2004-0099OC. [DOI] [PubMed] [Google Scholar]
- 57.King NE, Zimmermann N, Pope SM, et al. Expression and regulation of a disintegrin and metalloproteinase (ADAM) 8 in experimental asthma. Am J Respir Cell Mol Biol. 2004;31:257–65. doi: 10.1165/rcmb.2004-0026OC. [DOI] [PubMed] [Google Scholar]
- 58.Paulissen G, Rocks N, Quesada-Calvo F, et al. Expression of ADAMs and their inhibitors in sputum from patients with asthma. Mol Med. 2006;12:171–79. doi: 10.2119/2006-00028.Paulissen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsuno O, Miyazaki E, Nureki S, et al. Elevated soluble ADAM8 in bronchoalveolar lavage fluid in patients with eosinophilic pneumonia. Int Arch Allergy Immunol. 2007;142:285–90. doi: 10.1159/000097359. [DOI] [PubMed] [Google Scholar]
- 60.Matsuno O, Kumamoto T, Higuchi Y. ADAM8 in allergy. Inflamm Allergy Drug Targets. 2008;7:108–12. doi: 10.2174/187152808785107598. [DOI] [PubMed] [Google Scholar]
- 61.Kedda MA, Duffy DL, Bradley B, et al. ADAM33 haplotypes are associated with asthma in a large Australian population. Eur J Hum Genet. 2006;14:1027–36. doi: 10.1038/sj.ejhg.5201662. [DOI] [PubMed] [Google Scholar]
- 62.Noguchi E, Ohtsuki Y, Tokunaga K, et al. ADAM33 polymorphisms are associated with asthma susceptibility in a Japanese population. Clin Exp Allergy. 2006;36:602–8. doi: 10.1111/j.1365-2222.2006.02471.x. [DOI] [PubMed] [Google Scholar]
- 63.Werner M, Herbon N, Gohlke H, et al. Asthma is associated with single-nucleotide polymorphisms in ADAM33. Clin Exp Allergy. 2004;34:26–31. doi: 10.1111/j.1365-2222.2004.01846.x. [DOI] [PubMed] [Google Scholar]
- 64.Howard TD, Postma DS, Jongepier H, et al. Association of a disintegrin and metalloprotease 33 (ADAM33) gene with asthma in ethnically diverse populations. J Allergy Clin Immunol. 2003;112:717–22. doi: 10.1016/s0091-6749(03)01939-0. [DOI] [PubMed] [Google Scholar]
- 65.Blakey JD, Sayers I, Ring S, et al. Positionally cloned asthma susceptibility gene polymorphisms and disease risk in the British 1958 Birth Cohort. Thorax. 2009 doi: 10.1136/thx.2008.102053. published online, 22 February 2009. doi:10.1136/thx.2008.102053. [DOI] [PubMed] [Google Scholar]
- 66.Raby BA, Silverman EK, Kwiatkowski DJ, et al. ADAM33 polymorphisms and phenotype associations in childhood asthma. J Allergy Clin Immunol. 2004;113:1071–78. doi: 10.1016/j.jaci.2004.03.035. [DOI] [PubMed] [Google Scholar]
- 67.Lind DL, Choudhry S, Ung N, et al. ADAM33 is not associated with asthma in Puerto Rican or Mexican populations. Am J Respir Crit Care Med. 2003;168:1312–16. doi: 10.1164/rccm.200306-877OC. [DOI] [PubMed] [Google Scholar]
- 68.Tremblay K, Lemire M, Potvin C, Tremblay A, et al. Genes to diseases (G2D) computational method to identify asthma candidate genes. PLoS ONE. 2008;3:e2907. doi: 10.1371/journal.pone.0002907. published online 6 August 2008, doi:10.1371/journal.pone.0002907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Higuchi Y, Yasui A, Matsuura K, Yamamoto S. CD156 transgenic mice. Different responses between inflammatory types. Pathobiology. 2002;70:47–54. doi: 10.1159/000066003. [DOI] [PubMed] [Google Scholar]
- 70.Matsuno O, Miyazaki E, Nureki S, et al. Role of ADAM8 in experimental asthma. Immunol Lett. 2006;102:67–73. doi: 10.1016/j.imlet.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 71.Knolle MD, Cala L, Lonak SP, Owen CA. Anti-inflammatory role for ADAM8 in asthma. Am.J.Resp.Crit.Care Med. Abstracts issue. 2008:A978. [Google Scholar]
- 72.Puxeddu I, Pang YY, Harvey A, et al. The soluble form of a disintegrin and metalloprotease 33 promotes angiogenesis: implications for airway remodeling in asthma. J Allergy Clin Immunol. 2008;121:1400–6. doi: 10.1016/j.jaci.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 73.Sriramarao P, von Andrian UH, Butcher EC, et al. L-selectin and very late antigen-4 integrin promote eosinophil rolling at physiological shear rates in vivo. J Immunol. 1994;153:4238–46. [PubMed] [Google Scholar]
- 74.Berry MA, Hargadon B, Shelley M, et al. Evidence of a role of tumor necrosis factor α in refractory asthma. N Engl J Med. 2006;354:697–708. doi: 10.1056/NEJMoa050580. [DOI] [PubMed] [Google Scholar]
- 75.Kim H, Hwang JS, Woo CH, et al. TNF-α-induced up-regulation of intercellular adhesion molecule-1 is regulated by a Rac-ROS-dependent cascade in human airway epithelial cells. Exp Mol Med. 2008;40:167–75. doi: 10.3858/emm.2008.40.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang CC, Fu CL, Yang YH, et al. Adenovirus expressing interleukin-1 receptor antagonist alleviates allergic airway inflammation in a murine model of asthma. Gene Ther. 2006;13:1414–21. doi: 10.1038/sj.gt.3302798. [DOI] [PubMed] [Google Scholar]
- 77.Orlando S, Sironi M, Bianchi G, et al. Role of metalloproteases in the release of the IL-1 type II decoy receptor. J Biol Chem. 1997;272:31764–69. doi: 10.1074/jbc.272.50.31764. [DOI] [PubMed] [Google Scholar]
- 78.Hundhausen C, Misztela D, Berkhout TA, et al. The disintegrin-like metalloproteinase ADAM10 is involved in constitutive cleavage of CX3CL1 (fractalkine) and regulates CX3CL1-mediated cell-cell adhesion. Blood. 2003;102:1186–95. doi: 10.1182/blood-2002-12-3775. [DOI] [PubMed] [Google Scholar]
- 79.Tsou CL, Haskell CA, Charo IF. Tumor necrosis factor-α-converting enzyme mediates the inducible cleavage of fractalkine. J Biol Chem. 2001;276:44622–26. doi: 10.1074/jbc.M107327200. [DOI] [PubMed] [Google Scholar]
- 80.Rimaniol AC, Till SJ, Garcia G, et al. The CX3C chemokine fractalkine in allergic asthma and rhinitis. J Allergy Clin Immunol. 2003;112:1139–46. doi: 10.1016/j.jaci.2003.09.041. [DOI] [PubMed] [Google Scholar]
- 81.Wen LP, Fahrni JA, Matsui S, Rosen GD. Airway epithelial cells produce stem cell factor. Biochim Biophys Acta. 1996;1314:183–86. doi: 10.1016/s0167-4889(96)00138-3. [DOI] [PubMed] [Google Scholar]
- 82.Hartman M, Piliponsky AM, Temkin V, Levi-Schaffer F. Human peripheral blood eosinophils express stem cell factor. Blood. 2001;97:1086–91. doi: 10.1182/blood.v97.4.1086. [DOI] [PubMed] [Google Scholar]
- 83.Oliveira SH, Lukacs NW. Stem cell factor: a hemopoietic cytokine with important targets in asthma. Curr Drug Targets Inflamm Allergy. 2003;2:313–18. doi: 10.2174/1568010033483990. [DOI] [PubMed] [Google Scholar]
- 84.Ramenghi U, Ruggieri L, Dianzani I, et al. Human peripheral blood granulocytes and myeloid leukemic cell lines express both transcripts encoding for stem cell factor. Stem Cells. 1994;12:521–26. doi: 10.1002/stem.5530120508. [DOI] [PubMed] [Google Scholar]
- 85.Da Silva CA, Reber L, Frossard N. Stem cell factor expression, mast cells and inflammation in asthma. Fundam Clin Pharmacol. 2006;20:21–39. doi: 10.1111/j.1472-8206.2005.00390.x. [DOI] [PubMed] [Google Scholar]
- 86.Longley BJ, Tyrrell L, Ma Y, et al. Chymase cleavage of stem cell factor yields a bioactive, soluble product. Proc Natl Acad Sci U S A. 1997;94:9017–21. doi: 10.1073/pnas.94.17.9017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gallea-Robache S, Morand V, Millet S, et al. A metalloproteinase inhibitor blocks the shedding of soluble cytokine receptors and processing of transmembrane cytokine precursors in human monocytic cells. Cytokine. 1997;9:340–46. doi: 10.1006/cyto.1996.0174. [DOI] [PubMed] [Google Scholar]
- 88.Campbell E, Hogaboam C, Lincoln P, Lukacs NW. Stem cell factor-induced airway hyperreactivity in allergic and normal mice. Am J Pathol. 1999;154:1259–65. doi: 10.1016/S0002-9440(10)65377-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Oliveira SH, Hogaboam CM, Berlin A, Lukacs NW. SCF-induced airway hyperreactivity is dependent on leukotriene production. Am J Physiol Lung Cell Mol Physiol. 2001;280:L1242–L1249. doi: 10.1152/ajplung.2001.280.6.L1242. [DOI] [PubMed] [Google Scholar]
- 90.Edberg JC, Kimberly RP. Cell type-specific glycoforms of Fc gamma RIIIa CD16.: differential ligand binding. J Immunol. 1997;159:3849–57. [PubMed] [Google Scholar]
- 91.Tosi MF, Zakem H. Surface expression of Fcγ receptor III (CD16) on chemoattractant-stimulated neutrophils is determined by both surface shedding and translocation from intracellular storage compartments. J Clin Invest. 1992;90:462–70. doi: 10.1172/JCI115882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dransfield I, Buckle AM, Savill JS, et al. Neutrophil apoptosis is associated with a reduction in CD16 (Fc gamma RIII) expression. J Immunol. 1994;153:1254–63. [PubMed] [Google Scholar]
- 93.Davoine F, Lavigne S, Chakir J, et al. Expression of FcγRIII (CD16) on human peripheral blood eosinophils increases in allergic conditions. J Allergy Clin Immunol. 2002;109:463–69. doi: 10.1067/mai.2002.121952. [DOI] [PubMed] [Google Scholar]
- 94.Rivier A, Pene J, Rabesandratana H, et al. Blood monocytes of untreated asthmatics exhibit some features of tissue macrophages. Clin Exp Immunol. 1995;100:314–18. doi: 10.1111/j.1365-2249.1995.tb03670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lensmar C, Prieto J, Dahlen B, et al. Airway inflammation and altered alveolar macrophage phenotype pattern after repeated low-dose allergen exposure of atopic asthmatic subjects. Clin Exp Allergy. 1999;29:1632–40. doi: 10.1046/j.1365-2222.1999.00757.x. [DOI] [PubMed] [Google Scholar]
- 96.Weskamp G, Ford JW, Sturgill J, et al. ADAM10 is a principal ‘sheddase’ of the low-affinity immunoglobulin E receptor CD23. Nat Immunol. 2006;7:1293–98. doi: 10.1038/ni1399. [DOI] [PubMed] [Google Scholar]
- 97.Aberle N, Gagro A, Rabatic S, et al. Expression of CD23 antigen and its ligands in children with intrinsic and extrinsic asthma. Allergy. 1997;52:1238–42. doi: 10.1111/j.1398-9995.1997.tb02530.x. [DOI] [PubMed] [Google Scholar]
- 98.Williams J, Johnson S, Mascali JJ, et al. Regulation of low affinity IgE receptor (CD23) expression on mononuclear phagocytes in normal and asthmatic subjects. J Immunol. 1992;149:2823–29. [PubMed] [Google Scholar]
- 99.Daher S, Santos LM, Sole D, et al. Interleukin-4 and soluble CD23 serum levels in asthmatic atopic children. J Investig Allergol Clin Immunol. 1995;5:251–54. [PubMed] [Google Scholar]
- 100.Cernadas M, De Sanctis GT, Krinzman SJ, et al. CD23 and allergic pulmonary inflammation: potential role as an inhibitor. Am J Respir Cell Mol Biol. 1999;20:1–8. doi: 10.1165/ajrcmb.20.1.3299. [DOI] [PubMed] [Google Scholar]
- 101.Payet ME, Woodward EC, Conrad DH. Humoral response suppression observed with CD23 transgenics. J Immunol. 1999;163:217–23. [PubMed] [Google Scholar]
- 102.Karagiannis SN, Warrack JK, Jennings KH, et al. Endocytosis and recycling of the complex between CD23 and HLA-DR in human B cells. Immunology. 2001;103:319–31. doi: 10.1046/j.1365-2567.2001.01238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Aubry JP, Pochon S, Graber P, et al. CD21 is a ligand for CD23 and regulates IgE production. Nature. 1992;358:505–7. doi: 10.1038/358505a0. [DOI] [PubMed] [Google Scholar]
- 104.Hibbert RG, Teriete P, Grundy GJ, et al. The structure of human CD23 and its interactions with IgE and CD21. J Exp Med. 2005;202:751–60. doi: 10.1084/jem.20050811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lecoanet-Henchoz S, Plater-Zyberk C, Graber P, et al. Mouse CD23 regulates monocyte activation through an interaction with the adhesion molecule CD11b/CD18. Eur J Immunol. 1997;27:2290–94. doi: 10.1002/eji.1830270924. [DOI] [PubMed] [Google Scholar]
- 106.Aikawa T, Shimura S, Sasaki H, et al. Marked goblet cell hyperplasia with mucus accumulation in the airways of patients who died of severe acute asthma attack. Chest. 1992;101:916–21. doi: 10.1378/chest.101.4.916. [DOI] [PubMed] [Google Scholar]
- 107.Shim JJ, Dabbagh K, Ueki IF, et al. IL-13 induces mucin production by stimulating epidermal growth factor receptors and by activating neutrophils. Am J Physiol Lung Cell Mol Physiol. 2001;280:L134–L140. doi: 10.1152/ajplung.2001.280.1.L134. [DOI] [PubMed] [Google Scholar]
- 108.Lemjabbar H, Li D, Gallup M, et al. Tobacco smoke-induced lung cell proliferation mediated by tumor necrosis factor α-converting enzyme and amphiregulin. J Biol Chem. 2003;278:26202–07. doi: 10.1074/jbc.M207018200. [DOI] [PubMed] [Google Scholar]
- 109.Lemjabbar H, Basbaum C. Platelet-activating factor receptor and ADAM10 mediate responses to Staphylococcus aureus in epithelial cells. Nat Med. 2002;8:41–46. doi: 10.1038/nm0102-41. [DOI] [PubMed] [Google Scholar]
- 110.Deshmukh HS, Case LM, Wesselkamper SC, et al. Metalloproteinases mediate mucin 5AC expression by epidermal growth factor receptor activation. Am J Respir Crit Care Med. 2005;171:305–14. doi: 10.1164/rccm.200408-1003OC. [DOI] [PubMed] [Google Scholar]
- 111.Shao MX, Ueki IF, Nadel JA. Tumor necrosis factor α-converting enzyme mediates MUC5AC mucin expression in cultured human airway epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11618–23. doi: 10.1073/pnas.1534804100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sahin U, Weskamp G, Kelly K, et al. Distinct roles for ADAM10 and ADAM17 in ectodomain shedding of six EGFR ligands. J Cell Biol. 2004;164:769–79. doi: 10.1083/jcb.200307137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Davies DE, Wicks J, Powell RM, et al. Airway remodeling in asthma: new insights. J Allergy Clin Immunol. 2003;111:215–25. doi: 10.1067/mai.2003.128. [DOI] [PubMed] [Google Scholar]
- 114.Broide DH. Immunologic and inflammatory mechanisms that drive asthma progression to remodeling. J Allergy Clin Immunol. 2008;121:560–70. doi: 10.1016/j.jaci.2008.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Yu Q, Stamenkovic I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-β and promotes tumor invasion and angiogenesis. Genes Dev. 2000;14:163–76. [PMC free article] [PubMed] [Google Scholar]
- 116.Maeda S, Dean DD, Gay I, et al. Activation of latent transforming growth factor β1 by stromelysin 1 in extracts of growth plate chondrocyte-derived matrix vesicles. J Bone Miner Res. 2001;16:1281–90. doi: 10.1359/jbmr.2001.16.7.1281. [DOI] [PubMed] [Google Scholar]
- 117.Mohan S, Thompson GR, Amaar YG, et al. ADAM-9 is an insulin-like growth factor binding protein-5 protease produced and secreted by human osteoblasts. Biochemistry. 2002;41:15394–403. doi: 10.1021/bi026458q. [DOI] [PubMed] [Google Scholar]
- 118.Okada A, Mochizuki S, Yatabe T, et al. ADAM-12 (meltrin α) is involved in chondrocyte proliferation via cleavage of insulin-like growth factor binding protein 5 in osteoarthritic cartilage. Arthritis Rheum. 2008;58:778–89. doi: 10.1002/art.23262. [DOI] [PubMed] [Google Scholar]
- 119.Mochizuki S, Shimoda M, Shiomi T, et al. ADAM28 is activated by MMP-7 (matrilysin-1) and cleaves insulin-like growth factor binding protein-3. Biochem Biophys Res Commun. 2004;315:79–84. doi: 10.1016/j.bbrc.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 120.Martin J, Eynstone LV, Davies M, et al. The role of ADAM 15 in glomerular mesangial cell migration. J Biol Chem. 2002;277:33683–89. doi: 10.1074/jbc.M200988200. [DOI] [PubMed] [Google Scholar]
- 121.Millichip MI, Dallas DJ, Wu E, Dale S, et al. The metallo-disintegrin ADAM10 (MADM) from bovine kidney has type IV collagenase activity in vitro. Biochem Biophys Res Commun. 1998;245:594–98. doi: 10.1006/bbrc.1998.8485. [DOI] [PubMed] [Google Scholar]
- 122.Gronski TJ, Jr., Martin RL, Kobayashi DK, et al. Hydrolysis of a broad spectrum of extracellular matrix proteins by human macrophage elastase. J Biol Chem. 1997;272:12189–94. doi: 10.1074/jbc.272.18.12189. [DOI] [PubMed] [Google Scholar]
- 123.Chua F, Dunsmore SE, Clingen PH, et al. Mice lacking neutrophil elastase are resistant to bleomycin-induced pulmonary fibrosis. Am J Pathol. 2007;170:65–74. doi: 10.2353/ajpath.2007.060352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Taipale J, Koli K, Keski-Oja J. Release of transforming growth factor-β1 from the pericellular matrix of cultured fibroblasts and fibrosarcoma cells by plasmin and thrombin. J Biol Chem. 1992;267:25378–84. [PubMed] [Google Scholar]
- 125.Rajah R, Nachajon RV, Collins MH, et al. Elevated levels of the IGF-binding protein protease MMP-1 in asthmatic airway smooth muscle. Am J Respir Cell Mol Biol. 1999;20:199–208. doi: 10.1165/ajrcmb.20.2.3148. [DOI] [PubMed] [Google Scholar]
- 126.Cohen P, Noveral JP, Bhala A, et al. Leukotriene D4 facilitates airway smooth muscle cell proliferation via modulation of the IGF axis. Am J Physiol. 1995;269:L151–L157. doi: 10.1152/ajplung.1995.269.2.L151. [DOI] [PubMed] [Google Scholar]
- 127.Zhang HY, Phan SH. Inhibition of myofibroblast apoptosis by transforming growth factor β1. Am J Respir Cell Mol Biol. 1999;21:658–65. doi: 10.1165/ajrcmb.21.6.3720. [DOI] [PubMed] [Google Scholar]
- 128.Freyer AM, Johnson SR, Hall IP. Effects of growth factors and extracellular matrix on survival of human airway smooth muscle cells. Am J Respir Cell Mol Biol. 2001;25:569–76. doi: 10.1165/ajrcmb.25.5.4605. [DOI] [PubMed] [Google Scholar]
- 129.Rocks N, Paulissen G, Quesada CF, et al. Expression of a disintegrin and metalloprotease (ADAM and ADAMTS) enzymes in human non-small-cell lung carcinomas (NSCLC) Br J Cancer. 2006;94:724–30. doi: 10.1038/sj.bjc.6602990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Van Ganse E, Laforest L, Pietri G, et al. Persistent asthma: disease control, resource utilisation and direct costs. Eur Respir J. 2002;20:260–67. doi: 10.1183/09031936.02.02542001. [DOI] [PubMed] [Google Scholar]
- 131.Planagumà A, Kazani S, Marigowda G, et al. Airway lipoxin A4 generation and lipoxin A4 receptor expression are decreased in severe asthma. Am J Respir Crit Care Med. 2008;178:574–82. doi: 10.1164/rccm.200801-061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Haworth O, Cernadas M, Yang R, et al. Resolvin E1 regulates interleukin 23, interferon-γ and lipoxin A4 to promote the resolution of allergic airway inflammation. Nat Immunol. 2008;9:873–79. doi: 10.1038/ni.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Levy BD, Lukacs NW, Berlin AA, et al. Lipoxin A4 stable analogs reduce allergic airway responses via mechanisms distinct from CysLT1 receptor antagonism. FASEB J. 2007;21:3877–84. doi: 10.1096/fj.07-8653com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Levy BD, Kohli P, Gotlinger K, et al. Protectin D1 is generated in asthma and dampens airway inflammation and hyperresponsiveness. J Immunol. 2007;178:496–502. doi: 10.4049/jimmunol.178.1.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Parks WC, Wilson CL, Lopez-Boado YS. Matrix metalloproteinases as modulators of inflammation and innate immunity. Nat Rev Immunol. 2004;4:617–29. doi: 10.1038/nri1418. [DOI] [PubMed] [Google Scholar]
- 136.Shang T, Yednock T, Issekutz AC. α9β1 integrin is expressed on human neutrophils and contributes to neutrophil migration through human lung and synovial fibroblast barriers. J Leukoc Biol. 1999;66:809–16. doi: 10.1002/jlb.66.5.809. [DOI] [PubMed] [Google Scholar]
- 137.Berton G, Mocsai A, Lowell CA. Src and Syk kinases: key regulators of phagocytic cell activation. Trends Immunol. 2005;26:208–14. doi: 10.1016/j.it.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 138.Lee JH, Park HS, Park SW, et al. ADAM33 polymorphism: association with bronchial hyper-responsiveness in Korean asthmatics. Clin Exp Allergy. 2004;34:860–65. doi: 10.1111/j.1365-2222.2004.01977.x. [DOI] [PubMed] [Google Scholar]
- 139.Howard TD, Postma DS, Jongepier H, et al. Association of a disintegrin and metalloprotease 33 (ADAM33) gene with asthma in ethnically diverse populations. J Allergy Clin Immunol. 2003;112:717–22. doi: 10.1016/s0091-6749(03)01939-0. [DOI] [PubMed] [Google Scholar]
- 140.Schluesener HJ. The disintegrin domain of ADAM 8 enhances protection against rat experimental autoimmune encephalomyelitis, neuritis and uveitis by a polyvalent autoantigen vaccine. J Neuroimmunol. 1998;87:197–202. doi: 10.1016/s0165-5728(98)00080-0. [DOI] [PubMed] [Google Scholar]
- 141.Hinkle CL, Diestel S, Lieberman J, Maness PF. Metalloprotease-induced ectodomain shedding of neural cell adhesion molecule (NCAM) J Neurobiol. 2006;66:1378–95. doi: 10.1002/neu.20257. [DOI] [PubMed] [Google Scholar]
- 142.Naus S, Richter M, Wildeboer D, et al. Ectodomain shedding of the neural recognition molecule CHL1 by the metalloprotease-disintegrin ADAM8 promotes neurite outgrowth and suppresses neuronal cell death. J Biol Chem. 2004;279:16083–90. doi: 10.1074/jbc.M400560200. [DOI] [PubMed] [Google Scholar]
- 143.Kieran NE, Doran PP, Connolly SB, et al. Modification of the transcriptomic response to renal ischemia/reperfusion injury by lipoxin analog. Kidney Int. 2003;64:480–92. doi: 10.1046/j.1523-1755.2003.00106.x. [DOI] [PubMed] [Google Scholar]
- 144.Vrachnis N, Malamitsi-Puchner A, Samoli E, et al. Elevated mid-trimester amniotic fluid ADAM-8 concentrations as a potential risk factor for preterm delivery. J Soc Gynecol Investig. 2006;13:186–90. doi: 10.1016/j.jsgi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 145.Mahoney ET, Benton RL, Maddie MA, et al. ADAM8 is selectively up-regulated in endothelial cells and is associated with angiogenesis after spinal cord injury in adult mice. J Comp Neurol. 2009;512:243–55. doi: 10.1002/cne.21902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Wildeboer D, Naus S, Amy Sang QX, et al. Metalloproteinase disintegrins ADAM8 and ADAM19 are highly regulated in human primary brain tumors and their expression levels and activities are associated with invasiveness. J Neuropathol Exp Neurol. 2006;65:516–27. doi: 10.1097/01.jnen.0000229240.51490.d3. [DOI] [PubMed] [Google Scholar]
- 147.Valkovskaya N, Kayed H, Felix K, et al. ADAM8 expression is associated with increased invasiveness and reduced patient survival in pancreatic cancer. J Cell Mol Med. 2007;11:1162–74. doi: 10.1111/j.1582-4934.2007.00082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Fritzsche FR, Jung M, Xu C, et al. ADAM8 expression in prostate cancer is associated with parameters of unfavorable prognosis. Virchows Arch. 2006;449:628–36. doi: 10.1007/s00428-006-0315-1. [DOI] [PubMed] [Google Scholar]