Abstract
Several thieno-expanded purine nucleoside analogues were synthesized for use as tools in ongoing investigations into nucleic acid structure and function in our laboratories. The inclusion of the thiophene ring system in the nucleoside endows the purine scaffold with advantages not previously available in other reported expanded purines. The synthesis and preliminary biological studies are reported herein.
INTRODUCTION
A number of research groups1-5, including ours6-9, have designed and synthesized a number of varients of structurally unique unnatural nucleosides to study various aspects of nucleic acid structure and function. Each new foray into the expansion of the genetic alphabet allows for further investigation of just how absolute the requirements for base-pairing and stacking, helix stability and recognition by enzyme systems such as polymerases or other nucleoside-metabolizing enzymes involved in critical biological processes, really are.
A number of design strategies have focused on size or shape complementarities, or matching hydrogen-bonding interactions, pairing up complementary donor-acceptor patterns between unnatural bases. Recently, use of benzene-expanded purines such as Nelson Leonard’s lin-benzoadenosine10 has been explored, an approach we began to pursue some time ago beginning with the synthesis of a series of thieno-expanded tricyclic purines.7 In contrast to Leonard’s, and Kool’s11-15 more recent extension of Leonard’s work, and Matteucci’s linear systems16-18, use of a heteroaromatic spacer ring provides a number of advantages over the benzene spacers, including offering forth a less dramatic expansion of the helix due to the curvature of the nonlinear base pairing, while still retaining the hydrogen bonding elements involved in recognition and pairing.
In terms of stacking, high-level molecular dynamics calculations have indicated that inclusion of the heteroaromatic spacer ring will significantly increase the overall aromaticity and polarizability for these modified bases19-21, which in turn, should result in dramatic increases in stacking effects.
Parallel to those studies, the tricyclic ribose nucleosides were converted to their triphosphate analogues to explore requirements for polymerase recognition, since studies have shown that the presence of a heteratom increases the potential for incorporation by many polymerases.22,23 In that regard, preliminary results are discussed.
RESULTS AND DISCUSSION
The synthesis of the ribose tricyclic guanosine and adenosine have been reported.9,7 To obtain the 2′-deoxy from the ribose derivatives, a Barton deoxygenation was employed using standard conditions.24-26 Subsequent conversion of the tricyclics to the triphosphates27 gave the desired nucleotides in reasonable yields.
The triphosphates of the ribose analogues were initially evaluated as substrates/inhibitors with the two most commonly studied RNA-dependent RNA polymerases (RdRp), T7 and SP6. The tricyclic ribose GTP proved to be a good substrate but not a terminator for T7, and a poor substrate, but a moderate kinetic terminator of SP6 as shown in Table 1 on the next page. In addition, the tricyclic ribose GTP inhibited HCV1B NS5B RdRp (data not shown). Studies are currently underway with the 2′-deoxy analogues with various biologically significant DNA polymerases.
Table 1.
Polymerase assays
| Nucleotide | T7 RNA Polymerase (Km, μM) |
SP6 RNA Polymerase (Km, μM) |
|---|---|---|
| GTP | 0.61 | 0.41 |
| Tricyclic GTP | 3.1 | 5.8 |
| UTP after GTP | 0.72 | 0.96 |
| UTP after tri-GTP | 0.79 | 4.98 |
CONCLUSION
The Rd-RNA polymerases readily recognized the unnatural triphosphates and surprisingly, exhibited remarkable differences in recognition, thus providing impetus to examine more selective polymerases. Investigations with DNA polymerases are currently underway with the 2-deoxy analogues and those studies are currently underway and the results will be reported as they become available.
Fig 1.
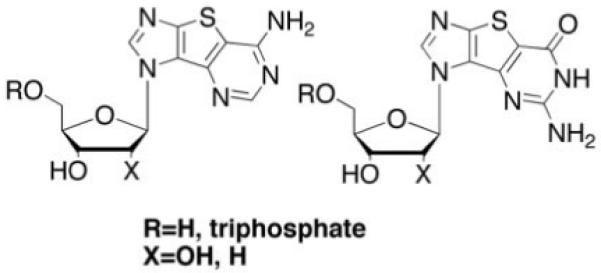
Hetero-expanded purine analogues.
Scheme 1.
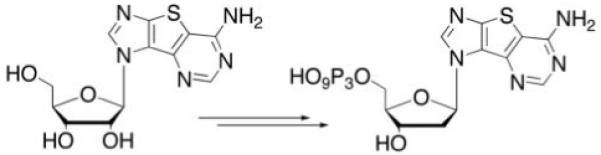
General synthesis overview.
ACKNOWLEDGEMENTS
The authors thank the National Institutes of Health RO1 GM073645 (KSR) for their kind support.
REFERENCES
- 1.Benner SA. Acc Chem Res. 2004;37:784–797. doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]
- 2.Kool ET. Acc Chem Res. 2002;35:936–943. doi: 10.1021/ar000183u. [DOI] [PubMed] [Google Scholar]
- 3.Krueger AT, Kool ET. Curr Opin Chem Biol. 2007;11:588–594. doi: 10.1016/j.cbpa.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Henry AA, Romesberg FE. Curr Opin Chem Biol. 2003;7:727–733. doi: 10.1016/j.cbpa.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Yu C, Henry AA, Schultz PG, Romesberg FE. Angew. Chem., Int. Ed. Engl. 2002;41:3841–3844. doi: 10.1002/1521-3773(20021018)41:20<3841::AID-ANIE3841>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 6.Seley KL, Salim S, Zhang L, O’Daniel PI. J. Org Chem. 2005;70:1612–9. doi: 10.1021/jo048218h. [DOI] [PubMed] [Google Scholar]
- 7.Seley KL, Januszczyk P, Hagos A, Zhang L, Dransfield DT. J Med Chem. 2000;43:4877–4883. doi: 10.1021/jm000326i. [DOI] [PubMed] [Google Scholar]
- 8.Seley KL, Mosley SL, Zeng F. Org Lett. 2003;5:4401–4403. doi: 10.1021/ol035696q. [DOI] [PubMed] [Google Scholar]
- 9.Seley KL, Zhang L, Hagos A, Quirk S. J Org Chem. 2002;67:3365–73. doi: 10.1021/jo0255476. [DOI] [PubMed] [Google Scholar]
- 10.Leonard NJ, Sprecker MA, Morrice AG. J Am Chem Soc. 1976;98:3987–3994. doi: 10.1021/ja00429a040. [DOI] [PubMed] [Google Scholar]
- 11.Krueger AT, Lu H, Lee AH, Kool ET. Acc Chem Res. 2007;40:141–50. doi: 10.1021/ar068200o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee AH, Kool ET. J Am Chem Soc. 2006;128:9219–30. doi: 10.1021/ja0619004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lynch SR, Liu H, Gao J, Kool ET. J Am Chem Soc. 2006;128:14704–11. doi: 10.1021/ja065606n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H, Gao J, Kool ET. J Am Chem Soc. 2005;127:1396–402. doi: 10.1021/ja046305l. [DOI] [PubMed] [Google Scholar]
- 15.Lee AH, Kool ET. J Am Chem Soc. 2005;127:3332–8. doi: 10.1021/ja0430604. [DOI] [PubMed] [Google Scholar]
- 16.Lin K-Y, Jones RJ, Matteucci MD. J Am Chem Soc. 1995;117:3873–3874. [Google Scholar]
- 17.Matteucci MD, von Krosigk U. Tetrahedron Lett. 1996;37:5057–5060. [Google Scholar]
- 18.Buhr CA, Matteucci MD, Froehler BC. Tetrahedron Letters. 1999;40:8969–8970. [Google Scholar]
- 19.Guckian KM, Schweitzer BA, Ren RX-F, Sheils CJ, Paris PL, Tahmassebi DC, Kool ET. J Am Chem Soc. 1996;118:8182–8183. doi: 10.1021/ja961733f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guckian KM, Schweitzer BA, Ren RX-F, Sheils CJ, Tahmassebi DC, Kool ET. J Am Chem Soc. 2000;122:2213–2222. doi: 10.1021/ja9934854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Daniel PI, Jefferson M, Wiest O, Seley-Radtke KL. J Biomol Struct Dyn submitted. doi: 10.1080/07391102.2008.10507243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morales JC, Kool ET. Nat Struct Biol. 1998;5:950–4. doi: 10.1038/2925. [DOI] [PubMed] [Google Scholar]
- 23.Hendrickson CL, Devine KG, Benner SA. Nucleic Acids Res. 2004;32:2241–50. doi: 10.1093/nar/gkh542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barton DHR, Jang DO, Jaszberenyl JC. Tetrahedron Lett. 1990;31:3991–3994. [Google Scholar]
- 25.Bennett SM, Nguyen-Ba N, Ogilvie KK. J Med Chem. 1990;33:2162–2173. doi: 10.1021/jm00170a019. [DOI] [PubMed] [Google Scholar]
- 26.Robins MJ, Wilson JS, Hansske F. J Am Chem Soc. 1983;105:4059–4065. [Google Scholar]
- 27.Ludwig J, Eckstein F. J Org Chem. 1989;54:631–635. [Google Scholar]


