Abstract
Leupaxin is a LIM-domain containing adapter protein belonging to the paxillin family that has been previously reported to be preferentially expressed in hematopoeitic cells. Herein, we identified leupaxin in a screen for FAK binding partners in aortic smooth muscle, show that leupaxin is enriched in human and mouse vascular smooth muscle, and that leupaxin expression is dynamically regulated during development. In addition, our studies reveal that leupaxin can undergo cytoplasmic/nuclear shuttling and functions as an SRF-cofactor in the nucleus. We found that leupaxin forms a complex with SRF, associates with CArG-containing regions of SM promoters, and that ectopic expression of leupaxin induces SM marker gene expression in both 10T1/2 cells and rat aortic smooth muscle cells (SMC). Subsequent studies indicated that enhanced FAK activity (induced by fibronectin or expression of constitutively active FAK) attenuates the nuclear accumulation of leupaxin and limits the ability of leupaxin to enhance SRF-dependent gene transcription. Thus, these studies indicate that modulation of the sub-cellular localization of SRF-cofactors is one mechanism by which extracellular matrix-dependent signals might regulate phenotypic switching of SMC.
Keywords: smooth muscle, differentiation, LIM proteins, adhesion, signal transduction
Introduction
Mature medial SMC express high levels of the SMC differentiation marker genes (ie. myosin heavy chain (SM-MHC), SM α-actin, SM22α among others) that contribute to the regulation of SMC contractility 1. Unlike cardiac and skeletal muscle, SMC never terminally differentiate and can transition to a synthetic phenotype characterized by decreased SMC marker gene expression, increased matrix production, and responsiveness to pro-growth and migratory signals. This unique plasticity is critical for proper vessel development, blood pressure homeostasis, and injury repair, but can also contribute to the development of various vascular pathologies 1. Thus, defining the signaling mechanisms that regulate SMC growth and differentiation will be important for understanding the processes that modulate vascular development and is critical for the design of agents that might regulate aberrant SMC responses in diseased vessels.
The transcription mechanisms that regulate SMC differentiation are starting to become clear. Serum response factor (SRF) binding to conserved CArG (CC(A/T)6GG) promoter elements is required for the expression of most SMC differentiation marker genes. It is well known that SRF activity is regulated through interactions with additional ubiquitous and cell-type-selective transcription factors or co-factors including the ternary complex factors (Elk-1, Sap-1, SAP-2/NET/ERP) the GATA factors, Nkx2.5, and the LIM domain proteins CRP1, CRP2, and FHL2 1. The myocardin factors (myocardin, MRTF-A, and MRTF-B) are particularly potent activators of SRF-dependent transcription 2-4. Their importance in the regulation of SMC differentiation is underscored by the lethal defects in SMC differentiation observed in myocardin-/- and MRTF-B-/- mice 5,6. The presence of multiple co-factors likely provides the opportunity for precise transcriptional control of the numerous SRF target genes that are known to regulate SMC growth, migration, and differentiation.
TGF-β, which promotes SMC differentiation 7,8, and PDGF-BB, which promotes phenotypic modulation, are important extrinsic regulators of SMC phenotype and genetic ablation of these genes resulted in defective vasculogenesis 9,10. In addition to these soluble factors, extracellular matrix (ECM) molecules also regulate SMC phenotype. Deletion of either FN, the α5 integrin FN receptor, or focal adhesion kinase (FAK) (the kinase that mediates α5-dependent signaling) result in extraembryonic and embryonic vessel defects leading to lethality in the mouse from E8.5 to E10 11-13. In vitro, FN (which is enriched in the developing vasculature) supports SMC proliferation and limits SMC differentiation, whereas the basement membrane components collagen type IV and laminin (which are more prominent in the mature vessel) promote SMC differentiation 14,15. However, it is currently unclear how integrin-dependent signals interface with the transcriptional machinery to regulate SMC phenotype.
Herein, we identified leupaxin, an understudied LIM protein in the paxillin family, as a FAK binding partner in SMC. We found that leupaxin is particularly abundant in SMC where it localizes both to focal adhesions and the nucleus, indicating that it may be a bi-functional adapter protein. Indeed, we found that nuclear localized leupaxin acts as an SRF-cofactor to enhance SMC differentiation, and that leupaxin undergoes regulated cytoplasmic-nuclear shuttling that is dependent on FAK activity. These data highlight the possibility that extrinsic signals can regulate the SMC gene profile by modulating the activation state of FAK and the localization of LIM-containing SRF cofactors.
Materials and Methods
See Supplemental Data section for a complete description of the reagents, tissue samples, DNA constructs, and general methods used for these studies.
Statistical Analysis
All promoter measurements were performed using the indicated reporter construct in parallel with a minimal thymidine kinase (TK-Luc) promoter. Data are presented as fold levels of SM-promoter activity over TK promoter activity (to rule out generic increases in gene transcription) and all data represent at least three separate experiments presented as mean +/- SEM. Means were compared by 2-tailed Students's t test or ANOVA (where indicated) and p<0.05 was considered statistically significant as indicated by an asterisk. All other data including Western analysis and ChIP assays are representative of at least three individual experiments.
Results
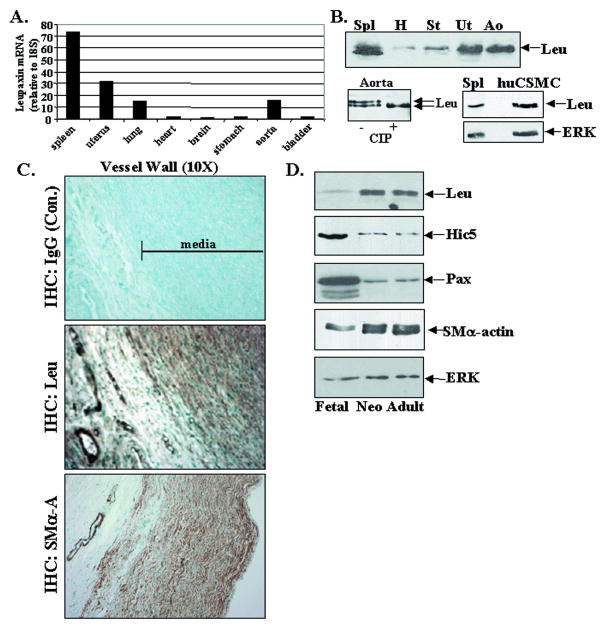
We identified leupaxin as a putative FAK binding partner in a yeast two-hybrid screen of an aortic smooth muscle cell library. This finding was somewhat surprising, because leupaxin was previously reported to be selectively expressed in lymphoid tissues as well as several cultured hematopoetic cell lines, osteoclasts, and a bone-derived cancer cell line 16,17. However, as our RT-PCR results demonstrate (Fig. 1A), high levels of leupaxin message were detected in several mouse tissues including spleen, aorta, lung, and uterus, while lower levels were observed in stomach, bladder, heart, and brain. Western analysis using a monoclonal antibody (Ab) that is completely specific for human leupaxin yielded a similar expression pattern and also that leupaxin runs as a doublet of approximately 45/47 kDa in some tissues (Fig 1B). Since a single mRNA species was identified by Northern analysis (not shown), the slower migrating form is likely due to post-translational modification. Indeed, the leupaxin mobility shift was reversed by treatment of human aortic lysates with calf intestinal alkaline phosphatase (CIP; Fig. 1B, bottom left). Leupaxin expression was also strong in cultured human coronary SMC (huCSMC) (Fig 1B, bottom right) and treatment of these cells with calyculin-A induced a mobility shift (data not shown), in further support of a phosphorylation-induced change in motility on SDS-PAGE. Importantly, the leupaxin antibody used does not recognize its closely related family members paxillin (68 kda) or Hic-5 (50 kDa), which are also expressed in huCSMC (Supplemental Fig 1A).
Figure 1. Leupaxin is expressed at high levels in arterial and visceral smooth muscle.
A. Leupaxin qRT-PCR was performed on adult mouse tissue. Data are represented as relative to 18S RNA control. B (top) Western analysis for leupaxin in adult human tissues (46-60 yrs; 50 μg each) including spleen (Spl), heart (H), stomach (St), uterus (UT), and aorta (Ao) (bottom left) Human aorta lysate was incubated in the presence (+) or absence (−) of calf intestinal alkaline phosphatase (10 units) for 20 min at 30°C. (bottom right) Levels of leupaxin in huCSMC compared to spleen. ERK is shown as a loading control C. Immunohistochemical staining for leupaxin and SMα-actin in human aorta (37 week pc) detected by DAB (brown) and counter-stained with methyl green. D. Western analysis of human thoracic aorta from fetal (37 week pc), neonatal (2 wk post-natal), and adult (55 yrs). ERK levels are shown as a loading control.
In accordance with the expression of leupaxin in huCSMC, immunohistochemical analysis of human aorta revealed strong leupaxin expression throughout the media as well as in the smooth muscle layers (but not endothelium) in the microvessels within the adventitia (Fig 1C; Supplemental Fig. 1B). Since several ECM/integrin signaling components that regulate SMC function have been shown to be developmentally regulated, we also examined leupaxin expression at several stages of human aortic development. Figure 1D demonstrates that leupaxin expression was relatively low in fetal human thoracic aorta (37 wk pc) but was much higher at P14 and in adult (55 yr), which contain more highly differentiated SMC as revealed by higher levels of SMα-actin. Interestingly, leupaxin was expressed in a reciprocal fashion relative to its family members, paxillin and Hic-5. Quantitative RT-PCR analysis for leupaxin message in thoracic aorta isolated from post-natal day 4 to adult (8weeks) mice revealed a similar striking increase in leupaxin expression in the more mature vessels (Supplemental Fig. 1C). Notably, the 8 week samples contained 6-fold higher levels of SM22 message when compared to day 4 vessels (not shown). Collectively, these data indicate that leupaxin is expressed strongly in SMC, and may play a previously un-recognized role during SMC maturation.
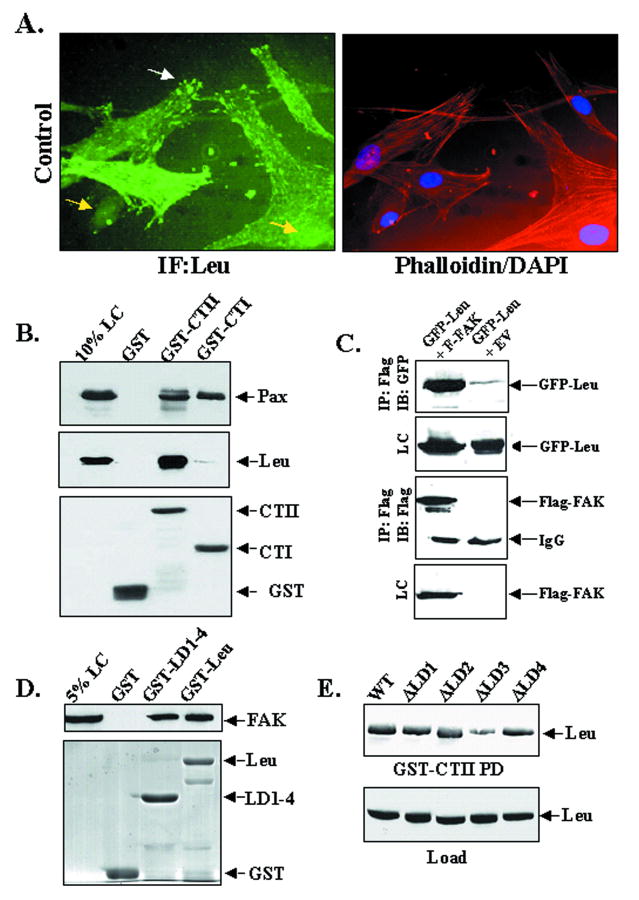
Given the high level of leupaxin expression in huCSMC, we utilized these cells to confirm an interaction between FAK and leupaxin. Leupaxin localizes within focal adhesions and co-localizes with FAK in these cells (Fig 2A and Supplemental Fig 2A). In addition, a GST fusion protein containing the entire FAK C-terminus (GST-CTII) but not GST alone strongly precipitated endogenous leupaxin from huCSMC lysates (Fig 2B). This interaction was subsequently confirmed in vivo by co-immunoprecipitation of a FAK-leupaxin complex from Cos 7 cells (Fig 2C). Somewhat surprisingly, a GST fusion protein containing the more C-terminal FAT domain of FAK (GST-CTI) efficiently precipitated paxillin but not leupaxin (Fig 2B). These results indicate that leupaxin association with FAK requires sequences N-terminal to the FAT domain, and perhaps, that leupaxin may not directly compete with paxillin and/or Hic-5 for FAK binding. Reciprocal mapping studies revealed that the N-terminal LD motifs in leupaxin were sufficient to precipitate FAK from cell lysates (Fig 2D) and subsequent deletion studies revealed that LD3 is the major site on leupaxin that directs binding to FAK (Fig 2E and Supplemental Fig 2B).
Figure 2. Leupaxin associates with FAK in huCSMC.
A HuCSMC cultured in 10% serum were co-stained with anti-leupaxin Ab, phalloidin, and DAPI (40 X) B. huCSMC lysates were incubated with GST or GST-FAK C-terminal fusion proteins (GST-CTI or GST-CTII) prior to Western analysis for paxillin (Pax) or leupaxin (Leu). C. GFP-tagged leupaxin was co-expressed in Cos7 cells with or without Flag-tagged FAK and immunoprecipitations (IP) with anti-Flag antibody were performed followed by immunoblotting with indicated antibodies. A 10% loading control (LC) is shown. D. Cos7 expressing Flag-FAK were incubated with GST or GST-Leu (full-length) or a N-terminal leupaxin fusion protein containing LD motifs 1-4 (GST-LD1-4) prior to Western analysis for FAK. E. Cos7 expressing GFP-Leupaxin variants with individual deletions of LD motifs 1-4 were incubated with GST or GST-CTII prior to Western analysis for GFP.
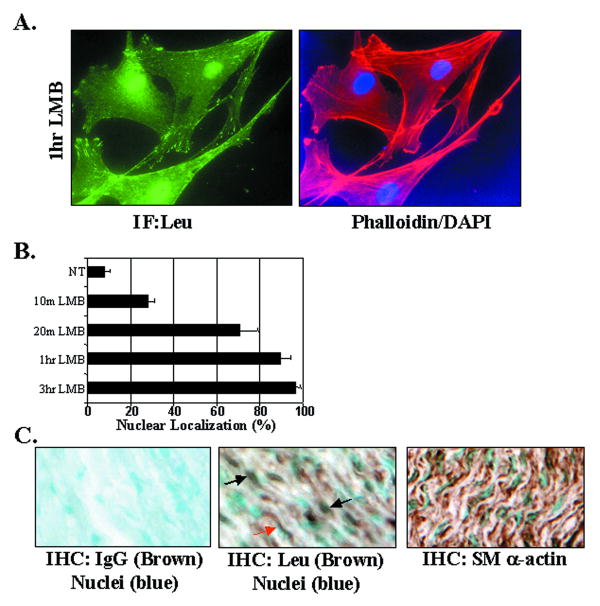
Interestingly, we also observed a subset of huCSMC (approximately 15% of cells maintained in serum) that exhibited both focal adhesion and nuclear-localized leupaxin suggesting that like several other LIM proteins, leupaxin shuttles between focal adhesion and nuclear compartments (Fig 2A). To test this more directly, we treated huCSMC with the CRM1-dependent nuclear export inhibitor, leptomycin B (LMB). LMB treatment for 20 min-1 hr induced a dramatic redistribution of endogenous leupaxin to the nuclear compartment (Fig 3A-B), while treatment for 3 h resulted in nearly complete nuclear localization (not shown). Importantly, immunohistochemical localization of leupaxin in the media of aortic tissue sections also revealed both nuclear and cytoplasmic staining (Fig 3B).
Figure 3. Leupaxin localizes within focal adhesions and the nucleus and undergoes Crm-1/exportin-dependent nuclear-cytoplasmic shuttling.
A,B. huCSMC cultured in 10% serum were treated with or without Leptomycin B (LMB; 5ng/ml) for indicated times prior to fixation. Cells were co-stained with anti-leupaxin Ab, phalloidin, and DAPI (40 X) and cells were scored for nuclear accumulation. Data represent 3 separate experiments (150-200 cells/time point) B. Subcellular localization of leupaxin or SMα-actin detected by DAB (brown) staining in medial smooth muscle. Sections were counter-stained with methyl green to demarcate the nuclei. Flag antibody was used for the IgG control (left panel). Black and red arrows demarcate nuclear and cytoplasmic leupaxin staining respectively.
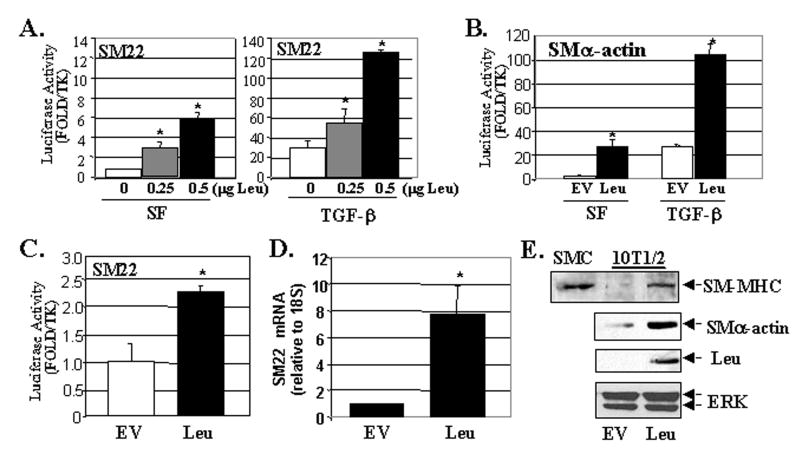
As noted above, several LIM domain proteins that exhibit nuclear accumulation have been implicated in the regulation of gene transcription 18. In fact, the smooth muscle-specific LIM proteins, CRP1 and CRP2, and the cardiac/SMC-selective LIM protein, FHL2, have been shown to interact with SRF and to regulate SRF-dependent gene expression 19,20. To test whether leupaxin can contribute to the regulation of transcription in SMC, we co-transfected flag-tagged leupaxin along with SM22 or SM α-actin promoter/luciferase constructs into multi-potential mouse 10T1/2 cells. As shown in figure 4, expression of leupaxin strongly increased SM22 and SM α-actin promoter activity in a dose-dependent fashion and enhanced the effects of TGF-β in this model. Expression of leupaxin also significantly up-regulated SM22 (Fig 4C) and SM α-actin promoter activity (data not shown) in primary rat aortic SMC (rASMC). The more modest effects of leupaxin in SMC are most likely due to the already high levels of SMC-specific transcriptional activity in primary SMC cultures. Importantly, ectopic expression of leupaxin in 10T1/2 cells induced the expression of endogenous SM gene transcription as assessed by quantitative RT-PCR for SM22 (Fig 4D) and by Western analysis for SM-MHC (the canonical smooth muscle-specific marker) and SM α-actin (Fig 4E).
Figure 4. Ectopic expression of leupaxin stimulates SM marker gene transcription.
A,B. 10T1/2 cells were co-transfected with the SM22- or SMα-actin luciferase constructs and Flag-leupaxin (Leu). Total DNA was normalized with empty vector, EV). Cells were serum starved (0.5% serum) for 24 hrs prior to treatment with vehicle or TGF-β (1ng/ml) overnight and luciferase activity was measured. The * indicates p<0.05 between groups (analyzed by ANOVA). C. Promoter-reporter assays in rASMC maintained in 10% serum measured 48 hrs following transfection. D, E Quantitiative RT-PCR for SM22 (D) or Western blot (E) of SMC (5 μg) or 10T1/2 cells (25 μg) transfected with EV or Leu for 48hrs under serum-starved conditions (0.5% serum). ERK is shown as a loading control.
We hypothesized that nuclear leupaxin, like CRP1/2, might regulate SMC-specific gene expression by interacting with SRF. In support of this idea, leupaxin failed to activate a SM α-actin promoter that contained mutations to all three SRF-binding CArG boxes (ABI; Fig 5A). In addition, we found a co-association of GFP-leupaxin and flag-SRF in reciprocal co-immunoprecipitation assays performed in Cos-7 cells (Fig 5B) and that GST-leupaxin interacted directly with 35S-labeled SRF translated in vitro (Fig 5C). We also used ChIP assays to demonstrate that endogenous leupaxin associated with the CArG-containing regions of the SM α-actin and SM-MHC promoters (but not with the c-fos promoter) in huCSMC grown in serum (Fig 5D). When combined with results from gel shift assays demonstrating that leupaxin did not associate directly with the SM α-actin CArGs (not shown), these results strongly suggest that leupaxin interacts with SRF in vivo. Interestingly, leupaxin was not found in association with CArG elements in serum-starved huCSMC, however under these conditions, TGF-β significantly promoted leupaxin association with CArG-containing region of the SMα-actin promoter, without effecting leupaxin expression levels (Fig 5E). These data corroborate the finding that leupaxin and TGF-β exhibit functional synergy in the promotion of SM marker gene expression. To directly test whether the effects of leupaxin on SMC differentiation marker gene required SRF, we expressed leupaxin in SRF-/- ES cells. As expected, expression of leupaxin did not enhance SM22 reporter gene expression in SRF -/- ES cells but this response could be rescued by co-expression of SRF (Supplemental Fig 3). Collectively, these data strongly support our hypothesis that leupaxin is recruited to the SMC-specific promoters through a direct interaction with SRF and that this leads to increased SMC differentiation marker gene expression.
Figure 5. Leupaxin interacts with SRF and induces SRF- and CArG-dependent gene transcription.
A. Promoter-reporter assay in rASMC using the SMα-actin promoter or a triple CArG mutant SM α-actin promoter (ABI Mutant) B. Cos7 cells were transfected with Flag-SRF and GFP-leupaxin or GFP and immunoprecipitation and Western analysis was performed with the indicated antibodies. A 10% lysate is shown as a loading control (LC). C. Pull-down assays using GST or GST-leupaxin with 35S labeled in vitro translated SRF. Complexes were electrophoresed and processed for autoradiography. 2% of the total 35S-SRF used is shown as a loading control. D,E. ChIP was performed on huCSMC grown in 10% serum (D) or serum starved (no serum) for 36 hr and treated with vehicle or TGF-β for 18 hr (E) using the indicated antibodies and primers. (bottom) Western analysis for leupaxin in vehicle and TGF-β treated huCSMC.
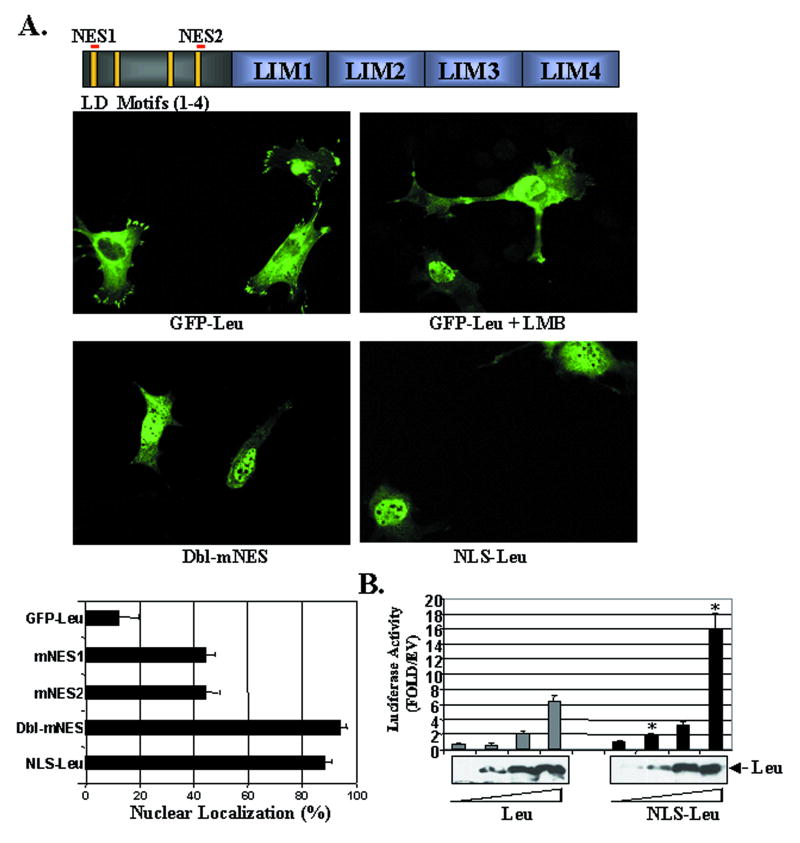
Our next goal was to test whether leupaxin nuclear/cytoplasmic shuttling was an important mechanism that regulated leupaxin's effects on SMC-specific transcription. The effects of LMB indicated that leupaxin likely contains a leucine-rich CRM-1-dependent nuclear export sequence (NES), and we identified two regions (aa 4-11 and aa 134-144) that conformed to consensus NES sequences identified in the LIM proteins, Trip6, Zyxin, LPP and Hic-5 (Fig 6A) 21. Four leucine to alanine mutations were made at each site (mNES1; L4,7,8,11 A and mNES2; L134,137,141,144A) separately and in combination (dbl-mNES) in the context of GFP-leupaxin. We utilized 10T1/2 cells and rat primary aortic SMC (rASMC) for these and subsequent localization experiments since the huCSMC proved to be remarkably resistant to transfection. Importantly, significant leupaxin mRNA is expressed in both 10T1/2 and rASMC (not shown), and the sub-cellular localization of GFP-leupaxin in these cells was virtually identical to that of flag-leupaxin and to the localization of endogenous leupaxin in huCSMC. (Fig 6A). Similar to our previous results, 15% of 10T1/2 cells exhibited nuclear accumulation of GFP-leupaxin while treatment with LMB for 1 h resulted in nuclear accumulation in nearly all cells (Fig 6A). While individual mutation of either NES1 or NES2 increased the percentage of cells with nuclear GFP-leupaxin (to approximately 45%), mutation of both sites resulted in nuclear localization in approximately 90% of transfected cells (Fig 6A and Supplemental Fig 4A). Similar results were observed in rASMC (not shown). Importantly, transfection of these constructs did not significantly affect cell morphology (Supplemental Fig 4B). These data provide strong evidence to support that leupaxin undergoes Crm-1 dependent export, and that the NES sequences identified are important for the rapid shuttling of leupaxin from the nucleus to the cytoplasm.
Figure 6. Regulation and function of nuclear-targeted leupaxin.
A (top) Schematic of leupaxin structure with alignment of assumptive NES1 and 2. (middle) GFP-leupaxin or GFP-leupaxin with both NES mutations (dbl-mNES) were transfected into 10T1/2 cells. Where indicated LMB was added 1 hr before fixation. (bottom) Quantification of nuclear leupaxin accumulation. Data represent at least 150 cells per condition collected from 3 separate experiments. B. Promoter-reporter assay in 10T1/2 cells transfected with increasing concentrations of Leu and NLS-Leu. Western blot reveals comparable expression levels of the two proteins.
Although the dbl mNES variant did exhibit nuclear-restricted expression, this construct contains disrupted LD1 and LD4 binding motifs that could affect normal leupaxin function. Thus to directly test the effects of nuclear localization on transcriptional activity we generated a leupaxin variant that was targeted to the nucleus by fusion of a triple NLS tag to the N-terminus of wild type leupaxin. This construct exhibited nearly complete nuclear localization in 100% of transfected 10T1/2 cells (Fig 6A) and SMC (data not shown) and was functional as shown by its ability to activate the SM22 promoter even more strongly than Wt leupaxin (Fig 6B). These data strongly support the idea that leupaxin's ability to induce gene expression is dependent on nuclear not focal adhesion localization. Furthermore, these data suggest that leupaxin's function as a transcriptional regulator can be modified by altering it's subcellular localization.
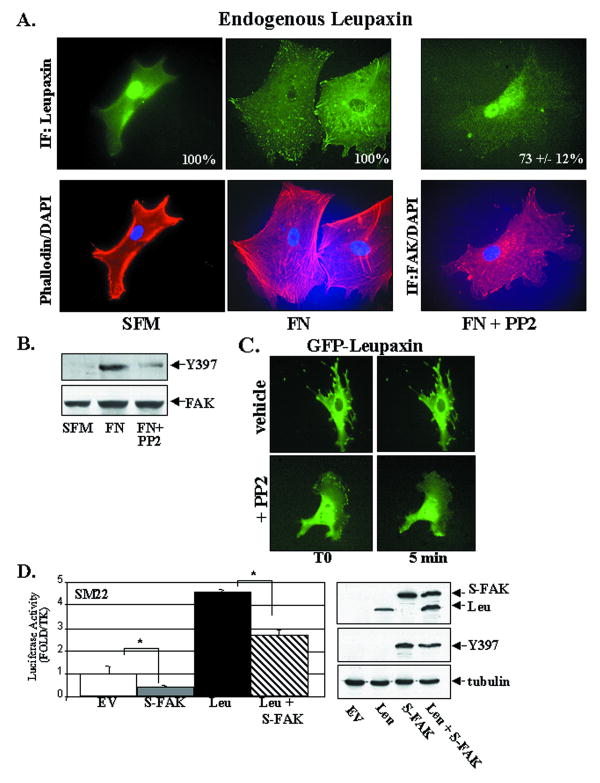
Our demonstration of a FAK-leupaxin interaction suggested that FAK may regulate leupaxin localization and/or transcriptional activity. We reasoned that enhanced FAK activity might lead to sequestration of leupaxin in focal adhesions, thus limiting its effects in the nucleus. To test this hypothesis, we examined endogenous leupaxin localization in huCSMC under different conditions that modulate FAK activity. huCSMC plated on tissue-culture plastic under serum-starved conditions exhibit low FAK activity (as assessed by auto-phosphorylation of FAK on Y397) and near complete restriction of leupaxin within the nucleus (Fig 7A, B). Interestingly, in serum-starved cells plated on FN for 90 min (that exhibit high FAK activity), leupaxin was exclusively associated with focal adhesions (Fig 7A). We next used a pharmacological approach to determine whether endogenous leupaxin shuttling was regulated by FAK/Src activity. We found that treatment with FAK/Src inhibitor, PP2, attenuated FAK activity and lead to remarkable nuclear accumulation of leupaxin in huCSMC plated on FN (Fig 7A-B).
Figure 7. FAK activity modulates leupaxin localization and function.
A,B serum-starved (no serum) huCSMC were plated on plastic (SFM) or fibronectin (FN) for 90 min in absence or presence of PP2 (10 μM) for the last 30 min. Cells were processed for immunocytochemistry (A) or Western analysis (B) using a phosporylation-site specific Y397FAK Ab or an antibody that recognizes total FAK levels. C. GFP-leupaxin expressing rASMC were pre-treated with vehicle or PP2 (10 μM) for 10 min prior to treatment with LMB at time 0 (T0). Fluorescent images were captured for 30 minutes (see Online Movies 1 and 2). Images shown are representative of 10 time-lapse movies for each condition. D. Λεφτ: Πρoμoτερ–ρεπoρτερ ασσαΨσ ιν 10T1/2 χελλσ εξπρεσσινγ Λευ ανδ/oρ Φλαγ–ΣυπερΦAK (ΣΦAK; 0.25μg each). Right: Western blot indicates levels of Flag-Leu, SFAK, and FAK activity.
We next utilized the GFP-tagged leupaxin construct to track leupaxin shuttling in real time in transfected SMC to determine whether FAK/Src activity regulates the rate of leupaxin translocation to the nucleus. To this end, we identified cells in which GFP-leupaxin was predominantly cytoplasmic, pre-treated the cells with PP2 or vehicle for 10 min and then performed time-lapse imaging immediately following LMB treatment. LMB-induced leupaxin nuclear accumulation was evident much earlier in PP2-treated SMC compared to vehicle-treated cells (Fig 7C; Online movies 1 and 2). Indeed, 90% of PP2 pre-treated cells exhibited marked leupaxin nuclear localization within 5 min following LMB treatment, while vehicle-treated cells required at least 20 min to exhibit significant nuclear accumulation.
In support of a functional significance of FAK-dependent leupaxin shuttling, we found that overexpression of a constitutively active FAK variant (termed SuperFAK; 22, promoted focal adhesion-associated leupaxin localization (similar to plating on FN) and partially reversed the marked leupaxin-induced SMC-marker gene transcription in 10T1/2 cells (Fig 7D). It should be noted that although both ectopic expression of SuperFAK (data not shown) and plating cells on FN resulted in pronounced focal adhesion localization in cells maintained in 10% serum, treatment of both populations of cells with LMB for 1 hr did result in substantial nuclear accumulation of leupaxin. Thus, even under conditions of enhanced FAK activity some nuclear shuttling of leupaxin does occur, which may (in part) account for the incomplete rescue of promoter activity observed following SuperFAK expression. Taken together, these results indicate that activation of FAK inhibits leupaxin-induced transcription by sequestering leupaxin to focal adhesions.
Discussion
Leupaxin is an understudied 43 kDa protein that was originally reported as having a lymphoid-restricted expression pattern 16. We show for the first time that leupaxin is also highly expressed in aorta and in cultured SMC. In addition, our studies reveal that leupaxin can undergo cytoplasmic/nuclear shuttling and functions as an SRF-cofactor in the nucleus. We found that leupaxin forms a complex with SRF, associates with CArG-containing regions of the SMC-specific promoters, and that ectopic expression of leupaxin induces SMC differentiation marker gene expression. Subsequent studies indicated that enhanced FAK activity attenuates the nuclear accumulation of leupaxin and limits the ability of leupaxin to enhance SRF-dependent gene transcription. Thus, these studies indicate that sequestration of leupaxin to focal adhesion plaques may be one mechanism by which ECM-dependent signals might regulate phenotypic switching of SMC.
Leupaxin belongs to a family of proteins including paxillin and Hic-5 that may share some overlapping cellular functions. Each of these proteins are comprised of four N-terminal LD motifs and four C-terminal LIM domains 21 and they share approximately 40% identity at the amino acid level. Leupaxin has also been reported to share several binding partners with paxillin such as Pyk2, FAK, Src, PTP-PEST and p95 paxillin kinase linker 16,17,23,24. Leupaxin was previously shown to co-localize with Pyk2 and FAK in the cortical F-actin domain in JY8 lymphoblasts and within podosomes of osteoclasts 16,23. Thus far, known functions for leupaxin include suppression of B-cell antigen receptor signaling in lymphoblasts 25, induction of bone resorption in osteoclasts 24, and induction of cell motility in bone-derived prostate cancer cells 17.
Treatment with the nuclear export inhibitor, leptomycin B causes retention of paxillin, Hic-5, and leupaxin in the nucleus, providing evidence that each of these family members may share a common function to coordinate cell adhesion status with specific changes in gene expression 21. While numerous LIM proteins have been shown to undergo nucleo-cytoplasmic shuttling, it is becoming clear that distinct mechanisms regulate their trafficking. Hic-5 demonstrates oxidant-sensitive nuclear export that was shown to be dependent on two cysteine residues located proximal to the canonical NES identified within the Hic-5 LD3 motif 26. Notably, Hic-5 appears to be unique in this regulation as neither paxillin nor leupaxin harbor similarly located cysteine residues. Recently, elegant studies by Tsujita et. al. indicate that nuclear shuttling of zyxin in cardiomyocytes is regulated in a cGMP and AKT-dependent fashion and that agonists that stimulate PKG (including estradiol and IGF-1) induce dramatic nuclear accumulation of zyxin and paxillin in these cells 27. On the other hand, the LIM only protein FHL2 has been shown to translocate to the nucleus in response to Rho A-dependent signals 28. While we have not observed Rho- or cGMP-dependent nuclear accumulation of leupaxin in huCSMC in these studies (not shown), we did find that leupaxin localization was regulated by the activation state of FAK in these cells. Interestingly, leupaxin was recently shown to be a substrate for Src in A20 B lymphoma cells 25 and we have shown that leupaxin is a direct substrate for FAK in vitro (unpublished data). Thus, we favor a model whereby activation of FAK and/or Src likely promotes phospho-tyrosine-dependent leupaxin protein interactions within focal adhesions, thus limiting the amount of leupaxin available to shuttle to the nucleus. It remains to be determined whether FAK activity may also regulate SM gene expression through cytoplasmic retention of other LIM-containing SRF co-factors.
Once in the nucleus, LIM proteins can participate in transcriptional control, a function likely dependent on the ability of these proteins to provide scaffolds for transcription factors and/or chromatin remodeling factors. Both paxillin and Hic-5 bind to steroid receptors and have been shown to co-activate androgen, glucocorticoid, and progesterone response genes by bridging an association with the nuclear matrix 29,30. In addition, Hic-5 has been shown to influence Sp1-dependent c-fos and p21 expression by inducing a complex between Sp1, SMAD3, and the histone acetyltransferase, p300 31. The finding that Hic-5 and leupaxin appear to be reciprocally expressed during development (with higher levels of Hic-5 observed in less differentiated SMC) coupled with evidence that Hic-5 regulates Sp1-dependent transcription, while leupaxin regulates SRF-dependent transcription, is particularly interesting with respect to SMC phenotypic switching. Although Sp1 can act as either a transcription enhancer in certain cell types, in SMC, Sp1 acts as a repressor of SMC marker gene transcription by interacting with G/C rich repressor elements contained in SM22 and SM-MHC promoters 32. Thus, it is possible that these two FAK binding partners have opposing functions with respect to SM gene transcription.
Although the paxillin family members may induce divergent gene regulation in SMC, several other LIM domain proteins besides leupaxin including CRP1/2, LPP, and FHL2 (which also cycle between focal adhesions and the nucleus) share the ability to influence SRF-mediated gene transcription 19,20,33. Although some of these LIM proteins may have partially overlapping functions, it is likely that additional regulatory pathways that alter the expression levels, localization, or protein binding interactions ultimately impart specific functions for each of these molecules.
In conclusion, we propose a model that leupaxin may play a dual role in SM function; aiding to coordinate multi-protein complexes in focal adhesions and in the nucleus. Since leupaxin localization is clearly modulated by FAK/Src activity in SMC, we postulate that ECM- and agonist-dependent regulation of FAK activity in vessels could impart precise control of leupaxin localization in order to balance the migratory and contractile capacities necessary for proper vasculogenesis during development and following vascular injury. Interestingly, recent studies in our laboratory indicate that FAK inactivation in SMC by homologous recombination promotes TGF-β induced SMC differentiation but attenuates PDGF-stimulated SMC motility (unpublished observaitons). Thus, it will be of future interest to determine to what extent these phenotypes are due to mis-localization of leupaxin. Although our data provide clear evidence of a role for nuclear leupaxin in promoting SRF-dependent gene transcription, future gene targeting studies will be necessary to determine under which circumstances leupaxin activity might be necessary for specific SMC functions.
Supplementary Material
Acknowledgments
The authors would like to thank Marisa Deburkarte (Lab Assistant, Dept. of Pathology, UNC) for excellent technical assistance and Vincent Moylan (Clinical Investigator, Dept. of Pathology, UNC) for human tissue procurement.
Sources of funding: This work was supported in part by grants from NIH-NHLBI (HL-081844 and HL-071054 to JMT; HL070953 to CPM) and the American Heart Association (0355776U to JMT; 0555476U to C.P.M; 0515329U to LSS).
Footnotes
Disclosures: none
References
- 1.Yoshida T, Owens GK. Molecular determinants of vascular smooth muscle cell diversity. Circ Res. 2005;96:280–91. doi: 10.1161/01.RES.0000155951.62152.2e. [DOI] [PubMed] [Google Scholar]
- 2.Wang D, Chang PS, Wang Z, Sutherland L, Richardson JA, Small E, Krieg PA, Olson EN. Activation of cardiac gene expression by myocardin, a transcriptional cofactor for serum response factor. Cell. 2001;105:851–62. doi: 10.1016/s0092-8674(01)00404-4. [DOI] [PubMed] [Google Scholar]
- 3.Chen J, Kitchen CM, Streb JW, Miano JM. Myocardin: a component of a molecular switch for smooth muscle differentiation. J Mol Cell Cardiol. 2002;34:1345–56. doi: 10.1006/jmcc.2002.2086. [DOI] [PubMed] [Google Scholar]
- 4.Lockman K, Hinson JS, Medlin MD, Morris D, Taylor JM, Mack CP. Sphingosine 1-phosphate stimulates smooth muscle cell differentiation and proliferation by activating separate serum response factor co-factors. J Biol Chem. 2004;279:42422–30. doi: 10.1074/jbc.M405432200. [DOI] [PubMed] [Google Scholar]
- 5.Li S, Wang DZ, Wang Z, Richardson JA, Olson EN. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc Natl Acad Sci U S A. 2003;100:9366–70. doi: 10.1073/pnas.1233635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Zhu X, Chen M, Cheng L, Zhou D, Lu MM, Du K, Epstein JA, Parmacek MS. Myocardin-related transcription factor B is required in cardiac neural crest for smooth muscle differentiation and cardiovascular development. Proc Natl Acad Sci U S A. 2005;102:8916–21. doi: 10.1073/pnas.0503741102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bobik A. Transforming growth factor-betas and vascular disorders. Arterioscler Thromb Vasc Biol. 2006;26:1712–20. doi: 10.1161/01.ATV.0000225287.20034.2c. [DOI] [PubMed] [Google Scholar]
- 8.Sinha S, Hoofnagle MH, Kingston PA, McCanna ME, Owens GK. Transforming growth factor-beta1 signaling contributes to development of smooth muscle cells from embryonic stem cells. Am J Physiol Cell Physiol. 2004;287:C1560–8. doi: 10.1152/ajpcell.00221.2004. [DOI] [PubMed] [Google Scholar]
- 9.Leveen P, Pekny M, Gebre-Medhin S, Swolin B, Larsson E, Betsholtz C. Mice deficient for PDGF B show renal, cardiovascular, and hematological abnormalities. Genes Dev. 1994;8:1875–87. doi: 10.1101/gad.8.16.1875. [DOI] [PubMed] [Google Scholar]
- 10.Goumans MJ, Zwijsen A, van Rooijen MA, Huylebroeck D, Roelen BA, Mummery CL. Transforming growth factor-beta signalling in extraembryonic mesoderm is required for yolk sac vasculogenesis in mice. Development. 1999;126:3473–83. doi: 10.1242/dev.126.16.3473. [DOI] [PubMed] [Google Scholar]
- 11.Bouvard D, Brakebusch C, Gustafsson E, Aszodi A, Bengtsson T, Berna A, Fassler R. Functional consequences of integrin gene mutations in mice. Circ Res. 2001;89:211–23. doi: 10.1161/hh1501.094874. [DOI] [PubMed] [Google Scholar]
- 12.George EL, Baldwin HS, Hynes RO. Fibronectins are essential for heart and blood vessel morphogenesis but are dispensable for initial specification of precursor cells. Blood. 1997;90:3073–81. [PubMed] [Google Scholar]
- 13.Ilic D, Kovacic B, McDonagh S, Jin F, Baumbusch C, Gardner DG, Damsky CH. Focal adhesion kinase is required for blood vessel morphogenesis. Circ Res. 2003;92:300–7. doi: 10.1161/01.res.0000055016.36679.23. [DOI] [PubMed] [Google Scholar]
- 14.Glukhova MA, Frid MG, Shekhonin BV, Balabanov YV, Koteliansky VE. Expression of fibronectin variants in vascular and visceral smooth muscle cells in development. Dev Biol. 1990;141:193–202. doi: 10.1016/0012-1606(90)90114-x. [DOI] [PubMed] [Google Scholar]
- 15.Glukhova M, Koteliansky V, Fondacci C, Marotte F, Rappaport L. Laminin variants and integrin laminin receptors in developing and adult human smooth muscle. Dev Biol. 1993;157:437–47. doi: 10.1006/dbio.1993.1147. [DOI] [PubMed] [Google Scholar]
- 16.Lipsky BP, Beals CR, Staunton DE. Leupaxin is a novel LIM domain protein that forms a complex with PYK2. J Biol Chem. 1998;273:11709–13. doi: 10.1074/jbc.273.19.11709. [DOI] [PubMed] [Google Scholar]
- 17.Sahu SN, Nunez S, Bai G, Gupta A. Interaction of Pyk2 and PTP-PEST with leupaxin in prostate cancer cells. Am J Physiol Cell Physiol. 2007;292:C2288–96. doi: 10.1152/ajpcell.00503.2006. [DOI] [PubMed] [Google Scholar]
- 18.Kadrmas JL, Beckerle MC. The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol. 2004;5:920–31. doi: 10.1038/nrm1499. [DOI] [PubMed] [Google Scholar]
- 19.Philippar U, Schratt G, Dieterich C, Muller JM, Galgoczy P, Engel FB, Keating MT, Gertler F, Schule R, Vingron M, Nordheim A. The SRF target gene Fhl2 antagonizes RhoA/MAL-dependent activation of SRF. Mol Cell. 2004;16:867–80. doi: 10.1016/j.molcel.2004.11.039. [DOI] [PubMed] [Google Scholar]
- 20.Chang DF, Belaguli NS, Iyer D, Roberts WB, Wu SP, Dong XR, Marx JG, Moore MS, Beckerle MC, Majesky MW, Schwartz RJ. Cysteine-rich LIM-only proteins CRP1 and CRP2 are potent smooth muscle differentiation cofactors. Dev Cell. 2003;4:107–18. doi: 10.1016/s1534-5807(02)00396-9. [DOI] [PubMed] [Google Scholar]
- 21.Hervy M, Hoffman L, Beckerle MC. From the membrane to the nucleus and back again: bifunctional focal adhesion proteins. Curr Opin Cell Biol. 2006;18:524–32. doi: 10.1016/j.ceb.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 22.Gabarra-Niecko V, Keely PJ, Schaller MD. Characterization of an activated mutant of focal adhesion kinase: ‘SuperFAK’. Biochem J. 2002;365:591–603. doi: 10.1042/BJ20020065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta A, Lee BS, Khadeer MA, Tang Z, Chellaiah M, Abu-Amer Y, Goldknopf J, Hruska KA. Leupaxin is a critical adaptor protein in the adhesion zone of the osteoclast. J Bone Miner Res. 2003;18:669–85. doi: 10.1359/jbmr.2003.18.4.669. [DOI] [PubMed] [Google Scholar]
- 24.Sahu SN, Khadeer MA, Robertson BW, Nunez SM, Bai G, Gupta A. Association of leupaxin with Src in osteoclasts. Am J Physiol Cell Physiol. 2007;292:C581–90. doi: 10.1152/ajpcell.00636.2005. [DOI] [PubMed] [Google Scholar]
- 25.Chew V, Lam KP. Leupaxin negatively regulates B cell receptor signaling. J Biol Chem. 2007;282:27181–91. doi: 10.1074/jbc.M704625200. [DOI] [PubMed] [Google Scholar]
- 26.Shibanuma M, Kim-Kaneyama JR, Ishino K, Sakamoto N, Hishiki T, Yamaguchi K, Mori K, Mashimo J, Nose K. Hic-5 communicates between focal adhesions and the nucleus through oxidant-sensitive nuclear export signal. Mol Biol Cell. 2003;14:1158–71. doi: 10.1091/mbc.02-06-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsujita Y, Muraski J, Shiraishi I, Kato T, Kajstura J, Anversa P, Sussman MA. Nuclear targeting of Akt antagonizes aspects of cardiomyocyte hypertrophy. Proc Natl Acad Sci U S A. 2006;103:11946–51. doi: 10.1073/pnas.0510138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller JM, Isele U, Metzger E, Rempel A, Moser M, Pscherer A, Breyer T, Holubarsch C, Buettner R, Schule R. FHL2, a novel tissue-specific coactivator of the androgen receptor. Embo J. 2000;19:359–69. doi: 10.1093/emboj/19.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujimoto N, Yeh S, Kang HY, Inui S, Chang HC, Mizokami A, Chang C. Cloning and characterization of androgen receptor coactivator, ARA55, in human prostate. J Biol Chem. 1999;274:8316–21. doi: 10.1074/jbc.274.12.8316. [DOI] [PubMed] [Google Scholar]
- 30.Kasai M, Guerrero-Santoro J, Friedman R, Leman ES, Getzenberg RH, DeFranco DB. The Group 3 LIM domain protein paxillin potentiates androgen receptor transactivation in prostate cancer cell lines. Cancer Res. 2003;63:4927–35. [PubMed] [Google Scholar]
- 31.Shibanuma M, Kim-Kaneyama JR, Sato S, Nose K. A LIM protein, Hic-5, functions as a potential coactivator for Sp1. J Cell Biochem. 2004;91:633–45. doi: 10.1002/jcb.10754. [DOI] [PubMed] [Google Scholar]
- 32.Wamhoff BR, Hoofnagle MH, Burns A, Sinha S, McDonald OG, Owens GK. A G/C element mediates repression of the SM22alpha promoter within phenotypically modulated smooth muscle cells in experimental atherosclerosis. Circ Res. 2004;95:981–8. doi: 10.1161/01.RES.0000147961.09840.fb. [DOI] [PubMed] [Google Scholar]
- 33.Gorenne I, Jin L, Yoshida T, Sanders JM, Sarembock IJ, Owens GK, Somlyo AP, Somlyo AV. LPP expression during in vitro smooth muscle differentiation and stent-induced vascular injury. Circ Res. 2006;98:378–85. doi: 10.1161/01.RES.0000202802.34727.fd. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.