Summary
The nuclear bile acid receptor FXR is critical for regulation of lipid and glucose metabolism. Here we report that FXR is a target of SIRT1, a deacetylase that mediates nutritional and hormonal modulation of hepatic metabolism. Lysine 217 of FXR is the major acetylation site targeted by p300 and SIRT1. Acetylation of FXR increases its stability but inhibits heterodimerization with RXRα, DNA binding, and transactivation activity. Down-regulation of hepatic SIRT1 increased FXR acetylation with deleterious metabolic outcomes. Surprisingly, in mouse models of metabolic disease, FXR interaction with SIRT1 and p300 was dramatically altered, FXR acetylation levels were elevated, and overexpression of SIRT1 or resveratrol treatment reduced acetylated FXR levels. Our data demonstrate that FXR acetylation is normally dynamically regulated by p300 and SIRT1 but is constitutively elevated in metabolic disease states. Small molecules that inhibit FXR acetylation by targeting SIRT1 or p300 may be promising therapeutic agents for metabolic disorders.
Introduction
Farnesoid X receptor (FXR) is a ligand-regulated transcription factor that belongs to a large superfamily of nuclear receptors (Mangelsdorf and Evans, 1995). Activated by physiological concentrations of bile acids, FXR regulates expression of numerous bile acid-responsive genes, mainly in the liver and intestine, to regulate cholesterol and bile acid homeostasis (Cariou and Staels, 2007; Kalaany and Mangelsdorf, 2006; Lee et al., 2006). Other studies have established that FXR is also a master regulator of lipid and glucose homeostasis (Lee et al., 2006; Ma et al., 2006) and is critically involved in liver regeneration (Huang et al., 2006), and protection of intestines from intestinal bacteria growth (Inagaki et al., 2006). Disruption of the FXR gene in transgenic mice is associated with metabolic diseases, including diabetes and hypercholesterolemia (Sinal et al., 2000). Interestingly, activation of FXR in diabetic mice improved metabolic outcomes by reducing serum glucose and lipid levels (Zhang et al., 2006). Although these critical roles of FXR in normal physiology and metabolic disease processes have been established, the molecular basis of how FXR activity is modulated in health and disease states remains largely unexplored.
Nuclear receptors collaborate with numerous transcriptional cofactors to effectively modulate transcription of their target genes (Rosenfeld et al., 2006). Transcriptional regulation by nuclear receptors, therefore, involves the recruitment of cofactors to target gene promoters that results in histone modification. In addition, these cofactors also modulate receptor activity by post-translational modification of the receptor itself, including acetylation and deacetylation. Previous studies have shown that the activities of regulatory proteins, such as PGC-1 , Foxo-1, LXR, and p53, are modulated by protein acetylation and deacetylation (Daitoku et al., 2004; Kitamura et al., 2005; Lerin et al., 2006; Li et al., 2007; Luo et al., 2001; Motta et al., 2004; Rodgers et al., 2005). We recently reported that p300 acetylates FXR, as well as histones at FXR target gene promoters (Fang et al., 2008), but the role of FXR acetylation in normal and metabolic disease states remains unclear.
A mammalian homologue of yeast sir2, SIRT1, regulates cellular metabolism such that aging and life longevity are affected (Guarente, 2007; Sinclair et al., 2006). SIRT1 is a NAD+-dependent deacetylase that removes acetyl groups from modified lysine residues in both histones and transcription factors (Sinclair et al., 2006). Recent studies demonstrate that SIRT1 plays an important role in the regulation of metabolism in response to hormonal and nutritional cues by modulating the activity of PGC-1α, a master metabolic regulator (Rodgers et al., 2008; Rodgers et al., 2005). For instance, while acetylation of PGC-1α by GCN5 acetylase decreased PGC-1α activity by altering its intra-nuclear distribution, deacetylation of PGC-1α by SIRT1 increased its activity (Lerin et al., 2006; Rodgers et al., 2005). Interestingly, activation of SIRT1 by a polyphenolic compound, resveratrol, in mouse models of metabolic disease reduced acetylation levels of PGC-1α and improved metabolic profiles (Baur et al., 2006; Lagouge et al., 2006).
Both FXR and SIRT1 are critically involved in liver metabolic regulation (Lee et al., 2006; Lerin et al., 2006; Rodgers and Puigserver, 2007; Zhang et al., 2004) and activation of these proteins in mouse models of metabolic disease improved metabolic outcomes (Baur et al., 2006; Lagouge et al., 2006; Zhang et al., 2006). Therefore, these previous studies, along with our recent findings that FXR is acetylated by p300 (Fang et al., 2008), prompted us to ask whether SIRT1 modulates the activity of FXR by deacetylation. Using molecular, cellular, and animal in vivo studies, we investigated the biological function of FXR acetylation in normal and metabolic disease states. Here, we demonstrate that FXR is a target of SIRT1 in metabolic regulation. Acetylation of FXR inhibits its activity and is dynamically regulated by p300 and SIRT1 under normal conditions. Down-regulation of hepatic SIRT1 by siRNA increased FXR acetylation with deleterious metabolic outcomes. Surprisingly, FXR acetylation levels were constitutively elevated in two mouse models of metabolic disorders, ob/ob mice and mice chronically fed a western style diet. Treatment with the SIRT1 activator, resveratrol, or adenoviral-mediated overexpression of SIRT1 substantially reduced FXR acetylation levels in these disease model mice. Our studies provide an intriguing correlation between elevated FXR acetylation by decreased SIRT1 activity, decreased FXR activities, and deleterious metabolic effects in hepatic metabolic disease.
Results
K217 in the hinge region of FXR is the major acetylation site
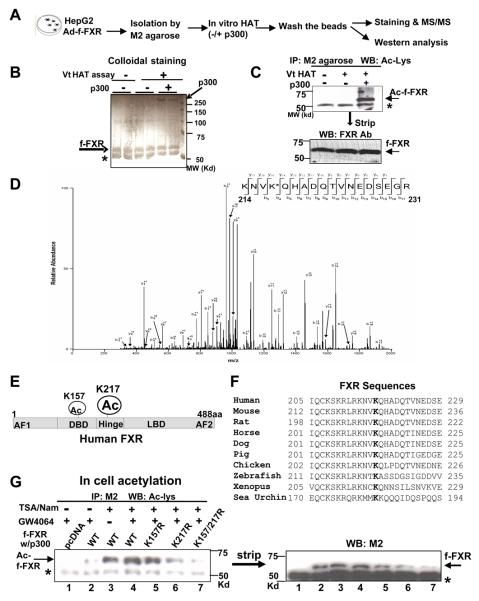
As the first step in defining the functional role of FXR acetylation, we identified lysine (K) residues acetylated using tandem mass spectrometry (MS/MS). Flag-tagged FXR was expressed in HepG2 cells and isolated flag-FXR was acetylated by p300 in vitro (Fig. 1A-C). The MS/MS analysis revealed that K217 in the hinge region was the major site acetylated and that K157 in the DNA binding domain was also acetylated (Fig. 1D, E, Fig. S1). K217 is highly conserved in FXR in vertebrates (Fig. 1F).
Fig. 1. The major site acetylated in FXR by p300 is K217.
A) Experimental outline. B, C) After incubation with p300, proteins were separated using SDS-PAGE and stained by colloidal Coomassie Blue (B) or acetylated flag-FXR was detected by western analysis (C). In C (lower panel), the membrane was stripped and reprobed with FXR antibody. An asterisk denotes IgG heavy chain. D) Tandem MS (MS/MS) spectrum of the FXR peptide showing acetylation at K217. E) A schematic diagram of acetylation sites in FXR is shown. F) Alignment of the FXR region containing K217 from various species. G) Cos-1 cells transfected with p300 and with flag-FXR (fFXR) plasmids were treated as indicated for 2 hr. Flag-FXR was immunoprecipitated and acetylated FXR was detected by western analysis.
To confirm these results, cells were cotransfected with plasmids for p300 and either FXR or one of the acetylation-defective FXR mutants, K157R and K217R. Acetylated FXR was detected by immunoprecipitation under stringent conditions with buffers containing SDS, followed by western analysis using an anti-acetyl lysine antibody. FXR acetylation levels were increased by treatment with deacetylase inhibitors, trichostatin A (TSA) and nicotinamide (Nam) (Fig. 1G, lanes 2 and 4), suggesting that FXR undergoes dynamic acetylation and deacetylation. Treatment with GW4064, a synthetic FXR agonist (Willson et al., 2001), modestly increased FXR acetylation (lanes, 3 and 4). FXR acetylation levels were substantially reduced in the K217R mutant, and nearly abolished in the K157/217R double mutant (lanes, 4-7). These results indicate that K217 of FXR is the major acetylation site by p300 and that K157 is also acetylated.
Acetylation of FXR increases its stability
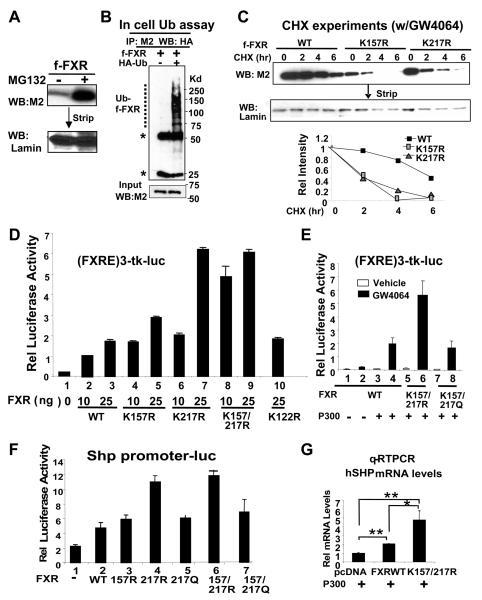
Since FXR protein levels of the acetylation mutants were markedly reduced (Fig. 1G), we tested whether acetylation of FXR affects its stability. Treatment with MG132, a proteasome inhibitor, dramatically increased FXR levels and resulted in its ubiquitination in vitro and in cells (Fig. 2A, B, Fig. S2, 3). The degradation rate was determined by monitoring the decrease of flag-FXR wild type or mutants after blocking protein synthesis by cycloheximide (CHX). The half-lives were 5-6 hr for ligand-activated flag-FXR wild type and about 2 hr for the K157R or K217R mutants (Fig. 2C), indicating that FXR acetylation increases its stability. These results suggest that FXR is a target of ubiquitin-proteasomal degradation and acetylation of FXR increases protein stability by inhibiting its degradation.
Fig. 2. FXR acetylation increases its stability but inhibits transactivation ability.
A) HepG2 cells infected with Ad-flag-FXR were treated with vehicle or MG132 and flag-FXR levels were detected. B) HepG2 cells were cotransfected with expression plasmids as indicated. Flag-FXR was immunoprecipitated from cell extracts and ubiquitinated flag-FXR was detected. C) Transfected Cos-1 cells were treated with CHX for the indicated times and flag-FXR levels in cell extracts were detected. D-F) HepG2 cells were cotransfected with indicated reporter plasmids, expression plasmids for p300 and FXR WT, or FXR mutants, as indicated. Cells were treated with 200 nM of GW4064 overnight. The values for firefly luciferase activities were normalized by dividing by β-galactosidase activities. G) HepG2 cells were cotransfected with plasmids as indicated and treated with GW4064. The mRNA levels were determined by q-RTPCR. Statistical significance was measured using the Student's t test. *, **, and NS indicate p<0.05, p<0.01, and statistically not significant, respectively, SEM (n=3).
Mutation of FXR acetylation sites increases transactivation
To determine whether acetylation of FXR affects its transactivation ability, the effects of mutation of K157 and/or K217 on FXR transactivation were examined. Overexpression of K157R and K217R mutants increased FXR activity compared to wild type and mutation of both sites synergistically increased FXR transactivation activity at the lower amounts of plasmids transfected (Fig. 2D, lanes, 2, 4, 6, 8). Mutation of K122 to arginine did not alter FXR acetylation levels (not shown) and had little effect on FXR transactivation (lanes, 3 and 10). In the absence of activation of FXR by GW4046, little transactivation was observed (fig. 2E). Mutation of K157 and K217 to glutamine residues (Q) to mimic acetylation resulted in transactivation by FXR similar to that of the wild type (Fig. 2E, lanes 4, 8). Similar results were observed in reporter assays using a natural SHP promoter-luc (Fig. 2F). With both reporters, the effects of the K217 mutant were greater than those of the K157 mutant.
We also examined the effects of FXR acetylation on expression of the endogenous SHP gene in HepG2 cells. The mRNA levels of SHP were significantly increased when the K157/217R mutant was coexpressed with p300 (Fig. 2G). These results indicate that FXR transactivation activity is decreased when FXR is acetylated.
Mutation of K157 and K217 increases binding of FXR/RXRα heterodimer to DNA
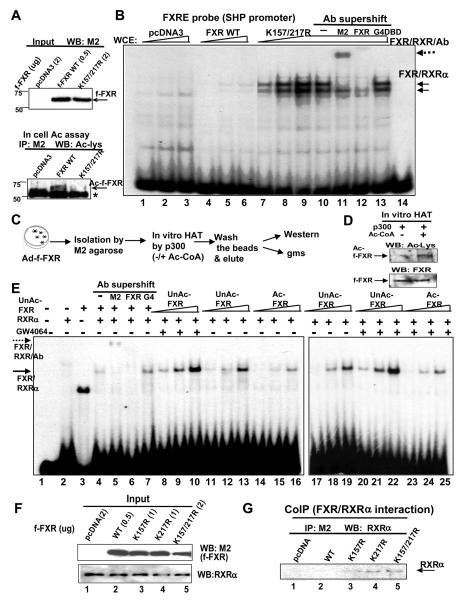
Since FXR activity was decreased by acetylation of FXR, we tested whether acetylation alters binding of the FXR/RXR heterodimer to the DNA. Cells were cotransfected with p300 along with different amounts of expression plasmids for flag-FXR or the K157/K217 mutant to yield similar protein expression levels (Fig. 3A). Acetylation of FXR was reduced in the K157/K217 mutant compared to wild type (Fig. 3A, lower panel). Antibody supershift gel mobility shift assays identified the FXR/RXR/DNA complexes (Fig. 3B, lanes 10-13). While little binding of wild type FXR was observed, robust DNA binding was detected with the K157/K217R mutant (lanes 4-6, 7-9). These results suggest that FXR acetylation inhibits binding of FXR/RXRα to DNA.
Fig. 3. Acetylation of FXR inhibits binding of FXR/RXRα to DNA.
A) flag-FXR (upper panel) or acetylated flag-FXR (lower panel) was detected in Cos-1 cell extracts. B) Increasing amounts of cell extracts were incubated with flag-RXRα and the radiolabeled oligonucleotide probe containing the FXRE from the SHP promoter, and complexes were detected using gel mobility shift (gms) assay. C) Experimental outline for the gms using flag-FXR acetylated in vitro. D, E) Purified flag-FXR was incubated with p300 (−/+ acetyl CoA). In D, acetylated flag-FXR was detected by western analysis. In E, increasing amounts of unacetylated or acetylated flag-FXR were incubated with purified RXRα and the complexes were detected. F, G) Cos-1 cells were cotransfected with expression plasmids as indicated with p300 plasmids and expressed protein levels were determined. Interaction between flag-FXR and RXRα was detected by CoIP assays.
To directly test the effect of acetylation of FXR on its DNA binding, purified flag-FXR was acetylated by p300 in vitro and acetylation of flag-FXR was confirmed (Fig. 3C, D). In gel shift assays, a single protein/DNA complex was detected which was supershifted by the M2 and FXR antibodies (Fig. 3E, lanes 4-7). Binding of unacetylated FXR/RXR was detected in a dose-dependent manner (lanes, 8-13, 17-22), whereas binding of acetylated FXR was substantially reduced (lanes, 14-16, 23-25). Addition of GW4064 resulted in increased DNA binding of the unacetylated FXR (lanes, 8-13, 17-22), but had little effect on binding of acetylated FXR (lanes 14-16, 23-25). These results demonstrate that binding of FXR/RXR to the DNA is inhibited when FXR is acetylated.
FXR acetylation inhibits FXR/RXRα heterodimerization
Since DNA binding of FXR/RXR could be impaired if formation of FXR/RXRα heterodimers is blocked, we examined the effects of mutation of K157 and K217 on the interaction of FXR with RXRα in cells coexpressing p300. RXRα was co-immunoprecipitated with the FXR acetylation mutants, whereas RXRα was not detected in FXR immunoprecipitates of wild type FXR (Fig. 3F, G, Fig. S4). These results indicate that FXR acetylation inhibits FXR/RXRα heterodimerization.
Since FXR acetylation decreases heterodimerization and DNA binding of FXR/RXR, then acetylation effects on its transactivation ability should be reduced in FXR fused to a Gal4 DNA binding domain (G4DBD). Binding to a Gal4 promoter reporter should be independent of heterodimerization. In contrast to the enhanced transactivation ability in (FXRE)3-tk-luc (Fig. 2D, E) or SHP promoter-luc (Fig. 2F), expression of G4DBD-K217R did not significantly increase its transactivation (Fig. S5). These results, together with gel shift assays and CoIP protein interaction studies, demonstrate that FXR acetylation inhibits FXR/RXRα heterodimerization, which accounts for, at least in part, impaired binding of FXR/RXR to the DNA, and reduced FXR transactivation ability.
SIRT1 is associated with FXR in mouse liver in vivo
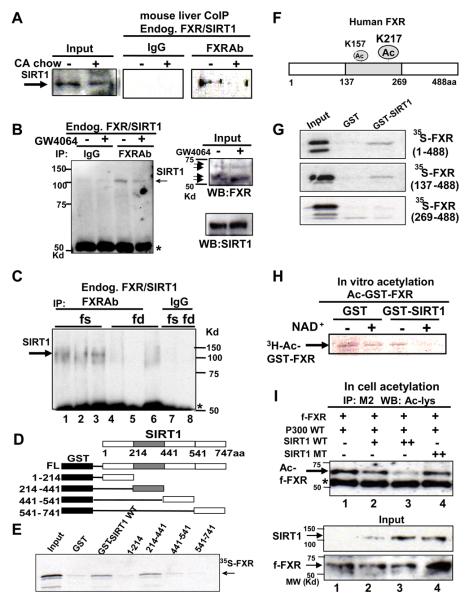
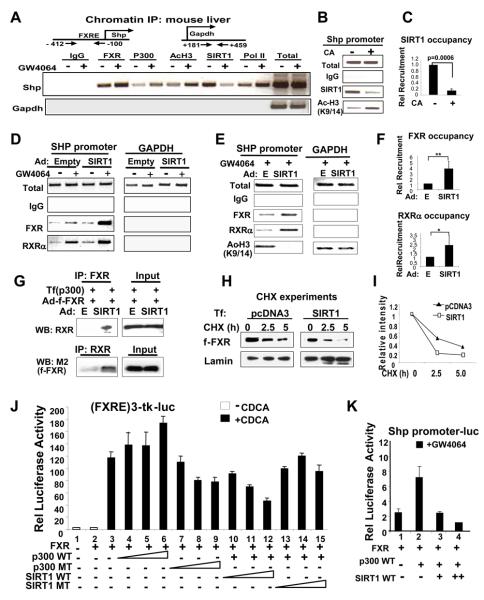
The level of acetylation of FXR is likely to be a balance between acetylation and deacetylation of the protein. The SIRT1 deacetylase is emerging as a master metabolic regulator (Guarente, 2007; Rodgers et al., 2008) and is a possible candidate for modulating FXR acetylation. We, therefore, examined whether SIRT1 associates with FXR in mouse liver. In CoIP assays, the amount of SIRT1 associated with flag-FXR was substantially reduced in mice fed cholic acid (CA), a primary bile acid and natural FXR agonist (Fig. S6). To determine if overexpression of flag-FXR resulted in non-specific interaction with SIRT1, we also examined the interaction between endogenous SIRT1 and endogenous FXR. CA feeding reduced the interaction of endogenous FXR with SIRT1 (Fig. 4A). Similarly, treatment with GW4046 reduced the interaction of FXR and SIRT1 in mouse liver (Fig. 4B) and in cultured cells (Fig. S7). In addition, the interaction of FXR with SIRT1 was substantially increased by fasting (Fig. 4C, Fig. S8). Conversely, FXR interaction with p300 was substantially increased after feeding (Fig. S9). These results demonstrate that FXR and SIRT1 interact in mouse liver and that the interaction is decreased by activation of FXR or in response to feeding.
Fig. 4. SIRT1 directly interacts with and deacetylates FXR.
A-C) Mice were fed normal or CA chow (+) for 6 hr (A) or treated with vehicle (−) or GW4064 (+) for 1 hr (B) or fasted (fs) overnight or refed (fd) after overnight fasting (C). CoIP studies were performed to examine FXR/SIRT1 interaction. D) Schematic diagrams of GST-SIRT1 full length (FL) and deletion mutants. The gray shaded area represents the sirtuin homology domain. E) 35S-FXR was synthesized in vitro, and GST pull down assays were performed. F) A schematic diagram of FXR acetylation sites. G) 35S-FXR full length wild type and mutants were synthesized, and binding to GST-FXR fusion proteins was determined. H) GST-FXR, which had been acetylated using p300 with 3H-acetyl CoA, was incubated with eluted GST or GST-SIRT1 and analyzed by SDS-PAGE followed by fluorography. I) Cos-1 cells were cotransfected with the indicated plasmids and treated with deacetylase inhibitors for 3 hr. Flag-FXR levels were immunoprecipitated and acetylated flag-FXR was detected by western analysis. Acetylated flag-FXR and IgG heavy chain are indicated by arrow and asterisk, respectively. SIRT1 and flag-FXR levels in input are shown.
SIRT1 directly interacts with and deacetylates FXR
The CoIP studies demonstrate that FXR and SIRT1 interact directly or indirectly within a complex. We next examined whether SIRT1 can bind directly to FXR using GST-pull downs. FXR interacted with full length GST-SIRT1 and with the GST-SIRT1 fragment (214-441) which contains the sirtuin homology domain, but not with fragments from other regions (Fig. 4D, E). Conversely, FXR and the fragment (137-488), but not (269-488), bound to GST-SIRT1 (Fig. 4F, G). The FXR region from residues (137-269) required for interaction with SIRT1 contains both K157/K217 acetylation sites. These results show that FXR interacts directly with SIRT1.
To test if SIRT1 can deacetylate FXR, in vitro deacetylation studies were performed. 3H-labeled acetylated GST-FXR levels were decreased by incubation with GST-SIRT1 in a NAD+-dependent manner (Fig. 4H, Fig. S10). To determine if SIRT1 decreases FXR acetylation levels in cells, increasing amounts of SIRT1 or a catalytically inactive SIRT1 mutant were cotransfected with expression plasmids for flag-FXR and p300. FXR acetylation levels were decreased by SIRT1, whereas levels were not decreased in cells cotransfected with the SIRT1 mutant (Fig. 4I). These results indicate that SIRT1 directly interacts with and deacetylates FXR.
FXR agonist treatment results in dissociation of SIRT1 from the Shp promoter
To determine whether FXR and SIRT1 interact at the promoters of FXR target genes in vivo, chromatin immunoprecipitation (ChIP) assays were performed in mouse liver using the Shp gene as a model. As reported (Fang et al., 2008; Goodwin et al., 2000; Lu et al., 2000), GW4064 treatment increased Shp mRNA levels about 8-fold (Fig. S11). Association of SIRT1 with the promoter was detected and GW4064 treatment resulted in dissociation of SIRT1 and recruitment of p300, consistent with the observed increases in histone H3 acetylation at K9/K14, a gene activation histone modification (Fig. 5A). Similarly, the amount of SIRT1 associated with the promoter was significantly reduced in mice fed CA-chow (Fig. 5B, C). These results indicate that the recruitment of SIRT1 to the Shp promoter is decreased by treatment with GW4064 or CA.
Fig. 5. SIRT1 and p300 reciprocally regulate FXR activity.
A-C) Mice were treated with vehicle (−) or GW4064 (+) for 3 hr (A) or fed CA chow for 6 hr (B). Livers were collected for ChIP assay. C) Band intensities were quantified using Image J and the values for control samples set to 1. The SEM is indicated and statistical significance was determined by the Student's t test, n=3. D-F) HepG2 cells were infected with control Ad-empty or Ad-SIRT1 and 24 h later, cells were treated with GW4064 for 1 hr and ChIP assays were performed. In F, relative recruitment of FXR and RXRα was determined as described in C. G) HepG2 cells were transfected with plasmids as indicated and then infected with adenoviral vectors as indicated. Cells were treated with GW4064 and CoIP assays were performed. H, I) Transfected HepG2 cells were further infected with Ad-flag-FXR and then treated with CHX for indicated times. J, K) HepG2 cells transfected with indicated plasmids were treated with FXR agonists overnight and reporter assays were performed.
SIRT1 increases association of FXR/RXRα with the promoter
Acetylation of FXR inhibited its interaction with RXRα and binding of FXR/RXRα heterodimer to the DNA (Fig. 3). Therefore, deacetylation of FXR by SIRT1 should increase binding of this heterodimer to the promoter. Indeed, adenoviral-mediated overexpression of flag-SIRT1 significantly increased association of FXR and RXRα with the SHP promoter chromatin in cells treated with GW4064 (Fig. 5D-F). Further, in CoIP assays, interaction of FXR with RXRα was substantially increased in cells overexpressing SIRT1 (Fig. 5G). Consistent with the histone deacetylation activity of SIRT1 (Vaquero et al., 2006), acetylated H3 levels were substantially decreased in cells overexpressing SIRT1 (Fig. 5E). These results demonstrate that SIRT1 increases FXR/RXRα interaction and binding of this heterodimer to the promoter chromatin.
SIRT1 decreases the stability of FXR
Mutation of K157 and K217 reduced acetylation of FXR and decreased its stability (Fig. 2C), therefore, deacetylation of FXR by SIRT1 should destabilize FXR. As expected, expression of SIRT1 increased the degradation rate of FXR (Fig. 5H, I, Fig. S12). These results suggest that SIRT1 deacetylates FXR, which increases the degradation of FXR in hepatocytes.
SIRT1 and p300 reciprocally modulate FXR transactivation
In previous cell-based reporter studies, we observed that p300 increased FXR transactivation of the SHP gene (Fang et al., 2008). Therefore, a deacetylase, such as SIRT1, should have the opposite effect. Consistent with previous studies, cotransfection of increasing amounts of expression plasmids for p300, but not of the inactive p300 mutant, increased FXR transactivation (Fig. 5J, lanes, 3-9). In contrast, increasing amounts of SIRT1 progressively decreased the p300-enhanced transactivation while an inactive SIRT1 mutant had smaller effects (lanes 6, 10-15). Similar results were observed with the natural Shp promoter-luc reporter (Fig. 5K). These data, together with acetylation studies and chromatin IP studies, suggest that p300 and SIRT1 reciprocally regulate FXR activity by modulating acetylation levels of both FXR and histones. Acetylation of histones at the promoter is likely responsible for increased gene transcription. In contrast, the increased acetylation of FXR by p300 may act to limit and reverse the increased transactivation by inhibiting the interaction with RXRα and binding to DNA.
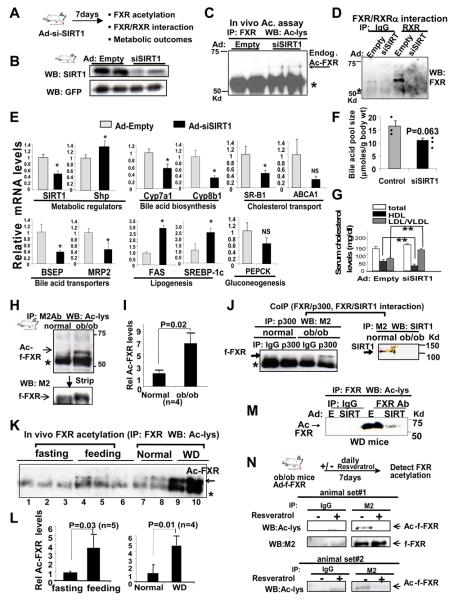
Down-regulation of SIRT1 increases FXR acetylation with deleterious metabolic outcomes
To directly determine whether acetylation levels of FXR in vivo are dependent on SIRT1, mice were infected with Ad-siSIRT1 to down-regulate SIRT1. Endogenous SIRT1 mRNA and protein levels were decreased by siSIRT1 expression (Fig. 6A, B, E). Acetylation levels of endogenous hepatic FXR were markedly elevated in mice infected with Ad-siSIRT1 (Fig. 6C). Furthermore, the interaction of endogenous FXR with RXRα in these mice was decreased as expected upon increased FXR acetylation (Fig. 6D). Also consistent with results from cell studies (Fig. 5), Shp mRNA levels were slightly, but significantly, increased in mice infected with Ad-siSIRT1 (Fig. 6E). Since overexpression of SIRT1 or SIRT1 activation by resveratrol, a polyphenolic SIRT1 activator, showed beneficial gene expression patterns and improved metabolic profiles (Baur et al., 2006; Lagouge et al., 2006; Rodgers and Puigserver, 2007), its down-regulation would be expected to have deleterious effects. Indeed, down-regulation of SIRT1 resulted in increased mRNA levels of SREBP-1c and FAS involved in hepatic lipogenesis, and decreased mRNA levels of SR-B1 and ABCA1 involved in cholesterol transport (Fig. 6E). We and others showed that PGC-1α is an important coactivator for regulation of the Cyp7a1 and Cyp8b1 genes (Bhalla et al., 2004; Miao et al., 2006; Shin et al., 2003), which encode key enzymes in hepatic bile acid biosynthesis. Consistent with these findings, mRNA levels of Cyp7a1 and Cyp8b1 were significantly decreased in mice infected with Ad-siSIRT1 (Fig. 6E). The mRNA levels of hepatic bile acid transporters, BSEP and MRP2, were significantly decreased. The bile acid pool size was also decreased as expected from these changes in gene expression (Fig. 6F). Serum VLDL and LDL levels were significantly elevated and HDL levels were markedly decreased, leading to a deleterious pro-artherogenic serum lipid profile (Fig. 6G). These results indicate that SIRT1 decreases FXR acetylation levels in vivo and further support the idea that decreased SIRT1 activity and elevated FXR acetylation correlate with deleterious metabolic outcomes.
Fig. 6. Elevated FXR acetylation levels in metabolic disease states.
A-G) Mice were infected with either Ad-empty or Ad-siSIRT1 which expresses SIRT1 siRNA. A) Experimental outlines. B) Levels of endogenous SIRT1 and exogenous GFP were detected. Results from two mice are shown. C) Endogenous FXR was immunoprecipitated and acetylated FXR was detected. Results from 2 mice are shown. D) CoIP assays to detect FXR/RXRα interaction. E) The mRNA levels of the indicated genes were determined by q-RTPCR. The mean and SEM (n=3) are shown. F) The total amount of bile acids in liver, gall bladder, and intestines was measured as described in Experimental Procedures. Dots indicate measurements from 3 mice in each group. G) Serum cholesterol levels were measured and the mean and SEM (n=3) are shown. H-J) Normal or ob/ob mice were infected with Ad-flag-FXR as described in Methods. H) Flag-FXR was immunoprecipitated from liver extracts and acetylated flag-FXR in the immunoprecipitates was detected. I) Band intensities of acetylated flag-FXR were quantified using Image J. The values from control mice were set to 1. The mean and SEM (n=4) are shown. J) CoIP assays were performed to detect FXR/p300 (left panel) and FXR/SIRT1 (right panel). K) Mice were fasted overnight or fasted overnight and then refed for 1 hr. Mice were fed either normal chow or WD chow for 16 weeks. Acetylated endogenous FXR levels were detected. L) Band intensities of acetylated FXR were quantified and the values from fasted mice or mice fed normal chow were set to 1. M) Acetylated endogenous FXR levels were detected in WD mice infected with Ad-empty or Ad-flag-SIRT1. N) The ob/ob mice injected with Ad-flag-FXR were treated daily with resveratrol for 1 week and livers were collected for acetylation assays as in Fig. 6H.
Elevated FXR acetylation levels in metabolic disease mice
To test whether acetylated FXR levels are altered in metabolic disease states, acetylation studies were performed in two mouse models of obesity and type II diabetes, the leptin-deficient ob/ob mice and mice fed chronic western-style diet (WD). Acetylation of FXR was substantially increased in the ob/ob mice although the acetylated FXR was barely separated from a strong non-specific band due to IgG (Fig. 6H, I). To reduce the amount of the IgG band, flag-FXR bound to M2 agarose was eluted using flag-peptide and again, the level of acetylated flag-FXR was substantially higher in the ob/ob mice (Fig. S13). Consistent with elevated FXR acetylation levels, the interaction of FXR with p300 was increased, whereas interaction with SIRT1 was decreased in ob/ob mice (Fig. 6J). Elevated FXR acetylation levels were also detected in mice chronically fed WD compared to mice fed normal chow (Fig. S14). Since acetylated FXR levels were elevated in two different mouse models of metabolic diseases, it is consistent with the idea that elevated FXR acetylation underlie metabolic disorders.
FXR acetylation levels in normal physiology and pathophysiology
Finally, we compared the modulation of FXR acetylation by normal physiological stimuli, fasting and feeding, and by metabolic disease conditions using WD mice as a model. FXR acetylation levels were slightly but significantly increased in mice refed after overnight fasting (Fig. 6K, L). Remarkably, FXR acetylation levels were dramatically elevated in WD mice compared to mice fed normal chow (Fig. 6K, L). Consistent with roles of SIRT1 in FXR deacetylation from cell studies (Fig. 4), hepatic overexpression of SIRT1 in these WD mice substantially reduced the FXR acetylation levels (Fig. 6M).
Treatment with resveratrol resulted in beneficial gene expression patterns and improved an overall metabolic outcome, partly due to deacetylation of PGC-1α in metabolic disease model mice (Baur et al., 2006; Lagouge et al., 2006). It is possible that activation of SIRT1 might also decrease the abnormally high levels of acetylated FXR in these disease models, which could contribute to the beneficial effects. To test this possibility, ob/ob mice were treated daily with resveratrol for one week and acetylated FXR levels and metabolic studies were performed. Treatment with resveratrol substantially decreased acetylated FXR levels in ob/ob mice (Fig. 6N). Further, treatment with resveratrol significantly decreased Shp mRNA levels and elevated bile acid pool sizes in WD mice (Fig. S15, S16). These results, together, demonstrate that acetylated FXR levels are dynamically regulated by p300 and SIRT1 under normal conditions but constitutively elevated in metabolic disease model mice and further suggest that elevated acetylated FXR levels reduce FXR activity, which is associated with deleterious outcomes in metabolic disease states.
Discussion
We demonstrate in these studies that acetylation of FXR normally dynamically modulates its activity and that abnormally elevated levels of acetylated FXR are present in mouse models of metabolic disease. The level of FXR acetylation is reciprocally regulated by the acetylase p300 and the deacetylase SIRT1. The major site of FXR acetylated by p300 is K217 within the hinge domain and acetylation was also detected at K157 within the DNA binding domain. Mutation of K157 and K217 resulted in decreased stability of FXR but increased heterodimerization with RXRα, binding to DNA, and transactivation activity indicating that acetylation of FXR increases its stability but inhibits its DNA binding and transactivation activity. Down-regulation of endogenous SIRT1 in mouse liver increased acetylation levels of endogenous FXR demonstrating that SIRT1 affected FXR acetylation in vivo. Interestingly, down-regulation of SIRT1 was associated with deleterious gene expression patterns and metabolic outcomes. Consistent with this observation, FXR acetylation levels were highly elevated in ob/ob mice and high fat western-style diet mice. Furthermore, activation of SIRT1 by treatment with resveratrol, which has been shown to improve metabolic outcomes (Baur et al., 2006; Lagouge et al., 2006), demonstrated reduced the elevated FXR acetylation in these metabolic disease mice. Our results suggest an intriguing connecting between elevated FXR acetylation and decreased FXR activity in animals with deleterious metabolic profiles.
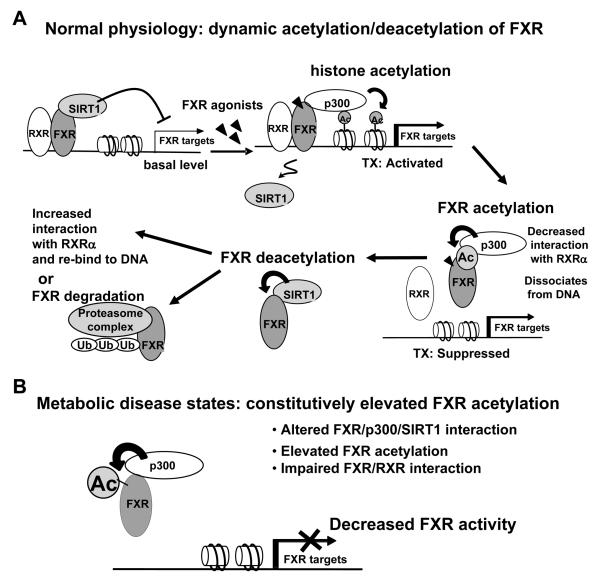
Based on results from the present and published studies (Fang et al., 2008), acetylation is likely to play a complex role in gene regulation by FXR (Fig. 7). Upon activation, FXR recruits p300 to target gene promoters and SIRT1 is dissociated, resulting in increased acetylation of FXR and histones. Acetylation of histones by p300 is associated with gene activation and is probably the major factor in the activation of the FXR target gene as we previously demonstrated (Fang et al., 2008). At the same time, however, acetylation of FXR itself inhibits the activity of FXR. This seemingly paradoxical effect may be important to limit or terminate the response to a stimulus response, which is essential in a dynamically regulated system. Once the acetylated FXR is released from the promoter, FXR is deacetylated by SIRT1, which then either interacts with RXRα and rebinds to the DNA as a heterodimer, or is degraded via the ubiquitin-proteasomal degradation pathway. In the absence of further stimulation, SIRT1 is recruited to the target genes by the unliganded FXR and histones are deacetylated so that gene expression remains at a low basal level. Importantly, acetylation and deacetylation of FXR appear to be a dynamic process under normal physiological conditions, so that the activity of FXR is tightly balanced by the opposing actions of p300 and SIRT1.
Fig. 7. A proposed model of FXR acetylation in normal and disease states.
A) In normal conditions, SIRT1 and FXR are associated with promoters of FXR target genes, such as the SHP gene. When FXR is activated by agonists, p300 is recruited and SIRT1 is dissociated, which results in increased acetylation of histone H3 and subsequently transcriptional activation. At the same time, the process for limiting the stimulated activity is initiated by acetylation of FXR by p300, which impairs FXR interaction with RXRα and DNA binding, resulting in dissociation of FXR from the promoter. SIRT1 deacetylates FXR, which decreases FXR acetylation levels. The deacetylated FXR is either degraded via the ubiquitin-proteasomal pathway or heterodimerizes with RXRα and re-binds to the target gene promoter with SIRT1, resulting in deacetylation of histones and low basal levels of gene expression. The activity of FXR is tightly balanced by the opposing actions of p300 and SIRT1 via FXR and histone acetylation. B) In contrast, in metabolic disease states, FXR acetylation levels are constitutively highly elevated, probably due to abnormally high p300 activity and low SIRT1 activity. Constitutively elevated acetylation of FXR inhibits FXR activity, at least in part, by inhibiting FXR interaction with RXRα and subsequently binding of FXR/RXR to the DNA. Deacetylation of the acetylated FXR may be required for its further activity in normal mice, but does not occur in the diseased mice because of low expression and activity of SIRT1.
In contrast to normal mice, in mice with abnormal metabolic profiles such as ob/ob mice, western diet mice, or mice expressing siSIRT1, the FXR acetylation levels are constitutively and highly elevated. The increased acetylation of FXR may be caused by low activity of SIRT1 since down-regulation of SIRT1 in normal mice led to deleterious metabolic outcomes. In metabolic disease states, continuously acetylated FXR would show impaired interaction with RXRα and DNA binding of FXR/RXRα, and subsequently decreased FXR transactivation of its metabolic target genes. This result suggests that the dynamic acetylation and deacetylation of FXR in normal mice may be required for activation of the genes, while continuously elevated acetylation in the diseased states blocks activation. Therefore, our studies provide a potential intriguing correlation between elevated FXR acetylation, decreased FXR activities, and deleterious metabolic effects in metabolic disease states.
The correlation of elevated FXR acetylation and metabolic disease suggests that activation of SIRT1 should have beneficial effects. Indeed, activation of SIRT1 by resveratrol has already been demonstrated to have beneficial metabolic effects in mouse models of metabolic disease by enhancing mitochondria function through activation of PGC-1α (Baur et al., 2006; Lagouge et al., 2006). Our observation that activation of resveratrol or overexpression of SIRT1 results in decreased acetylation of FXR in disease model mice suggests that effects of resveratrol on FXR, in addition to PGC-1α, may also contribute to improved metabolic outcomes. These results are consistent with our previous studies showing that down-regulation of p300, which should reduce acetylation of FXR, altered expression of metabolic target genes involved in lipoprotein and glucose metabolism, such that beneficial lipid and glucose profiles would be expected (Fang et al., 2008). In addition to the direct effects of SIRT1 on FXR, SIRT1 may also indirectly increase FXR activity by activating PGC-1α (Baur et al., 2006; Rodgers et al., 2008). PGC-1α has been shown to coactivate FXR transactivation and increase FXR expression (Zhang et al., 2004). It is possible that SIRT1-mediated PGC-1α coactivation and FXR deacetylation synergistically regulate hepatic FXR activity under physiological conditions.
It has been demonstrated that bile acids, not only activate FXR signaling by binding to the ligand binding domain of FXR, but also activate upstream cellular kinase signaling pathways, such as protein kinase B (PKB, Akt), PKC, JNK, and ERK kinases, in hepatocytes (Dent et al., 2005; Gineste et al., 2008; Hylemon et al., 2009; Miao et al., 2009). We recently demonstrated that bile acids increase the stability of SHP, a well known FXR target, by inhibiting ubiqutin-proteasomal degradation in an ERK-dependent manner (Miao et al., 2009). A recent study showed that inhibition of PKC impaired ligand-mediated regulation of FXR activity of its target genes by blocking FXR interaction with PGC-1α (Gineste et al., 2008). Interestingly, one of the two reported PKC sites in FXR, S154, is located near K157, an acetylation site in FXR. It will be, therefore, interesting to know whether phosphorylation of FXR by PKB or PKC affects acetylation of FXR and whether impaired signaling pathways are associated with elevated FXR acetylation in metabolic disease states.
Although K217 was identified as the major acetylation site for FXR, acetylation at K157 was also observed. Recent studies demonstrated that acetylation of p53 at different lysine residues affected different biological processes, for example, acetylation of p53 at K120 in the DNA binding domain by Tip60 acetylase is crucial for apoptosis but is dispensable for cell cycle arrest (Tang et al., 2006). Likewise, acetylation at K157 and K217 in FXR may have different functional outcomes and may selectively play a role in regulation of subsets of target genes involved in different metabolic pathways, such as cholesterol/bile acids, fatty acid, and glucose. It will be, therefore, important to determine whether acetylation in the disease models is generally increased or if acetylation at specific sites is increased and whether FXR acetylated at different sites is recruited in a gene-selective manner.
In this study, we demonstrate for the first time that FXR acetylation is tightly and reciprocally regulated by p300 and SIRT1 and is critical for activation of FXR target genes in response to FXR signaling under normal conditions and further, this dynamic regulation of FXR acetylation is disrupted in metabolic disease model mice, resulting in constitutively elevated FXR acetylation. We propose that small molecules that inhibit FXR acetylation by increasing SIRT1 or decreasing p300 activity may be promising therapeutic agents for treatment of metabolic disorders, such as, fatty liver, diabetes, and obesity.
Experimental Procedures
Materials
Antibodies against FXR, RXR, p300, GFP, lamin, tubulin, and RNA polymerase II were purchased from Santa Cruz Biotech. M2 antibody was purchased from Sigma and SIRT1 and acetylated H3 (K9/K14) antibodies were from Upstate Biotech. Acetyl lysine antibody was purchased from Cell Signaling, Inc. GW4046 was a kind gift from T. M. Willson. TSA, nicotinamide (Nam), and resveratrol were purchased from Sigma, Inc.
Plasmid and adenoviral vector constructs
Acetylation defective- and mimic-flag-FXR mutants were constructed by site-directed mutagenesis (Stratagene, Inc.) and confirmed by sequencing. Flag-FXR in this manuscript refers to 3flag-human FXR. Ad-flag-FXR has been previously described (Fang et al., 2008).
Animal experiments
BALB/c male mice were fed with chow supplemented with 0.5% CA (Harland Teklad TD05271) for 3 to 6 hr or treated with GW4064 (30 mg/kg in corn oil) by intraperitoneal injection and 1-3 hr later, livers were collected for further analyses. BALB/c mice were fasted overnight or refed for 1 hr after overnight fasting and livers were collected for further analyses. About 8-12 week old ob/ob mice or congenic C57BL/6J mice were tail vein injected with Ad-flag-FXR and then daily and orally treated with resveratrol (50 mg/kg body wt) for 1 week. BALB/c mice were fed normal chow or high fat western style chow for 16 weeks and then infected with control Ad-empty or Ad-flag-FXR or daily and orally treated with resveratrol for 1 week. Recombinant adenovirus was injected via the tail vein of mice as previously described (Fang et al., 2007; Miao et al., 2009). All the animal use and adenoviral protocols were approved by the Institutional Animal Care and Use and Institutional Biosafety Committees at University of Illinois at Urbana-Champaign and were in accordance with National Institutes of Health guidelines.
Tandem mass (MS/MS) spectrometry analysis
Flag-FXR was expressed in HepG2 cells (3 plates of 15 cm per group) by adenoviral infection. Flag-FXR was purified by binding to M2 agarose and subjected to MS/MS analysis as described previously (Miao et al., 2009).
Acetylation and deacetylation assays
HepG2 cells (ATCC HB8065) were maintained in Dulbecco's modified Eagle's medium (DMEM)/F12 (1:1) medium. Cos-1 cells were maintained in DMEM medium. Cells were infected with Ad-flag-FXR as previously described (Fang et al., 2008). For acetylation assays in cells, Cos-1 cells were cotransfected with expression plasmids of flag-FXR along with p300. Cells were treated with 200 nM of GW4064, 500 nM of TSA and 10 mM of Nam for 2-5 hr. Flag-FXR or endogenous FXR in cells or mouse liver was immunoprecipitated from freshly prepared cell or liver extracts at 4°C for 2 hr in buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 5 mM EDTA, 1% NP40, 0.1 % SDS). Acetylated FXR was detected by western analyses. For in vitro assays, acetylated flag-FXR or GST-flag-FXR was incubated with purified GST or GST-SIRT1 in the presence of 50 μM NAD+ in deacetylation buffer (Tris-HCl, pH 8.8, 5 % glycerol, 50 mM NaCl, 4 mM MgCl2, 1 mM DTT) at 37°C for 1 hr as previously described (Fang et al., 2008).
Protein interaction assays
Chromatin IP (ChIP), coimmunoprecipitation (CoIP), and GST pull down assays were performed as described previously (Fang et al., 2007; Kemper et al., 2004; Miao et al., 2009). Gel mobility shift assays were done as described (Bhalla et al., 2004; Miao et al., 2006) with some modifications. Briefly, a 26-mer oligonucleotide containing the FXRE sequence present in the SHP promoter was labeled with γ-32P and gel purified. Increasing amounts of flag-FXR or Cos-1 whole cell extracts were incubated with purified flag-RXRα and incubated at RT for 10 min. The complexes were analyzed using nondenaturing acrylamide gel electrophoresis.
Quantification of mRNA
RNA was isolated from liver or cultured cells and the levels of mRNA were determined by quantitative reverse transcriptase-PCR (qRT-PCR) as previously described (Miao et al., 2009).
Bile acid pool size and serum cholesterol levels measurement
The total amount of bile acids from gall bladder, liver, and entire intestines was measured by colorimetric analysis (Trinity Biotech). Serum cholesterol levels were measured using a HDL and LDL/VLDL cholesterol quantification kit (Biovision, Inc).
Supplementary Material
Acknowledgments
*We are grateful to P. Puigserver for Ad-siSIRT1 and Ad-flag-SIRT1, P. Edwards for FXR deletion constructs, M. Leid for GST-SIRT1 constructs, R. Sato for plasmids CMV-3 flag-FXR and G4DBD-FXR, W. Gu and T. Finkel for plasmids for SIRT1 wt and mutant, M. Ananthanarayanan for (FXRE)3-tk-luc, Yoon K. Lee for the Shp-luc reporter plasmid, and T. Imamura for HA-Ub plasmid. Special thank Dr. T. M. Willson for providing GW4064. We also thank B. Kemper for helpful discussions. This study was supported by NIH grants CA103867 and CA124760 to C.M.C. and NIH DK062777, NIH DK80032, and ADA basic research award to J.K.K. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract N01-CO-12400 to T.D.V.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla S, Ozalp C, Fang S, Xiang L, Kemper JK. Ligand-activated pregnane X receptor interferes with HNF-4 signaling by targeting a common coactivator PGC-1alpha. Functional implications in hepatic cholesterol and glucose metabolism. J Biol Chem. 2004;279:45139–45147. doi: 10.1074/jbc.M405423200. [DOI] [PubMed] [Google Scholar]
- Cariou B, Staels B. FXR: a promising target for the metabolic syndrome? Trends Pharmacol Sci. 2007;28:236–243. doi: 10.1016/j.tips.2007.03.002. [DOI] [PubMed] [Google Scholar]
- Daitoku H, Hatta M, Matsuzaki H, Aratani S, Ohshima T, Miyagishi M, Nakajima T, Fukamizu A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc Natl Acad Sci U S A. 2004;101:10042–10047. doi: 10.1073/pnas.0400593101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent P, Fang Y, Gupta S, Studer E, Mitchell C, Spiegel S, Hylemon PB. Conjugated bile acids promote ERK1/2 and AKT activation via a pertussis toxin-sensitive mechanism in murine and human hepatocytes. Hepatology. 2005;42:1291–1299. doi: 10.1002/hep.20942. [DOI] [PubMed] [Google Scholar]
- Fang S, Miao J, Xiang L, Ponugoti B, Treuter E, Kemper JK. Coordinated recruitment of histone methyltransferase G9a and other chromatin modifying enzymes in SHP-mediated regulation of hepatic bile acid metabolism. Mol Cell Biol. 2007;27:1407–1424. doi: 10.1128/MCB.00944-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S, Tsang S, Jones R, Ponugoti B, Yoon H, Wu SY, Chiang CM, Willson TM, Kemper JK. The P300 acetylase is critical for ligand-activated farnesoid X receptor (FXR) induction of SHP. J Biol Chem. 2008 doi: 10.1074/jbc.M803531200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gineste R, Sirvent A, Paumelle R, Helleboid S, Aquilina A, Darteil R, Hum DW, Fruchart JC, Staels B. Phosphorylation of farnesoid X receptor by protein kinase C promotes its transcriptional activity. Mol Endocrinol. 2008;22:2433–2447. doi: 10.1210/me.2008-0092. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol Cell. 2000;6:517–526. doi: 10.1016/s1097-2765(00)00051-4. [DOI] [PubMed] [Google Scholar]
- Guarente L. Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol. 2007;72:483–488. doi: 10.1101/sqb.2007.72.024. [DOI] [PubMed] [Google Scholar]
- Huang W, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J, Dong B, Huang X, Moore DD. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 2006;312:233–236. doi: 10.1126/science.1121435. [DOI] [PubMed] [Google Scholar]
- Hylemon PB, Zhou H, Pandak WM, Ren S, Gil G, Dent P. Bile acids as regulatory molecules. J Lipid Res. 2009;50:1509–1520. doi: 10.1194/jlr.R900007-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T, Moschetta A, Lee YK, Peng L, Zhao G, Downes M, Yu RT, Shelton JM, Richardson JA, Repa JJ, et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc Natl Acad Sci U S A. 2006;103:3920–3925. doi: 10.1073/pnas.0509592103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaany NY, Mangelsdorf DJ. LXRS and FXR: the yin and yang of cholesterol and fat metabolism. Annu Rev Physiol. 2006;68:159–191. doi: 10.1146/annurev.physiol.68.033104.152158. [DOI] [PubMed] [Google Scholar]
- Kemper J, Kim H, Miao J, Bhalla S, Bae Y. Role of a mSin3A-Swi/Snf chromatin remodeling complex in the feedback repression of bile acid biosynthesis by SHP. Mol Cell Biol. 2004;24:7707–7719. doi: 10.1128/MCB.24.17.7707-7719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura YI, Kitamura T, Kruse JP, Raum JC, Stein R, Gu W, Accili D. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2005;2:153–163. doi: 10.1016/j.cmet.2005.08.004. [DOI] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Lee FY, Lee H, Hubbert ML, Edwards PA, Zhang Y. FXR, a multipurpose nuclear receptor. Trends Biochem Sci. 2006;31:572–580. doi: 10.1016/j.tibs.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1alpha. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang S, Blander G, Tse JG, Krieger M, Guarente L. SIRT1 deacetylates and positively regulates the nuclear receptor LXR. Mol Cell. 2007;28:91–106. doi: 10.1016/j.molcel.2007.07.032. [DOI] [PubMed] [Google Scholar]
- Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell. 2000;6:507–515. doi: 10.1016/s1097-2765(00)00050-2. [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Ma K, Saha PK, Chan L, Moore DD. Farnesoid X receptor is essential for normal glucose homeostasis. J Clin Invest. 2006;116:1102–1109. doi: 10.1172/JCI25604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- Miao J, Fang S, Bae Y, Kemper JK. Functional inhibitory cross-talk between car and HNF-4 in hepatic lipid/glucose metabolism is mediated by competition for binding to the DR1 motif and to the common coactivators, GRIP-1 and PGC-1alpha. J Biol Chem. 2006;281:14537–14546. doi: 10.1074/jbc.M510713200. [DOI] [PubMed] [Google Scholar]
- Miao J, Xiao Z, Kanamaluru D, Min G, Yau PM, Veenstra TD, Ellis E, Strom S, Suino-Powell K, Xu HE, Kemper JK. Bile acid signaling pathways increase stability of Small Heterodimer Partner (SHP) by inhibiting ubiquitin-proteasomal degradation. Genes Dev. 2009;23:986–996. doi: 10.1101/gad.1773909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- Shin DJ, Campos JA, Gil G, Osborne TF. PGC-1alpha activates CYP7A1 and bile acid biosynthesis. J Biol Chem. 2003;278:50047–50052. doi: 10.1074/jbc.M309736200. [DOI] [PubMed] [Google Scholar]
- Sinal C, Tohkin M, Miyata M, Ward J, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell. 2000;102:731–744. doi: 10.1016/s0092-8674(00)00062-3. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Lin SJ, Guarente L. Life-span extension in yeast. Science. 2006;312:195–197. doi: 10.1126/science.312.5771.195d. author reply 195-197. [DOI] [PubMed] [Google Scholar]
- Tang Y, Luo J, Zhang W, Gu W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell. 2006;24:827–839. doi: 10.1016/j.molcel.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willson TM, Jones SA, Moore JT, Kliewer SA. Chemical genomics: functional analysis of orphan nuclear receptors in the regulation of bile acid metabolism. Med Res Rev. 2001;21:513–522. doi: 10.1002/med.1023. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Castellani LW, Sinal CJ, Gonzalez FJ, Edwards PA. Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha) regulates triglyceride metabolism by activation of the nuclear receptor FXR. Genes Dev. 2004;18:157–169. doi: 10.1101/gad.1138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Lee FY, Barrera G, Lee H, Vales C, Gonzalez FJ, Willson TM, Edwards PA. Activation of the nuclear receptor FXR improves hyperglycemia and hyperlipidemia in diabetic mice. Proc Natl Acad Sci U S A. 2006;103:1006–1011. doi: 10.1073/pnas.0506982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.