Summary
In Drosophila, miRNA strands are predominantly sorted into AGO1 to regulate seed-matched target transcripts, while their partner miRNA* strands are thought to be mostly degraded. Here we report that Drosophila Argonautes exhibit different strand preferences for miRNA duplexes, and that in particular, many miRNA* species accumulate in the RNAi effector AGO2. AGO2-loaded miRNA* species require canonical RNAi factors for their accumulation, are efficiently 3′ modified, and are preferentially active on extensively-matched target transcripts. Differential miRNA/miRNA* sorting profiles are correlated with specific central mismatches. In vitro assays revealed an active role for Watson-Crick basepairing at positions 9 and 10 in promoting strand selection by AGO2, with little reciprocal effect on strand selection by AGO1. We conclude that miRNA strand selection and sorting are actually linked processes that stem from distinct loading preferences of AGO proteins, and that independent sorting of duplex strands is a general feature of Drosophila microRNA genes.
Keywords: microRNA*, strand selection, small RNA sorting, AGO1, AGO2
Introduction
microRNAs (miRNAs) are endogenous ~21–24 nucleotide (nt) RNAs that generally function as post-transcriptional repressors, and they collectively regulate diverse aspects of development and physiology (Flynt and Lai, 2008). Canonical primary miRNA transcripts in animals bear local inverted repeats that are cleaved by the nuclear RNAse III enzyme Drosha to yield ~60–70 nt pre-miRNA hairpins, which are cleaved in the cytoplasm by the Dicer RNAse III enzyme to yield a ~22 nt RNA duplex (Kim et al., 2009). One of the strands is preferentially incorporated into an Argonaute protein and guides it to imperfectly-paired targets for repression, while its partner miRNA* strand accumulates to a lower level and is presumed to be mostly degraded.
miRNA biogenesis and function is related to RNA interference (RNAi) induced by small interfering RNAs (siRNAs) (Farazi et al., 2008; Okamura and Lai, 2008; Zhou et al., 2008). The specific selection of mature miRNAs and guide strand siRNAs is governed by thermodynamic asymmetry of precursor duplexes, as the small RNA whose 5′ end resides in the less-structured duplex end is favored for incorporation into an Argonaute protein (Figure 1A) (Khvorova et al., 2003; Schwarz et al., 2003). Nevertheless, Drosophila has substantially separate molecular pathways for miRNAs and siRNAs. While miRNAs are processed by Dicer-1 (Dcr-1) and mostly incorporated into Argonaute1 (AGO1) complex, siRNAs are processed by Dcr-2 and sorted into AGO2. Mutations in Dcr-2 and AGO2 abolish RNAi, but do not strongly affect development or physiology (Lee et al., 2004; Okamura et al., 2004). On the other hand, mutants of Dcr-1 and AGO1 are lethal (Kataoka et al., 2001; Lee et al., 2004), reflecting essential roles for miRNA genes during fly development (Flynt and Lai, 2008).
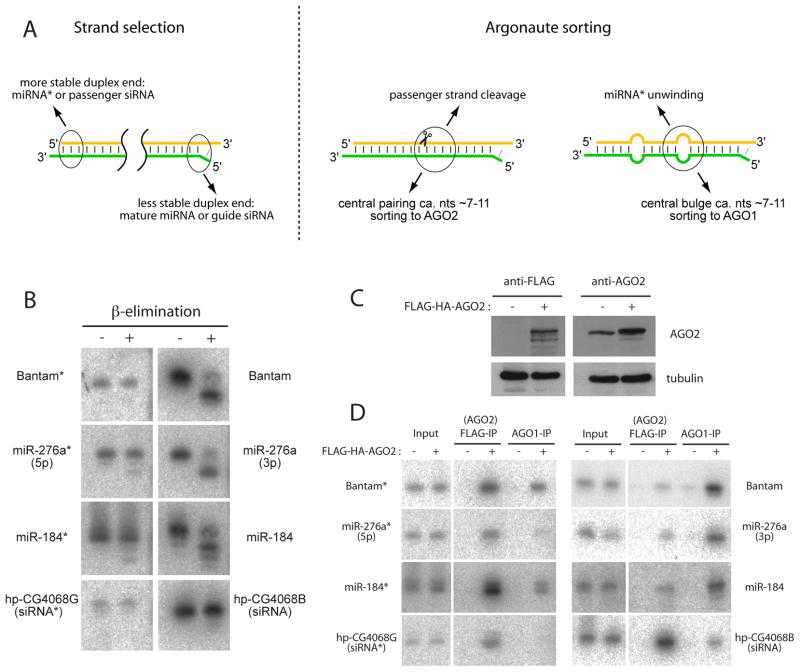
Figure 1. Sorting of miRNA* species into mature AGO2 complexes.
(A) Previously proposed rules for the selection and sorting of guide strand miRNAs and siRNAs. For a given siRNA or miRNA duplex, the strand whose 5′ end inhabits the less structured duplex end is favored for selection as the guide strand (Khvorova et al., 2003; Schwarz et al., 2003). In Drosophila, centrally paired nucleotides were proposed to promote sorting of the duplex to AGO2, followed by cleavage of the passenger strand. Conversely, centrally bulged nucleotides was proposed to promote sorting to AGO1 (Forstemann et al., 2007; Tomari et al., 2007), followed by unwinding and ejection of the miRNA* strand.
(B) Total RNA samples were subjected to β-elimination followed by Northern blotting for the indicated small RNAs. miRNA species such as bantam, miR-276a, miR-184 are sensitive to β-elimination and migrate more quickly following this treatment. siRNAs such as hp-CG4068B and hp-CG4068G (which derive from a single precursor duplex), as well as the partner star strands of all these miRNAs, are resistant to β-elimination.
(C) S2-R+ cells stably transfected with FLAG-HA-AGO2 plasmid exhibit only a slight increase in AGO2 levels, as assayed by either anti-FLAG antibody (left) or anti-AGO2 antibody (right).
(D) AGO2 and AGO1 complexes were sequentially purified from S2-R+ or S2-R+/FLAG-HA-AGO2 cells, using anti-FLAG or anti-AGO1 antibodies, and their associated small RNAs were assayed by Northern blotting. Input lanes (10% of the sample used for immunoprecipitation) demonstrate that the levels of small RNAs are not markedly altered by the presence of AGO2 plasmid. bantam*, miR-276a*, miR-184*, hp-CG4068B and hp-CG4068G predominantly load AGO2, while miR-276a, miR-184 and mature bantam predominantly load AGO1.
Separation of the miRNA and siRNA pathways is believed to require active sorting mechanisms that direct different RNAs into appropriate Argonaute complexes. Strict sorting of small RNAs was directly confirmed by largescale sequencing of small RNAs purified with AGO1 and AGO2 (Czech et al., 2008). In vitro tests indicated that sorting can occur post-dicing in Drosophila, and pointed to the importance of central mismatches in precursor duplexes (Figure 1A) (Forstemann et al., 2007; Tomari et al., 2007). Perfect duplex complementarity correlated with sorting to AGO2 as siRNAs, while the presence of unpaired nucleotides in miRNA duplexes were proposed to promote sorting to AGO1. Indeed, a single central bulge involving positions 7–11 was sufficient to redirect an otherwise fully-paired siRNA duplex to AGO1 (Tomari et al., 2007). Reciprocally, an endogenous miRNA duplex (mir-277) with extensive pairing exhibited substantial sorting to AGO2 (Forstemann et al., 2007).
In contrast to the routing of miRNA strands and siRNA guide strands, whose appropriate sorting is relevant to understanding miRNA biology and optimizing siRNA efficacy, the sorting of miRNA* strands has received little attention to date. However, as profiling of small RNA populations has become increasingly precise (Czech et al., 2008; Ruby et al., 2007; Seitz et al., 2008), it has become apparent that different miRNA* species accumulate to distinct and characteristic levels. A substantial proportion of miRNA* strands are subject to stringent sequence constraint, reside in AGO1 complexes, and possess gene regulatory activity, indicating that miRNA* strands are often used as endogenous regulatory species (Okamura et al., 2008b). Therefore, miRNA* strands have fates other than default degradation.
It is plausible that the behavior of miRNA* strands is linked to the sorting pathways that act on their partner miRNA strands, such that miRNA* sorting simply reflects the consequences of miRNA sorting. However, we show in this study that several miRNA* species, unlike their partner mature miRNAs, are preferentially incorporated into AGO2 complex and are 3′ modified. Similar to endogenous siRNAs (endo-siRNAs) in Drosophila (Czech et al., 2008; Ghildiyal et al., 2008; Kawamura et al., 2008; Okamura et al., 2008a), these miRNA* species require canonical RNAi factors for their accumulation and preferentially repress extensively paired targets rather than seed-matched targets.
Although miRNA strand selection by AGO1 is generally consistent with thermodynamic asymmetry, AGO2 strand preference differed substantially from that of AGO1. Notably, we observed substantial preference of miRNA* sorting to AGO2, which correlated strongly with mismatches at position 9 and 10. In vitro assays demonstrated facile redirection of AGO2 strand selection by altering position 9 pairing, without effect on AGO1 strand selection. These data indicate that independent routing of partner miRNA and miRNA* strands is due to the distinct strand preferences of Drosophila Argonautes, revealing unexpected complexity of small RNA biogenesis pathways.
Results
miRNA* species with modified 3′ ends
We previously reported that many miRNA* strands are present in AGO1 complex in Drosophila embryos and cultured cells (Okamura et al., 2008b). However miR-276a* and miR-184* caught our attention because they co-precipitated poorly with AGO1, relative to their amount in total RNA. Such behavior might be explained by atypically slow unwinding of their precursor duplexes during maturation of the AGO1 complex. Alternatively, these miRNA* strands might be located in a complex other than AGO1. To distinguish these possibilities, we checked for 3′ modification. HEN1 methyltransferase modifies single stranded siRNAs in AGO2 and piRNAs in PIWI-class Argonaute complexes, which therefore carry 2′-O-methylation at their 3′ ends (Horwich et al., 2007; Saito et al., 2007). In contrast, double-stranded duplexes and mature miRNAs in AGO1 complex have predominantly free 2′ and 3′ –OH at their 3′ ends.
The β-elimination assay increases mobility of small RNAs with free 2′ and 3′ –OH group at their 3′ termini (Hutvagner et al., 2001). We observed that the miRNAs bantam, miR-276a and miR-184 all exhibited increased mobility following this treatment (Figure 1B), supporting the previous conclusion that a majority of the pool of mature Drosophila miRNAs have free 3′ ends and sort to AGO1. We also tested the endo-siRNAs hp-CG4068B and G, which are known to complex with AGO2, and they were resistant to β-elimination as previously reported (Kawamura et al., 2008; Okamura et al., 2008a). When we analyzed miR-276a* and miR-184*, which were not efficiently co-precipitated with AGO1 (Okamura et al., 2008b), we found that they were nearly completely resistant to β-elimination like endo-siRNAs (Figure 1B). Bantam* was similarly β-resistant. Therefore, some small RNAs derived from Drosophila miRNA genes are efficiently modified at their 3′ termini.
miRNA* species that are preferentially sorted into AGO2 complex
Because small RNAs in AGO2 complex are modified at their 3′ ends (Horwich et al., 2007; Pelisson et al., 2007), we tested whether these miRNA* species associated with AGO2. To do so, we generated S2-R+ cells that were stably transfected with FLAG-HA-AGO2 plasmid (Czech et al., 2008). These cells slightly overexpressed AGO2 protein, as judged by Western blotting using anti-AGO2 and anti-FLAG antibodies (Figure 1C), but the moderate increase in AGO2 did not substantially affect the steady levels of endo-siRNAs or miRNAs in these cells (Figure 1D, see input lanes). Therefore, these cells should represent an appropriate setting to study small RNA sorting.
We purified AGO2 and AGO1 complexes from these cells using anti-FLAG and anti-AGO1 antibodies. Consistent with previous reports, endo-siRNAs and miRNAs were highly enriched in AGO2 and AGO1 complexes, respectively (Figure 1D). In contrast to their partner mature miRNAs, miR-276a*, miR-184* and bantam* were highly enriched in AGO2 complex, with enrichment comparable to that of endo-siRNAs (Figure 1D). Together with the β-elimination experiments, we conclude that the star strands of mir-276a, mir-184, and bantam are preferentially sorted into AGO2 like endo-siRNAs, even though they derive from the miRNA pathway. Notably, the specific incorporation of only one duplex strand into AGO2 was not observed in previous studies. For example, the two strands of the endo-siRNA duplex hp-CG4068B/G accumulate asymmetrically, but both strands are sorted to AGO2 (Figure 1D). Therefore, the rules for strand selection by different Drosophila Argonautes are more complex than previously anticipated.
The accumulation of AGO2-loaded miRNA* species requires RNAi factors
In light of the efficient loading of many miRNA* species into AGO2, we tested whether their biogenesis required miRNA or RNAi factors. We depleted a panel of small RNA biogenesis factors by soaking aliquots of S2R+ cells with cognate dsRNAs for 8 days. We observed reduction of mature miRNAs (bantam) and hpRNA-derived siRNAs (hp-CG4068B) upon knockdown of the expected miRNA or RNAi factors (Czech et al., 2008; Kawamura et al., 2008; Okamura et al., 2008a), indicating their functional depletion (Figure 2A).
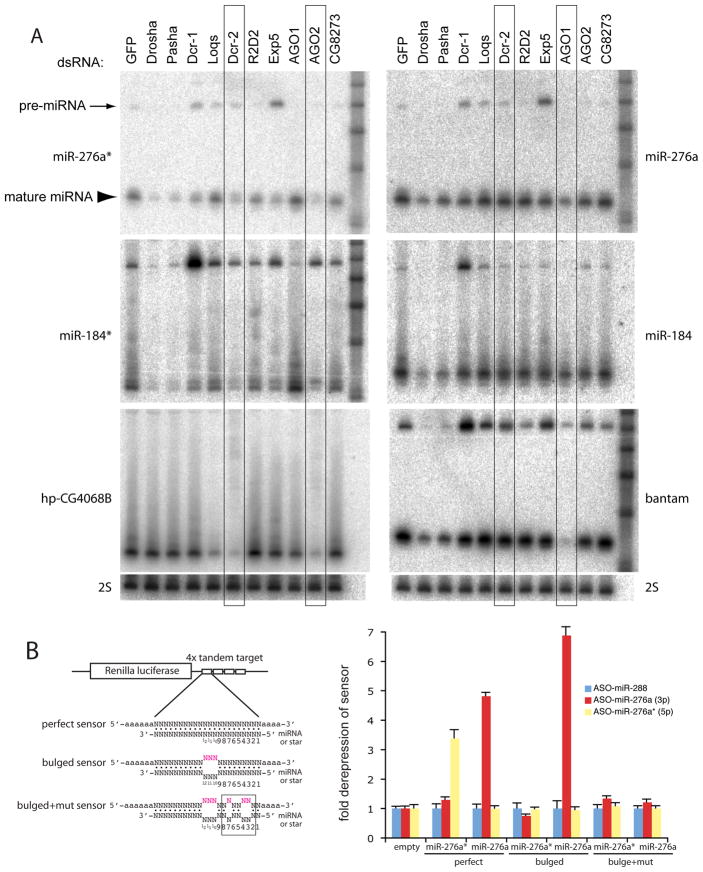
Figure 2. Distinct biogenesis and activity of miRNA and miRNA* species.
(A) Accumulation of miRNA* species requires both miRNA and RNAi factors. The indicated factors were depleted in S2-R+ cells, and small RNAs were assayed by Northern blot. These experiments represent sequential probing and stripping of two master membranes, used to analyze miR-276a/miR-276a*/bantam and miR-184/miR-184*/hp-CG4068. Membranes were re-exposed to verify complete stripping of old probes, and were finally probed for 2S rRNA to confirm equal loading (bottom). The appropriate behavior of bantam and hp-CG4068B across the dsRNA panel verified the efficacy of knockdowns (Czech et al., 2008; Okamura et al., 2008a). These experiments showed that miRNA* species share requirements for Drosha, Pasha, Exp5 and Dcr-1 with their partner miRNA species. However, the accumulation of miR-276a* and miR-184* is decreased upon depletion of AGO2, while the accumulation of miR-276a and miR-184 is specifically decreased upon depletion of AGO1. The accumulation of the various miRNA and miRNA* species is comparably less dependent on Loqs than is the endo-siRNA hp-CG-4068B.
(B) AGO2-loaded miRNA* species exhibit greater target stringency than AGO1-loaded miRNA species. Renilla luciferase sensors bearing four tandem perfect or bulged targets for the indicated miRNA (cloned in psiCHECK2, which contains an internal control firefly luciferase gene); bulged+mut targets also contained 3 mutations in seed pairs (shown is the miR-276a mutant pattern). These were co-transfected with 2′OMe antisense oligos (ASO) and derepression of sensor activity was measured. For each construct, two sets of quadruplicate transfections were done on 2 different days and the data were pooled. Values were normalized to sensor activity in the presence of non-cognate ASO against miR-288, and mean values ± S.D. are shown. Both miR-276a and miR-276a* perfect sensors were derepressed by cognate ASO; however, only miR-276a bulged sensor could be derepressed.
In contrast to mature miR-276a and miR-184, their partner miRNA* species were sensitive not only to loss of the miRNA factor Dcr-1, but also to loss of the canonical RNAi factor Dcr-2 (Figure 2A, “Dcr-2”-boxed lanes). In addition, we observed that these miRNA* species exhibited specific dependence on AGO2 instead of AGO1 for normal accumulation, consistent with the biochemical evidence for the residence of these miRNA* species in AGO2 (Figure 2A). However, the accumulation of miRNA* species was more similar to miRNA strands in that they lacked the strong requirement for loqs observed for most endo-siRNAs (Figure 2A, hp-CG4068B blot). In summary, the biogenesis of these miRNA* species depends on a unique combination of canonical miRNA and RNAi factors, not previously observed for mature miRNAs or endo-siRNAs.
AGO2-loaded miRNA* species are poorly active on seed targets
Unlike AGO1 complex that can suppress target expression by seed base-pairing, AGO2 was suggested to be poorly active on target mRNAs with imperfect complementarity to guide small RNAs (Forstemann et al., 2007). Because miR-276a* and miR-184* were preferentially sorted to AGO2 complex, one might predict that these small RNAs would fail to suppress bulged targets as efficiently as canonical miRNAs.
We tested this using luciferase sensors with perfectly complementary target sites or target sites with central bulges (Figure 2B). To avoid possible mis-sorting of overexpressed miRNA hairpins, we measured the activity of endogenous miRNA and miRNA* species by introducing 2′-O-methyl antisense oligo nucleotides (2Ome-ASOs) and assaying for sensor de-repression. As shown in a previous report (Okamura et al., 2008b), antisense inhibitors of mature and star strands of miR-276a were able to de-repress both cognate perfect targets confirming that both strands of the duplex are active in S2 cells (Figure 2B). The bulged sensor for the mature strand of miR-276a was also de-repressed by cognate ASO, but further mutation of its seed match resulted in a complete loss of target regulation, as expected for a typical miRNA. We constructed a similar set of sensors for miR-184, which was similarly active on perfect and bulged targets, but not on a bulged target with seed mismatches (Supplementary Figure 1).
In contrast, 2Ome-ASOs against miR-276a* did not substantially de-repress bulged sensor activity (Figure 2B). miR-184* similarly showed little activity on its bulged sensor, even though clear de-repression of its perfect sensor by 2Ome-ASO was observed (Supplementary Figure 1). These results are not simply due to different levels of mature and star strands, at least in the case of mir-276, since perfect sensors of both mir-276a strands were similarly de-repressed by cognate inhibitors. We conclude that there is a functional consequence of miRNA* sorting to AGO2, namely the restriction of their target activity on seed-type binding sites. We note that translational repression by AGO2-RISC was recently recognized using in vitro extracts (Iwasaki et al., 2009). Lack of substantial translational regulation by miRNA*-programmed AGO2 in our cell-based assay might be explained if translational repression by AGO2 is reversible and relatively weaker (Iwasaki et al., 2009), while AGO1 mediated repression is usually associated with deadenylation and mRNA destabilization.
The strands of synthetic miRNA/miRNA* duplexes can be independently sorted
The cloning of small RNAs from total RNA inevitably yields a variety of related sequences, whose relationship to the RNAs detected by hybridization cannot be explicitly assigned. Conceivably, different cleavage positions of miRNA precursors by Drosha or Dcr-1 might yield a mixture of duplexes with distinct strand selection and/or sorting preferences, which would confound the interpretation of independent sorting of strands from any specific miRNA/miRNA* duplex.
To exclude this possibility, we developed an in vitro assay using synthetic small RNA duplexes (Figure 3A). We labeled the 5′ end of mature miR-276a or star with 32P, and then annealed them to unlabeled partner strands. The labeled duplexes were incubated with lysate prepared from S2-R+ cells expressing FLAG-HA-AGO2 for one hour. AGO2 and AGO1 complexes were then sequentially purified using anti-FLAG and anti-AGO1 antibodies, and the immunoprecipitated small RNAs were analyzed on native acrylamide gels. Although the input RNAs were completely double-stranded, the co-purified small RNAs were predominantly single stranded. Since the labeled RNA recovered from supernatant were mostly double stranded (Figure 3B–E, “supernatant” lanes), we conclude that single stranded small RNAs co-precipitated with Argonaute complexes represent genuine small RNAs in mature complexes, as opposed to non-specific interactions of fortuitously unwound RNAs to sticky Argonaute proteins.
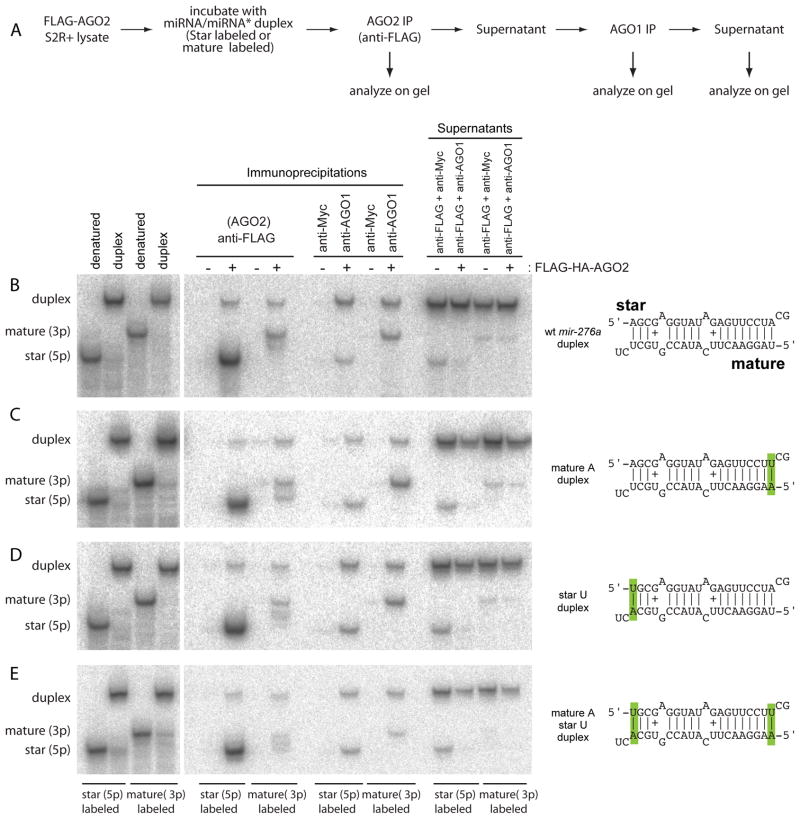
Figure 3. Distinct loading preferences of the strands of synthetic miRNA duplexes.
(A) In vitro sorting assay. Synthetic duplexes carrying one labeled strand were incubated with lysate from S2-R+ cells stably transfected with FLAG-HA-AGO2 (Figure 1C). AGO2 and AGO1 complexes were sequentially purified and analyzed along with the final supernatants on native gels; 2% of input and 2.5% of supernatant relative to the IP lanes were loaded. Note that miRNA and miRNA* species of similar length exhibit distinct mobility in this matrix.
(B) Wild-type mir-276a duplex shows preferential sorting of star strand to AGO2 and mature strand to AGO1.
(C) Mutation of the 5′ U of miR-276a into 5′ A does not substantially alter its sorting pattern.
(D) Mutation of the 5′ A of miR-276a* into 5′ U does not substantially alter its sorting pattern.
(E) Double 5′ alterations do not affect AGO2 strand selection, but result in preferred incorporation of star strand into AGO1 over the mature strand.
Under this experimental protocol, unwound miR-276a* strand co-precipitated more efficiently with AGO2 than its partner mature strand (Figure 3B). On the other hand, AGO1 preferred the miR-276a strand (Figure 3B), consistent with in vivo results (Figure 1D). We confirmed this result using a synthetic miR-184/miR-184* duplex, for which AGO1 and AGO2 also exhibited different strand preference (Supplementary Figure 2). These experiments demonstrate that the strands of defined small RNA duplexes can be independently sorted to different Argonaute complexes.
The influence of 5′ nucleotide on strand selection by AGO1 and AGO2
In plants, the 5′ nucleotide of a small RNA is not only strongly correlated with the choice of Argonaute host, but can actively determine its sorting (Mi et al., 2008). Animal mature miRNAs exhibit a substantial preference for 5′ uridine, whereas miRNA* species do not exhibit this bias. Largescale sequencing of Drosophila AGO1- and AGO2-associated RNAs verified that the former are highly enriched in 5′ U, while the latter are not (Czech et al., 2008), with certain populations of AGO2-resident RNAs exhibiting 5′ C bias (Ghildiyal et al., 2008). Therefore, one might hypothesize that AGO1 simply prefers RNAs with 5′ U and that AGO2 selects against such RNAs. This might be plausible since the mir-276a, mir-184 and bantam loci we studied in depth, like most miRNAs, encode 5′ U only on their mature strands.
We performed in vitro loading assays of a mir-276a duplex in which the mature strand was altered to 5′ A, while maintaining its structure. The strands of this variant duplex were still reciprocally sorted (Figure 3C), indicating that the lack of 5′ U was not sufficient to promote sorting of miR-276a to AGO2, nor did AGO2 lose its ability to distinguish the star strand when both duplex RNAs bore 5′ A. Similarly, a variant duplex in which both duplex strands bore 5′ U maintained its reciprocal sorting (Figure 3D). However, when we simultaneously changed mature miR-276a to 5′ A and miR-276a* to 5′ U, we reproducibly observed a reversal in the strand selection by AGO1, which now showed preference for the star strand (Figure 3E). Still, AGO2 preferred the star strand of this variant duplex. In summary, strand selection by AGO1 can be dominantly altered by 5′ nt identity, without changing relative end energies, and the strand preference of AGO2 is not adequately explained by either duplex end energy or by lack of 5′ U.
Distinct preferences of AGO1 and AGO2 with respect to miRNA and star strands
Based on our detailed analyses of selected miRNAs, we wished to determine how broadly the principle of independent sorting of miR/miR* strands applies in Drosophila. Hannon and colleagues reported largescale data on small RNA sequences obtained from AGO1 and AGO2 complexes immunoprecipitated from S2-NP cells (Czech et al., 2008). In order to obtain an appropriate baseline for interpreting the relative enrichment of depletion of small RNA reads in these IP libraries, we analyzed the total small RNA content of S2-NP cells using Illumina sequencing. We prepared 18–28 nt RNAs and generated 4,310,254 small RNA reads that perfectly matched the Drosophila melanogaster genome release 5. We then isolated reads that matched the miRNA and miRNA* strands of miRbase version 11 from the S2-NP total RNA, S2-NP-AGO1 and S2-NP-AGO2 libraries (Supplementary Table 1).
Our S2-NP total RNA dataset contained 3,819,484 reads matching 100 known miRNA loci, with the content of individual genes ranging over 6 orders of magnitude. We focused our analysis on genes with >100 miRNA reads and >10 miRNA* reads in this dataset. The star read requirement was instated so that inferred miRNA:miRNA* ratios were based on a reasonable sampling of data; however, few (8/46) genes failed this criterion due to our deep library coverage. We then compared the miRNA:miRNA* ratios in total RNA with those obtained from AGO1- and AGO2-IP libraries.
If AGO1 and AGO2 had identical strand preference, we would expect most values to be ~1, with deviations from 1 being due to either cloning/sequencing bias or perhaps small RNAs not in mature complexes. However, we instead saw that most miRNAs were enriched in AGO1 while a majority of miRNA* species were enriched in AGO2. This indicated distinct strand preferences of miRNA duplexes by the two Argonautes. When plotted on a gene-by-gene basis, ~2/3 of miRNA genes exhibited enrichment in AGO2 relative to total RNA (Figure 4A). Collectively, we observed significantly different distribution patterns between AGO1 and AGO2 (Figure 4B), with AGO2 in S2-NP cells carrying more miRNA* reads than miRNA reads (Figure 4C and Table S2). Note that some genes exhibiting star-enrichment in AGO2 had more partner miRNA reads in AGO2, because the number of miRNA reads was often much greater than miRNA* reads (Supplementary Table 1). For example, even though there were 7335 reads of mature bantam and 2238 reads of bantam* in AGO2, this represented ~50 fold enrichment of bantam* in AGO2 in light of the fact that bantam* represents <0.63% of mature bantam in total RNA. We observed similar enrichment of miRNA* strands in AGO2-IP data from female ovaries (Figure 4C) (Czech et al., 2008), indicating that this is a genuine feature of miRNA* sorting in the animal.
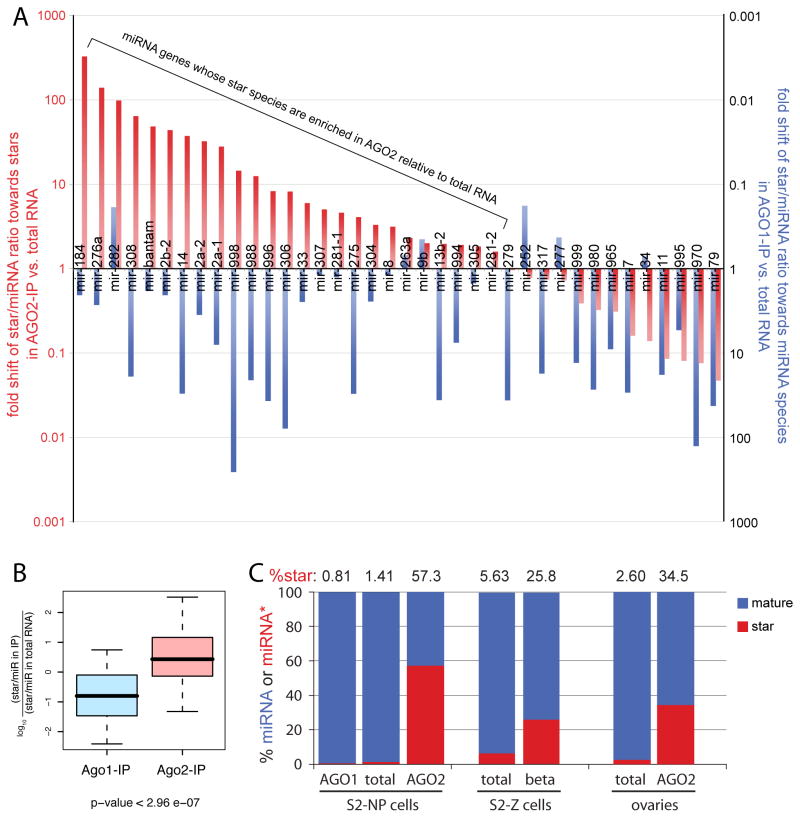
Figure 4. Distinct miRNA duplex strand preferences of AGO1 and AGO2.
(A) We sampled 3.82 million miRNA reads from S2-NP cells, and selected the 38 miRNA genes that generated at least 100 miRNA reads and 10 miRNA* reads for further analysis. In red, we plotted the fold enrichment of the reported ratio of miRNA*:miRNA reads in AGO2 (Czech et al., 2008) relative to total S2-NP RNA (this study). In blue, we plotted the fold enrichment of the miRNA:miRNA* ratio in AGO1 relative to total S2-NP RNA. Ratios deviating from 1 are unexpected unless AGO1 and AGO2 have different strand preferences; however, most miRNAs are enriched in AGO1 and many miRNA* species are enriched in AGO2 relative to total RNA. For genes exhibiting star-enrichment (>1) in AGO2, the AGO2-IP library might contain more mature strand than star reads, due to the highly asymmetric nature of miRNA:star populations in total RNA (see also Supplementary Table 1).
(B) Log values of the miR* enrichment factors in AGO2 and AGO1 complexes were plotted. Box plots follow Tukey’s standard conventions: A rectangle represents inter-quartile range of the first to the third quartiles, a bold horizontal line shows the median, whiskers connected to the rectangle indicate the largest and smallest values. P-values were calculated by the two-sided Wilcoxon test. There is a highly significant difference between the miR* enrichment factors in AGO2 vs. total RNA and AGO1 vs. total RNA.
(C) miRNA and miRNA* content of total RNA and AGO1-IP and AGO2-IP from cells and ovaries (Czech et al., 2008) shows tremendous enrichment of miRNA* species in AGO2. Similar trends were observed in β-eliminated samples of S2 cells (Seitz et al., 2008).
The IP assay by itself does not distinguish between immature duplexes in Argonautes and functionally matured RISC. This may be tested using beta-eliminated libraries, since Hen1 modifies single stranded species in AGO2 (Horwich et al., 2007). The beta-elimination reaction impairs standard 3′ linker ligation, thereby depleting for AGO1-loaded species and enriching for methylated miRNA or miRNA* species in AGO2. Indeed, we observed a strong enrichment for miRNA* species in S2 beta-eliminated RNA data reported by Zamore and colleagues (Seitz et al., 2008) (Figure 4C and Table S2). In summary, these largescale analyses demonstrate that AGO1 and AGO2 exhibit distinct preferences for the partner strands of most miRNA duplexes, and that a general attribute of AGO2 is the preferred selection of miRNA* strands relative to AGO1.
The pairing status of nucleotides 9 and 10 correlates with sorting profiles
Although strand selection by miRISC and siRISC were thought to be governed by the same thermodynamic rule (Khvorova et al., 2003; Schwarz et al., 2003; Tomari et al., 2004), our genomewide analyses clearly demonstrated distinct loading preferences of partner miRNA and miRNA* species into AGO1 and AGO2. To search for additional factors that influence strand selection and sorting, we analyzed the pairing status across all miRNA/miRNA* duplexes; G:U pairs were considered as a distinct category. To ensure that their companion sorting ratios were meaningful, we used only those miRNA or star sequences with >30 reads in total S2-NP data and >10 AGO1- and AGO2-IP reads; in total, 46 mature strands and 32 star sequences.
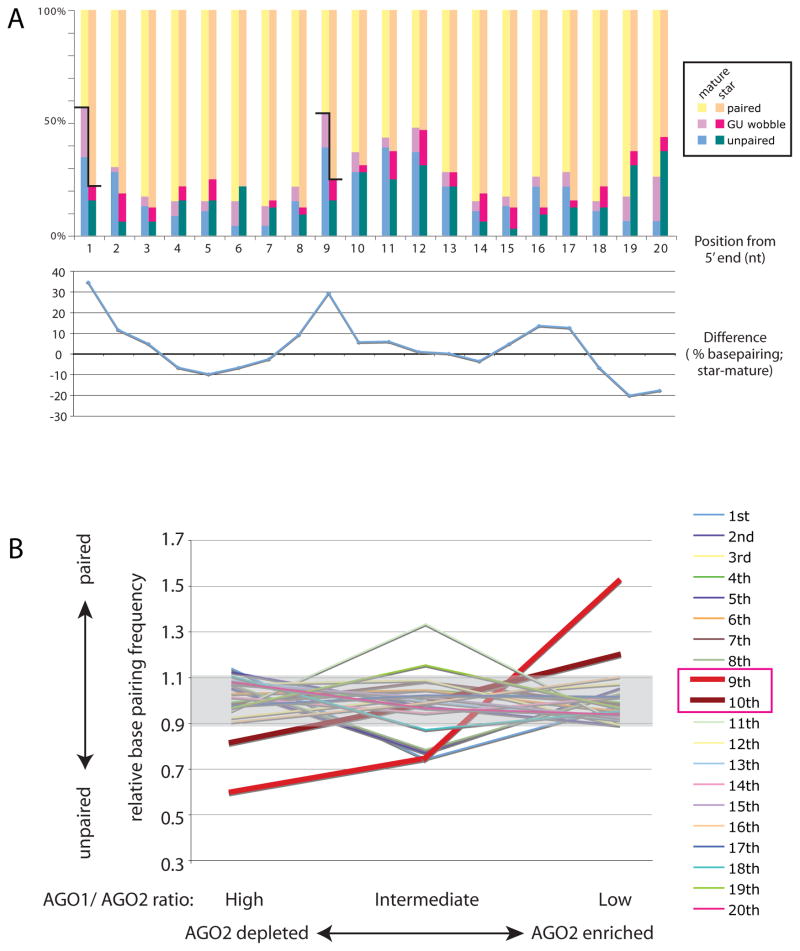
As expected, the most substantial difference between the collective pairing status of miRNAs and miRNA* species occurred at the 5′ nucleotide, reflecting the strong influence of thermodynamic asymmetry on strand selection by AGO1 (Figure 5A and Table S3). The second-most substantial difference occurred at position 9 from the 5′ end, with unpaired or G:U status correlating with AGO1 loading and paired status correlating with AGO2 loading. We analyzed pairing status with respect to the degree of enrichment in AGO1 or AGO2. This clearly showed that, by far, the pairing status of the 9th position was most correlated with differential sorting into the Argonautes; the 10th position also showed a limited correlation that was evident in this largescale analysis (Figure 5B and Table S3).
Figure 5. Mispairing at positions 9 and 10 correlates with sorting profiles.
(A) The pairing status of well-expressed miRNA duplexes in S2-NP cells was analyzed. Upper chart: Frequency of basepairing. Bars indicate % frequency of paired nucleotides at each position. Yellow: canonical basepairing (G-C or U-A), Pink: G-U wobble pair, Green: unpaired. Lower chart: Differences of % basepairing frequency between star and mature strands were plotted. A strong discrepancy between mature and star pairing status at their 5′ nucleotides was observed, correlating with the thermodynamic asymmetry rule for AGO1 strand selection. Another highly discrepant position was found at the nucleotide #9, suggesting a previously unrecognized constraint on the miRNA structures.
(B) Correlation between sorting and 9–10th mismatches. miRNA genes were grouped according to the ratio of reads in AGO1-IP and AGO2-IP libraries (Czech et al., 2008). We considered three categories of AGO1/AGO2 ratio, high (>15), intermediate (5–15) and low (<5). Relative basepairing frequency was calculated by basepairing frequency at each position in the given group of genes divided by basepairing frequency at the position in the all genes. Clear correlation was observed at the position 9, suggesting the pairing status at this position affects the sorting trends. The position 10 also exhibits modest but recognizable correlation. Note that these correlations do not distinguish between attraction to AGO1 and repulsion by AGO2; however, we designate a high AGO1/AGO2 ratio as representing depletion from AGO2 on the basis of in vitro sorting assays (Figure 6).
Pairing at nucleotides 9 and 10 actively determines strand selection by AGO2
The observation of 9th position as a sensitive point for differential AGO loading of miRNA strands was reminiscent of previous observations that central mismatches of positions 7–11 strongly determined AGO partitioning of model duplexes (Tomari et al., 2007); in fact, greatest sensitivity was observed at position 9. However, the effect of central structure on duplex strand preference was not determined in that study. We directly tested this using the in vitro sorting assay. We first analyzed the sorting of a miR-276 duplex in which the order of bulged positions was reversed with respect to the endogenous duplex (Figure 6A, B). Interestingly, AGO1 still preferred the mature strand of this variant duplex, but the preference of AGO2 was completely reversed and now loaded only the mature strand. Therefore, the strand selection of AGO2 is very sensitive to internal duplex structure, and its loading properties can be altered without affecting those of AGO1.
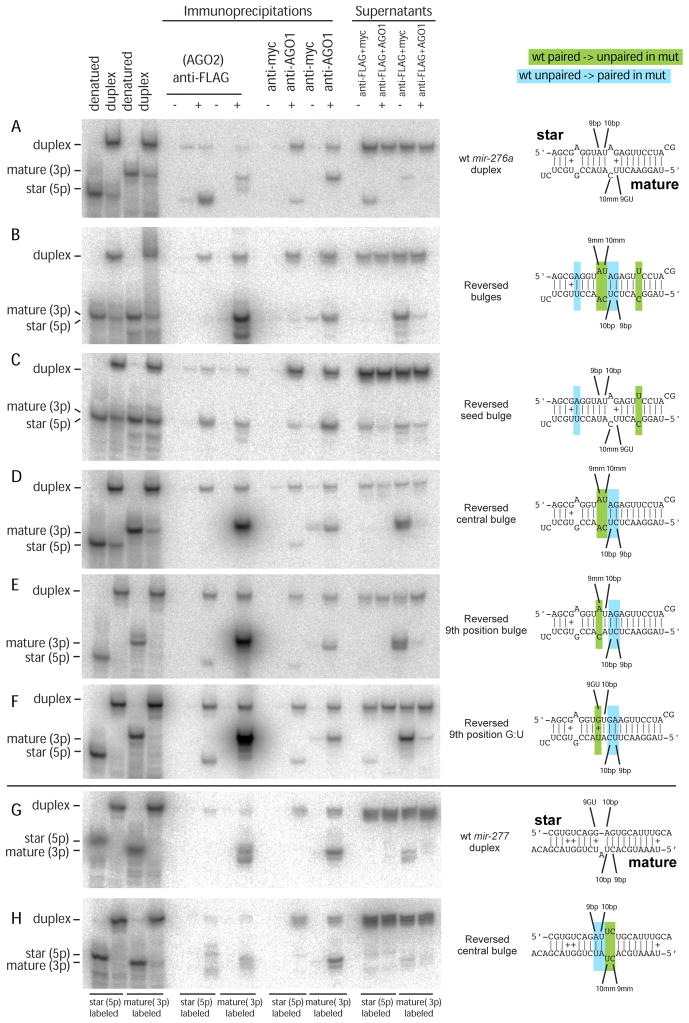
Figure 6. AGO2 strand selection is directed by duplex structure at positions 9/10.
(A) Wild-type mir-276a duplex shows preferred sorting of mature strand to AGO1 and star strand to AGO2.
(B) Complete reversal of the order of bulged nucleotides in mir-276a duplex induces AGO2 to select the mature strand exclusively; AGO1 still maintains preference for mature strand as well.
(C) Specific reversal of the miR-276a* seed bulge does not alter strand preference.
(D) Specific reversal of central bulges reverses the strand preference of AGO2.
(E) A single 9th position bulge of the star strand induces AGO2 to select mature miR-276a.
(F) A single 9th position G:U pair of the star strand induces AGO2 to select mature miR-276a.
(G) Wild-type mir-277 duplex shows that both AGO1 and AGO2 select mature miR-277. Note that our bioinformatics analysis showed that 9/10th positions correlated best with sorting profiles when gaps were not counted as nucleotide positions (Figure 5); thus, the 10th nucleotide of miR-277* was annotated as paired.
(H) Continuous pairing at the 9th and 10th positions of miR-277* redirects the star strand into AGO2. Note that mature strand miR-277 has gone from being fully paired to fully unpaired at 9th and 10th positions, but this did not alter strand selection by AGO1.
We probed this further with additional variants. In the endogenous miR-276a duplex, the mature strand is unpaired at positions 9, 10 and 16, whereas the star strand is unpaired at positions 5, 11 and 12. To test whether a seed bulge was relevant for AGO2 strand selection, we reversed the miR-276a* seed bulge to be present in the mature strand; however, this maintained preferred star sorting to AGO2 (Figure 6C). To test whether this was attributable to the position of the centrally paired nucleotides, we reversed the position of the central bulges (Figure 6D). This duplex showed complete selection of the mature strand by AGO2. Given that position 9 showed the greatest correlation for differential miRNA duplex sorting, we tested a miR-276a duplex variant with a single star mismatch at position 9, maintaining a paired 10th position (Figure 6E). This also yielded predominant selection of the mature strand by AGO2. Finally, as our computational analysis suggested that 9th position G:U pairing might be functionally similar to mispairing with respect to AGO2 strand selection, we tested an additional variant. Indeed, introduction of a single G:U pair at position 9 of miR-276a* resulted in nearly complete selection of miR-276a by AGO2 (Figure 6F). We conclude that Watson-Crick pairing at position 9 can actively dictate strand selection by AGO2, and that G:U or mispairing at this position are inhibitory to AGO2 strand selection.
We recognize that the end energies of mir-276a duplex are relatively similar, so that this particular duplex might in principle be especially sensitive to central structure. We therefore wished to confirm our results with a different miRNA duplex. Zamore and colleagues reported mir-277 as an unusual locus for which a substantial amount of mature strand is sorted to AGO2 (Forstemann et al., 2007); this duplex is clearly asymmetric with the 5′ end of mature miR-277 residing in the less-structured end. We subjected mir-277 duplex to in vitro sorting assay, and confirmed that AGO2 selected mature miR-277 exclusively. Inspection of this duplex showed that the miR-277* strand has G:U at position 9 (Figure 6G), consistent with rejection of the star strand by AGO2. We tested this by mutating the miR-277* strand to reverse its pattern of central bulges, now paired at 9th and 10th positions of the star strand and unpaired at 9th and 10th positions of the mature strand (Figure 6H). This reversed the behavior of AGO2, which now accepted a substantial amount of miR-277*. This reciprocal redirection strongly supports our proposal that AGO2 strand selection is actively directed by the duplex pairing status of positions 9 and 10, even in the context of a thermodynamically asymmetric duplex.
Studies of siRNA biogenesis indicated that the star strand of the precursor duplex represents the first target of guide strand-programmed cleavage. This is functionally relevant since the introduction of non-cleavable phosphorothiorate (PS) linkages at the scissile phosphodiester bond (Schwarz et al., 2004) impedes RISC maturation (Leuschner et al., 2006; Matranga et al., 2005; Miyoshi et al., 2005; Rand et al., 2005). We tested whether passenger strand cleavage was similarly important for maturation of miRNA*-loaded AGO2 complex by introducing PS linkage between nucleosides 10 and 11 of mature miR-276a, thereby blocking its potential cleavage by miR-276a*. However, unwound miR-276a* still accumulated efficiently in AGO2 (Supplementary Figure 3). This suggests that passenger strand cleavage of miRNA duplexes is not absolutely required for the maturation of miRNA*-loaded AGO2-RISC.
Central duplex structure is less critical for strand selection by AGO1
It is relevant to note that in none of these manipulations, which redirected AGO2 preference from star to mature (for mir-276) or from mature to star (for mir-277), did we observe a reversal of strand selection by AGO1. Moreover, it was clearly evident that while pairing at positions 9/10 are critical for star strand selection by AGO2, pairing at these positions of the mature miRNA was not reciprocally unfavored for loading into AGO1. For example, in the mir-276a duplexes in Figure 6B, D, E and F all are fully paired at the 9th and 10th positions of mature miR-276a. Nevertheless, mature miR-276a was selected by AGO1 from all of these duplex variants, as in the endogenous situation where miR-276a contains G:U 9 and mismatch 10. In addition, full pairing at positions 9 and 10 of mature miR-277 (Fig. 6G), as well as full mismatching at these positions (Fig. 6H), were both compatible with nearly complete selection of mature miR-277 by AGO1. We infer from these data that AGO1 selects its strand primarily on the basis of the thermodynamic rules and on 5′ nucleotide, and is less actively influenced by central duplex structure.
Discussion
Because higher eukaryotes usually have multiple Argonaute proteins with distinct functions (Carmell et al., 2002; Farazi et al., 2008), it follows that the sorting of small RNAs to the appropriate effectors is critical for their function. Several mechanisms for small RNA sorting have been reported in different organisms. In Arabidopsis, sorting is actively influenced by the 5′ nt of the small RNA, with AGO1 preferring 5′ U, AGO2 and AGO4 preferring 5′ A, and AGO5 preferring 5′ C (Mi et al., 2008). In C. elegans, miRNA and siRNA sorting are influenced by RNA precursor structures and the protein machineries that process them (Jannot et al., 2008; Steiner et al., 2007). Mammals have 4 Argonaute-class proteins, only one of which is endowed with Slicer activity (Liu et al., 2004; Meister et al., 2004). Although initial studies suggested that mammalian miRNAs are similarly apportioned to the different Argonautes, recent immunoprecipitation studies suggest potential differences in their miRNA cargoes (Azuma-Mukai et al., 2008; Ender et al., 2008; Su et al., 2009). It also remains to be seen whether the recently-discovered mammalian endo-siRNAs are sorted differently from miRNAs (Babiarz et al., 2008; Tam et al., 2008; Watanabe et al., 2008).
In Drosophila, predominant sorting of mature miRNAs and exogenous siRNAs to AGO1 and AGO2 complexes, respectively, was reported earlier (Miyoshi et al., 2005). Our studies allow us to infer a substantial pathway that preferentially sorts miRNA* species into AGO2. We hypothesize that Drosophila miRNA genes are in fact finely tuned to generate specific amounts of miRNA and miRNA* species in both AGO1 and AGO2 complexes. Whilst the incorporation of miRNA* species in AGO1 has generated modest target pools, adding to those of conventional miRNA binding sites, the biological basis of either miRNA or miRNA* loading into AGO2 remains mysterious. Despite biochemical evidence for programming of AGO2 by endogenous miRNAs and miRNA* species (Czech et al., 2008; Forstemann et al., 2007; Ghildiyal et al., 2008; Kawamura et al., 2008; Seitz et al., 2008; this work), there are hardly any perfectly complementary targets apparent in the transcriptome. We searched for imperfect targets of many miRNA* species with strong AGO2 preference using general criteria that identified plant miRNA targets, for which extensive complementarity is the rule (Rhoades et al., 2002). Although there were some candidate targets with only a few mismatches and G:U pairs, we also observed similar numbers of hits to sense strands of mRNAs (Supplementary Table 4). However, the lack of clear enrichment for antisense matches does not rule out that some might be functional targets of miRNA*-programmed AGO2 complex, and relatively little is known about the parameters for recognition of imperfect targets by AGO2. In any case, the broad sorting of miRNA* species into AGO2 suggests that this feature is under evolutionary selection, perhaps either to regulate some endogenous or viral targets, or alternatively to suppress their ability to repress transcripts via miRNA-like seed matching.
Distinct mechanisms for miRNA and miRNA* strand selection by Argonautes
Distinct AGO preferences of partner miRNA and miRNA* species were not anticipated, since the thermodynamic rules for guide strand loading into miRISC and siRISC were thought to be the same (Khvorova et al., 2003; Schwarz et al., 2003; Tomari et al., 2004). While we cannot exclude that the two strands of an individual duplex are sent to different Argonautes, we envision that independent sorting occurs across a population of a given duplex. If the loading complexes for AGO1 and AGO2 have different strand preferences, duplexes that can bind both loading factors would appear to exhibit independent sorting (Figure 7).
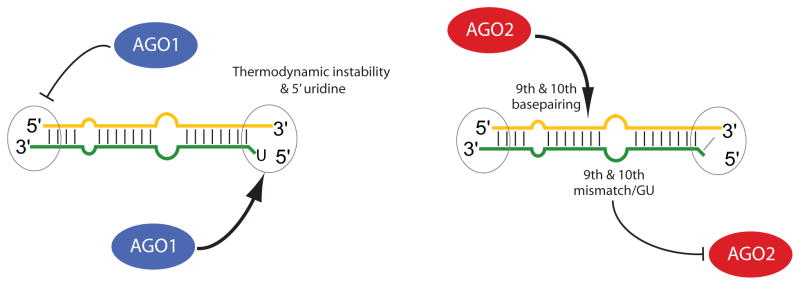
Figure 7. Model for differential miRNA strand selection by AGO1 and AGO2.
AGO1 strand selection is influenced by 5′ uridine in addition to the previously proposed thermodynamic rule, while it is insensitive to the positions of central mismatches. In contrast, AGO2 strand selection is highly sensitive to 9th and 10th basepairing status. Drosophila miRNA genes are generally configured to access distinct strand preferences of AGO1 and AGO2, thereby yielding the seemingly independent sorting of miRNA and miRNA* species.
We used in vitro assays to distinguish duplex features that actively influence strand selection and sorting. The apparent insensitivity of AGO1 strand preference to central mismatch positions (Figure 6) is consistent with the thermodynamic asymmetry rule. However, we could reverse the strand selection by AGO1 by making reciprocal changes in the duplex 5′ ends so that only the star strand had 5′ U (Figure 3). Therefore in addition to preferring the less-structured duplex end, AGO1 also detectably prefers 5′ U, perhaps underlying the propensity of mature miRNAs to have 5′ U.
On the other hand, our studies of the small RNAs associated with AGO1 and AGO2 indicated substantial differences in their strand selection from individual duplexes, suggesting that AGO2 is not solely governed by thermodynamic asymmetry. Although in vitro sorting assays did not show a strong influence of 5′ nucleotide, they clearly demonstrated that AGO2 strand selection could be reversed by manipulating central duplex structure. In particular, we found that pairing at position 9 and 10 strongly promoted strand selection by AGO2 (Figure 6). In all of these manipulations, strand selection by AGO1 was largely unaffected, indicating that strand selection and sorting to AGO2 are controlled actively and independently of the behavior of AGO1. Therefore in addition to thermodynamic duplex asymmetry (Schwarz et al., 2003; Tomari et al., 2004), our data reveal a powerful influence of central structure on strand selection by Drosophila AGO2.
Altogether, our study unifies the phenomena of strand selection and sorting of miRNAs in Drosophila, providing evidence that they stem from the distinct strand preferences of the Argonautes (Figure 7). Moreover, the structural and sequence features of Drosophila miRNA genes appear to have evolved in concert with the properties of AGO1 and AGO2 to yield differential sorting of miRNA duplex strands, implying that this arrangement is of biological utility.
Materials and Methods
Northern blotting and β-elimination
Northern blotting was performed as described (Okamura et al., 2007) using total RNA from S2-R+ cells or cells that had been treated with two consecutive 4-day treatments with previously described dsRNAs (Okamura et al., 2008a). 5′ 32P labeled LNA or DNA oligonucleotide probes were used to detect small RNAs. Total RNA was subjected to β-elimination as described (Horwich et al., 2007).
Immunoprecipitation
S2-R+ cells were stably transfected with a genomic construct of FLAG-HA-tagged AGO2. Cells were lysed in IP buffer (30 mM HEPES-KOH (pH 7.3), 150 mM KOAc, 2 mM MgOAc, 5 mM DTT, 0.1% NP40) and cleared by a centrifugation at 10,000g for 10 min at 4 degrees. FLAG-M2 conjugated beads (Sigma) or Gamma Bind G Beads (GE Healthcare) bound with rabbit anti-myc (Santa Cruz) or rabbit anti-AGO1 (AbCam) were used for immunoprecipitation. Beads were incubated with lysate for 1–1.5h at 4 degrees and supernatant from 1st IP was used for 2nd IP. Co-precipitated RNA was extracted by a Phenol/Chloroform/Isoamyl alcohol extraction and precipitated with EtOH.
Luciferase assay
A modified psiCHECK plasmid was used to construct perfect or bulged targets for miRNA or miRNA* species. The complete list of oligos used for sensor cloning is listed in the Supplementary Table 5. The sensor plasmids were co-transfected with 100nM 2′-O-methyl antisense oligonucleotides. Three days after transfection, luciferase activity was measured with Dual-Glo Luciferase assay system (Promega). Details were described before (Okamura et al., 2007).
In vitro sorting assay
S2 lysate was prepared as described (Okamura et al., 2008a). We prepared duplexes of 32P-labeled oligos annealed to unlabeled phosphorylated partner oligos. 1nM (~1–2×105 cpm) labeled duplexes were incubated for 1h in 80 μl in vitro RNAi reactions and the reactions were stopped by adding chilled 320 μl IP buffer. IP was performed as described above. Co-precipitated RNA was extracted by phenol/chloroform followed by EtOH precipitation. Excess of unlabeled small RNA identical to labeled strand was added after phenol/chloroform extraction to prevent re-annealing. The RNA oligos used for this assay was listed in Supplementary Table 5.
Small RNA library analysis
Small RNAs (18–28nt) were purified from S2-NP cells and cloned as described (Czech et al., 2008), and subjected to a single lane of sequencing on the Illumina 1G platform (BC Genome Sciences Centre). The reads were deposited at NCBI Gene Expression Omnibus under sample GSM371638. We also analyzed published AGO1-IP (GSM280088) and AGO2-IP (GSM280087) libraries from S2-NP cells, total RNA (GSM280082) and AGO2-IP (GSM280086) libraries from ovaries (Czech et al., 2008), and total RNA and beta-eliminated RNA from S2 cells (GSE9389; 0, 1 and 2 linker mismatch datasets) (Seitz et al., 2008).
Small RNAs were clipped of 3′ linkers and string-matching was used to identify known miRNA and miRNA* reads (Ruby et al., 2007). Star/miR enrichment analysis (Figure 4A and B) was done for 38 genes with >100 miRNA reads and >10 miRNA* reads in S2-NP total library. For miR*/miR ratio analysis (Figure 4C), all the non-redundant reads mapped on miRNA hairpins were counted. For Figure 5A and B, all miRNA or miRNA* species with >30 reads in the total RNA library and >10 reads in the combined AGO1-IP and AGO2-IP datasets. Mature and star strands were considered separately; 46 mature + 32 star strands were analyzed. No reads were recorded for miR-306* and miR-994* in AGO1, or for miR-252*, miR-11* and miR-970*in AGO2; we arbitrarily assigned them values of 0.5 so that real number miRNA:miRNA* ratios could be calculated. mir-2 genes were removed because the vast majority had multiple blast hits, complicating their assignment to any particular duplex; mir-281-1 and -2 genes were kept because both have a similar structure; miR-13b reads were all assigned to 13b-2 because 13b-1 had no star reads in S2 cells (the other members of the mir-13b-1, mir-13a mir-2c operon also had very few reads). 5′ nucleotided positions were determined by the most frequently cloned sequences in NP library; Gaps were not considered as nucleotides. Cutoffs for the three expression groups were high (AGO1/AGO2>15), medium (15≥AGO1/AGO2>5) and low (AGO1/AGO2≤5).
Supplementary Material
Acknowledgments
We thank Gregory Hannon for FLAG-HA-AGO2 plasmid. Illumina sequencing was performed at the BC Genome Sciences Centre. Haruhiko and Mikiko Siomi provided AGO2 antibody. K.O. was supported by the Charles Revson Foundation. E.C.L. was supported by the Sidney Kimmel Cancer Foundation, the Alfred Bressler Scholars Fund and the US National Institutes of Health (R01-GM083300 and U01-HG004261).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azuma-Mukai A, Oguri H, Mituyama T, Qian ZR, Asai K, Siomi H, Siomi MC. Characterization of endogenous human Argonautes and their miRNA partners in RNA silencing. Proc Natl Acad Sci U S A. 2008;105:7964–7969. doi: 10.1073/pnas.0800334105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiarz JE, Ruby JG, Wang Y, Bartel DP, Blelloch R. Mouse ES cells express endogenous shRNAs, siRNAs, and other Microprocessor-independent, Dicer-dependent small RNAs. Genes Dev. 2008;22:2773–2785. doi: 10.1101/gad.1705308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel J, Sachidanandam R, et al. An endogenous siRNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ender C, Krek A, Friedlander MR, Beitzinger M, Weinmann L, Chen W, Pfeffer S, Rajewsky N, Meister G. A human snoRNA with microRNA-like functions. Mol Cell. 2008;32:519–528. doi: 10.1016/j.molcel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Farazi TA, Juranek SA, Tuschl T. The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development. 2008;135:1201–1214. doi: 10.1242/dev.005629. [DOI] [PubMed] [Google Scholar]
- Flynt AS, Lai EC. Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet. 2008;9:831–842. doi: 10.1038/nrg2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, Zamore PD. Endogenous siRNAs Derived from Transposons and mRNAs in Drosophila Somatic Cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli A, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Iwasaki S, Kawamata T, Tomari Y. Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol Cell. 2009;34:58–67. doi: 10.1016/j.molcel.2009.02.010. [DOI] [PubMed] [Google Scholar]
- Jannot G, Boisvert ME, Banville IH, Simard MJ. Two molecular features contribute to the Argonaute specificity for the microRNA and RNAi pathways in C. elegans. Rna. 2008;14:829–835. doi: 10.1261/rna.901908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka Y, Takeichi M, Uemura T. Developmental roles and molecular characterization of a Drosophila homologue of Arabidopsis Argonaute1, the founder of a novel gene superfamily. Genes Cells. 2001;6:313–325. doi: 10.1046/j.1365-2443.2001.00427.x. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada T, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct Roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA Silencing Pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi K, Tsukumo H, Nagami T, Siomi H, Siomi MC. Slicer function of Drosophila Argonautes and its involvement in RISC formation. Genes Dev. 2005;19:2837–2848. doi: 10.1101/gad.1370605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008a;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The Mirtron Pathway Generates microRNA-Class Regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Lai EC. Endogenous small interfering RNAs in animals. Nat Rev Mol Cell Biol. 2008;9:673–678. doi: 10.1038/nrm2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol. 2008b;15:354–363. doi: 10.1038/nsmb.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelisson A, Sarot E, Payen-Groschene G, Bucheton A. A novel repeat-associated small interfering RNA-mediated silencing pathway downregulates complementary sense gypsy transcripts in somatic cells of the Drosophila ovary. J Virol. 2007;81:1951–1960. doi: 10.1128/JVI.01980-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110:513–520. doi: 10.1016/s0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, Lai EC. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Seitz H, Ghildiyal M, Zamore PD. Argonaute loading improves the 5′ precision of both MicroRNAs and their miRNA strands in flies. Curr Biol. 2008;18:147–151. doi: 10.1016/j.cub.2007.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner FA, Hoogstrate SW, Okihara KL, Thijssen KL, Ketting RF, Plasterk RH, Sijen T. Structural features of small RNA precursors determine Argonaute loading in Caenorhabditis elegans. Nat Struct Mol Biol. 2007;14:927–933. doi: 10.1038/nsmb1308. [DOI] [PubMed] [Google Scholar]
- Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, Hannon GJ. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Du T, Zamore PD. Sorting of Drosophila small silencing RNAs. Cell. 2007;130:299–308. doi: 10.1016/j.cell.2007.05.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Zhou R, Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol Cell. 2008;32:592–599. doi: 10.1016/j.molcel.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.