Abstract
The mouse cytomegalovirus (CMV), a β-herpesvirus, exploits its large (~230 kb) double-stranded DNA genome for both essential and non-essential functions. Among the non-essential functions are those that offer the virus selective advantage in eluding both the innate and adaptive immune responses of the host. Several non-essential genes of MCMV are thought to encode MHC-I-like genes and to function as immunoevasins. To understand further the evolution and function of these viral MHC-I (MHC-Iv) molecules, X-ray structures of several of them have been determined, confirming the overall MHC-I-like structure, but also elucidating features unique to this family. Future efforts promise to clarify the nature of the molecular ligands of these molecules, their evolution in the context of the adapting immune response of the murine host, and by analogy the evolution of the host response to human CMV as well.
Keywords: MHC recognition, NK cells, T cells, immunoevasins, X-ray crystallography, MHC-Iv molecules, cytomegalovirus
Introduction
Viruses engage in an interpretive dance with the cells and hosts that they infect. As viruses, they are not free-living organisms but are dependent upon metabolic functions provided by their hosts. As genetic beings, their primary mission is to preserve the viral genome and transmit it to future generations, and they can accomplish this successfully through a variety of mechanisms. Vertebrate viruses encounter resistance from the host in the form of the inflammatory response and innate and adaptive immunity, which form an overlapping network of cellular and molecular reactions to the perturbations induced by viral infection. The virus exploits its rapid life cycle to generate mutants, which allow escape from the repression of the immune system. The host immune system matures and evolves, refining its repertoire of responses. The virus is controlled, to a degree; its mutants then escape again.
This evolutionary play is enacted by all parasites and all hosts. Viruses, as examples of relatively simple cellular parasites, may be as simple as the phage that infect bacteria, in which lytic infection gives rise to a burst of progeny that serve as a source of mutants to combat cellular variation and resistance of the bacterial host. Some bacterial viruses establish a persistent infection that leads to long-term production of viral particles that can proceed to infect neighboring bacteria. They may also enter a cryptic state known as lysogeny (e.g. bacteriophage λ), in which the phage genome integrates into the host chromosome and, repressed, replicates in step with the host. Appropriate environmental conditions lead to derepression and excision of the viral genome and entry into a lytic phase. Among the viruses that affect vertebrates, those with relatively small coding capacity (such as the RNA viruses rhinovirus (~7 kb for human rhinovirus A) or influenza A (~13 kb total for 8 segments)), rely on extensive antigenic variation to escape the host immune response. Vertebrate viruses with larger DNA genomes, such as the pox- or herpesviruses, follow a long-term strategy, and often manage to co-exist with their hosts for life. The advantage of the larger genomes of these DNA viruses is that in addition to encoding essential functions for virus replication, survival, and maturation, they may encode a number of proteins that can interact directly with molecules involved in the host immune response. They thus can tip the balance in favor of the virus, resulting in the establishment of persistent infection or of latency. Since the host inflammatory/immune system has evolved to generate antibodies, natural killer (NK) cells, and antigen-specific T cells against foreign viruses or their products, the virus has correspondingly evolved many different mechanisms to achieve its survival. One general scheme used by the latent/persistent viruses (such as herpes simplex) is to sequester themselves from the host by infecting non-permissive cell types that act as reservoirs for their genetic material. In such cellular reservoirs, little or no gene transcription occurs until the viral genome is reactivated, allowing entry into a new maturation phase and further viral spread (2). In addition to viral genes involved in establishing latency, other pathways of viral interference include the blockade of antigen presentation, evasion of NK cell responses, disruption of cytokine signaling networks, inhibition of apoptosis in virus-infected cells, and evasion of antibody and complement responses (3, 4). The herpesvirus family provides examples of virus-encoded proteins that target each of these different pathways. Recently, the human cytomegalovirus has been shown to modulate the host immune response via a microRNA that down regulates a stress-induced molecule seen by the host (5, 6).
The herpesvirus family, which includes α, β, and γ- herpesvirinae, serves as a prime example of the many immunoevasive strategies that such large DNA viruses employ. The prototypic human cytomegalovirus (HCMV), for which some 50% to 90% of humans are seropositive (7), causes no clinical disease in the healthy, but serious and fatal disease may result from primary infection or activation in immunocompromised patients (such as transplant recipients, AIDS patients, and newborns). The murine CMV (MCMV) serves as a valid experimental model for the human disease, and is particularly valuable in that specific viral genes can be targeted in deletion mutants. Specifically, non-essential genes of MCMV have been studied extensively for their effects on T cell and NK cell recognition. MCMV causes acute, persistent and latent infection in mice (8). The 230 kb genome, encoding about 170 unique open reading frames (ORF), is organized such that its central portion directs the synthesis of molecules crucial for essential viral functions such as virus replication. This region is conserved among CMVs of different species. The termini of the genome are considerably more divergent and, in the mouse, consist of two sets of genes that function in immunoevasion. On the left arm of the genome are the genes of the m02 family, consisting of m02 through m16, and one member of the m145 family, m17, and toward the right arm are the other members of the m145 family, comprised of m145, m146, m150 through m155, m157, and m158 (9). Early on, it was noted that the sequence and biochemical similarities of the product of the MCMV gene m144, which maps proximal to m145, though technically not considered a member of the m145 family, may be considered a distant relative (10).
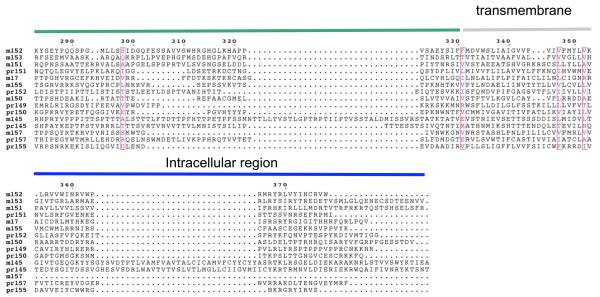
Each of the genes of the m145 family encodes a likely signal peptide, a transmembrane region (except for the m157 protein which has a glycosyl-phosphatidyl-inositol (GPI) linkage site instead), and several sites for N-linked glycosylation. The rat CMV encodes a homologous and expanded family of such genes. However all members of the mouse m145 family are not conserved. Rat homologs with both sequence and positional homology are r145, r149, r150, r151, r152, r155 and r157 (11). An interesting feature of the mouse m145 protein family is that eight members (nine including m144) are predicted to adopt an MHC-I-like fold despite no significant sequence similarity to MHC-I molecules (12). A multiple sequence alignment of the predicted MHC-I-like m145 proteins and their rat homologs (including signal peptide, transmembrane and intracellular domains) is shown in Figure 1. The viral proteins share low levels of amino acid similarity (< 30%) and there are very few residues conserved in all family members. The predicted extracellular regions of the proteins are most similar, whereas the N-terminal (signal peptide) and C-terminal (predicted intracellular domain) regions show very little conservation. Some cysteine residues, involved in disulfide bonds, are conserved. These are indicated in the figure. Several important immune evasion functions of MCMV are attributed to members of the m145 family.
Figure 1.
Amino acid sequence alignment of mouse and rat members of the m145 family. Encoded amino acid sequences of the mouse and rat indicated proteins were aligned with ClustalW and the alignments were further arranged based on the crystallographic structures of m153 and m157. Conserved cysteines are aligned, and disulfide bond linkages are indicated by dashed lines. Indicated numbering is based on the full translated sequence of m152, beginning with the first methionine. Disulfides are numbered indicated by stars and the numbering is according to the amino acid sequence of the mature m153 protein.
Evidence for MCMV interference with the immune response
Among the strategies employed by MCMV to avoid the host immune response are to interfere with the activation of NK cells and to subvert the complex process of antigen presentation (13, 14). Although m144 was identified as an MHC-I-like molecule, the mechanism for its biological effects has been unclear. Farrell et al (15) showed that a mutant MCMV with a disrupted m144 gene was less virulent during acute infection and that this was reversed by NK cell depletion of the host. m144 expression in human cells (16) as well as in a mouse cell line (10) revealed resistance to NK cell cytotoxicity. Thus, it has been hypothesized that m144 binds an inhibitory NK cell surface receptor. To date, however, no NK cell or other ligand for m144 has been identified.
Of the m145 family, four members have been shown to interfere with the NK cell response. m145, m152, and m155 have all been shown to contribute to the downregulation of specific host cell encoded molecules that serve as ligands for the activating NK cell receptor NKG2D (17-20). In addition, m157 can bind an NK inhibitory receptor expressed in some mouse strains (12, 21). Cells exposed to various types of cellular stress, including but not limited to temperature, oncogenic transformation, or viral infection, express higher levels of Rae1 (of some 5 isoforms), H60, and MULT-1 (22), and m145, m152, and m155 have an antagonistic effect on the surface expression of MULT-1, Rae1, and H60 respectively. Subtleties of the relative effects of m152 on the different isoforms of Rae1 are currently under active investigation. It is interesting to note that Rae1 (whose structure has been determined crystallographically, (23)), H60 and MULT-1 are also related to MHC-I in structure, but only have the amino terminal α1 and α2 domains of the MHC-I fold. Another MCMV encoded protein interferes with NKG2D ligand expression. The viral FcRγ receptor encoded by the m138 gene was recently shown to reduce the surface expression levels of H60 and MULT-1 in MCMV infected cells (24). This study revealed distinct modes of interaction of the FcRγ with H60 and MULT-1 and emphasizes the diversity of Fc receptor function. MCMV m157 has been identified as a ligand for the NK cell activating receptor, Ly49H (12, 21), which is expressed in mouse strains that are resistant to MCMV (such as C57BL/6). Ly49H expression accounts for the MCMV resistance locus, Cmv1, which maps to the NK cell gene complex (NKC) on mouse chromosome 6. In addition, in some MCMV sensitive mouse strains, such as 129/J, an m157 ligand on NK cells, Ly49I, an inhibitory receptor, has been identified (12, 21). It would thus appear that the virus first was selected to express m157 as a ligand for an NK inhibitory receptor (such as Ly49I) providing a selective advantage to those viruses that could deliver a negative signal to the host NK cells. The host, under the selective pressure of the virus, then generated a variant of Ly49I (such as Ly49H) that then could bind m157 as an activating receptor. This is the situation in mouse strains that are resistant to MCMV. Finally, the virus can generate mutants that have escaped the recognition of the activating receptor. Indeed, in an experiment where MCMV was serially passaged through resistant B6 (Ly49H+) mice, several virus clones could be isolated that contained mutations in the m157 gene. The mutations all either blocked the cell surface expression of m157, or failed to bind to Ly49H (25). These results indicate that the virus quickly adapts under selective pressure from the host innate immune system.
MCMV also encodes several proteins that target antigen presentation by MHC class I molecules. Three of these are unrelated to MHC-I and the fourth, m152, is similar to MHC-I in amino acid sequence. Such molecules have been called VIPRs -Viral proteins that Interfere with antigen Presentation- to distinguish them from immunoevasins that function via different mechanisms. The m04 and m06 genes encode gp34 and gp48 respectively and they affect MHC-I trafficking in different ways. m04/gp34 associates with MHC-I in the ER and these complexes are transported to the cell surface where the proteins remain associated (26). m06/gp48 redirects MHC-I molecules to lysosomes, where they are subsequently degraded. m06/gp48 has a dileucine motif in its cytoplasmic tail that facilitates binding to adaptor proteins AP-1A and AP-3A and is sorted into the lysosomal pathway. Both m04 and m06 belong to the m02 gene family of MCMV. Other m02 family members have not been characterized functionally. The m152 gene that encodes the gp40 glycoprotein was the first MCMV immunoevasin to be described (27). Initial observations showed that an epitope derived from the immediate early (IE) protein pp89 was presented during IE MCMV gene expression but not when early genes were expressed (28, 29). Further studies revealed that the virus blocks antigen presentation by retaining the tri-molecular MHC-I complex (heavy chain, β2-microglobulin (β2-m) and peptide) in the ER/cis-Golgi compartment (30). The region affecting MHC-I transport was mapped to the HindIII E fragment of the genome (31). m152 was identified as the gene responsible for MHC-I downregulation (32) and the lumenal domain of m152 was implicated in promoting the retention of MHC-I molecules (33). There is as yet no published evidence of a stable m152/MHC-I interaction and the exact mechanism of retention remains a mystery.
The biological consequences of the concerted action of m04, m06 and m152 are not well understood. A complex picture has emerged from studies utilizing engineered deletion viruses specific for single immunoevasins or combinations of them. These three viral genes exhibit different efficiencies in targeting different MHC-I alleles (34). Early studies indicated that m04 could inhibit CTL lysis of MCMV infected cells (35). It was proposed that m04 prevents the TCR from recognizing MHC-I, thereby reducing CTL activity. m04 was thus considered a negative regulator of antigen presentation (36). More recently a comprehensive analysis of the effect of m04 on antigen presentation both in vitro and in vivo has been undertaken (37). These experiments provided important new insight, namely that m04 instead acts as a positive regulator of antigen presentation. Cells that were infected with an m04/m06 deletion mutant MCMV successfully evaded CTL stimulation and lysis. Thus, m152 expression is sufficient to block antigen presentation and evade CTL attack. However, when m04 was expressed in combination with m152, the infected cells became susceptible to CTL lysis, suggesting that m04 restores antigen presentation by escorting MHC-I to the cell surface. Wild type MCMV-infected cells, where m04, m06 and m152 are expressed in combination, were able to evade CTL responses. Therefore m06 counters the positive effect of m04 and allows escape from immune recognition (37).
It was assumed that VIPR function was essential for MCMV to establish persistent infection in mice. However, a deletion mutant for m04, m06 and m152 did not differ in its ability to establish acute, persistent and latent infection in adult C57BL/6 mice. Furthermore the size and effector memory phenotype of the CD8+ T cell response elicited after infection with the triple mutant virus was comparable to the response against wild type virus (38). MCMV infection in salivary gland acinar epithelial cells cannot be controlled by CD8+ T cells and is only cleared by CD4+ T cells. The effect of deleting all three VIPRs on virus replication in the salivary gland was recently tested. Mice infected with the m04/m06/m152 triple knockout virus had 10-fold lower salivary gland titers than mice infected with wild type MCMV. The virus titers could be restored to the levels reached in a wild type MCMV infection by depleting CD8+ T cells (39). Taken together, it seems that the MCMV VIPRs affect antigen presentation to CD8+ T cells in the salivary gland epithelial cells and may be important to sustain virus production in the salivary glands and promote transmission to new hosts.
MCMV also encodes two molecules that influence chemokine function during infection. MCK-2, a CC chemokine homolog, promotes dissemination of the virus to the salivary glands (40) and M33, a viral chemokine receptor, has been shown to be very important for successful virus replication in the salivary glands (41). MCMV can infect dendritic cells and encodes the modB7-2 protein that selectively downregulates CD86 in antigen presenting cells (42). Other viral products that modulate the complement pathway (43) and prevent apoptosis in infected cells have also been described (44).
In addition to the proteins indicated above that interfere with the host immune response, recent papers have begun to address the possibility that virus-encoded microRNAs might target sequences in host genes that are involved in virus resistance. Stern-Ginossar et al (5) identified a microRNA of the HCMV, hcmvmiUL112 RNA, that downregulates the expression of MICB, a stress-induced molecule which is also a ligand for the activating NK cell receptor, NKG2D (6). A similar mechanism that might also function in the MCMV, is another means by which the virus can modulate its interactive dance.
Do MHC-I-like genes of CMV encode MHC-I like molecules?
Major questions raised by the initial analysis of the DNA and encoded protein sequences of the mouse and human CMVs included: 1) are the molecules predicted to be MHC-I-like by bioinformatic analysis truly MHC-I-like in biochemical characteristics and molecular structure? and 2) do such molecules bind ligands that would explain their selective advantage for the virus? Both Arase et al (21) and Smith et al (12) addressed the second concern with respect to the Cmv1 locus that encodes Ly49H. Our laboratory has taken a multifaceted approach to address these questions for the mouse CMVs. We have cloned the cDNAs of the predicted MHC-I-like genes, analyzed their expression following transfection into several cell types, and determined the structure of two representative members, m144 (45) and m153 (46). In addition, the structure of m157 has also recently been reported (47), and the structure of a distantly related MHC-I-like molecule from HCMV, UL18, has also been described (48).
Expression at the cell surface of m145 family members
The normal expression of classical MHC-I molecules at the cell surface is dependent on at least two additional components of the cellular machinery: the presence of the light chain of the MHC-I complex, β2-m, and the availability of peptides, initially processed by the proteasome in the cytoplasm and delivered to the endoplasmic reticulum via TAP, the transporter associated with antigen processing. We first asked whether members of the m145 family were expressed as cell surface protein on transfection of COS-7 cells (1). Using expression constructs encoding amino-terminal FLAG-Tag epitopes, we showed that m17, M37, m144, m145, m151, m152, m153, and m155 were expressed with varying efficiency (Figure 2). Both m152 and m155, which are expressed poorly, or not at all at the cell surface, are expressed well when assessed by intracellular staining (Figure 2B). Since the MHC-I fold was predicted for all members of the MCMV m145 family as well as m144 and M37 (12), we determined whether m144, m145, m151, m153 and M37 share the requirements of classical MHC-I molecules for β2m and peptide for stable expression. Thus, we transfected N-terminally FLAG-tagged constructs into a cell line deficient in β2m (R1E (49)) as well as its β2m sufficient counterpart (R1.1) (Figure 3). The surface expression of all five proteins was equivalent in both cell types indicating that none of them require β2m for stable surface expression (1, 45, 46). As reported elsewhere, for m144, although the molecule copurifies with β2m, in such transfection experiments, there is no absolute requirement of β2m for cell surface expression (45). Also, we explored the requirement for TAP for cell surface expression of M37, m145, m151, and m153 by transfection experiments with RMA (TAP+) and RMA-S (TAP−) cells (Figure 4). Clearly, the viral proteins do not require normal TAP function for stable cell surface expression.
Figure 2.
Cell surface expression of MCMV proteins related to MHC-I molecules. A) pIRES-GFP vectors encoding N-terminally FLAG-tagged constructs of the indicated proteins were transfected into COS-7 cells. Surface expression was evaluated 48 h after transfection by staining with the anti-FLAG M2 monoclonal antibody and anti-mouse IgG1-PE followed by FACS analysis. B) Intracellular staining of COS-7 cells transfected with m152- or m155-pIRES-GFP using the same antibodies as in A.
Figure 3.
Surface expression of MCMV MHC-I-like proteins is independent of β2m. R1.1 (β2m+) and R1E (β2m−) cells were transfected with the indicated FLAG-tagged viral genes in pIRES-GFP or H-2Dd-pIRES-GFP as a control. Eighteen hours post transfection surface levels of the viral proteins and H-2Dd were determined. The percentage surface expression is indicated in bold in the upper right quadrant.
Figure 4.
TAP-independent cell surface expression of M37, m145, m151 and m153. RMA (TAP+) and RMA-S (TAP−) cells were transfected with the indicated FLAG-tagged viral genes in pIRES-GFP or H-2Dd-pIRES-GFP as a control. Eighteen hours post transfection surface levels of m153 and H-2Dd were determined. The number of GFP positive cells indicates transfection efficiency. The percentage surface expression is shown in the upper right quadrant in bold (percentage of GFP+ cells that stain for either FLAG-M2 or H-2Dd). The results are representative of at least two independent experiments.
Structures of MHC-Iv molecules
Although amino acid sequence alignments, and bioinformatic evaluation suggested that members of the m145 family are structurally similar to MHC-I molecules, indeed the only definitive approach is to determine the molecular structure of several representative molecules in three dimensions, using the technique of X-ray crystallography. Our laboratory has determined the structures of m144 and of m153, and the structure of m157 has also recently been reported (47). We will summarize these structures here and indicate the salient differences.
The structure of m144
To determine the structure of m144, we first engineered the extracellular domain of the mature molecule for expression as inclusion bodies in E. coli, determined conditions for its refolding in the presence of the light chain, β2–m, and, following purification, identified conditions under which diffraction quality crystals formed. One crystal diffracted synchrotron radiation to 1.9 Å, and a complete data set was obtained (45). Solved by molecular replacement, the structure was further refined by standard methods, and provided the structure illustrated in Figure 5. The overall structure of the m144/β2-m complex revealed the canonical MHC-I-fold, with an α1α2 domain unit consisting of two anti-parallel α-helices supported on a platform of anti-parallel β-sheets. One conserved disulfide bond (connecting Cys 98 to Cys 128) and one unique disulfide (Cys 28 to Cys 70) stabilize the structure. The α3 immunoglobulin-like domain is structurally similar to that of bona fide MHC-I molecules, although the “hinge” angle that it makes with the α1α2 domain unit is extreme as compared to those of other MHC-I molecules. The molecular contacts of the m144 heavy chain to the β2m subunit are largely conserved. Nevertheless, the amino acid residues of genuine MHC-I molecules that make contact with the CD8 coreceptor on T cells (residues of the α3 domain “acidic loop”, residues 222 to 229 of classical MHC-I molecules) are not conserved, consistent with the view that m144 does not interact with CD8.
Figure 5.
Structure of m144. A ribbon diagram of the X-ray structure of m144 (PDB code 1U58) is illustrated, with the domains of the protein labelled. The region of the missing density at the amino terminal end of the α2 helix is sketched in as a cartoon.
One remarkable feature of the m144 structure is that a stretch of 13 amino acids that exists in the protein (residues 115 to 127), is absent from the electron density map. We conclude that this region, a region that one would expect to be the amino terminal part of the α2 helix, exhibits remarkable flexibility in the crystal. Although one interpretation of such apparent flexibility is that this is merely a mobile region of the molecule that failed to assume a consistent position in space in the crystallographic unit cell, an alternative is that such flexibility implies that this is a binding site for a ligand. Indeed, there is a developing concept that unstructured regions in proteins play an important role in their adaptability as binding partners in important cellular signaling pathways (50-52). Finally, in m144, no electron density was visualized in the cleft region between the α1 and α2 domains, and no space for a missing peptide was apparent in this region either. This is consistent with the finding that m144 fails to require peptide for in vitro refolding, and that no peptides copurified with m144 expressed from cultured cells (53).
Structure of m153
To address further the question as to whether other members of the m145 family are structurally related to MHC-I molecules, we explored the recombinant expression, purification, and crystallization of several additional members of the family. Despite the successful expression of m17, m90, m150, m151, M37, m153, m155, and m159 in E. coli as inclusion bodies, we were unsuccessful in refolding any of these proteins following solubilization. Several of these (M37, m151, and m153) were then expressed as proteins from secreted Drosophila S2 cells (1), and one of these, m153, was selected for further study because of the high level of protein expression and the lack of apparent aggregation in size exclusion chromatography. In addition, m153 serves as a representative of the group, or sub-family, which includes, m150, m151, and m152. These proteins are 41 to 44% similar to each other with respect to their amino acid sequence and m152 and m153 have 28% amino acid identity. m152 has two known functions that bestow a selective advantage on those virus isolates that express this molecule--the downregulation of host cell MHC-I expression and the downregulation of host cell Rae-1 expression.
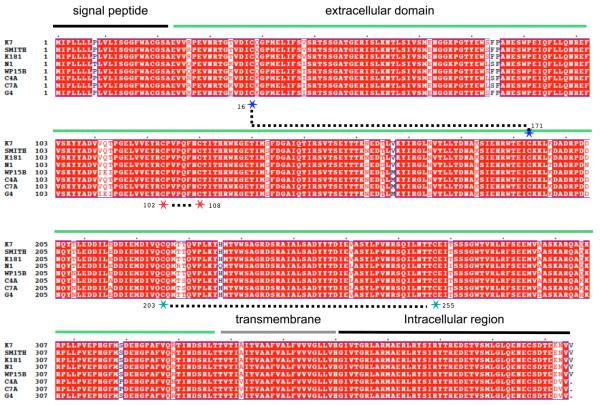
m153 has (as of this writing) no known biological function, but analysis of the gene and encoded protein sequences of a number of wild isolates of MCMV indicates that the encoded protein is highly conserved, suggesting that it is under selection for a particular function. The encoded m153 protein sequences from six different virus isolates as compared to the laboratory Smith and K181 strains are presented in Figure 6.
Figure 6.
Multiple amino acid sequence alignment of laboratory and wild isolates of MCMV m153 encoded proteins. The sequences of two laboratory strains, Smith and K181, are compared with six additional wild isolates. Numbering is from the amino terminus of the unprocessed translation product, and domain locations are indicated. Cysteine residues involved in disulfide bonds are indicated by stars, and are numbered according to the mature protein sequence.
The biochemical characterization of the recombinant m153 protein initially suggested that the extracellular domain of the molecule formed a non-covalent dimer. This was confirmed by analytical ultracentrifugation analysis, pull-down experiments, and fluorescence complementation analysis in transfectant cells (1, 46). The three-dimensional crystal structure, solved at 2.4 Å resolution using selenomethionine-substituted protein for phasing by the method of single anomalous dispersion (SAD), offers more detailed visualization not only of the overall protein fold, but of the nature of the dimerization interface. Once the detailed atomic interactions at the interface were defined crystallographically, site-directed mutagenesis studies and analysis of the resulting protein confirmed the nature of the dimer interface (1, 46).
The overall structure of the m153 homodimer revealed a single homodimer in the crystallographic asymmetric unit, with two individual chains associated non-covalently (see Figure 7A-F). The structure reveals the basic features of the MHC-I fold, two anti-parallel α-helices representing the α1 and α2 domains, supported on a platform of anti-parallel β-strands, and a relatively independent immunoglobulin-like α3-domain. Novel features include the lack of a β2-m subunit, the narrow juxtaposition of the helices that in other MHC-I molecules define the peptide binding cleft, the extended amino-terminal region joined by a disulfide bond to the H2b helix (Figure 7A), and tight non-covalent dimer assembly (Figure 7D-F). The lack of a binding cleft is emphasized in a surface representation of the molecule (Figure 7C), which shows the lack of space to accommodate a ligand. The most provocative aspect of the m153 structure, however, is that the non-covalent dimer appears to form a unique binding site poised for interaction with some ligand. Several experimental strategies are currently being employed in efforts to identify a functional ligand that binds m153, and that would explain the conservation of the m153 sequence in the genomes of the wild mouse-derived cytomegaloviruses.
Figure 7.
X-ray structures of m153 and m157 reveal novelty, similarities, and differences among members of the m145 family of proteins. Structures of m153 (PDB id: 2O5N)(panels A to F) and m157 (PDB id: 2NYK) (panels G to I) are shown in either ribbon (A, B, D, E, G, H, I) or surface (C, F) representation). The m153 monomer is shown in A to C, and the homodimer in E to F. m157 is displayed in G and H. I shows a superposition of m153 and m157. All molecular illustrations were prepared with PyMOL http://www.pymol.org (55).
Structure of m157
m157 is one of several members of the m145 family for which molecular ligands are known (12, 21), and recently the X-ray structure of m157 was determined (47). As shown in the amino acid alignment of Figure 1, m157, though clearly related to m153, lacks the amino terminal disulfide bond common to m152, m153, m151, m17, m155, pr151, and pr152. m157 clearly has a peptide-free, β2-m-free MHC-I-like structure, but lacks the remarkable homodimerization observed for m153 (Figure 7G-H). Its amino terminal residues form an α–helix that extends away from the rest of the molecule in a unique configuration. In addition, electron density corresponding to amino acids 132 to 146 was not observed in the structure, a region very similar to the region of m144 that lacked electron density. We would suggest that this again indicates that this region, located at the amino terminal end of the α2-helix, may prove to be an unstructured region that is involved in ligand binding. The structure of m157 also provides a valuable framework for understanding a set of m157 mutants that escape NK cell resistance or that are incapable of stimulating Ly49H+-reporter cells (25, 54). Two possible regions, either the β-sheet floor of the α1α2-domain or an area that seems close to the novel α0 helix have been proposed to serve as the Ly49H or Ly49I129 binding sites on m157 (47).
Conclusions
The past several years bear witness to the determination of the nucleotide sequences of the entire genomes of numerous organisms including a number of medically important viruses. Initial analysis of these nucleotide sequences establish likely ORFs and further bionformatic analysis of the ORFs has supported the prediction that in the mouse cytomegalovirus there are a number of genes that encode molecules with structural similarity to classical MHC-I molecules. Functional studies of these MHC-Iv molecules have led to the conclusion that several of them perform valuable immunological functions to allow the successful propagation of the virus. Experiments surveying the expression of the ORFs and the nature of the proteins they encode lead to several notable conclusions. It is clear from transfection experiments, that most of the m145 family members are effectively expressed at the cell surface, even in the absence of β2-m and access to peptide (via TAP function). Structure studies from our laboratory and others have definitively shown that several representative members of the MHC-Iv molecule family, m144, m153, and m157, are structurally related to classical MHC-I molecules, though they also have distinctive features. Further studies promise to define the molecular function, ligands, and sites of interaction of these interesting molecules, and perhaps to shed additional light on the selective forces that help pathogen and host survive and continue their evolutionary dance.
Acknowledgements
This work was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases.
Footnotes
This review is based in part on a Doctoral Dissertation submitted to the Faculty of Health Sciences, University of the Witwatersrand, Johannesburg, South Africa, for the degree of Doctor of Philosophy by Janet Mans (1)
REFERENCES
- 1.Mans J. Characterization of Mouse Cytomegalovirus MHC-I Homologs. University of the Witwatersrand; Johannesburg: 2008. [Google Scholar]
- 2.Steiner I, Spivack JG, Deshmane SL, Ace CI, Preston CM, Fraser NW. A herpes simplex virus type 1 mutant containing a nontransinducing Vmw65 protein establishes latent infection in vivo in the absence of viral replication and reactivates efficiently from explanted trigeminal ganglia. J Virol. 1990;64:1630–8. doi: 10.1128/jvi.64.4.1630-1638.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finlay BB, McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767–82. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 4.Tortorella D, Gewurz BE, Furman MH, Schust DJ, Ploegh HL. Viral subversion of the immune system. Annu Rev Immunol. 2000;18:861–926. doi: 10.1146/annurev.immunol.18.1.861. [DOI] [PubMed] [Google Scholar]
- 5.Stern-Ginossar N, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–81. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stern-Ginossar N, et al. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat Immunol. 2008;9:1065–73. doi: 10.1038/ni.1642. [DOI] [PubMed] [Google Scholar]
- 7.Sissons JG, Carmichael AJ, McKinney N, Sinclair JH, Wills MR. Human cytomegalovirus and immunopathology. Springer Semin Immunopathol. 2002;24:169–85. doi: 10.1007/s00281-002-0104-0. [DOI] [PubMed] [Google Scholar]
- 8.Sweet C. The pathogenicity of cytomegalovirus. FEMS Microbiol Rev. 1999;23:457–82. doi: 10.1111/j.1574-6976.1999.tb00408.x. [DOI] [PubMed] [Google Scholar]
- 9.Rawlinson WD, Farrell HE, Barrell BG. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–49. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cretney E, Degli-Esposti MA, Densley EH, Farrell HE, Davis-Poynter NJ, Smyth MJ. m144, a murine cytomegalovirus (MCMV)-encoded major histocompatibility complex class I homologue, confers tumor resistance to natural killer cell-mediated rejection. J Exp Med. 1999;190:435–44. doi: 10.1084/jem.190.3.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vink C, Beuken E, Bruggeman CA. Complete DNA sequence of the rat cytomegalovirus genome. J Virol. 2000;74:7656–65. doi: 10.1128/jvi.74.16.7656-7665.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith HR, et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc Natl Acad Sci U S A. 2002;99:8826–31. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lodoen MB, Lanier LL. Viral modulation of NK cell immunity. Nat Rev Microbiol. 2005;3:59–69. doi: 10.1038/nrmicro1066. [DOI] [PubMed] [Google Scholar]
- 14.Pinto AK, Hill AB. Viral interference with antigen presentation to CD8+ T cells: lessons from cytomegalovirus. Viral Immunol. 2005;18:434–44. doi: 10.1089/vim.2005.18.434. [DOI] [PubMed] [Google Scholar]
- 15.Farrell HE, et al. Inhibition of natural killer cells by a cytomegalovirus MHC class I homologue in vivo. Nature. 1997;386:510–4. doi: 10.1038/386510a0. [DOI] [PubMed] [Google Scholar]
- 16.Kubota A, Kubota S, Farrell HE, Davis-Poynter N, Takei F. Inhibition of NK cells by murine CMV-encoded class I MHC homologue m144. Cell Immunol. 1999;191:145–51. doi: 10.1006/cimm.1998.1424. [DOI] [PubMed] [Google Scholar]
- 17.Hasan M, et al. Selective down-regulation of the NKG2D ligand H60 by mouse cytomegalovirus m155 glycoprotein. J Virol. 2005;79:2920–30. doi: 10.1128/JVI.79.5.2920-2930.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krmpotic A, et al. NK cell activation through the NKG2D ligand MULT-1 is selectively prevented by the glycoprotein encoded by mouse cytomegalovirus gene m145. J Exp Med. 2005;201:211–20. doi: 10.1084/jem.20041617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lodoen MB, Abenes G, Umamoto S, Houchins JP, Liu F, Lanier LL. The cytomegalovirus m155 gene product subverts natural killer cell antiviral protection by disruption of H60-NKG2D interactions. J Exp Med. 2004;200:1075–81. doi: 10.1084/jem.20040583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lodoen M, et al. NKG2D-mediated natural killer cell protection against cytomegalovirus is impaired by viral gp40 modulation of retinoic acid early inducible 1 gene molecules. J Exp Med. 2003;197:1245–53. doi: 10.1084/jem.20021973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–6. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 22.Cerwenka A, Lanier LL. NKG2D ligands: unconventional MHC class I-like molecules exploited by viruses and cancer. Tissue Antigens. 2003;61:335–43. doi: 10.1034/j.1399-0039.2003.00070.x. [DOI] [PubMed] [Google Scholar]
- 23.Li P, McDermott G, Strong RK. Crystal structures of RAE-1beta and its complex with the activating immunoreceptor NKG2D. Immunity. 2002;16:77–86. doi: 10.1016/s1074-7613(02)00258-3. [DOI] [PubMed] [Google Scholar]
- 24.Lenac T, et al. The herpesviral Fc receptor fcr-1 down-regulates the NKG2D ligands MULT-1 and H60. J Exp Med. 2006;203:1843–50. doi: 10.1084/jem.20060514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voigt V, et al. Murine cytomegalovirus m157 mutation and variation leads to immune evasion of natural killer cells. Proc Natl Acad Sci U S A. 2003;100:13483–8. doi: 10.1073/pnas.2233572100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kleijnen MF, et al. A mouse cytomegalovirus glycoprotein, gp34, forms a complex with folded class I MHC molecules in the ER which is not retained but is transported to the cell surface. EMBO J. 1997;16:685–94. doi: 10.1093/emboj/16.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krmpotic A, Messerle M, Crnkovic-Mertens I, Polic B, Jonjic S, Koszinowski UH. The immunoevasive function encoded by the mouse cytomegalovirus gene m152 protects the virus against T cell control in vivo. J Exp Med. 1999;190:1285–96. doi: 10.1084/jem.190.9.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Del Val M, Munch K, Reddehase MJ, Koszinowski UH. Presentation of CMV immediate-early antigen to cytolytic T lymphocytes is selectively prevented by viral genes expressed in the early phase. Cell. 1989;58:305–15. doi: 10.1016/0092-8674(89)90845-3. [DOI] [PubMed] [Google Scholar]
- 29.Reddehase MJ, Rothbard JB, Koszinowski UH. A pentapeptide as minimal antigenic determinant for MHC class I-restricted T lymphocytes. Nature. 1989;337:651–3. doi: 10.1038/337651a0. [DOI] [PubMed] [Google Scholar]
- 30.del Val M, et al. Cytomegalovirus prevents antigen presentation by blocking the transport of peptide-loaded major histocompatibility complex class I molecules into the medial-Golgi compartment. J Exp Med. 1992;176:729–38. doi: 10.1084/jem.176.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thale R, Szepan U, Hengel H, Geginat G, Lucin P, Koszinowski UH. Identification of the mouse cytomegalovirus genomic region affecting major histocompatibility complex class I molecule transport. J Virol. 1995;69:6098–105. doi: 10.1128/jvi.69.10.6098-6105.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziegler H, et al. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity. 1997;6:57–66. doi: 10.1016/s1074-7613(00)80242-3. [DOI] [PubMed] [Google Scholar]
- 33.Ziegler H, Muranyi W, Burgert HG, Kremmer E, Koszinowski UH. The luminal part of the murine cytomegalovirus glycoprotein gp40 catalyzes the retention of MHC class I molecules. EMBO J. 2000;19:870–81. doi: 10.1093/emboj/19.5.870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pinto AK, Munks MW, Koszinowski UH, Hill AB. Coordinated function of murine cytomegalovirus genes completely inhibits CTL lysis. J Immunol. 2006;177:3225–34. doi: 10.4049/jimmunol.177.5.3225. [DOI] [PubMed] [Google Scholar]
- 35.Kavanagh DG, Gold MC, Wagner M, Koszinowski UH, Hill AB. The multiple immune-evasion genes of murine cytomegalovirus are not redundant: m4 and m152 inhibit antigen presentation in a complementary and cooperative fashion. J Exp Med. 2001;194:967–78. doi: 10.1084/jem.194.7.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lilley BN, Ploegh HL. Viral modulation of antigen presentation: manipulation of cellular targets in the ER and beyond. Immunol Rev. 2005;207:126–44. doi: 10.1111/j.0105-2896.2005.00318.x. [DOI] [PubMed] [Google Scholar]
- 37.Holtappels R, et al. Cytomegalovirus encodes a positive regulator of antigen presentation. J Virol. 2006;80:7613–24. doi: 10.1128/JVI.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gold MC, et al. Murine cytomegalovirus interference with antigen presentation has little effect on the size or the effector memory phenotype of the CD8 T cell response. J Immunol. 2004;172:6944–53. doi: 10.4049/jimmunol.172.11.6944. [DOI] [PubMed] [Google Scholar]
- 39.Lu X, Pinto AK, Kelly AM, Cho KS, Hill AB. Murine cytomegalovirus interference with antigen presentation contributes to the inability of CD8 T cells to control virus in the salivary gland. J Virol. 2006;80:4200–2. doi: 10.1128/JVI.80.8.4200-4202.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fleming P, et al. The murine cytomegalovirus chemokine homolog, m131/129, is a determinant of viral pathogenicity. J Virol. 1999;73:6800–9. doi: 10.1128/jvi.73.8.6800-6809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis-Poynter NJ, et al. Identification and characterization of a G protein-coupled receptor homolog encoded by murine cytomegalovirus. J Virol. 1997;71:1521–9. doi: 10.1128/jvi.71.2.1521-1529.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loewendorf A, Kruger C, Borst EM, Wagner M, Just U, Messerle M. Identification of a mouse cytomegalovirus gene selectively targeting CD86 expression on antigen-presenting cells. J Virol. 2004;78:13062–71. doi: 10.1128/JVI.78.23.13062-13071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nomura M, et al. Mechanism of host cell protection from complement in murine cytomegalovirus (CMV) infection: identification of a CMV-responsive element in the CD46 promoter region. Eur J Immunol. 2002;32:2954–64. doi: 10.1002/1521-4141(2002010)32:10<2954::AID-IMMU2954>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 44.Menard C, et al. Role of murine cytomegalovirus US22 gene family members in replication in macrophages. J Virol. 2003;77:5557–70. doi: 10.1128/JVI.77.10.5557-5570.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Natarajan K, et al. Crystal structure of the murine cytomegalovirus MHC-I homolog m144. J Mol Biol. 2006;358:157–71. doi: 10.1016/j.jmb.2006.01.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mans J, et al. Cellular expression and crystal structure of the murine cytomegalovirus major histocompatibility complex class I-like glycoprotein, m153. J Biol Chem. 2007;282:35247–58. doi: 10.1074/jbc.M706782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Adams EJ, et al. Structural elucidation of the m157 mouse cytomegalovirus ligand for Ly49 natural killer cell receptors. Proc Natl Acad Sci U S A. 2007;104:10128–33. doi: 10.1073/pnas.0703735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang Z, Bjorkman PJ. Structure of UL18, a peptide-binding viral MHC mimic, bound to a host inhibitory receptor. Proc Natl Acad Sci U S A. 2008;105:10095–100. doi: 10.1073/pnas.0804551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allen H, Fraser J, Flyer D, Calvin S, Flavell R. Beta 2-microglobulin is not required for cell surface expression of the murine class I histocompatibility antigen H-2Db or of a truncated H-2Db. Proc Natl Acad Sci U S A. 1986;83:7447–51. doi: 10.1073/pnas.83.19.7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dunker AK, Uversky VN. Signal transduction via unstructured protein conduits. Nat Chem Biol. 2008;4:229–30. doi: 10.1038/nchembio0408-229. [DOI] [PubMed] [Google Scholar]
- 51.Khan AN, Lewis PN. Unstructured conformations are a substrate requirement for the Sir2 family of NAD-dependent protein deacetylases. J Biol Chem. 2005;280:36073–8. doi: 10.1074/jbc.M508247200. [DOI] [PubMed] [Google Scholar]
- 52.Verkhivker GM. Protein conformational transitions coupled to binding in molecular recognition of unstructured proteins: deciphering the effect of intermolecular interactions on computational structure prediction of the p27Kip1 protein bound to the cyclin A-cyclin-dependent kinase 2 complex. Proteins. 2005;58:706–16. doi: 10.1002/prot.20351. [DOI] [PubMed] [Google Scholar]
- 53.Chapman TL, Bjorkman PJ. Characterization of a murine cytomegalovirus class I major histocompatibility complex (MHC) homolog: comparison to MHC molecules and to the human cytomegalovirus MHC homolog. J Virol. 1998;72:460–6. doi: 10.1128/jvi.72.1.460-466.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.French AR, et al. Escape of mutant double-stranded DNA virus from innate immune control. Immunity. 2004;20:747–56. doi: 10.1016/j.immuni.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 55.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific LLC.; San Carlos, CA, USA: [Google Scholar]