Abstract
Reliable prediction of protein-ligand docking pose requires proper account of induced fit effects. Treating both the ligand and the protein as flexible molecules is still challenging because many degrees of freedom are involved. Peptides are one type of ligands that are particularly difficult to study because of their extreme flexibility. In this study, we tested a molecular dynamics-based simulated-annealing cycling protocol in docking peptides to four protein kinases and two phosphatases using two implicit-solvent models: a distance-dependent dielectric model (ε(r)=4r) and a version of the Generalized Born model termed GBMV. We found that the simpler ε(r)=4r model identified docking pose better than the more expensive GBMV model. In addition, rescoring structures obtained from one implicit-solvent model with the other identified good docking poses for all six systems studied. Including protein energy in scoring also improved results.
Keywords: simulated annealing, distance-dependent dielectric model, generalized Born model, protein kinase A, insulin receptor protein kinase, insulin-like growth factor 1 receptor kinase, cyclic dependent protein kinase 2, protein tyrosine phosphatase 1B, YopH in Yersinia pestis
1. Introduction
Reliable prediction of protein-ligand docking structure remains a challenging problem. Peptide ligands present one of the more difficult cases because of their high flexibility, requiring extensive configurational sampling. Even for the rigid-protein model, it is demanding to handle flexible ligands with over 15–20 rotatable bonds, which is typical even for short peptides. This difficulty increases when the protein receptor is also flexible and a peptide ligand only binds to a small subset of the many possible conformations of the protein.
Nevertheless, an increasing number of research groups have employed various strategies to include protein flexibility in docking. For example, one can dock a ligand to an ensemble of receptor conformations obtained from molecular dynamics simulation, from normal mode calculation, from crystal structures with various ligands bound, from NMR structures, and from structures constructed with different combination of side-chain conformations in the binding site1–14. One can also “enlarge” the docking pocket in a protein by softening the van der Waals (vdw) potential (ref15 gives an example) or by offsetting vdw radii (e.g., reference16) to approximate the effects of protein flexibility.
More realistic approaches directly simulated the coupled structural change of the protein and the ligand. These approaches allowed protein movement in different ways. For example, the program FLIPDOCK17 used a Flexibility Tree (FT) data structure to choose a small number of variables to describe the key structural changes required by the protein to accommodate the ligand. On the other hand, the program FITTED18 employed a genetic algorithm to generate diverse protein structures.
Molecular dynamics-based methods provide another useful approach. Unlike many docking potentials solely designed for docking rigid or flexible ligands to rigid proteins, molecular dynamics-based methods take advantage of force fields already designed for simulating protein motion. One approach by Mangoni et al19 facilitated the docking of a flexible ligand to a flexible protein by assigning a high temperature to the translational motion of the center-of-mass of the ligand and only allowed the binding pocket of the protein to move. Another study by Nakajima et al20 employed the multi-canonical molecular dynamics simulation method to dock a flexible peptide to the SH3 domain of Abl tyrosine kinase. In their work, they only allowed the side chains in the ligand-binding pocket to move. On the other hand, our earlier work permitted full flexibility of every side chain but restrained the α carbons to suitable experimental reference structures to reduce computational time and to avoid sampling unphysical structures due to the use of non-optimal force fields21–23. This model have successfully docked small organic ligands to protein kinases and phosphatases21–23. This study further explores its application to the more challenging problem of peptide-protein docking.
One way to facilitate extensive configurational search is to use implicit- rather than explicit-solvent models. Researchers have now developed numerous implicit-solvent models, including simple distance-dependent dielectric models, generalized Born models, and Poisson-Boltzmann models. (Gilson and Zhou recently provided a good review of these models24.) Distance-dependent dielectric and Generalized Born models are cheaper to use and were first models to consider in this study. The ε(r) = r model, where r is the distance between two atoms in a molecular system, was the first distance-dependent dielectric model employed in molecular dynamics simulation of proteins. Later on, dielectric function that increased more rapidly with distance has become popular; ε(r) = 4r model is one of them. This and other distance-dependent dielectric models are now widely used in programs such as CHARMM25, DOCK26; FITTED18 and others21–23,27–31.
A more complex model is the Generalized Born model first proposed by Still and co-workers32. One form of their model calculates the electrostatic contribution to solvation energy by:
| (1) |
where εp represents the low dielectric constant inside a solute such as a protein, εw is the dielectric constant of water, and qi and qj are the charges of atoms i and j, respectively. There are various ways of calculating the effective Born radii αi and αj33–42, among them is the Generalized Born using Molecular Volume (GBMV)39,40 model which we used in this study. Because Generalized Born models are more expensive than simple distance-dependent dielectric functions to use, they are still not commonly employed in molecular docking, especially in larger scale virtual screening. On the other hand, it has been used to refine and rescore docking pose. For example, Lee and Sun recently reported43 that further optimizing and rescoring docking poses obtained from the DOCK program could improve the identification of correct docking pose in a test set of 79 protein-ligand complexes.
In this work, we focused on evaluating the ε(r) = 4r and the GBMV models in docking peptides to protein kinases and phosphatases. We used both models to perform docking directly and to rescore docking poses obtained from the other model. The main goal was to find out whether these models or a mixture of them could identify good docking pose for protein-peptide systems.
Section 2 describes the simulated annealing cycling protocol that we applied successfully to dock small molecules to protein kinases and phosphatases earlier. It also describes how we set up the systems and presents several energy models we used to identify docking pose. Section 3 details the results and discusses the performance of different models. Section 4 concludes with our major findings.
2. Methods
2.1. Simulated annealing cycling protocol
We used a novel simulated annealing cycling protocol described previously for protein-ligand docking.21,23 This protocol was found to consume less computational time than regular replica-exchange and simulated annealing methods if the goal was solely to find good docking pose.22 Each simulated annealing (SA) cycling simulation was performed as follows:
Assigned initial velocities at 500 K.
Allowed the system to evolve for 1 ps in the NVT ensemble. We used the Nosé-Hoover chain method44 to maintain constant temperature.
At the end of each 1-ps simulation, we reduced the temperature by half.
Looped back to step 3 if the temperature was above 5 K, to step 1 otherwise.
We continued the above loop until a prescribed simulation length was reached. Typically, each simulated annealing cycle approximately covered the following temperatures: 500 K, 250 K, 125 K, 62.5 K, 31.2 K, 15.6 K, 7.8 K, and 3.9 K. The end of each simulated-annealing cycle identified a structure near a local or the global energy minimum. By performing many such short simulated annealing cycles in a molecular dynamics simulation, one could identify many energy minima. If the energy and solvation model was sufficiently realistic, one could identify the correct docking pose from the lowest-energy structures, which we showed previously to be the case in docking small organic molecules to protein kinases22,23 and phosphatases21. In the simulation performed here, we ran twenty independent trajectories starting from the same structure and temperature but initiated atomic velocities with different random number generating seeds. By using two starting structures, we ran a total of forty trajectories for each protein-peptide system. We modified the MMTSB Toolkit45 to use with the CHARMM package25 to perform the simulated annealing docking just described.
2.2. System set up
We chose six protein-peptide test systems in this study: 1) protein tyrosine phosphatase 1B (PTB1B) and a phosphorylated tetrapeptide Ac-DEpYL--NH246; 2) protein tyrosine phosphatase YopH of Yersinia pestis and a mimic of a phosphorylated hexapeptide Ac-DADE-F2Pmp-L-NH247; 3) the activated form of the catalytic domain of the insulin receptor protein kinase (PTK) and a hexapeptide with the sequence GDYMNM48; 4) insulin-like growth factor 1 receptor kinase (IGF) and an octapeptide with the sequence GEYVNIEF49; 5) cyclic AMP-dependent kinase or protein kinase A (PKA) and a fragment of its peptide inhibitor PKI containing residues 14 through 2450; 6) cyclin dependent protein kinase 2 (CDK2), with cyclin A bound, and a recruitment 9-mer peptide derived from E2F151. Entries 1PTT46, 1QZ052, 1IR348, IK3A49, 1FMO50 and 1H2451 in the Protein Data Bank provided the co-crystal structures for these protein-peptide systems.
For the YopH-peptide system, we included six residues of the peptide in our simulation although only four residues could be observed in the crystal structure (in PDB entry 1QZ0). In addition, we included three crystal water molecules in the binding pocket that facilitate the interactions between the phosphotyrosine residue of the peptide and the protein. As this study focused on evaluating whether current force fields and the two solvation models were good enough for identifying good docking pose, we constrained these water molecules near their experimental structures during docking to reduce sampling time. In principle, one could allow these waters to move throughout the docking process but this would require longer simulation time to find good docking pose, especially when the starting structure was far away from the best docking pose. For the PTK-peptide system, the hexapeptide GDYMNM in the crystal structure was used. Its N-terminus was acetylated and its C-terminus was in the N-methyl amide form. We replaced the ATP analog in the crystal structure by ATP in the simulation. For the IGF-peptide system, we used the octapeptide GEYVNIEF in the crystal structure and blocked it similarly as the hexapeptide above. We also replaced the ATP analog by ATP. In addition, the coordinates of eight missing residues (A1069-A1076) in PDB entry 1K3A were obtained from the corresponding residues in PDB entry 1IR3 after superposition of their protein structures. Because three of these eight residues were different in these two protein structures, we only used the coordinates of the backbone atoms for these three residues from PDB entry 1IR3 and we used CHARMM25 to build the side chains. For the PKA-peptide system, we only used eleven residues, 14 through 24 with the sequence GRTGRRNAIHD, of PKI in the docking study to reduce computational time, even though the crystal structure contains a longer segment from residues 5 to 24. In the simulation, we also kept the adenosine in the crystal structure as it might facilitate protein-ligand recognition. For the CDK2-peptide system, we used the nine-residue peptide PVKRRLDLE and we only included the B chain of CDK2 from the crystal structure in the simulation.
During the simulation, we applied RMSD-restraint to keep the proteins, along with the additional water molecules, adenosine, or ATP, close to their crystal structures, as done previously21,22. Only the α carbons of the proteins, the water oxygens in the YopH-peptide system, the adenosine in the PKA-peptide system, and ATP in the PTK-peptide and IGF-peptide systems were restrained. These restraints prevented sampling of unrealistic structures far from experiment - due to the limitation of current force fields and solvation models - and reduced computational costs. Yet, the weak restraints permitted ample conformational flexibility of the proteins to accommodate a more diverse set of small organic molecules and peptides that would have been excluded by rigid-protein docking models.
Prior to each molecular dynamics simulation, we performed 500 steps of steepest descent energy minimization of a protein-peptide complex to remove bad contacts. During the energy minimization, the α carbons, the water oxygens, and the heavy atoms of adenosine and ATP were held fixed. On the other hand, for the IGF-peptide system, we also allowed the α carbons of the eight residues that we modeled in to relax.
We started each peptide-docking simulation either from the X-ray structure or with the peptide in the extended conformation on the protein surface near its binding site. We generated the extended structure of each peptide by the CHARMM program25. Twenty trajectories of simulated annealing cycling simulations were performed for each of the two starting structures so that forty trajectories resulted. Because each trajectory lasted 2 ns, the aggregate simulation time for each protein-peptide system covered 80 ns. We used a time step of 2 fs in these simulations. For the simulations using the GBMV model, we ran each trajectory for only 1 ns rather than 2 ns because it consumed significantly more computational time.
The simulated annealing cycling docking simulations were conducted with the CHARMM param27 force field53,54. In the simulation using the ε(r) = 4r model, we used a non-bonded cutoff distance of 14 Å, a switching function for the electrostatic interactions that began at 10 Å and ended at 12 Å, and a shifting function for the Lennard-Jones potential. We also used this ε(r) = 4r model in the energy minimization preceding each molecular dynamics simulation. In rescoring docking pose with the GBMV39,40 model, we used the GBMV1 parameters of Chocholoušová and Feig41 and we changed the above cutoff distances to 12 Å, 8 Å and 10 Å respectively.
In the simulation using the GBMV39–41 model, we also used a non-bonded cutoff distance of 12 Å and a switching function applied between 8 Å and 10 Å. However, subsequent rescoring with the ε(r) = 4r model changed these cutoff distances back to 14 Å, 10 Å and 12 Å respectively.
Different simulation models required different CPU times on a dual core-dual processor cluster node with 2.8GHz Intel Xeon EM64T processors. Table I compares the CPU time of the two simulation models for the six protein-peptide systems studied. The flexible-protein/GBMV model was about an order of magnitude more expensive than the flexible-protein/ε(r) = 4r model, even though the length of the former simulation was only half of the latter. It took more than six months using twenty CPUs to perform the GBMV simulations for the six protein-peptide systems studied here.
Table I.
CPU hours for six protein-peptide docking using two implicit-solvent models.
| Model | ε(r)=4r Forty 2-ns trajectories | GBMV Forty 1-ns trajectories |
|---|---|---|
| System | ||
| PTP-phosphorylated peptide | 1255.02 | 14738.52 |
| YopH-phosphorylated peptide mimic | 1142.88 | 13311.20 |
| PTK-peptide | 1203.75 | 15061.04 |
| IGF-peptide | 1208.44 | 14980.75 |
| PKA-peptide | 1497.91 | 17653.82 |
| CDK-peptide | 1097.17 | 13163.07 |
2.3 Energy models
We tested several energy models for selecting good docking pose:
Energy Model I: total energy of the whole system.
Energy Model II: total energy minus the protein (including additional waters, adenine, or ATP, if present) energy.
Energy Model III: Energy of whole system minus energy of protein (together with additional waters, adenine, or ATP if present) and peptide.
3. Results and Discussions
Table II shows the closest structures to experiment sampled by the simulated annealing cycling simulations using the ε(r) = 4r model. Obviously, simulations staring from the X-ray structure sampled more structures near the crystal structures (within about 1.5 Å) than simulations starting from the extended conformation of the peptide on the protein surface. Nevertheless, four out of the six systems (the YopH-peptide, PTK-peptide, IGF-peptide, and PKA-peptide systems) found structures closer to ~2.14 Å (RMSD of all heavy atoms: RMSDheavy) of experiment when the simulations were started from the protein surface. The other two systems - PTP-peptide and CDK-peptide –found structures up to only about 4 Å from experiment. Therefore, it will take longer simulation time to find good docking poses if one starts simulations with the extended conformation of the peptides on the protein surface.
Table II.
Best docking poses from six protein-peptide docking simulations with the ε(r)=4r model.
| Property | Smallest peptide RMSDheavy (Å) | Smallest peptide RMSDbackbone (Å) | Smallest peptide RMSDbackbone+pTyr#(Å) | |
|---|---|---|---|---|
| Simulation | ||||
| PTP-Peptide | X-ray* | 1.16 | 0.77 | 0.75 |
| Extended^ | 3.75 | 1.05 | 3.13 | |
| YopH-Peptide | X-ray | 0.60 | 0.31 | 0.32 |
| Extended | 1.30 | 0.56 | 1.06 | |
| PTK-Peptide | X-ray | 1.00 | 0.77 | |
| Extended | 1.53 | 0.98 | ||
| IGF-Peptide | X-ray | 0.95 | 0.61 | |
| Extended | 1.47 | 0.80 | ||
| PKA-Peptide | X-ray | 0.98 | 0.78 | |
| Extended | 2.14 | 1.43 | ||
| CDK-Peptide | X-ray | 1.05 | 0.68 | |
| Extended | 4.09 | 2.96 | ||
Simulations starting from the X-ray structure
Simulations starting with the peptide ligand on the protein surface and with an extended conformation
RMSDbackbone+pTyr: RMSD calculated by the backbone atoms of the peptide and the heavy atoms of the phosphotyrosine residue.
Bold: RMSD > 3 Å.
Table III shows the best peptide-protein docking poses obtained with the GBMV model. In this case, only two out of the six systems, PTK-peptide and IGF-peptide, found docking poses within about 1.6 Å (RMSDheavy) of experiment if the simulations were started from the extended form of the peptides on the protein surface. The other four systems failed to find docking poses within 3 Å (RMSDheavy) of experiment within the duration of the 40-ns simulation. Because this study focused on examining whether the CHARMM force field along with two implicit-solvent models could properly identify the experimental co-crystal structures, we started half of the trajectories from the crystal structures, instead of from the extended conformation on the protein surface - to ensure that we sampled the region near the experimental structures well without running much longer simulation.
Table III.
Best docking poses from six protein-peptide docking simulations using the GBMV model.
| Property | Smallest peptide RMSDheavy (Å) | Smallest peptide RMSDbackbone (Å) | Smallest peptide RMSDbackbone+pTyr#(Å) | |
|---|---|---|---|---|
| Simulation | ||||
| PTP-Peptide | X-ray* | 1.35 | 0.74 | 0.60 |
| Extended^ | 6.31 | 2.32 | 6.93 | |
| YopH-Peptide | X-ray | 0.73 | 0.57 | 0.48 |
| Extended | 6.49 | 2.74 | 6.43 | |
| PTK-Peptide | X-ray | 0.88 | 0.49 | |
| Extended | 1.58 | 0.92 | ||
| IGF-Peptide | X-ray | 0.91 | 0.44 | |
| Extended | 1.65 | 0.76 | ||
| PKA-Peptide | X-ray | 1.15 | 0.73 | |
| Extended | 3.78 | 2.15 | ||
| CDK-Peptide | X-ray | 1.18 | 0.64 | |
| Extended | 4.37 | 2.59 | ||
Simulations starting from the X-ray structure
Simulations starting with the peptide ligand on the protein surface and with an extended conformation
RMSDbackbone+pTyr: RMSD calculated by the backbone atoms of the peptide and the heavy atoms of the phosphotyrosine residue.
Bold: RMSD > 3 Å.
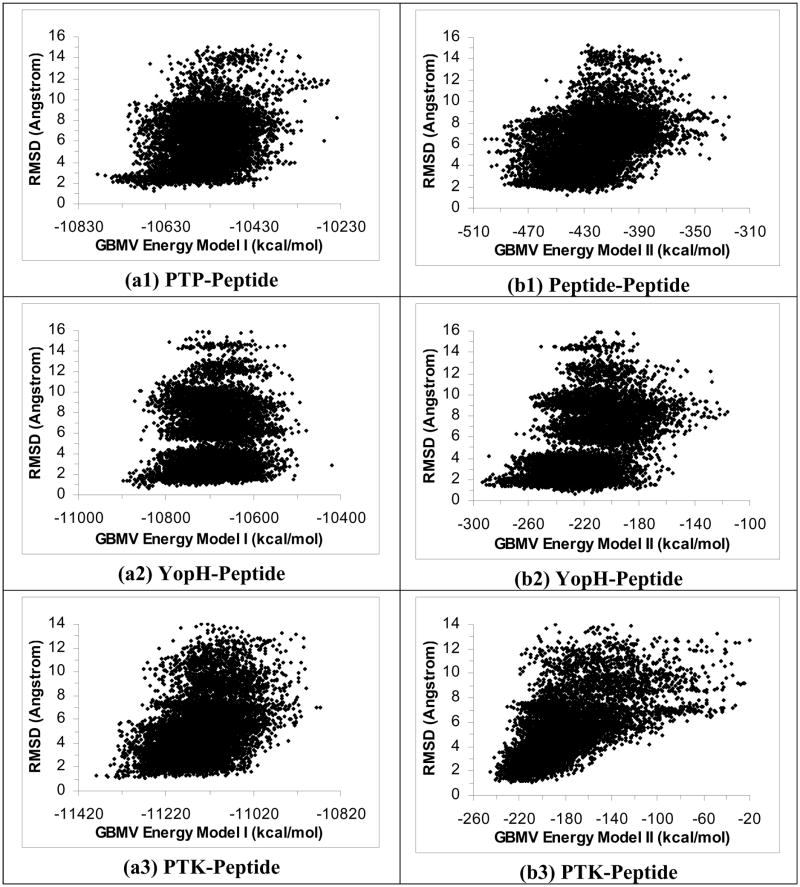
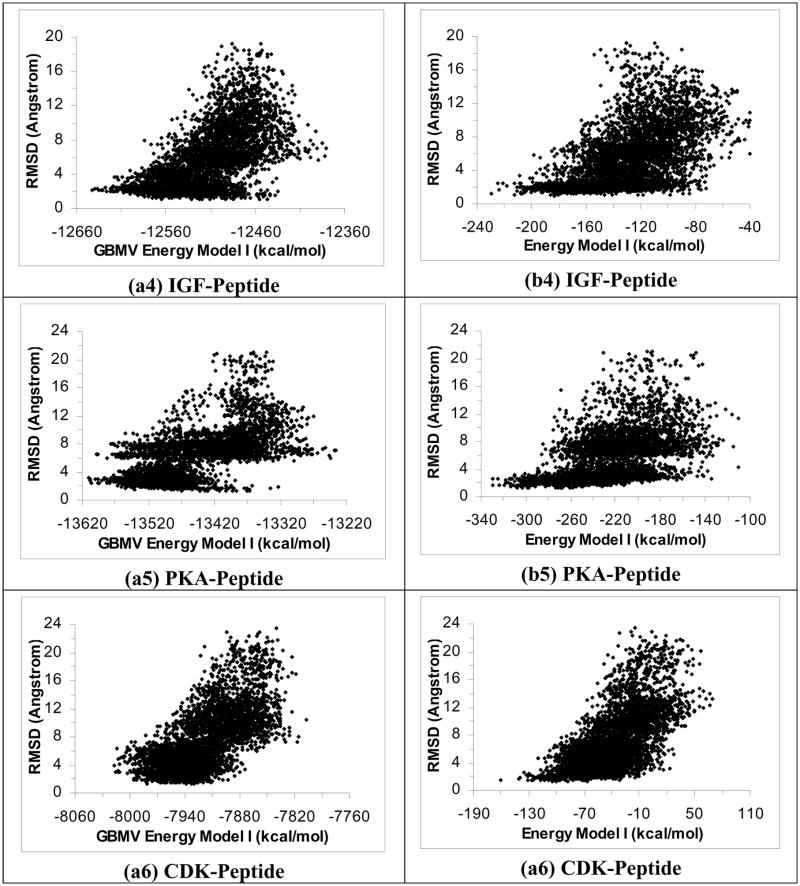
Figures 1a1 to 1a6 plot RMSDheavy versus energy for the six protein-peptide systems using the ε(r) = 4r model and Energy Model I (total energy). We only used structures near local energy minima obtained below 5 K in the plots. These plots show that this energy model identified good docking poses within ~3 Å of experiment for five of the six systems. The structures with the lowest energy were within 3.13 Å, 1.57 Å, 3.38 Å, 2.78 Å and 2.42 Å (RMSDheavy) of experiment for the PTP-, YopH-, PTK-, IGF-, and PKA-peptide systems respectively. On the other hand, this energy model failed to identify good docking pose for the CDK-peptide system; the lowest-energy structure was 6.30 Å (RMSDheavy) away from experiment.
Figure 1.
Plots of RMSD from experiment for the peptide heavy atoms versus energy for 40 2-ns simulated annealing cycling simulations using the ε(r)=4r model. Half of the trajectories were started from the crystal structure and half from the extended conformation on the protein surface. These plots include only structures near local energy minima obtained below 5 K. a1 to a6 are the results for the PTP-peptide, YopH-peptide, PTK-peptide, IGF-peptide, PKA-peptide, and CDK-peptide systems respectively using Energy Model I. b1 to b6 are similar except using Energy Model II to score the structures. (Energy Model I used the total energy of the whole system; Energy Model II excluded the energy of the protein.)
Figures 1b1 to 1b6 show similar results when we re-scored the structures with Energy Model II, which included only the peptide energy and the protein-peptide interaction energy but not the protein energy. Although we previously found this model to work well for selecting good docking poses for small organic ligands21–23, it did not work well for peptide ligands. The lowest-energy poses for the PTP-, PTK-, IGF-, PTK-, and CDK-peptide systems deviated significantly from their corresponding crystal structures - with RMSDheavy equaled to 7.35 Å, 8.29 Å, 7.35 Å, 8.93 Å and 8.41 Å, respectively. The YopH-peptide worked a bit better but the lowest-energy pose still deviated from the crystal structure by 4.04 Å. Further excluding the ligand energy (Energy Model III) did not work either (the plots resembled Figures 1b1 to 1b6 and are not shown). Therefore, it seems important to include protein energy in scoring protein-peptide structures.
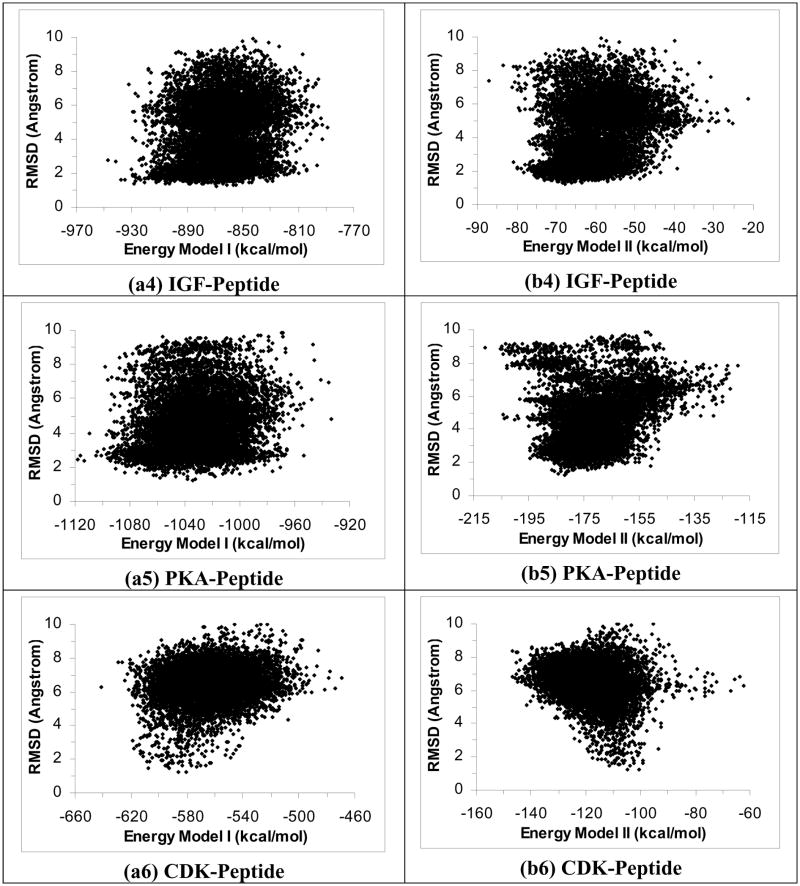
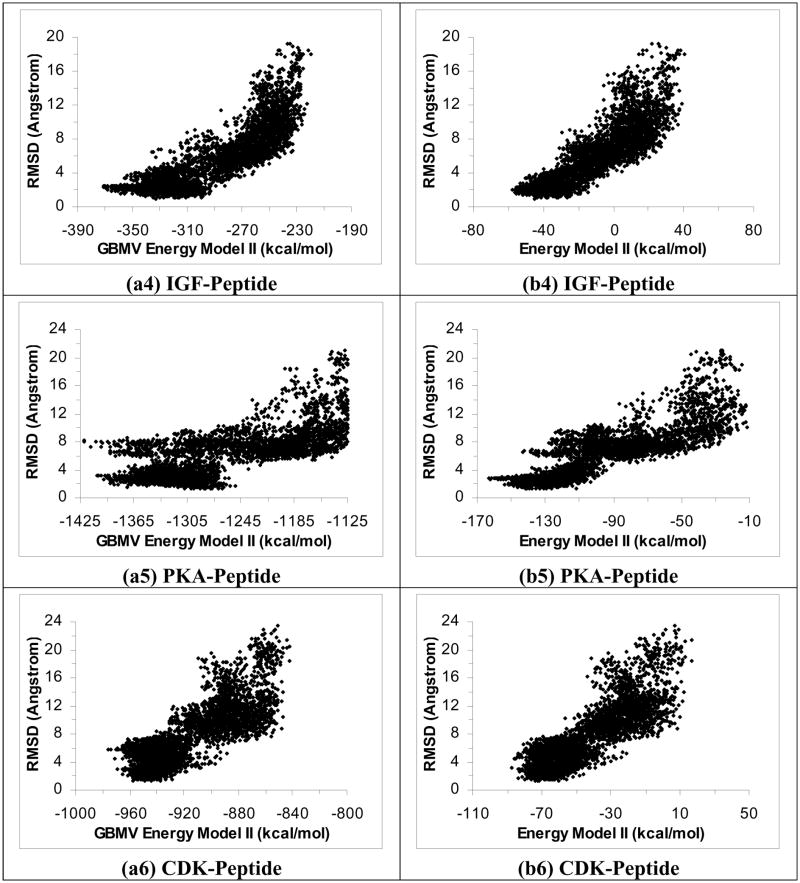
To test whether rescoring results from the ε(r) = 4r model with the GBMV model could improve results, we plotted RMSDheavy versus energy for the six protein-peptide systems using GBMV Energy Model I (Figures 2a1 to 2a6). These plots show that rescoring with the GBMV model significantly improved the identification of good docking poses. The lowest-energy docking poses were within 2.76 Å, 1.34 Å, 1.27 Å, 1.66 Å, 1.48 Å and 1.28 Å of experiment for the PTP-, YopH-, PTK-, IGF-, PKA-, and CDK-peptide systems respectively. On the other hand, rescoring with GBMV Energy Model II also notably improved the identification of experimental docking poses than the original ε(r) = 4r Energy Model II but it did not work as well as rescoring with GBMV Energy Model I (Figures 2b1 to 2b6); we obtained good docking poses for only four out of the six systems: YopH-, PTK-, IGF-, and PKA-peptide systems with RMSDheavy equaled 1.70 Å, 1.92 Å, 1.63 Å, and 2.45 Å respectively for their lowest energy structures. In contrast, the lowest-energy poses for the PTP-peptide and the CDK-peptide deviated significantly from their corresponding crystal structures - with RMSDheavy reaching 6.42 Å and 6.14 Å respectively.
Figure 2.
Rescoring results in Figure 1 with the GBMV model. a1 to a6 used GBMV Energy Model I and b1 to b6 used GBMV Energy Model II.
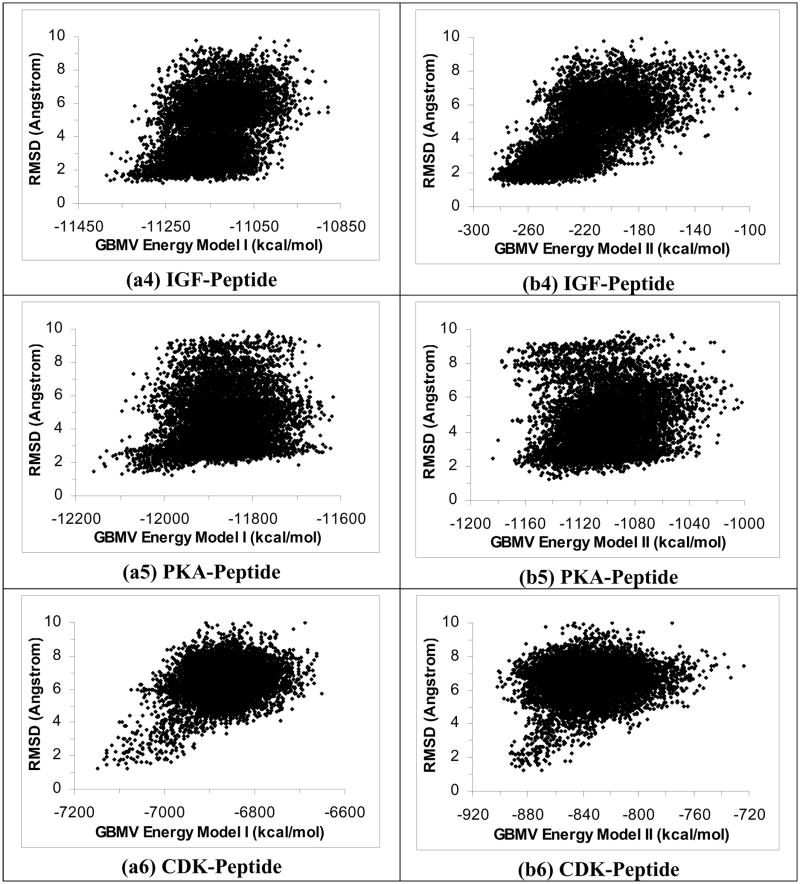
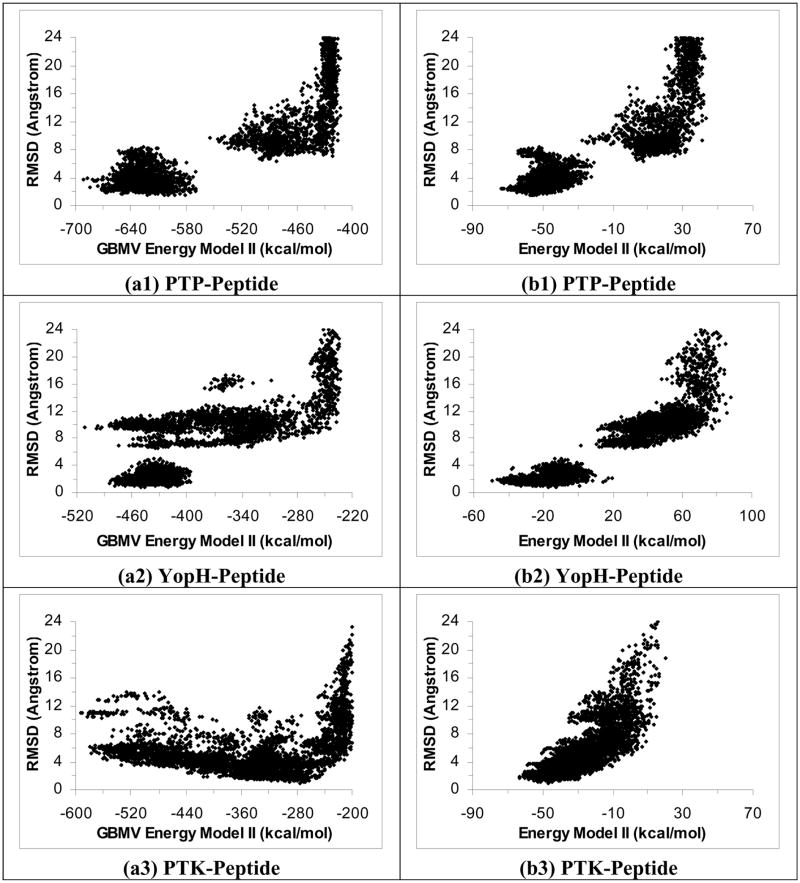
We performed similar studies by running the simulated annealing cycling simulation with the GBMV model instead of the ε(r)=4r model. Figures 3a1 to 3a6 plot RMSDheavy versus energy for the six systems using Energy Model I. These plots show that the GBMV model could identify good docking poses for the PTP-peptide and IGF-peptide systems to within 2.72 Å (RMSDheavy) and 2.10 Å of the crystal structures respectively. It worked a bit less well for the PKA-peptide and CDK-peptide systems, with the lowest-energy structures deviated from experiment by 3.09 Å and 3.96 Å (RMSDheavy) respectively. And it failed to identity docking poses closer than 8 Å for the other two systems: YopH-peptide and PTK-peptide. Interestingly, rescoring the structures from the GBMV simulation with the ε(r)=4r model managed to identify good docking poses for all the six systems (Figures 3b1 to 3b6). The lowest-energy rescoring poses were within 1.98 Å, 1.21 Å, 2.42 Å, 1.19 Å, 1.62 Å and 1.47 Å of experiment (RMSDheavy) for the PTP-, YopH-, PTK-, IGF-, PKA- and CDK-peptide systems respectively.
Figure 3.
Plots of RMSD from experiment for the peptide heavy atoms versus energy for 40 1-ns simulated annealing simulations using the GBMV model. a1 to a6 are the results for the PTP-peptide, YopH-peptide, PTK-peptide, IGF-peptide, PKA-peptide, and CDK-peptide systems respectively, using GBMV Energy Model I. b1 to b6 are for rescoring the results with the ε(r)=4r model using Energy Model I.
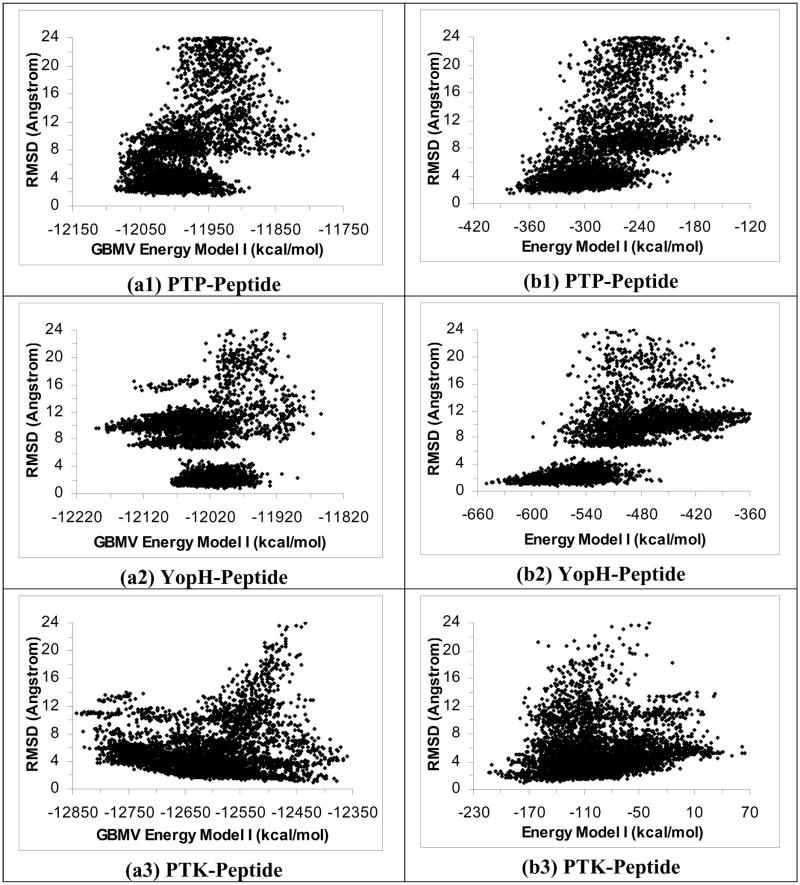
We further examined excluding the protein energy in the GBMV model (Energy Model II) for scoring but obtained even poorer results. Figures 4a1 to 4a6 show that this energy model failed to identify good docking pose except for the IGF-peptide system, whose lowest-energy docking structure was within 2.37 Å (RMSDheavy) of experiment. On the other hand, GBMV Energy Model II could identify docking poses to 3.79 Å (RMSDheavy) at best for the PTP-peptide system, 5.76 Å for the CDK-peptide system, and more than 8 Å for the remaining 3 systems. However, rescoring these structures using the other solvation model, ε(r)=4r (Energy Model II), again could identify good docking poses for all six systems (Figures 4b1 to 4b6). The lowest-energy rescoring poses were within 2.36 Å, 1.73 Å, 1.76 Å, 2.04 Å, 2.66 and 3.17 Å of experiment (RMSDheavy) for the PTP-, YopH-, PTK-, IGF-, PKA- and CDK-peptide systems respectively.
Figure 4.
Repeat Figure 3 by using Energy Model II, i.e., excluding protein energy in scoring.
The above results show that the ε(r)=4r model performed better than the GBMV model for peptide-protein docking when each model was used alone. In addition, Energy Model I (total energy) performed the best. Interestingly, no matter which solvation model was used in running the simulated annealing cycling simulation, rescoring structures with the other solvation model always yielded better results, with good docking poses identified for all the six systems studied here. Table IV summarizes the results for the six protein-peptide systems using the ε(r)=4r model for docking and several energy models for scoring. It is clear that scoring the structures obtained from ε(r)=4r simulated annealing cycling simulation with the GBMV model always gave good docking poses for Energy Model I. Table V shows that scoring structures obtained from the GBMV simulations with the ε(r)=4r model also identified good docking poses for all six systems if Energy Model I and Energy Model II were used. These results suggest the following practical strategy to identify good docking pose in protein-peptide docking: First use the cheaper ε(r)=4r model to dock a peptide to a protein kinase or phosphatase and then score the resulting structures with the GBMV model to identify docking pose using Energy Model I, i.e., total energy of the system.
Table IV.
RMSD of the best docking pose identified by different energy models. The docking simulations were run with the ε(r)=4r model.
| System | PTP- Peptide | YopH- Peptide | PTK- Peptide | IGF- Peptide | PKA- Peptide | CDK- Peptide | |
|---|---|---|---|---|---|---|---|
| Energy and solvation model | |||||||
| I | ε(r)=4r | 3.13* | 1.57 | 3.38 | 2.78 | 2.42 | 6.30 |
| 1.92^ | 0.69 | 2.50 | 2.09 | 2.17 | 5.05 | ||
| GBMV | 2.76 | 1.34 | 1.27 | 1.66 | 1.48 | 1.28 | |
| 1.88 | 0.67 | 1.12 | 1.33 | 1.02 | 0.78 | ||
| II | ε(r)=4r | 7.35 | 4.04 | 8.29 | 7.35 | 8.93 | 8.41 |
| 4.31 | 2.01 | 8.28 | 6.52 | 8.62 | 5.89 | ||
| GBMV | 6.42 | 1.70 | 1.92 | 1.63 | 2.45 | 6.14 | |
| 3.84 | 1.10 | 1.37 | 1.25 | 2.25 | 5.06 | ||
| III | ε(r)=4r | 7.54 | 3.39 | 9.14 | 7.35 | 8.93 | 7.84 |
| 4.48 | 1.36 | 9.04 | 6.52 | 8.62 | 4.92 | ||
| GBMV | 5.29 | 4.21 | 1.83 | 1.99 | 8.08 | 6.21 | |
| 3.13 | 2.12 | 1.51 | 1.46 | 8.13 | 5.06 | ||
Upper: RMSDheavy in Å for the best-scoring pose; lower: similar but using only the backbone atoms or backbone and heavy atoms of the phosphotyrosine residue in the YopH and PTP systems. The phosphotyrosine residue was included in the latter systems because it was an important component for recognizing the proteins.
Table V.
RMSD of the best docking pose identified by different energy models. The simulations were run by using the GBMV model.*
| System | PTP- Peptide | YopH- Peptide | PTK- Peptide | IGF- Peptide | PKA- Peptide | CDK- Peptide | |
|---|---|---|---|---|---|---|---|
| Energy and solvation model | |||||||
| I | GBMV | 2.72 | 9.69 | 10.95 | 2.10 | 3.09 | 3.96 |
| 1.72 | 9.41 | 10.48 | 1.70 | 2.30 | 3.13 | ||
| ε(r)=4r | 1.98 | 1.21 | 2.42 | 1.19 | 1.62 | 1.47 | |
| 1.10 | 0.70 | 1.60 | 0.88 | 1.23 | 0.70 | ||
| II | GBMV | 3.79 | 9.63 | 10.89 | 2.37 | 8.14 | 5.76 |
| 2.26 | 9.55 | 10.37 | 1.91 | 7.53 | 4.54 | ||
| ε(r)=4r | 2.36 | 1.73 | 1.76 | 2.04 | 2.66 | 3.17 | |
| 1.49 | 0.78 | 1.69 | 1.39 | 2.38 | 2.84 | ||
| III | GBMV | 3.79 | 9.62 | 5.99 | 2.37 | 7.93 | 5.66 |
| 2.26 | 9.53 | 5.56 | 1.92 | 7.60 | 4.77 | ||
| ε(r)=4r | 2.44 | 1.81 | 1.89 | 2.85 | 2.66 | 5.68 | |
| 1.43 | 1.16 | 1.60 | 2.23 | 2.36 | 4.79 | ||
See footnote of Table IV.
We mentioned above that the GBMV model had more difficulty finding the correct docking pose when we started simulations with the extended conformations of the peptides on the protein surface. To analyze this in more detail, we compared the range of peptide structures sampled by the GBMV and the ε(r)=4r models. Table VI shows that for all the six systems, the GBMV simulations sampled structures further away from the crystal structures in comparison to the ε(r)=4r model. (Note that only the first 1 ns of the trajectories in the ε(r)=4r model was used for comparison because the GBMV trajectories lasted only 1 ns, instead of 2 ns.) This is consistent with earlier studies that protein-peptide structures were less stable in molecular dynamics simulations using Generalized Born models30.
Table VI.
Range of RMSDheavy (in Å) from experiment for structures sampled from the docking simulations starting from the extended conformation, using two different implicit-solvent models.
| Traj. Num. | PTP-Peptide | YopH-Peptide | PTK-Peptide | |||
|---|---|---|---|---|---|---|
| ε(r)=4r | GBMV | ε(r)=4r | GBMV | ε(r)=4r | GBMV | |
| 1 | 5.74–11.64 | 7.31–38.21 | 1.47–10.36 | 12.22–24.24 | 3.83–13.15 | 4.33–12.86 |
| 2 | 4.09–13.37 | 9.77–47.66 | 6.20–10.29 | 8.02–12.40 | 4.03–9.40 | 3.80–7.39 |
| 3 | 5.59–10.85 | 7.34–35.08 | 5.28–10.52 | 8.11–11.59 | 2.75–6.78 | 2.18–6.82 |
| 4 | 5.60–9.22 | 9.09–31.86 | 7.92–10.89 | 7.72–12.14 | 3.74–9.88 | 5.38–25.61 |
| 5 | 5.97–10.37 | 7.73–41.23 | 7.54–10.87 | 7.70–15.86 | 3.75–6.58 | 2.84–6.04 |
| 6 | 6.09–9.98 | 7.85–27.99 | 6.63–11.96 | 10.93–25.11 | 4.00–12.01 | 5.68–14.13 |
| 7 | 5.33–12.91 | 9.45–56.61 | 5.56–9.95 | 7.74–13.51 | 3.89–8.77 | 2.86–5.28 |
| 8 | 3.86–8.50 | 7.04–11.14 | 5.56–9.41 | 6.61–9.71 | 4.88–12.13 | 5.21–21.21 |
| 9 | 5.12–8.20 | 7.47–18.35 | 7.81–11.38 | 9.35–12.79 | 3.89–7.34 | 1.58–8.47 |
| 10 | 6.02–10.78 | 7.49–15.05 | 5.82–9.45 | 7.08–9.78 | 3.74–8.07 | 4.38–18.33 |
| 11 | 4.72–10.53 | 8.31–51.80 | 5.98–11.67 | 9.31–12.12 | 3.24–6.95 | 3.09–5.08 |
| 12 | 4.28–9.61 | 7.82–34.60 | 9.45–14.45 | 8.22–13.42 | 4.28–7.97 | 3.23–5.35 |
| 13 | 3.75–8.67 | 6.92–13.16 | 6.58–10.93 | 6.49–9.70 | 1.54–6.26 | 2.79–7.17 |
| 14 | 5.41–9.41 | 7.04–13.14 | 7.75–11.58 | 7.30–10.60 | 4.74–10.96 | 2.53–9.01 |
| 15 | 4.87–8.24 | 9.43–41.63 | 8.45–14.05 | 8.04–11.13 | 3.85–7.10 | 3.79–10.44 |
| 16 | 6.17–10.39 | 6.31–10.63 | 5.32–9.71 | 8.30–12.65 | 5.64–13.00 | 4.40–13.42 |
| 17 | 7.50–16.60 | 7.47–14.73 | 8.49–11.26 | 10.06–29.85 | 3.60–8.29 | 5.20–16.70 |
| 18 | 6.42–10.31 | 8.09–37.94 | 5.59–9.81 | 8.95–13.01 | 3.77–9.58 | 3.23–7.78 |
| 19 | 5.12–8.20 | 8.56–31.65 | 7.39–13.79 | 9.73–12.53 | 3.95–7.70 | 3.99–7.74 |
| 20 | 6.02–10.78 | 9.21–37.95 | 8.17–11.91 | 8.60–12.38 | 4.81–8.31 | 4.36–16.05 |
| Traj. Num. | IGF-Peptide | PKA-Peptide | CDK-Peptide | |||
| ε(r)=4r | GBMV | ε(r)=4r | GBMV | ε(r)=4r | GBMV | |
| 1 | 4.30–8.78 | 2.41–7.66 | 3.20–6.90 | 6.36–9.69 | 5.72–8.02 | 7.10–17.02 |
| 2 | 4.67–8.05 | 4.53–15.74 | 4.68–6.51 | 6.94–13.66 | 6.24–8.94 | 6.74–11.56 |
| 3 | 4.39–7.35 | 4.82–14.13 | 5.22–8.19 | 5.92–8.71 | 5.23–7.59 | 7.85–15.51 |
| 4 | 4.26–7.22 | 5.76–10.04 | 3.85–6.88 | 7.00–16.64 | 4.11–7.58 | 7.26–18.65 |
| 5 | 4.34–9.71 | 1.65–7.28 | 5.66–8.27 | 5.51–10.85 | 5.24–8.47 | 7.79–13.31 |
| 6 | 4.53–7.42 | 5.28–14.59 | 5.23–7.50 | 4.88–9.82 | 4.89–6.78 | 8.26–20.34 |
| 7 | 4.55–8.21 | 5.20–12.34 | 5.30–8.76 | 5.26–9.38 | 5.22–8.96 | 7.78–13.76 |
| 8 | 4.50–8.17 | 6.43–17.10 | 3.24–6.92 | 5.20–10.07 | 5.55–8.53 | 7.86–17.92 |
| 9 | 4.56–8.84 | 5.71–12.60 | 3.91–6.90 | 5.39–8.37 | 4.09–8.18 | 8.32–44.49 |
| 10 | 4.28–9.21 | 4.34–13.01 | 4.19–6.90 | 6.20–14.77 | 5.53–7.96 | 4.37–10.20 |
| 11 | 3.86–7.65 | 1.72–10.38 | 4.47–7.47 | 5.30–19.05 | 5.30–8.59 | 7.85–22.40 |
| 12 | 4.16–6.14 | 3.67–8.12 | 5.11–7.94 | 6.75–15.95 | 5.15–8.69 | 7.93–14.18 |
| 13 | 4.55–7.12 | 6.07–13.13 | 3.33–6.56 | 6.54–22.24 | 4.80–7.99 | 7.63–46.25 |
| 14 | 4.27–7.79 | 2.95–9.97 | 4.65–8.16 | 6.49–8.94 | 5.88–9.28 | 4.65–12.21 |
| 15 | 4.65–8.46 | 3.33–13.53 | 3.31–6.30 | 5.64–9.11 | 5.80–8.39 | 5.90–13.60 |
| 16 | 1.72–6.09 | 6.01–11.67 | 4.40–8.01 | 5.71–8.26 | 5.55–8.54 | 4.80–9.92 |
| 17 | 4.52–6.46 | 6.16–15.21 | 4.05–7.05 | 5.24–15.83 | 6.16–10.50 | 5.82–10.32 |
| 18 | 4.71–8.33 | 6.40–19.77 | 4.53–6.97 | 6.11–10.74 | 5.19–7.73 | 8.81–16.17 |
| 19 | 3.61–6.71 | 1.80–8.83 | 4.49–7.98 | 5.75–8.71 | 5.06–8.23 | 6.51–11.41 |
| 20 | 4.59–9.07 | 5.66–13.04 | 3.20–7.54 | 3.78–12.00 | 5.44–8.23 | 4.39–9.80 |
Bold: RMSD < 6 Å. Bold: RMSD > 12 Å for PTP-Peptide system.
Bold: RMSD < 6 Å. Bold: RMSD > 12 Å for YopH-Peptide system.
Bold: RMSD < 5 Å. Bold: RMSD > 10 Å for PTK-Peptide system.
Bold: RMSD < 5 Å. Bold: RMSD > 10 Å for IGF-Peptide system.
Bold: RMSD < 5 Å. Bold: RMSD > 10 Å for PKA-Peptide system.
Bold: RMSD < 6 Å. Bold: RMSD > 12 Å for CDK-Peptide system.
4. Conclusions
Previously, we applied a molecular dynamics-based simulated annealing cycling protocol to dock small organic molecules to flexible models of protein kinases and phosphatases. In this work, we extended this approach to docking peptides to similar proteins. We ran simulations with the CHARMM force field using either the ε(r)=4r or the GBMV implicit-solvent model but we scored the structures with several energy models: total energy (Energy Model I), total minus protein energy (Energy Model II), and protein-peptide interaction energy only (Energy Model III) within the same solvation model and with the other solvation model. We obtained the best results by running docking simulation with one solvation model and scoring the structures with the other solvation model using the total energy of the system (Energy Model I). This strategy worked for all six systems studied here. If one used only one solvation energy model in running the simulated annealing cycling docking simulation and in scoring, the cheaper ε(r)=4r model performed better than the GBMV model and it was better to use the total energy of the system. In addition, using one solvation energy model alone could not identify the correct docking pose for all six systems.
Therefore, taking computational efforts into account, the best model tested here involved running docking simulation with the cheaper ε(r)=4r model and scoring the resulting structures with the GBMV model using the total energy of the system (Energy Model I).
Acknowledgments
This research was supported by a Research Award from the University of Missouri-Saint Louis, a Research Board Award from the University of Missouri System, the National Cancer Institute, and the National Institute of Allergy and Infectious Diseases. We also thank the University of Missouri Bioinformatics Consortium and the University of Missouri-Saint Louis Information Technology Services for providing computational resources.
References
- 1.Leach AR, Lemon AP. Proteins-Structure Function and Genetics. 1998;33:227. doi: 10.1002/(sici)1097-0134(19981101)33:2<227::aid-prot7>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 2.Leach AR. Journal Molecular Biology. 1994;235:345. doi: 10.1016/s0022-2836(05)80038-5. [DOI] [PubMed] [Google Scholar]
- 3.Knegtel RMA, Kuntz ID, Oshiro CM. Journal of Molecular Biology. 1997;266:424. doi: 10.1006/jmbi.1996.0776. [DOI] [PubMed] [Google Scholar]
- 4.Meng EC, Shoichet BK, Kuntz ID. Journal of Computational Chemistry. 1992;13:505. [Google Scholar]
- 5.Claussen H, Buning C, Rarey M, Lengauer T. Journal of Molecular Biology. 2001;308:377. doi: 10.1006/jmbi.2001.4551. [DOI] [PubMed] [Google Scholar]
- 6.Kramer B, Rarey M, Lengauer T. Proteins: Structure, Function, and Bioinformatics. 1999;37:228. doi: 10.1002/(sici)1097-0134(19991101)37:2<228::aid-prot8>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 7.Lin JH, Perryman AL, Schames JR, McCammon JA. Journal of the American Chemical Society. 2002;124:5632. doi: 10.1021/ja0260162. [DOI] [PubMed] [Google Scholar]
- 8.Lin JH, Perryman AL, Schames J, McCammon JA. Biopolymers. 2003;68:47. doi: 10.1002/bip.10218. [DOI] [PubMed] [Google Scholar]
- 9.Frembgen-Kesner T, Elcock AH. Journal of Molecular Biology. 2006;359:202. doi: 10.1016/j.jmb.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 10.Zacharias M, Sklenar H. Journal of Computational Chemistry. 1999;20:287. [Google Scholar]
- 11.Keseru GM, Kolossvary I. Journal of the American Chemical Society. 2001;123:12708. doi: 10.1021/ja0160086. [DOI] [PubMed] [Google Scholar]
- 12.Cavasotto CN, Kovacs JA, Abagyan RA. Journal of the American Chemical Society. 2005;127:9632. doi: 10.1021/ja042260c. [DOI] [PubMed] [Google Scholar]
- 13.Perryman AL, Lin JH, McCammon JA. Chemical Biology & Drug Design. 2006;67:336. doi: 10.1111/j.1747-0285.2006.00382.x. [DOI] [PubMed] [Google Scholar]
- 14.Gracia L, Speidel JA, Weinstein H. New computational approaches for NMR-based drug design: a protocol for ligand docking to flexible target sites. FROM PHYSICS TO BIOLOGY: The Interface between Experiment and Computation: Bifi 2006 II International Congress; Zaragoza, Spain. 2006. [Google Scholar]
- 15.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Journal of Medicinal Chemistry. 2006;49:534. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 16.Mizutani M, Takamatsu Y, Ichinose T, Nakamura K, Itai A. Proteins: Structure Function and Bioinformatics. 2006;63:878. doi: 10.1002/prot.20931. [DOI] [PubMed] [Google Scholar]
- 17.Zhao Y, Sanner MF. Proteins: Structure Function and Bioinformatics. 2007;68:726. doi: 10.1002/prot.21423. [DOI] [PubMed] [Google Scholar]
- 18.Corbeil CR, Englebienne P, Moitessier N. Journal of Chemical Information and Modeling. 2007;47:435. doi: 10.1021/ci6002637. [DOI] [PubMed] [Google Scholar]
- 19.Mangoni M, Roccatano D, Di Nola A. Proteins: Structure, Function, and Genetics. 1999;35:153. doi: 10.1002/(sici)1097-0134(19990501)35:2<153::aid-prot2>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 20.Nakajima N, Higo J, Kidera A, Nakamura H. Chemical Physics Letters. 1997;278:297. [Google Scholar]
- 21.Huang Z, Wong CF. Biophysical Journal. 2007;93:4141. doi: 10.1529/biophysj.107.113860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang Z, Wong CF, Wheeler RA. Proteins: Structure, Function, and Genetics. 2008;71:440. doi: 10.1002/prot.21781. [DOI] [PubMed] [Google Scholar]
- 23.Huang Z, Wong CF. Journal of Computational Chemistry. 2009;30:631. doi: 10.1002/jcc.21090. [DOI] [PubMed] [Google Scholar]
- 24.Gilson MK, Zhou HX. Annual Review of Biophysics and Biomolecular Structure. 2007;36:21. doi: 10.1146/annurev.biophys.36.040306.132550. [DOI] [PubMed] [Google Scholar]
- 25.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. Journal of Computational Chemistry. 1983;4:187. [Google Scholar]
- 26.Ewing TJA, Makino S, Skillman AG, Kuntz ID. Journal of Computer-Aided Molecular Design. 2001;15:411. doi: 10.1023/a:1011115820450. [DOI] [PubMed] [Google Scholar]
- 27.Grosdidier S, Pons C, Solernou A, Fernández-Recio J. Proteins: Structure, Function, and Bioinformatics. 2007;69:853. doi: 10.1002/prot.21796. [DOI] [PubMed] [Google Scholar]
- 28.Bueno M, Camacho CJ. Proteins: Structure, Function, and Bioinformatics. 2007;69:786. doi: 10.1002/prot.21722. [DOI] [PubMed] [Google Scholar]
- 29.Glick M, Robinson DD, Grant GH, Richards WG. Journal of the American Chemical Society. 2002;124:2337. doi: 10.1021/ja016490s. [DOI] [PubMed] [Google Scholar]
- 30.Wang T, Wade RC. Proteins: Structure, Function, and Genetics. 2003;50:158. doi: 10.1002/prot.10248. [DOI] [PubMed] [Google Scholar]
- 31.Moitessier N, Therrien E, Hanessian S. Journal of Medicinal Chemistry. 2006;49:5885. doi: 10.1021/jm050138y. [DOI] [PubMed] [Google Scholar]
- 32.Still WC, Tempczyk A, Hawley RC, Hendrickson T. Journal of the American Chemical Society. 1990;112:6127. [Google Scholar]
- 33.Schaefer M, Karplus M. Journal of Physical Chemistry. 1996;100:1578. [Google Scholar]
- 34.Qiu D, Shenkin PS, Hollinger FP, Still WC. Journal of Physical Chemistry A. 1997;101:3005. [Google Scholar]
- 35.Dominy BN, Brooks CL., III Journal of Physical Chemistry B. 1999;103:3765. [Google Scholar]
- 36.Tsui V, Case DA. Biopolymers. 2001;56:275. doi: 10.1002/1097-0282(2000)56:4<275::AID-BIP10024>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 37.Onufriev A, Bashford D, Case DA. Journal of Physical Chemistry B. 2000;104:3712. [Google Scholar]
- 38.Jayaram B, Sprous D, Beveridge DL. Journal of Physical Chemistry B. 1998:102. [Google Scholar]
- 39.Lee MS, Feig M, Salsbury FR, Jr, Brooks CL., III Journal of Computational Chemistry. 2003;24:1348. doi: 10.1002/jcc.10272. [DOI] [PubMed] [Google Scholar]
- 40.Lee MS, Salsbury FR, Jr, Brooks CL., III Journal of Chemical Physics. 2002;116:10606. [Google Scholar]
- 41.Chocholousova J, Feig M. Journal of Computational Chemistry. 2006;27:719. doi: 10.1002/jcc.20387. [DOI] [PubMed] [Google Scholar]
- 42.Mongan J, Simmerling C, McCammon JA, Case DA, Onufriev A. Journal of Chemical Theory and Computation. 2006;3:156. doi: 10.1021/ct600085e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee MR, Sun Y. Journal of Chemical Theory and Computation. 2007;3:1106. doi: 10.1021/ct6003406. [DOI] [PubMed] [Google Scholar]
- 44.Martyna GJ, Klein ML, Tuckerman ME. Journal of Chemical Physics. 1992;97:2635. [Google Scholar]
- 45.Feig M, Karanicolas J, Brooks CL., III Journal of Molecular Graphics and Modelling. 2004;22:377. doi: 10.1016/j.jmgm.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 46.Jia Z, Barford D, Flint AJ, Tonks NK. Science. 1995;268:1754. doi: 10.1126/science.7540771. [DOI] [PubMed] [Google Scholar]
- 47.Narayana N, Diller TC, Koide K, Bunnage ME, Nicolaou KC, Brunton LL, Xuong N-H, Ten Eyck LF, Taylor SS. Biochemistry. 1999;38:2367. doi: 10.1021/bi9820659. [DOI] [PubMed] [Google Scholar]
- 48.Hubbard SR. The EMBO Journal. 1997;16:5573. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Favelyukis S, Till JH, Hubbard SR, Miller WT. Nature Structural Biology. 2001;8:1058. doi: 10.1038/nsb721. [DOI] [PubMed] [Google Scholar]
- 50.Narayana N, Cox S, Shaltiel S, Taylor SS, Xuong NH. Biochemistry. 1997;36:4438. doi: 10.1021/bi961947+. [DOI] [PubMed] [Google Scholar]
- 51.Lowe ED, Tews I, Cheng KY, Brown NR, Gul S, Noble MEM, Gamblin SJ, Johnson LN. Biochemistry. 2002;41:15625. doi: 10.1021/bi0268910. [DOI] [PubMed] [Google Scholar]
- 52.Phan J, Lee K, Cherry S, Tropea JE, Burke J, Terrence R, Waugh DS. Biochemistry. 2003;42:13113. doi: 10.1021/bi030156m. [DOI] [PubMed] [Google Scholar]
- 53.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Jr, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M. Journal of Physical Chemistry B. 1998;102:3586. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 54.MacKerell AD, Jr, Feig M, Brooks CL., III Journal of Computational Chemistry. 2004;25:1400. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]