Abstract
In addition to having a Cx43 ortholog, the zebrafish genome also contains a Cx43-like gene, Cx40.8. Here, we investigate the expression of cx40.8 in zebrafish fins and the function of Cx40.8 in HeLa cells. We find that cx40.8 is present in the same population of dividing cells as cx43. Unlike Cx43, dye coupling assays suggest that Cx40.8 only inefficiently forms functional gap junction channels. However, co-transfection reveals that Cx40.8 can co-localize with Cx43 in gap junction plaques, and that the resulting plaques contain functional gap junction channels. Together, these data suggest the possibility that Cx40.8 may functionally interact with Cx43 to regulate cell proliferation in vivo.
Keywords: connexin, Cx40.8, Cx43, gap junction, zebrafish, short fin
1. Introduction
Connexins, the subunits of gap junction channels, are part of a large multigene family consisting of about 20 genes in mammals [1]. Connexins are integral membrane proteins that consist of four transmembrane domains, two extracellular loops, one cytoplasmic loop, and cytoplasmic amino- and carboxy- termini. Six connexin proteins oligomerize to form one connexon (or hemichannel), and the docking of two connexons at the plasma membranes of neighboring cells forms a single gap junction channel. Gap junction channels typically aggregate into plaques containing about 100–1000 channels, allowing the passage of ions and small molecules (< 1200 Da) between adjacent cells. Interestingly, most tissues express a unique assortment of 2–7 connexin genes, suggesting that distinct homomeric and heteromeric gap junction channels contribute to cell-cell coupling [reviewed in 2]. Further, channel composition can determine metabolite selectivity and specificity [3–5]. Therefore, one possibility is that regulated hetero-oligomerization of different connexin isotypes may in turn regulate tissue function.
Gap junctional intercellular communication (GJIC) is necessary for normal tissue development, evidenced by the identification of mutations in connexins causing such disease phenotypes as deafness, skin disorders, peripheral neuropathies, and cataracts [2,6,7]. In particular, missense mutations in human CX43 cause the craniofacial and limb skeletal malformations associated with oculodentodigital dysplasia (ODDD) [8]. ODDD is transmitted as an autosomal dominant disease, suggesting that mutant forms of Cx43 may hetero-oligomerize with wild-type Cx43, perhaps poisoning Cx43 gap junction channels [reviewed in 2,9]. Mutations in zebrafish cx43 also cause skeletal phenotypes, including short bony fin ray segments associated with the short fin phenotype [10,11]. In addition to the sofb123 allele that has reduced mRNA and protein levels [12,13], three non-complementing ENU-induced alleles were shown to cause missense mutations (sof j7e1 codes Cx43-F30V, sof j7e2 codes Cx43-P191S, and sof j7e3 codes Cx43-F209I). Each missense allele can form gap junction plaques, but channels exhibit aberrant ionic coupling properties [11]. Indeed, recent studies on the zebrafish cx43 mutations reveal a correlation between segment length, level of proliferation, and GJIC [11,13]. Therefore, the zebrafish fin represents an excellent model system to understand how changes in GJIC may lead to important developmental phenotypes, including bone growth.
The zebrafish caudal fin consists of 16–18 bony rays, and fin length depends on the number and size of numerous segments [14]. Fins also have the capacity for rapid growth during regeneration. Amputation is immediately followed by wound healing, and the regeneration blastema is next established [reviewed in 15,16]. The blastema is a specialized structure composed primarily of proliferating cells, and is required for outgrowth. Dividing cells differentiate to contribute to all the tissues of the fin. Expression of cx43 is identified both in the proliferating cells of the blastema and in differentiated cells surrounding fin ray joints [10]. Further, all of the sof alleles exhibit reduced levels of cell proliferation. This and other data suggests that Cx43-based GJIC may be required to establish the population of dividing cells [13]. Therefore, it is important to understand how Cx43-mediated coupling is regulated in this tissue.
The zebrafish genome includes a large connexin gene family (n=37) [17], almost twice the size of human (n=21) and mouse (n=20) families. Phylogenetic analysis revealed that 23 of the zebrafish connexins were a result of the additional whole genome duplication in the teleost lineage [18], while the remaining 14 may have arisen by individual gene duplication events and subsequent divergence [17]. Here we describe the zebrafish cx43-like gene, cx40.8. The cx40.8 gene shares 79% amino acid (Figure 1) and 80% nucleotide identity with cx43. Given the close phylogenetic relationship, Cx40.8 may be compatible to form heteromeric channels with Cx43 and directly modify Cx43-based GJIC. Indeed, heteromeric gap junctions often exhibit distinct channel properties from either homomeric form [3,19–21]. Here, we provide an initial characterization of Cx40.8 expression and function. Results from these and continued studies may provide an endogenous model system to understand how closely related connexins regulate GJIC, and/or how missense alleles may modify wild-type activity.
Figure 1.
Alignment of Cx43-related connexins. Amino acid alignments of human (Hs) Cx43, zebrafish (Dr) Cx43, and zebrafish (Dr) Cx40.8 were completed using ClustalW. The stars indicate amino acid identity, 2 dots indicates strong conservation, 1 dot indicates moderate conservation, and dashes indicate gaps. The arrow identifies the start of the carboxy-tail.
2. Materials and Methods
Fish maintenance
The fish used in this study were derived from the C32 strain and were raised in a 14 light:10 dark photoperiod at 25°C [22].
In situ hybridization
The coding sequence for cx40.8 was amplified (F-GAATCTCGAGATGGGTGACTGGAGCGCACTGG; R-GAATGTCGACTGGATGTCAAGATCATCCGGCC), TA-cloned into pGEMT, and sequenced. Digoxigenin-labeled antisense probe for cx40.8 was generated. Tissue was fixed overnight with 4% paraformaldehyde in PBS and stored in 100% methanol at −20°C. Gradual aqueous washes were completed in methanol/PBST. Tissue was treated with 5 µg/ml proteinase K (5 min for embryos; 45 min for fins) and re-fixed for 20 min. Prehybridization (50 % formamide, 5X SSC, 10 mM citric acid, 0.1 % Tween20) occurred for 1 hour at 65°C, and hybridization in the presence of digoxigenin-labeled antisense probes was completed overnight. Gradual washes into 0.2X SSC were followed by gradual washes into PBST. Anti-digoxigenin Fab fragments (pre-absorbed against zebrafish tissue) were used at 1:5000 overnight. Extensive washes in PBST followed by three short washes in staining buffer (100 mM Tris, 9.5, 50 mM MgCl2, 100 mM NaCl, 0.1 % Tween20, pH 9.0) were completed. Tissue was next transferred to staining solution (staining buffer plus 0.22 mg/ml NBT and 0.175 mg/ml BCIP) and development proceeded until purple color was observed.
qRT-PCR analysis
Trizol reagent (Gibco) was used to isolate mRNA from 5 dpa regenerating fins (5–10 fins were pooled) and first strand cDNA was prepared using oligodT(12–15) and reverse transcriptase. Dilutions of template cDNA were prepared (1:5, 1:50, 1:500, 1:5000). Oligos were designed for cx40.8 using Primer Express software (F-ATTACAATGACTGCTCGGCCC; R-TGCTCATTAGCCTGCTTGTCG). Oligos for cx43 and internal standard ker4 were described previously [12]. All amplicons were amplified independently using the Power SYBR green PCR master mix (Applied Biosystems). Samples were run in triplicate on the ABI7300 Real Time PCR system and the average cycle number (CT) was determined for each amplicon. Delta CT (ΔCT) values represent normalized cx43 or cx40.8 levels with respect to keratin4. Delta Delta CT (ΔΔCT) values represent expression levels of the test sample (sofb123) minus the expression levels of the calibrator sample (wild-type) for either cx43 or cx40.8. The fold-change was calculated using the double ΔCT method (i.e. using the equation 2 −ΔΔCT). Three independent experiments were completed.
Detection of mitotic cells using H3P
This method identifies mitotic cells by detecting a phosphorylated form of serine-10 (H3P) that is present only during mitosis [23]. Following processing for cx40.8 in situ hybridization, the fins were washed in PBS then treated with 1mg/mL collagenase in PBS for 45 mins at room temperature and blocked using a 0.5% BSA in PBS solution with 0.1% Triton-X. A rabbit antibody against anti-phosphohistone H3 (H3P, Upstate Biotechnology) was diluted 1:100 in block and fins were incubated overnight at 4°C. Following a series of washes in block, fins were incubated with an anti-rabbit antibody conjugated to Alexa-546 (Molecular Probes, diluted 1:200 in block) for 2.5 h at room temperature. Washes were performed in block followed by cryosectioning to detect cell doubly-labeled for cx40.8 and H3P. All H3P-positive cells within the distal-most 250µm were identified as cx40.8 positive or negative [13].
Plasmid Construction and HeLa Cell Transfection
The cx40.8 coding sequence was excised from pGEMT using XhoI and SalI restriction endonucleases engineereed into the primers used for the initial amplification (see above under in sity hybridization), and subcloned into p-mApple or pEGFP-N1 (Clontech). For cx43 constructs, the coding sequence from Cx43-N1-EGFP (described in [11]) was subcloned into the EcoRI site of p-mApple. Plasmid DNA was prepared using the MiniPrep Plasmid Purification kit (Qiagen).
HeLa cells were stored at 5% CO2 and 37°C and grown in tissue culture dishes with minimal essential media supplemented with 10% FBS, antibiotics, and antimycotics (Invitrogen). The cells were seeded onto poly-L-lysine coverglasses, incubated overnight, then transfected for 3 hours using either 2µg of Cx40.8-EGFP-N1 or Cx40.8-mApple using Superfect (Qiagen). For double transfections, a combination of 1µg Cx40.8-EGFP-N1 and 1µg Cx43-mApple was co-transfected using the same procedure. Transfected cell pairs were evaluated by confocal microscopy (Zeiss).
Dye coupling assays
Following transfection (t = 21h), transfected cell pairs were identified and a single cell within the pair was injected with propidium iodide (PI, MW = 668.4) using the Eppendorf FemtoJet microinjector and Eppendorf InjectMan N12 micromanipulator. Dye transfer to the non-injected cell of the pair was indicative of functional gap junction channels, and the percentage of cell pairs that showed dye transfer was calculated for each cell culture dish. A total of 25 cell pairs were evaluated for Cx40.8-EGFP and for doubly-transfected cells. The average and standard deviation from 3 trials for each experiment was determined. Statistical significance was calculated using the student’s t-test.
3. Results and Discussion
Cx40.8 is expressed in the same population of cells as Cx43
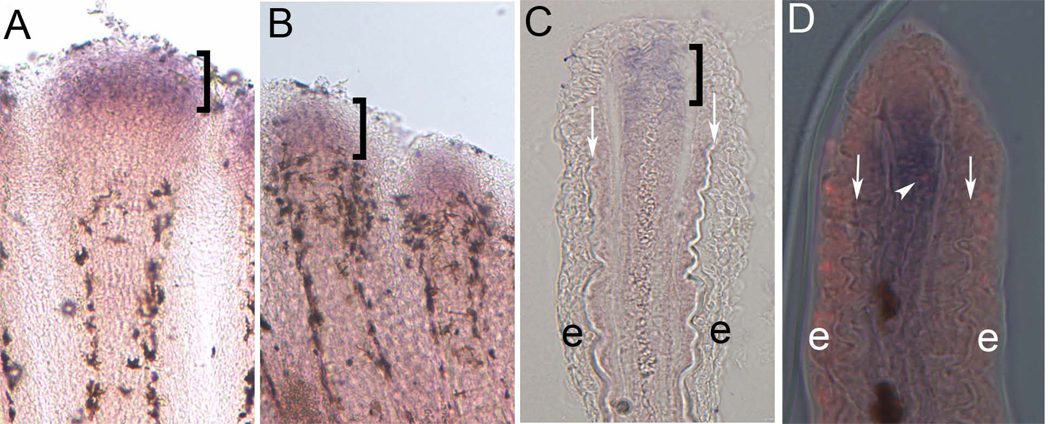
First, we examined cx40.8 expression in regenerating wild-type fins by in situ hybridization. Following whole mount staining and cryosectioning, we found that cx40.8 is expressed in the distal mesenchymal compartment (Figure 2A,C), consistent with the location of proliferating cells and similar to the location of cx43 expression [10]. Given the high percentage of nucleotide similarity between cx40.8 and cx43, it was necessary to rule out the possibility that the above observations were due to cross-hybridization of the cx40.8 probe with the cx43 mRNA. Previously, we found that cx43 is undetectable in short fin (sofb123) regenerating fins by ISH [10]. If the cx40.8 probe hybridizes to cx43 (i.e. in addition to or instead of hybridizing to the cx40.8 mRNA), we would expect that cx40.8 might also fail to be detected in sofb123fins. Instead the cx40.8 expression pattern in sofb123 (Figure 2B) is similar to the wild-type pattern, indicating that the cx40.8 probe identifies the correct target sequence and does not rely on the presence of cx43 mRNA for detection.
Figure 2.
Expression of cx40.8 mRNA in zebrafish regenerating fins is consistent with cx43 expression and the compartment of proliferating cells. (A) ISH of cx40.8 in 5 dpa wild-type regenerating fins. (B) ISH of cx40.8 in 5 dpa sofb123 regenerating fins. (C) Cryosection of stained wild-type regenerating fin showing cx40.8 expression is located in the distal mesenchymal compartment. Brackets identify cx40.8-positive cells. (D) Cryosection of stained wild-type fin showing both cx40.8 and co-localization with an H3P-positive cell. Arrows point to bone matrix. e, epidermis.
The expression domain of cx43 and cx40.8 is consistent with the population of proliferating cells in the regeneration blastema. Previous studies revealed that 89% of the H3P-positive mitotic cells in this region also express cx43 [13]. To determine if cx40.8 is also expressed in actively dividing cells, regenerating fins were processed for cx40.8 ISH and H3P detection. Cryosectioning of doubly-stained fins revealed that 80.6% of mesenchymal H3P-positive cells also expressed cx40.8 (n = 31 H3P cells, Figure 2D). The finding that cx40.8 and cx43 are expressed in the same compartment, together with the finding that cx40.8 is expressed in mitotic cells, suggests that cx40.8 is expressed in the same population of proliferating cells as cx43.
Expression levels of cx40.8 are unchanged in sofb123
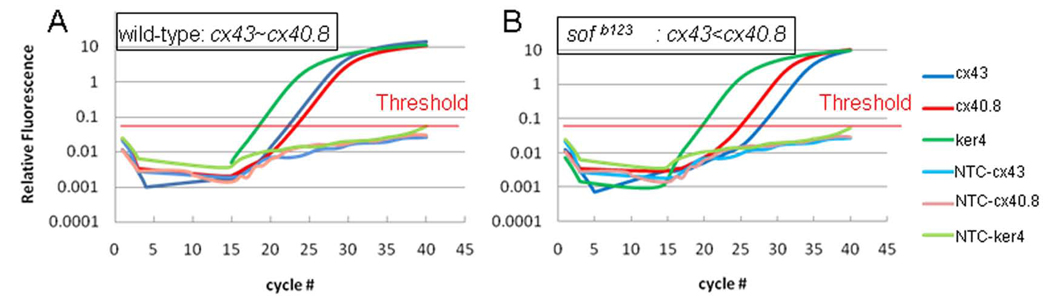
Since cx40.8 and cx43 appear to be expressed in the same cells, the expression levels of these genes may be co-regulated. To test this possibility, qRT-PCR was used to compare the relative levels of cx43 and cx40.8 mRNA in regenerating wild-type and sofb123 fins. First we determined the relative expression of cx43 and cx40.8 in wild-type regenerating fins and found expression at similar levels (Figure 3A). Next, we compared cx40.8 levels in wild-type and sofb123 and found no substantial difference in expression levels of cx40.8 (Figure 3 and Table 1). At the same time, we followed cx43 levels (previous studies revealed that cx43 levels are reduced 15–20-fold in sofb123 compared to wild-type [12]). As before, we found that cx43 levels are significantly reduced in sofb123 (Figure 3). Since the level of cx40.8 is unchanged in sofb123 while cx43 is significantly reduced, the relative levels of cx40.8 and cx43 are drastically different between wild-type and sofb123. Together, these data indicate that the transcription of cx40.8 is not affected by reduced cx43 mRNA. Moreover, the relative expression cx40.8 is almost equal to cx43 in wild-type fins, but it is 8–10 fold higher than cx43 in sofb123 regenerating fins.
Figure 3.
Relative expression levels of cx40.8 and cx43 in regenerating fins. Data from representative wild-type (A) and sofb123 (B) fins are shown. (A) qRT-PCR of cx43 and cx40.8 levels relative to the internal standard, ker4. The amplicons for cx43 and cx40.8 are detected at similar cycle numbers (CT), suggesting similar expression levels in wild-type. (B) In sofb123, the cx40.8 amplicon is detected at similar levels as in wild-type fins (see also Table 1), but cx43 is detected much later (suggesting reduced mRNA levels in sofb123 fins). NTC, no transcript controls for each gene do not permit amplification.
Table 1.
Expression levels of cx40.8 are similar in wild-type and in sofb123 regenerating fins.
| ΔΔCT values | fold-change | |
|---|---|---|
| cx40.8 | 0.575 | 0.671 |
| 0.53 | 0.692 | |
| 0.005 | 0.996 |
ΔΔCT values represent the relative expression of normalized cx40.8 mRNA in sofb123 compared with wild-type. Fold-change values close to 1 suggests that the relative level of cx40.8 is similar between wild-type and sofb123.
Cx40.8 fails to associate in gap junction plaques and does not form channels efficiently
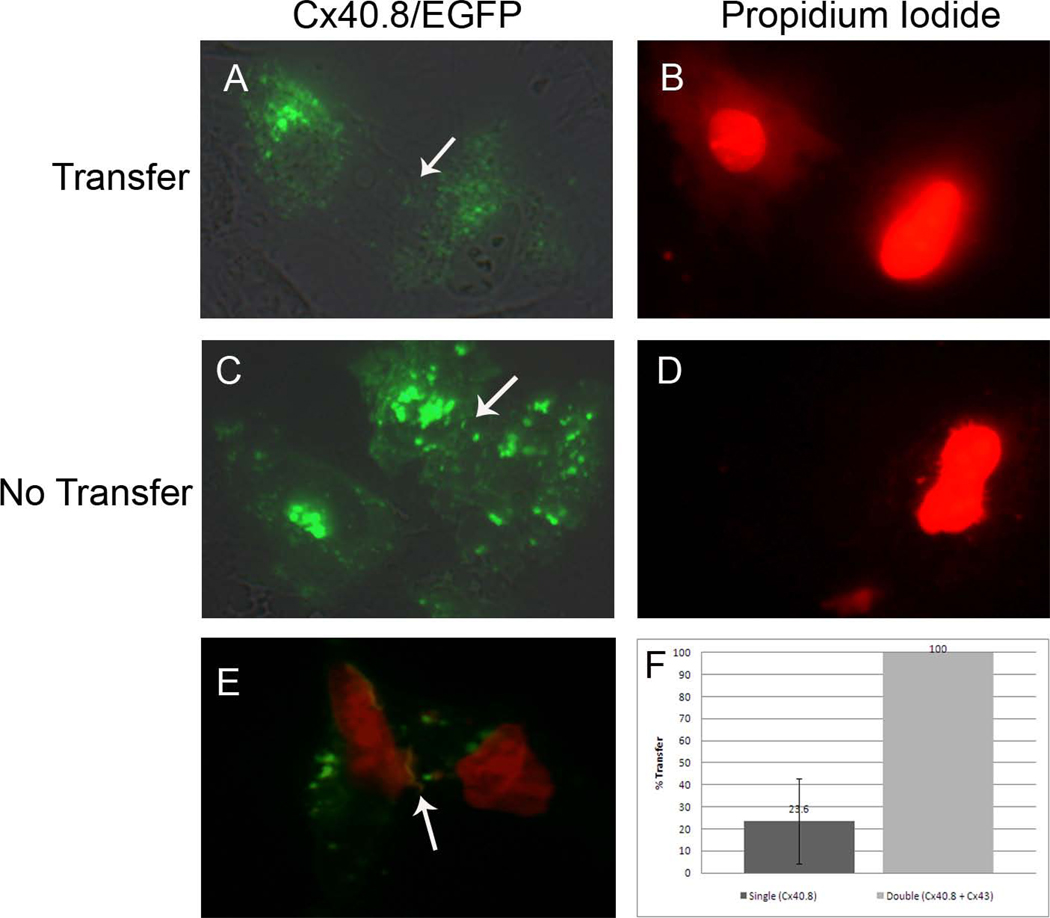
Next, we sought to determine if Cx40.8 functions as a gap junction channel. HeLa cells transfected with an EGFP-tagged form of Cx40.8, Cx40.8-EGFP, revealed that Cx40.8 is localized primarily intracellularly and does not form detectable gap junction plaques at the plasma membrane (Figure 4A,C). One possibility is that the EGFP-tagged zebrafish connexin protein is not trafficked normally using this heterologous system. While we cannot eliminate this possibility for Cx40.8-EGFP, previous studies have shown that zebrafish Cx43-EGFP is localized properly to the plasma membrane in gap junction plaques. Further, zebrafish Cx43-EGFP readily forms gap junction channels evidenced by transfer of the small dye propidium iodide in about 80 % of the injected cell pairs [11]. To determine if Cx40.8 channels might be detected similarly, HeLa cells were transfected with Cx40.8-EGFP, and individual cells from transfected cell pairs were injected with propidium iodide. We found that 23.6 % (+/− 19.4 ) of cell pairs demonstrated dye transfer, suggesting that functional gap junction channels were formed, while most of the cell pairs fail to share dye (Figure 4). Since it is possible to detect dye transfer, we suggest that Cx40.8 does not readily form gap junction channels, but channels that do form appear capable of GJIC.
Figure 4.
Cx40.8 does not readily form gap junction channels when transfected alone. (A,B) An example of positive dye transfer in Cx40.8-EGFP transfected cell pairs. (C,D) An example of failure of dye transfer in Cx40.8-EGFP transfected cell pairs. Arrows point to plasma membranes of adjacent cells. (E) An example of positive dye transfer in HeLa cells co-transfected with Cx40.8-EGFP and Cx43-mAPPLE. Red and green images were overlaid, and the red image is slightly underexposed (compared to B and D) to insure visualization of the yellow gap junction plaque containing both connexins (arrow). (F) Quantitative analysis of positive dye transfer in singly-transfected Cx40.8-EGFP HeLa cells and doubly transfected HeLa cells. The data sets are significantly different (p < 0.001).
Cx40.8 co-localizes with Cx43 in gap junction plaques
HeLa cell transfections showed that Cx40.8 is located intracellularly, albeit with the ability to form some gap junction channels at the plasma membrane. In an endogenous setting where Cx40.8 and Cx43 are co-expressed, Cx40.8 may have the opportunity to hetero-oligomerize or otherwise associate with Cx43. We investigated the possibility of an interaction between Cx40.8 and Cx43 by co-transfecting differentially tagged forms of Cx43 and Cx40.8.
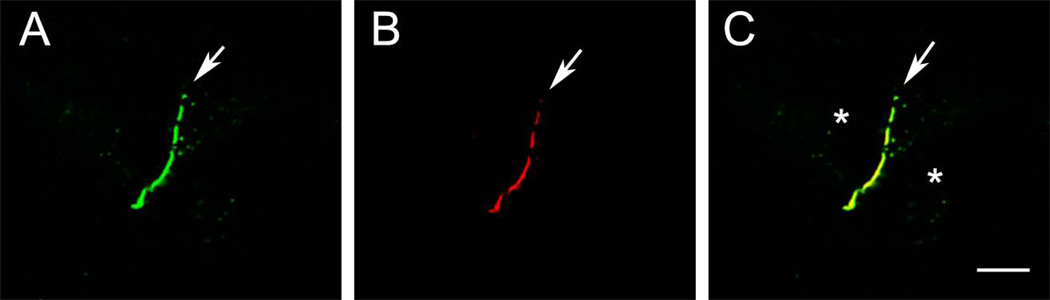
We generated Cx43-mApple and Cx40.8-mApple constructs and found that they localized similarly to their EGFP counterparts. Next, we demonstrated that Cx40.8-EGFP and Cx40.8-mApple co-transfections resulted in yellow intracellular signal and Cx43-EGFP and Cx43-mApple co-transfections resulted in yellow plaques using confocal microscopy (not shown). Then, we co-transfected equal concentrations of Cx43-mApple with Cx40.8-EGFP, mimicking wild-type conditions (switching the fluorescent tags does not alter the results). Using confocal microscopy, cell pairs containing both green and red signals were first analyzed for the presence of Cx43 gap junction plaques. Cx43 plaques were then assessed for the presence of Cx40.8. Interestingly, among doubly-transfected cell pairs, all plaques containing Cx43 also contain Cx40.8 (n=15 cell pairs, Figure 5). This co-localization reveals that Cx40.8 (expressed mostly intracellularly when transfected alone) can be found in gap junction plaques when co-expressed with Cx43 and may suggest that Cx40.8 can hetero-oligomerize with Cx43. At this time, we cannot rule out the possibility that homomeric Cx40.8 channels more efficiently associate among homomeric Cx43 channels within the plaques. However, it is clear that the presence of Cx43 enhances the ability of Cx40.8 to localize in gap junction plaques at the plasma membrane.
Figure 5.
Cx43 and Cx40.8 can associate together at the plasma membrane in HeLa cells. Cells were co-transfected with both Cx40.8-EGFP (A) and Cx43-mApple (B). Both connexins co-localize in gap junction plaques (C). Asterisks identify each cell in this cell pair.
Next we determined if the plaques containing both Cx43 and Cx40.8 permit dye transfer. Cells were co-transfected with Cx43-mApple and Cx40.8-EGFP prior to propidium iodide injection. Double-transfected cell pairs were identified by Cx40.8-EGFP positive plaques (i.e. since Cx40.8 is not found in plaques in singly-transfected cells). Interestingly, all of the injected cell pairs showed dye transfer (Figure 4). At least two explanations are possible for the increased level of dye transfer in the double transfections over the Cx40.8-EGFP single transfections. First, it is apparent that Cx40.8 does not inhibit Cx43 channel function, so one possibility is that Cx40.8-Cx43 heteromeric channels are capable of transferring propidium iodide. Therefore, we do not rule out the hypothesis that Cx40.8 might directly modify the channel properties of Cx43 and enhance or alter the coupling of particular small molecules (although further studies are required to demonstrate that channel properties differ among Cx40.8-channels, Cx43-channels, and heteromeric forms). Alternatively, the observed positive dye coupling may be permitted by homomeric forms of Cx43 and Cx40.8 gap junction channels. Indeed, Cx43 homomeric gap junction channels transfer propidium iodide efficiently [11], and such channels may exist within the plaques. Moreover, since co-transfection of Cx43 and Cx40.8 causes an increased association of Cx40.8 in gap junction plaques, and since we conclude above that Cx40.8 channels are capable of dye transfer, double transfection may simply increase the efficiency of bringing Cx40.8 to the plasma membrane where it can function as a typical channel. Future analyses using higher resolution imaging and ionic coupling assays are required to distinguish these possibilities.
Conclusions
Zebrafish Cx40.8 represents the closest connexin relative to Cx43. While the cx40.8 gene is not found in mammalian genomes, the ODDD phenotypes may be caused by mutant forms of Cx43 modifying the function of wild-type Cx43 protein. Indeed, this study reveals that while cx40.8 is expressed similar to cx43 in vivo, its function may be distinct. The Cx40.8 protein does not contribute to gap junction plaques efficiently on its own, but does in combination with Cx43. Therefore, when expression levels are similar (as in wild-type fins), Cx40.8 may have the opportunity to interact functionally with Cx43. However, when cx40.8 is expressed at much higher levels than cx43 (as in sofb123 fins), Cx40.8 may not be found at the plasma membrane. Therefore, it is possible that the reduced cell proliferation phenotype of sof mutants may be attributable to the absence of Cx40.8 at the plasma membrane, and/or due to the failure of Cx40.8-modification of Cx43 gap junction channels.
Acknowledgments
The authors thank Rebecca Jefferis for care of the zebrafish, Sarah Neilsen for the H3P and cx40.8 co-localization studies, as well as Dr. Jutta Marzillier for her assistance with qRT-PCR. This research was funded by the NICHD (R01HD047737 to MKI).
List of Abbreviations
- ODDD
Oculodentaldigital dysplasia
- GJIC
Gap junction intercellular communication
- ISH
In situ hybridization
- H3P
Histone-3-phosphate
- dpa
days post amputation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Structured summary:
MINT-7266123:
cx40.8 (genbank_protein_gi:68354404) and cx43 (uniprotkb:O57474) colocalize (MI:0403) by fluorescence microscopy (MI:0416)
References
- 1.Sohl G, Willecke K. Gap junctions and the connexin protein family. Cardiovasc Res. 2004;62:228–232. doi: 10.1016/j.cardiores.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 2.Laird DW. Life cycle of connexins in health and disease. Biochem J. 2006;394:527–543. doi: 10.1042/BJ20051922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bevans CG, Kordel M, Rhee SK, Harris AL. Isoform composition of connexin channels determines selectivity among second messengers and uncharged molecules. J Biol Chem. 1998;273:2808–2816. doi: 10.1074/jbc.273.5.2808. [DOI] [PubMed] [Google Scholar]
- 4.Elfgang C, Eckert R, Lichtenberg-Frate H, Butterweck A, Traub O, Klein RA, Hulser DF, Willecke K. Specific permeability and selective formation of gap junction channels in connexin-transfected HeLa cells. J Cell Biol. 1995:805–817. doi: 10.1083/jcb.129.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldberg GS, Valiunas V, Brink PR. Selective permeability of gap junction channels. Biochim Biophys Acta. 2004;1662:96–101. doi: 10.1016/j.bbamem.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 6.Gerido DA, White TW. Connexin disorders of the ear, skin, and lens. Biochim Biophys Acta. 2004;1662:159–170. doi: 10.1016/j.bbamem.2003.10.017. [DOI] [PubMed] [Google Scholar]
- 7.White TW, Paul DL. Genetic diseases and gene knockouts reveal diverse connexin functions. Annu Rev Physiol. 1999;61:283–310. doi: 10.1146/annurev.physiol.61.1.283. [DOI] [PubMed] [Google Scholar]
- 8.Paznekas WA, et al. Connexin 43 (GJA1) mutations cause the pleiotropic phenotype of oculodentodigital dysplasia. Am J Hum Genet. 2003;72:408–418. doi: 10.1086/346090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laird DW. Closing the gap on autosomal dominant connexin-26 and connexin-43 mutants linked to human disease. J Biol Chem. 2008;283:2997–3001. doi: 10.1074/jbc.R700041200. [DOI] [PubMed] [Google Scholar]
- 10.Iovine MK, Higgins EP, Hindes A, Coblitz B, Johnson SL. Mutations in connexin43 (GJA1) perturb bone growth in zebrafish fins. Dev Biol. 2005;278:208–219. doi: 10.1016/j.ydbio.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 11.Hoptak-Solga AD, Klein KA, Derosa AM, White TW, Iovine MK. Zebrafish short fin mutations in connexin43 lead to aberrant gap junctional intercellular communication. FEBS Lett. 2007;581:3297–3302. doi: 10.1016/j.febslet.2007.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sims K, Jr., Eble DM, Iovine MK. Connexin43 regulates joint location in zebrafish fins. Dev Biol. 2009;327:410–418. doi: 10.1016/j.ydbio.2008.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoptak-Solga AD, Nielsen S, Jain I, Thummel R, Hyde DR, Iovine MK. Connexin43 (GJA1) is required in the population of dividing cells during fin regeneration. Dev Biol. 2008;317:541–548. doi: 10.1016/j.ydbio.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iovine MK, Johnson SL. Genetic analysis of isometric growth control mechanisms in the zebrafish caudal Fin. Genetics. 2000;155:1321–1329. doi: 10.1093/genetics/155.3.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poss KD, Keating MT, Nechiporuk A. Tales of regeneration in zebrafish. Dev Dyn. 2003;226:202–210. doi: 10.1002/dvdy.10220. [DOI] [PubMed] [Google Scholar]
- 16.Akimenko MA, Mari-Beffa M, Becerra J, Geraudie J. Old questions, new tools, and some answers to the mystery of fin regeneration. Dev Dyn. 2003;226:190–201. doi: 10.1002/dvdy.10248. [DOI] [PubMed] [Google Scholar]
- 17.Eastman SD, Chen TH, Falk MM, Mendelson TC, Iovine MK. Phylogenetic analysis of three complete gap junction gene families reveals lineage-specific duplications and highly supported gene classes. Genomics. 2006;87:265–274. doi: 10.1016/j.ygeno.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Amores A, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 19.Cao F, Eckert R, Elfgang C, Nitsche JM, Snyder SA, DF Hu, Willecke K, Nicholson BJ. A quantitative analysis of connexin-specific permeability differences of gap junctions expressed in HeLa transfectants and Xenopus oocytes. J Cell Sci. 1998;111(Pt 1):31–43. doi: 10.1242/jcs.111.1.31. [DOI] [PubMed] [Google Scholar]
- 20.Bukauskas FF, Angele AB, Verselis VK, Bennett MV. Coupling asymmetry of heterotypic connexin 45/ connexin 43-EGFP gap junctions: properties of fast and slow gating mechanisms. Proc Natl Acad Sci U S A. 2002;99:7113–7118. doi: 10.1073/pnas.032062099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moreno AP. Biophysical properties of homomeric and heteromultimeric channels formed by cardiac connexins. Cardiovasc Res. 2004;62:276–286. doi: 10.1016/j.cardiores.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 22.Westerfield M. The Zebrafish Book: A guide for the laboratory use of zebrafish (Brachydanio rerio) Eugene, OR: University of Oregon Press; 1993. [Google Scholar]
- 23.Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]