Abstract
Resistance or susceptibility of the snail host Biomphalaria glabrata to Schistosoma mansoni is determined by the genetics of both the snail and parasite. Although Mendelian genetics governs adult resistance to infection, juvenile resistance and susceptibility are complex traits. In this study, suppression subtractive hybridization was used to construct forward and reverse cDNA libraries to identify genes involved in the immediate response of juvenile resistant (BS-90), non-susceptible (LAC2) snails, and susceptible (NMRI) snails after early exposure to S. mansoni. Expressed Sequence Tags (ESTs) were generated from the repertoire of enriched transcripts. In resistant snails, several ESTs corresponded to transcripts involved in immune regulation/defense response. While no defense related transcripts were found among juvenile susceptible snail ESTs, we detected transcripts involved in negative regulation of biological process/morphogenesis /proliferation. Differential gene expression and temporal regulation of representative transcripts were compared among snails pre- and post-exposure to either normal or attenuated miracidia using quantitative real time RT-PCR. Results showed that several transcripts, such as fibrinolytic C terminal domain, cytidine deaminase, macrophage expressed gene 1, protein kinase C receptor, anti- microbial peptide; theromacin and Fas remained up- regulated regardless of whether or not snails were exposed to normal or attenuated miracidia. While ESTs related to C- type lectin and low- density lipoprotein receptor were induced only by exposure to normal miracidia. By comparing changes in gene expression between resistant and susceptible juvenile snails responding either to normal or attenuated parasites, we can conclude that the transcription of genes associated with the intra-dermal penetration process of the snail host by invading miracidia may need to be taken into account when assessing differential gene expression between resistant and susceptible strains of B. glabrata in relation to S. mansoni exposure.
Keywords: juvenile Biomphalaria glabrata, suppression subtractive hybridization (SSH), resistant, susceptible, Schistosoma mansoni, differential gene- regulation
1. Introduction
Schistosomiasis is a snail- borne trematode infection of humans, domestic and wild animals in different parts of Asia, Africa, the Middle East, South America and the Carribbean. The causative schistosome parasite is acquired trans- cutaneously while swimming or wading in contaminated waters; all other trematodes infect only via ingestion. Approximately 200 million people in 74 countries [1, 2] are affected; 120 million of these are symptomatic and 20 million have severe disease [3]. The risk of infection is spreading as new dams are constructed in endemic areas. Control of schistosomiasis, in particular that caused by S. japonicum has been severely hindered by the fact that several non- human mammalian species, including domesticated animals e.g. cattle, pigs, equines and sheep [4] serve as zoonotic carriers of this infection. Transmission reduction to solve public health problems and agricultural concerns associated with schistosomes have traditionally relied on the use of molluscicides and mass chemotherapy to curtail infections in the snail and human population, respectively. However, due to persisting conditions of poor health infrastructure, lack of access to clean water and poverty, re- infections in humans still poses a challenge for the long-term control of schistosomiasis. It is hoped that vaccines, and better diagnosis of human and snail infections will help alleviate some of these challenges. However, until these become available, alternative strategies, including blocking parasite transmission in the snail host have been considered. Several studies have been conducted in recent years to begin to understand the molecular basis of the snail- parasite interaction and to identify genes that may be involved in rendering snails either susceptible or resistant to infection.
It is now clear that the molecular basis of the intermediate snail host-schistosome relationship is complex, involving a variety of genes associated with receptor recognition [5], cell adhesion [6, 7], immune regulation [5, 8–10], proteolytic enzymes [11], and enzymes involved in detoxification reactions (e.g. anti- oxidants) [12, 13] that are modulated by soluble parasite factor(s) in the snail host [14–16]. A stress response, manifested by the modulation of genes encoding the stress response protein such as heat shock protein 70 (Hsp 70), may also underlie the snail- host/ parasite encounter [8, 17].
Schistosome infection of the snail host, unlike vertebrate immune responses that have both adaptive and innate systems depends solely on non-self recognition by the snail’s innate defense system (IDS). While parasites develop successfully into sporocysts and cercariae in susceptible/ compatible snails, they are destroyed by both cellular and humoral (hemocytes and hemolymph) components of the IDS within a short time after infection in a resistant/ incompatible host [18].
Previous studies have used compatible/ incompatible snail stocks to begin to elucidate the molecular mechanisms underlying compatibility issues in snail- host/ trematode infections. Thus, from numerous studies where transcriptome profiles (some using SSH- cDNA cloning strategies) of susceptible and resistant stocks have been compared, many genes that are differentially expressed between these two stocks of B. glabrata, either to S. mansoni or Echinostoma caproni infections, have been identified [9, 10, 15, 19–21]. These include genes encoding calcium- binding proteins and glycolytic fibrinogen related protein (FREPs) [22–26]. Similar comparative studies, based on proteomics of either snail or parasite protein extracts are also beginning to reveal key molecules (such as mucin- like proteins from both parasite and snail) that may play a role in snail/ schistosome compatibility [27, 28]. Despite abundant emerging molecular information, very little is known about which snail genes to specifically target to develop transmission - blocking strategies for the eventual goal of disease control.
Most studies aimed towards deciphering differences in gene regulation between resistant/ incompatible and susceptible/ compatible snails during the snail/ schistosome encounter have focused mainly on this relationship in adult, but not juvenile snails. Age dependent variability in B. glabrata susceptibility to S. mansoni has been well documented with results showing that juvenile snails (even within the same stock) are, in general, more vulnerable than their adult counterparts to infection [29–31]. How this biological phenomenon impacts the dynamics of disease transmission is not yet known. It is reasonable, however, to assume that if intervention tools to block transmission in B. glabrata are to one day become a practical reality, then it would be essential to determine the full complexity of the snail host transcriptome (that is up- regulated by infection) not only in adults but also in juvenile snails (both resistant and susceptible), especially during the early stages of schistosome infection.
Thus, in the present study we constructed suppression subtractive hybridization (SSH) cDNA libraries to determine the immediate response- specific transcripts from susceptible (NMRI), non- susceptible (LAC2), and resistant (BS-90) B. glabrata juvenile snails following early exposure (5 h) to miracidia. Our rationale was to identify genes that are differentially up regulated during early S. mansoni infection of the three stocks with the view that these transcripts, in susceptible snails may be involved in miracidia transformation/ development, while, in non-susceptible and resistant snails early response genes may play a role in miracidia destruction before they differentiate into primary sporocysts.
In addition, in the aforementioned studies regarding parasite- induced changes in gene- expression profiles of either resistant or susceptible snails, transcriptional regulation was investigated entirely in relation to normal miracidia but not to attenuated parasites. Because studies have shown that gamma- irradiated miracidia, while successful in penetrating the snail, fail to develop in B. glabrata [32, 33], we further evaluated transcriptional regulation of cloned, representative enriched resistant and susceptible- specific transcripts before and after exposure of these snails to either normal or live attenuated miracidia. We hoped by this analysis to differentiate between transcripts that are specifically up- regulated by infection from those that are induced because of the intra-dermal penetration of miracidia into the snail.
Expressed sequence tags (ESTs) generated from the recombinant SSH clones would then reflect a repertoire of enriched transcripts that are representative of these snail stocks (resistant and susceptible) in response to infection. We tested this hypothesis by constructing SSH libraries from exposed juvenile resistant BS-90 (jx-BS-90) snails, exposed juvenile non- susceptible LAC2 (jx-LAC2) snails, and exposed juvenile susceptible NMRI (jx-NMRI) snails. Using this strategy, we report here that in response to normal S. mansoni infection, transcripts encoding genes involved in the innate defense system, such as homologs of the fibrinogen (Fg) C- terminal domain, C- type lectin, low-density lipoprotein receptor (LDLR), and mucin were all up- regulated early during infection (within 5 h) in the susceptible (NMRI) snail. In resistant and non- susceptible snails (BS-90 and LAC2, respectively), transcripts related to genes involved in parasite killing (phagocytosis, anti- microbial response and enzyme, e.g. macrophage expressed gene 1 [Mpeg1], Fas, theromacin and cytidine deaminase), and protein kinase C receptor (PKCR), were all up- regulated shortly (within 5 h) after infection. We also report that some of the resistant and susceptible- specific SSH- ESTs that were up- regulated shortly after exposure to normal miracidia either remained induced or were down regulated in response to live, attenuated miracidia indicating that transcriptional regulation of genes involved, specifically, in the parasite invasion/penetration process, and not parasite development in the snail host may also influence changes in gene expression observed between resistant and susceptible stocks of B. glabrata upon exposure by S. mansoni.
2. Materials and Methods
2.1 Biomphalaria glabrata stocks and parasite exposure
Juvenile B. glabrata snails (4–5 mm in diameter) used in this study belonged to the BS-90 (resistant) and LAC2 (non-susceptible) stocks selected for their 100% and 75% resistance at any age, respectively and the NMRI (susceptible) strain that is 100% susceptible to the NMRI strain of S. mansoni. The LAC2 stock was derived [34] from the susceptible NMRI snails that failed to accommodate infection. Parasite- negative progeny snails were then selected from each successive generation by self- fertilization. LAC2 snails used for this study were selected for the non- susceptible phenotype for nineteen generations (F19). Prior to all experiments, snails were incubated overnight in sterile distilled water containing 100 µg/ml ampicillin at room temperature in an attempt to reduce contaminating resident bacterial in the snails. Individual snails were exposed to either normal or attenuated miracidia (10 miracidia/ snail) for different time points (5, 10, 24, 48 h). Attenuated miracidia were obtained by irradiating miracidia immediately after hatching using a Mark 1 Cesium-137 irradiator (JL Shepherd, San Fernando, CA) for 9.2 min delivering 20,000 Rads at the rate of 2,156 Rads/min. Following parasite exposure (to either normal or attenuated parasites), snails were snap frozen in liquid nitrogen and kept at −80°C until required. The time periods of post- exposure were chosen to capture the snail’s response after haemocyte contact with the invading miracidia prior to sporocyst encapsulation (24– 48 h) [18]. The penetration behavior of miracidia for each of the resistant and susceptible snail stocks was monitored for both normal and irradiated parasites, and shown not to differ [17].
2.2 RNA extraction and cDNA synthesis
Total RNA was extracted from whole juvenile snails (5 h after exposure to 10 miracidia) or S. mansoni adult worms using RNA Bee as previously described [19]. Trace amounts of contaminating genomic DNA were removed by treating all RNA samples with DNase I prior to use for cDNA synthesis according to the manufacturer’s instructions (Promega, WI). First strand cDNA was synthesized from pre- heated RNA (70°C, 2 min) (2 µg) with 1 µM of poly dT15 (Roche Molecular Systems Inc, PA) by reverse transcription (Superscript reverse transcriptase II) in 40 µl reaction volume following the manufacturer’s instructions (Invitrogen Life Technologies, CA). The first strand cDNA template was either processed immediately for second strand PCR or frozen at −20°C until required. cDNA processed without reverse transcriptase in the reaction was incubated in parallel as negative control to monitor for any residual contaminating genomic DNA in RNA samples.
2.3 Construction of subtractive resistant- and susceptible- specific transcripts cDNA libraries
Libraries were constructed by suppression subtractive hybridization (SSH) with the Clontech PCR-Select™ cDNA Subtraction kit (BD Biosciences, CA) [35]. Four libraries were constructed as follows: resistant juvenile BS-90 snail (tester) was subtracted from the susceptible juvenile NMRI (driver) snail (forward subtracted library resulting in susceptible snail enriched transcripts) and the reciprocal subtraction (reverse subtracted library resulting in resistant snail enriched transcripts). In addition, the non-susceptible juvenile LAC2 snail (tester) transcripts subtracted from susceptible juvenile NMRI snail (driver) RNA and its reciprocal (forward and reverse subtracted libraries for LAC2 and NMRI) were also constructed. For the above libraries, cDNA was synthesized from pooled poly A enriched RNA (isolated from 15 similarly sized snails within a short time, 5 h post exposure to normal miracidia). Library construction including all steps of suppression subtractive hybridization, adaptor ligation, and selective amplification were performed following the protocol of the manufacturer. Briefly, double strand tester cDNA was digested with Rsa I, and the digested tester cDNA split into two groups and linked to either adapter I or adapter 2R. The subtraction hybridization reaction was performed by annealing excess driver cDNA using adaptor ligated tester cDNA as follows: cDNA samples were denatured and incubated at 68°C for 8 h. After the first hybridization, the two samples were mixed together and the hybridization repeated with freshly denatured driver cDNA overnight at 68°C. The two rounds of hybridization generated a normalized population of tester specific cDNAs possessing different adaptors at each end. After filling in the ends, two rounds of PCR amplifications were performed to enrich for the desired cDNA containing both adapters. The optimized amplification for the first and second rounds of PCR, to increase representation and reduce redundancy of subtracted cDNAs, was performed for 27 and 10 cycles, respectively.
In the BS-90/ NMRI library, SSH was performed to produce a cDNA library enriched in resistant specific transcripts. The tester sample was the cDNA population from BS-90 and the driver sample was the cDNA population from the NMRI stock. In the NMRI/ BS-90 library, the tester and driver samples was reversed and SSH was performed to produce a cDNA library enriched in susceptible specific transcripts. In the LAC2/ NMRI and NMRI/ LAC2 libraries, the tester and driver samples were reversed as above to produce cDNA libraries enriched with non-susceptible- specific and susceptible-specific transcripts, respectively.
2.4 Cloning of the subtracted cDNAs
For each SSH procedure, the final amplification product was cloned into the TOPO®-TA cloning vector (Invitrogen Life Technologies, CA) and E. coli DH5α was transformed with the individual ligation products. Transformed bacteria in 395 µl of SOC medium were incubated for 1 h at 37°C with continuous shaking before plating onto agar plates containing 100 µg/ ml ampicillin, 100 mM IPTG and 100 mg/ ml X-gal overnight at 37°C. Ten random white clones selected from each library were screened by PCR using universal M13 primers (forward and reverse) to check for the presence, and sizes of inserts prior to picking single colonies into 96-well microtiter plates containing 150 µl of super broth medium supplemented with 100 µg/ ml ampicillin. The cultures of enriched recombinant clones were then grown overnight without shaking at 37°C.
2.5 Single pass sequencing and nucleotide sequence analysis
Plasmid DNA from 282, 417 and 981 clones of the subtracted BS-90, LAC2 and NMRI libraries respectively were purified and sequenced using the universal M13 reverse primer at the Institute of Genomic Research (TIGR, now the J. Craig Venter Institute), Rockville, MD. Base calling was performed using PHRED software with the quality cut-off set as PHRED 2.0. Raw sequences were then imported into Vector NTI Advance™ 10 software (Invitrogen Life Technologies, CA) and subjected to trimming of vector sequences and adapter sequences (for subtractive hybridization) using default setting. EST sequences representing contamination from bacterial, yeast and fungal sources were identified using BLASTN algorithm and removed from further analyses. ESTs were aligned and assembled into contigs using Vector NTI Advance™ 10 software when the criterion of a minimum identity of 95% over 50 bp was met. When an EST could not be assembled with others in a contig, it remained as a “singleton”. The contigs and the singletons should thus correspond to sequences of unique genes. The consensus sequences of the contigs and the sequences of the singlets were compared to the sequences in GenBank’s non- redundant (nr) using the BLASTX algorithms. The cut- off for sequence similarity was E- value < 10−4 for all analyses.
2.6 Biological process classification
Gene ontology (GO) term annotation and function- based analysis [36] of unique sequences were performed using AmiGO software [37]. The putative biological functions of sequences were assessed using GO based on BLAST analysis. GO terms for a given biological process were from sequence similarities using the application filter biological process parameter. From these annotations, pie charts were made using 3rd level GO.
2.7 Qualitative RT-PCR and SYBR Green quantitative real time RT-PCR
RNA isolated from individual juvenile snails (size matched) either unexposed (0 h) or exposed for different time points (5,10, 24, 48 h) to normal or irradiated (20 Krads) miracidia was used to synthesize first strand cDNA that was subsequently used for monitoring the transcription of SSH- ESTs relative to the housekeeping gene, myoglobin of B. glabrata. Using commercially purchased (Operon Biotechnologies, AL) gene specific primers (GSPs) for Q-RT-PCR designed using Primer-Blast algorithm in GenBank (Ta = 58°C) showing no cross hybridization to S. mansoni (Table 3) we performed second strand PCR as previously described [20]. Amplified material was analysed by 1.2% TBE- agarose electrophoresis (data not shown).
Table 3.
List of primers used for real time PCR and their GenBank Accession numbers
| Transcript | 5'primer | 3'primer | fragment length (bp) | Accession no. |
|---|---|---|---|---|
| Calcium binding protein (CaBP) | 5'-TTTATCATGGATCTATTGTGGGAAC-3' | 5'-CGTGATTATGCAACATCCTGTG-3' | 410 | GH716778 |
| Chitinase | 5’-ACTTCCCTTGTCCACATGTCTGTCC-3’ | 5’-TGATCAAAGCAGGGGCTCAGGT-3’ | 204 | GH716922 |
| Cathepsin B1 | 5'-AGCAACACCATTCCACATC-3' | 5'-ATAGCCTCCGTTACATCC-3' | 420 | EU035711 |
| Cytidine deaminase (CDA) | 5’-TCCCAAACTCTAAGAGAA-3’ | 5’-CGAGGTACATTAGTCCAT-3’ | 54 | DQ117977 |
| DEC_1 | 5’-ATACACGAAGTGTGGCTTGCATGTG-3’ | 5’-ACTACGCCAGGACCCATACCTTCA-3’ | 451 | GH717041 |
| Fas | 5’-GAAATATCATTCATTCTGCCCCGG-3’ | 5’-CGACGCCAAAAGTCAGCTCACTT-3’ | 159 | GH716733 |
| Ferritin3 | 5'-CTCTCCCACACTGTACCTATC-3' | 5'-CGGTCTGCATCTCGTTTTC-3' | 236 | GH716790 |
| Fibrinogen (Fg) C-terminal domain | 5’-AACCGTTAAGATTGACGAAGCCACA-3’ | 5’-GAAAGAGGCACTGCCTTCAACTTCA’3’ | 235 | GH716248 |
| Glutathione S-transferase (GST) | 5'-CAGCTGTTGGTCAACAATTTG-3' | 5'-CGAGCTAAATACGTTGCTATAGC-3' | 151 | GH716560 |
| Heat shock protein 40 (HSP-40) | 5'-GTTCCTTCTGGAGCAGATGA-3' | 5'-AAACCCCATGCTAAGGTCTC-3' | 152 | GH716707 |
| Heat shock protein 70 (HSP-70)3 | 5'-AGGCGTCGACATTCAGGTCTA-3' | 5'-TGGTGATGTTGTTGGTTTTACCA-3' | 199 | L44127 |
| Immunoglobulin (Ig) | 5’-AAAGACCTCTTACTGCGGCTCGAC-3’ | 5’-CATTTGTTTCACGTGCTTGGCTG-3’ | 219 | GH716944 |
| Low density lipoprotein receptor (LDLR) | 5’-ACATCCCAGTTCATCTGAAGCGTCT-3’ | 5’-ATTGCCACAAACAGGAGTGCACTG-3’ | 350 | GH716919 |
| C-type lectin | 5’-ACTTGGGACAGGGTTGAATGAAGG-3’ | 5’-TTTCAGGAGGGGTCACAATTTTGAA-3’ | 214 | GH716225 |
| Macrophage expressed gene 1 (Mpeg1) | 5’-TCCAAATGGAACTGATGACAACGC-3’ | 5’-TTGGTGTGTTGCAACTGAGCGAG-3’ | 199 | GH716015 |
| Melanogenic peroxidase | 5’-TCTTCAACGAATGGGTCAACATCG-3’ | 5’-AGATCGTTGGTTCTTCACCCACAGA-3’ | 213 | GH716074 |
| Mucin | 5'-GCCCCTGCACCCTGTTT-3' | 5'-ATGAGCTTCCACACACTGAGAAGT-3' | 110 | CAC83675.1 |
| Ornithine decarboxylase | 5'-AGGAATGTCGTCTGCCTGAGA-3' | 5'-TTGATTTCATCCCAGATATCTTGACT | 150 | GH716341 |
| Protein kinase C receptor (PKCR) | 5’-TTGATGCTCTTGTCACGAGATGCA-3’ | 5’-CATGGCCATGGACATTTTGTTAGTG-3’ | 212 | GH716153 |
| Serine protease | 5'-GTGAAGAATTTGGCGGGAGA-3' | 5'-GCGCTGAAGTGGACGATCA-3' | 239 | GH716785 |
| Theromacin | 5'-CGGGATGTTTCGAAGATTGG-3' | 5'-CGTTGTGGACAATTTAGACTTGGT-3' | 150 | GH716927 |
with the exception of Cathepsin B, Ferritin and HSP-70 regulation (Myers et al., 2008 and Ittiprasert et al., 2009)
with the exception of Ferritin (manuscript in preparation)
To confirm elevated transcription of selected SSH-ESTs (soon after parasite penetration) SYBR green quantitative real-time RT- PCR was used to determine transcriptional regulation (pre- and post- exposure) of genes that were classified as being resistant- specific, susceptible specific, and shared (those ESTs detected in both resistant and susceptible specific snail libraries). Thus, 21 transcripts (Table 3) from these 3 categories were selected for quantitative analysis. Quantitative real time RT-PCR was performed using total RNA from nine individual juvenile snails for each time point. Reverse transcription and PCR using one step Full Velocity SYBR Green Q-RT- PCR master mix (Stratagene, CA) was performed as previously described [11]. Reactions were performed in a one- step format with the first stand cDNA synthesis and PCR amplification done in triplicate. Primers specific for B. glabrata housekeeping gene, myoglobin (F: GATGTTCGCCAATGTTCCC; R: AGCGATCAAGTTTCCCCAG) [20] were also used in parallel amplifications to monitor the transcription of this housekeeping relative to that of SSH- ESTs. At the beginning of the study a validation experiment was performed to test amplification efficiencies of the transcripts of interest (ESTs) and myoglobin using corresponding GSPs. Validation experiments were performed using four different RNA template dilutions to confirm that amplification efficiencies (optimized for each primer) were equal between genes of interest and myoglobin (data not shown). The RT-PCR reactions were performed using an Applied Biosystems 7300 Real Time PCR System (Applied Biosystem, Foster City, CA) as described by Myers et al., 2008. The reaction was performed in a one-step format with 80 ng of DNase treated (RQ1 kit, Promega, WI) total RNA. First strand cDNA synthesis and PCR amplification were performed in triplicate using FullVelocity SYBR Green QRT-PCR Master mix (Stratagene, TX) according to the manufacturer’s instructions. The amplification protocol included an initial incubation at 48°C for 45 min for cDNA synthesis and a 95°C initial denaturation for 10 min followed by 40 cycles with 95°C denaturation for 10 sec, and annealing/ amplification at 58°C for 1min. The fluorescent signal was detected at the end of the amplification step of each cycle. For each assay, S. mansoni RNA samples were used in parallel to confirm that the products were snail specific (data not shown) and not parasite derived. Fold changes were calculated by the comparative delta-delta -Ct method (2−ΔΔCt) [38]. Transcript levels corresponding to the genes of interest were normalized relative to myoglobin expression. P- values were calculated by comparing the delta- Ct value (N=9) for each group using a Student’s t-test in order to determine if the differential expression of the specific enriched transcripts between the unexposed and exposed groups (5– 48 h) were significant.
3. Results
3.1 Identification of resistant and susceptible enriched transcripts using the SSH cloning strategies
To identify strain- specific transcripts from either resistant or susceptible snails that may be involved in parasite destruction or survival respectively, the subtracted cDNAs from all SSH libraries cloned as described above, were sequenced. From single-pass direct sequencing ESTs were obtained from between 58- 68% of the clones. Results summarized in Table 1 shows the success rate of sequencing. Thus, from 480 transcript enriched- clones from the juvenile exposed resistant BS-90 (jxBS-90) snails, 282 (58.75%) yielded useful data. In addition, from the 960 and 1,440 enriched clones from the juvenile exposed non- susceptible LAC2 (jxLAC2) and susceptible NMRI (jxNMRI) snails useful information was obtained from 417 (43.44%) and 981 (68.13%) clones, respectively. Average insert sizes from the SSH libraries were as follows: 197 bp for the SSH- juvenile exposed resistant BS-90 (jxBS-90), 244 bp for the SSH- juvenile exposed non- susceptible LAC2 (jxLAC2), and 302 bp for the juvenile exposed susceptible NMRI (jxNMRI) libraries. The average insert size values were calculated from insert size ranges shown in Table 1.
Table 1.
Characteristics of the SSH libraries: number of clones, insert size ranges and number of clones (ESTs) from each library showing either significant sequence matching information, or not, by BLASTX analysis.
| SSH libraries | insert size range (bp) |
Average insert size (bp) |
number of clones |
available ESTs |
Number of clones |
||||
|---|---|---|---|---|---|---|---|---|---|
| BLASTX < 10−4 | BLASTX ≥ 10−4 | ribosomal protein |
hypothetical/ unknown |
no hits | |||||
| juvenile exposed- BS-90 | 73–865 | 197 | 282 | 58.75% | 15.96% | 13.47% | 4.61% | 14.54% | 51.42% |
| juvenile exposed- LAC | 64–788 | 244 | 417 | 43.44% | 35.25% | 11.51% | 24.94% | 2.39% | 25.89% |
| juvenile exposed- NMRI | 64–735 | 302 | 981 | 68.13% | 28.75% | 23.34% | 9.38% | 4.79% | 33.74% |
SSH sequences were compared to those in the public domain (GenBank) using BLASTX with an E-value of less than 1×10−4 considered as significant. From this analysis, results showed several enriched sequences (between 25.89–51.42%) with no significant matches to nucleotide sequences currently deposited in both the GenBank non-redundant database and B. glabrata trace reads, 2.39–14.54% matched hypothetical/ unknown proteins, and 4.61–24.94% showed sequence homology to ribosomal proteins (Table 1).
3.2 Analysis of nucleotide sequences generated from resistant and susceptible SSH cDNA libraries
To evaluate the degree of redundancy and sequence diversity among transcripts enriched using this cloning strategy, we performed multiple sequence alignment analysis of the ESTs generated from the libraries using the Vector NTI software (Invitrogen Life Technologies, CA) [39]. The results revealed 23 contigs of which the six highly redundant contigs, containing 120 ESTs, showed significant similarity to various genes involved in protein binding. Twelve contigs (in total 72 ESTs) had sequences that showed significant similarity to genes involved in biological processes and cell component proteins and 5 contigs containing a total of 103 ESTs showed significant similarity to ribosomal protein. No contig corresponding to ESTs that were classified as ‘no hit’ was detected.
3.3 Biological process classification based on GO assignment
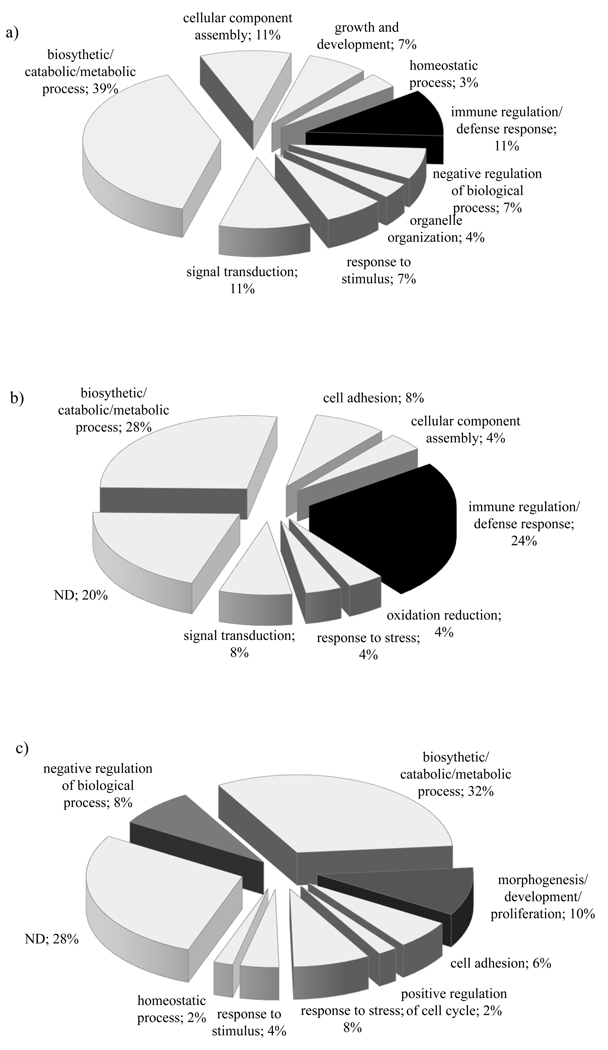
To determine the putative function of sequences categorized either as shared (common) by all the libraries, or specific to the resistant or susceptible enriched libraries we used (as mentioned above) the GO classification scheme (April 2009 release of GO database, Gene Ontology Consortium). Thus, SSH sequences that showed significant similarity to several annotated proteins with functions involved in cellular component assembly, catabolic/ metabolic processes, signal transduction, homeostatic processes, cell adhesion, cell cycle regulation, development/ proliferation, and immune regulation/ defense response were identified. Graphical representations of sequences that were identified as shared or specific for the resistant or susceptible libraries are shown in Figure 1. Analysis of transcripts identified as shared (Fig. 1a) showed most sequences corresponded to genes that function in the snail’s biosynthetic/ catabolic/ metabolic process (39%), cellular component assembly (11%), immune regulation/defense response (11%), signal transduction (11%), growth and development (7%), response to stimulus (7%), negative regulation of biological process (7%) organelle organization (4%), and the homeostatic process (3%). From resistant and susceptible snail - specific SSH-ESTs (Fig. 1b and 1c), we were unable to assign any biological information by GO analysis for 20% and 28% of the sequences, respectively. Analysis of remaining ESTs from the resistant- specific category (Fig. 1b) showed sequences with putative biological function in cell adhesion (8%), signal transduction (8%), stress response (4%), oxidation-reduction (4%), and cellular component assembly (4%). Several resistant specific transcripts were found to be involved in the snail’s immune regulation/ defense response e.g. programmed cell death and endocytosis (24%) (Fig.1b, and Table 2). While none of these defense related transcripts were detected in the susceptible- specific category of SSH-ESTs (Fig 1c), we identified transcripts that are involved in morphogenesis/ development/ proliferation (10%), negative regulation of biological process (8%), response to stress (8%), response to stimulus (4%), cell adhesion (6%) homeostatic process (2%) and positive regulation of the cell cycle (2%) (Fig. 1c, and Table 2).
Figure 1.
Pie chart representations depicting relative percentages of deduced putative gene functions according to their biological process category by Gene ontology of transcripts falling into a) shared b) resistant specific and c) susceptible- specific categories. Note the highly percentage of SSH- ESTs with defense response in resistant, but not in susceptible category of enriched transcripts. *ND = no biological data available
Table 2.
Conclusion of biological process of shared, non- susceptible and susceptible specific transcripts analysed by gene ontology. Note that individual transcripts can have multiple functions.
| Blastx homology (best e-value) | Biological process |
|---|---|
| Shared snail transcripts | |
| actin; B. glabrata (3e-57) | GO:0052171 : growth or development during symbiotic interaction, GO:0044408 : growth or development of symbiont on or near host |
| alpha/ beta-tubulin; Maconellicoccus hirsutus (4e-81) | GO:0006996 : organelle organization, GO:0006928 : cell motion |
| ATP synthase subunits; B. glabrata (2e-18) | GO:0065003 : macromolecular complex assembly, GO:0022607 : cellular component assembly |
| beta-1, 4-endoglucanase; Ampullaria crossean (3e-44) | GO:0009057 : macromolecule catabolic process, GO:0005975 : carbohydrate metabolic process |
| calcium binding protein; B. glabrata (2e-18) | GO:0007166 : cell surface receptor linked signal transduction |
| cathepsin L; Hymeniacidon perlevis (1e-32) | GO:0009057 : macromolecule catabolic process, GO:0006508 : proteolysis |
| cellulase; Haliotis discus (3e-25) | GO:0033692 : cellular polysaccharide biosynthetic process, GO:0009250 : glucan biosynthetic process |
| chitinase; Homo sapiens (8e-24) | GO:0016052 : carbohydrate catabolic process, GO:0043283 : biopolymer metabolic process |
| cytochrome b, c, III ; B. glabrata (6e-83) | GO:0006091 : generation of precursor metabolites and energy, GO:0015980 : energy derivation by oxidation of organic compounds, GO:0006793 : phosphorus metabolic process |
| cytochrome c oxidase subunit I, II, III; B. glabrata (6e-83) | GO:0006091 : generation of precursor metabolites and energy, GO:0015980 : energy derivation by oxidation of organic compounds, GO:0006793 : phosphorus metabolic process |
| DEC-1; Lymnaea stagnalis (1e-29) | GO:0048646 : anatomical structure formation involved in morphogenesis, GO:0031324 : negative regulation of cellular metabolic process |
| DEC-3; Lymnea stagnalis (4e-34) | ND |
| elongation factor 1-gamma/ 2; Aurelia aurita (6e-47) | GO:0044249 : cellular biosynthetic process, GO:0044260 : cellular macromolecule metabolic process, GO:0006414 : translational elongation |
| endo-beta-1,4-glucanase; Ampullaria crossean (7e-33) | GO:0009057 : macromolecule catabolic process, GO:0005975 : carbohydrate metabolic process |
| ezrin/radixin/moesin; B. glabrata (7e-42) | GO:0050794 : regulation of cellular process, GO:0007165 : signal transduction |
| ferritin; Haemaphysalis longicornis (2e-70) | GO:0042592 : homeostatic process, GO:0051235 : maintenance of location |
| fibrinogen C-terminal domain like; Homo sapiens (2e-15) | GO:0048518 : positive regulation of biological process ,GO:0002682 : regulation of immune system process |
| goose-type lysozyme; Chlamys farreri (7e-37) | GO:0016052 : carbohydrate catabolic process, GO:0006810 : transport |
| gram negative bacteria binding protein; B. glabrata (3e-33) | GO:0006952 : defense response, |
| haemoglobin; B. glabrata (1e-125) | GO:0044249 : cellular biosynthetic process, GO:0050794 : regulation of cellular process |
| Fas; immunoGlobulin-like Cell adhesion Molecule; Caenorhabditis elegans (7e-17) | GO:0050794 : regulation of cellular process, GO:0010467 : gene expression |
| laminin receptor; Pinctada fucata (1e-49) | GO:0048513 : organ development, GO:0009888 : tissue development, GO:0048731 : system development |
| C-type lectin; Danio rerio (2e-7) | GO:0048518 : positive regulation of biological process ,GO:0002682 : regulation of immune system process |
| mitochondrial ATP synthase alpha subunit; Aedes aegypti (6e-54) | GO:0065003 : macromolecular complex assembly, GO:0022607 : cellular component assembly |
| mitochondrial ATP synthase; Aedes aegypti (2e-16) | GO:0065003 : macromolecular complex assembly, GO:0022607 : cellular component assembly |
| mucin 5; Homo sapiens (4e-5) | GO:0042221 : response to chemical stimulus, GO:0009991 : response to extracellular stimulus |
| putative mucin-like protein; Aedes aegypti (3e-4) | GO:0042221 : response to chemical stimulus, GO:0009991 : response to extracellular stimulus |
| serine protease alpha/beta; B. glabrata (4e-44) | GO:0009057 : macromolecule catabolic process, GO:0019538 : protein metabolic process |
| serine proteinase inhibitor; Methanosarcina acetivorans (4e-11) | GO:0048519 : negative regulation of biological process, |
| Resistant-specific snail transcripts | |
| BS-90 specific transcripts | |
| activated protein kinase C receptor; Pagrus major (5e-45) | GO:0050794 : regulation of cellular process, GO:0044093 : positive regulation of molecular function, GO:0007165 : signal transduction |
| carboxypeptidase B precursor; Aedes polynesiensis (3e-10) | GO:0009057 : macromolecule catabolic process, GO:0044238 : primary metabolic process |
| CD63 antigen (melanoma 1 antigen); Xenopus spp. (7e-11) | GO:0006897 : endocytosis, GO:0048518 : positive regulation of biological process, GO:0032879 : regulation of localization, |
| countin; Dictyostelium discoideum (6e-17) | GO:0016337 : cell-cell adhesion |
| cytidine deaminase; B. glabrata (2e-6) | GO:0006139 : nucleobase, nucleoside, nucleotide and nucleic acid metabolic process, GO:0043094 : cellular metabolic compound salvage |
| galectin 4-like protein transcript variant; Halio discus hannai (3e-27) | GO:0007155 : cell adhesion |
| heat shock protein 40; Homo sapiens (2e-7) | GO:0006950 : response to stress, GO:0009266 : response to temperature stimulus |
| macrophage expressed gene 1; Homo sapiens (1e-14) | GO:0002472 : macrophage antigen processing and presentation |
| melanogenic peroxidase; Sepia officinalis (2e-10) | GO:0006897 : endocytosis, GO:0016192 : vesicle-mediated transport |
| microfibrillar- associated protein 4; Bos taurus (2e-11) | ND |
| ornithine decarboxylase antizyme; Euprymna scolopes (3e-56) | GO:0006519 : cellular amino acid and derivative metabolic process, GO:0034641 : cellular nitrogen compound metabolic process, GO:0044249 : cellular biosynthetic process |
| putative enoyl-CoA hydratase; Ralstonia spp. (6e-8) | GO:0008152 : metabolic process |
| tyrosine 3- monooxygenase/ tryptophan 5 monoooxigenase; Gallus gallus (6e-20) | GO:0044248 : cellular catabolic process, GO:0034754 : cellular hormone metabolic process |
| LAC2 specific transcripts | |
| antimicrobial protein Achacin precursor; Aplysia punctata (5e-5) | GO:0017144 : drug metabolic process, GO:0017000 : antibiotic biosynthetic process |
| carboxypeptidase A2; Strongylocentrotus purpuratus (1e-13) | GO:0009057 : macromolecule catabolic process, GO:0044238 : primary metabolic process |
| CHK1 checkpoint homolog; S. pombe (2e-16) | GO:0050794 : regulation of cellular process, GO:0007165 : signal transduction, GO:0033554 : cellular response to stress |
| Chondroitin sulfate proteoglycan; Xenopus laevis (3e-13) | GO:0044249 : cellular biosynthetic process, GO:0044262 : cellular carbohydrate metabolic process |
| CPA 4 protein; Strongylocentrotus purpuratus (4e-14) | GO:0003674 : molecular function; GO:0003824 : catalytic activity |
| deoxyribonuclease I; Rattus norvegicus (2e-15) | GO:0012501 : programmed cell death, GO:0022411 : cellular component disassembly |
| ependymin-related protein; Aplysia californica (8e-11) | ND |
| NADH dehydrogenase subunit 5; B. glabrata (6e-11) | GO:0055114 : oxidation reduction, O:0006091 : generation of precursor metabolites and energy |
| paramyosin; S. japonicum (1e-5) | GO:0022607 : cellular component assembly, GO:0048468 : cell development, GO:0030154 : cell differentiation |
| proprotein convertase aPC6C isoform; Branchiostoma californiense (3e-11) | ND |
| putative toxin; Aplysia californai (1e-7) | ND |
| selectin 1; B. glabrata (2e-5) | GO:0051707 : response to other organism, GO:0006952 : defense response, GO:0016337 : cell-cell adhesion |
| Susceptible-specific snail transcripts | |
| NMRI minus both BS-90 and LAC2 | |
| cathepsin B; Araneus ventricosus (6e-44) | GO:0006950 : response to stress, GO:0009611 : response to wounding, GO:0009057 : macromolecule catabolic process |
| cystatin type 2; B. glabrata (4e-5) | GO:0044092 : negative regulation of molecular function |
| low density lipoprotein receptor; Gallus gallus(2e-27) | GO:0009059 : macromolecule biosynthetic process, GO:0043112 : receptor metabolic process |
| myosin light chain 1; Aedes aegypti (5e-35) | GO:0048513 : organ development, GO:0003012 : muscle system process |
| NMRI minus BS-90 | |
| cadmium-metallothionein; Arianta arbustorum[terrestrial snail] (1e-4) | GO:0010035 : response to inorganic substance |
| carbonyl reductase; Danio rerio 1 (2e-22) | GO:0051186 : cofactor metabolic process , GO:0055086 : nucleobase, nucleoside and nucleotide metabolic process, GO:0032504 : multicellular organism reproductio |
| fatty acid binding protein H8- isoform; Cryodraco antarcticus (6e-5) | GO:0006629 : lipid metabolic process |
| galactosylceramidase like; Monodelphis domestica (2e-33) | GO:0016052 : carbohydrate catabolic proces |
| heat shock protein 70; Callinectes sapidus (4e-52) | GO:0010035 : response to inorganic substance, GO:0006950 : response to stress |
| prediced; brain protein; Homo sapiens(1e-9) | ND |
| putative zinc-dependent alcohol dehydrogenase; Lysiphlebus testaceipes (1e-13) | GO:0006066 : cellular alcohol metabolic process |
| selenoprotein Pb precursor (6e-4) | GO:0006950 : response to stress |
| solute carrier family 25; mitochondrial carrier; Danio rerio (8e-5) | ND |
| translationally controlled tumor protein; Lateolabrax japonicus (2e-12) | GO:0048519 : negative regulation of biological process, GO:0060548 : negative regulation of cell death |
| tyrosinase; Sepia officinalis (3e-5) | GO:0008283 : cell proliferation |
| NMRI minus LAC2 | |
| adenosine deaminase; Aedes aegypti (5e-14) | GO:0048519 : negative regulation of biological process, GO:0060548 : negative regulation of cell death |
| adenosyl homocysteinase; Stenotrophomonas sp. (8e-16) | GO:0006519 : cellular amino acid and derivative metabolic process, GO:0044248 : cellular catabolic process |
| adhesin lipoprotein (9e-6) | GO:0007155 : cell adhesion |
| alginase; Haliotis discus (3e-4) | GO:0005975 : carbohydrate metabolic process |
| alpha amylase; Crassostrea gigas (2e-15) | GO:0005975 : carbohydrate metabolic process |
| aminoglycoside-3: adenyltransferase; synthetic construct (1e-119) | GO:0044249 : cellular biosynthetic process, GO:0034641 : cellular nitrogen compound metabolic process |
| atrial gland-specific antigen precursor; Aplysia californica (3e-35) | ND |
| calreticulin 3; Mus musculus (8e-5) | GO:0048518 : positive regulation of biological process, GO:0045787 : positive regulation of cell cycle |
| carboxypeptidase D; Mus musculus (8e-8) | GO:0009057 : macromolecule catabolic process, GO:0043283 : biopolymer metabolic process |
| caveolin; Xenopus laevis (1e-16) | ND |
| crystal structure of Rhogdi ; Aedes aegypti (2e-20) | ND |
| cys-rich cocoon protein; Theromyzon rude (4e-6) | ND |
| developmentally regulated albumen gland; B.glabrata (9e-31) | ND |
| dopamine beta hydroxylase-like protein; Aplysia californica (6e-4) | GO:0007267 : cell-cell signaling, GO:0050877 : neurological system process |
| elastase 2A; Homo sapiens (6e-10) | GO:0080134 : regulation of response to stress, GO:0031348 : negative regulation of defense response |
| ependymin-related protein; Aplysia californica (8e-11) | ND |
| eukaryotic translation initiation factor 4A; Homo sapiens (3e-33) | GO:0010035 : response to inorganic substance, GO:0051789 : response to protein stimulus |
| fasciclin-like protein; Aplysia californai (2e-8) | GO:0044262 : cellular carbohydrate metabolic process, GO:0010382 : cellular cell wall macromolecule metabolic process |
| Flp recombinase; Saccharomyces cerevisiae (6e-8) | ND |
| fucosidase, alpha L 1 tissue; Danio rerio (5e-66) | GO:0016052 : carbohydrate catabolic proces |
| glutamine synthetase; Caenorhabditis elegans (2e-24) | GO:0006519 : cellular amino acid and derivative metabolic process |
| glutathione- S- transferase; Haliotis discus discus (1e-14) | GO:0006970 : response to osmotic stress, GO:0051186 : cofactor metabolic process |
| insulin responsive sequence DNA binding protein; Homo sapiens (3e-5) | GO:0016486 : peptide hormone processing, GO:0043283 : biopolymer metabolic process |
| lactase-phlorizin hydrolase; Strongylocentrotus purpuratus (8e-8) | GO:0008152 : metabolic process |
| legumain; Danio rerio (4e-14) | GO:0009057 : macromolecule catabolic process, GO:0019538 : protein metabolic process |
| lipophorin receptor; Leucophaea maderae (8e-27) | ND |
| mantle gene2; Pinctada fucata (2e-20) | GO:0009653 : anatomical structure morphogenesis, GO:0048513 : organ development, GO:0048731 : system development |
| matrilin; B. glabrata (1e-9) | ND |
| phospholipase A2; Mus musculus (4e-6) | GO:0016042 : lipid catabolic process, GO:0050794 : regulation of cellular process, GO:0007165 : signal transduction, GO:0031323 : regulation of cellular metabolic proces |
| poly A binding protein; Apis mellifera (4e-5) | ND |
| putative tumor suppressor; Suberites domunc (3e-12) | ND |
| signal sequence receptor; Strongulocentrotus purpuratus (1e-8) | GO:0042221 : response to chemical stimulus |
| surfactant protein D; Homo sapiens (5e-6) | GO:0042592 : homeostatic process |
| theromacin; Theromyzon tessulatum (5e-5) | ND |
| thiamine pyrophosphate enzyme-like TPP binding protein; Burkholderia phytofirmans (7e-8) | GO:0008150: biological process; ND |
| xylanase; Clostridium thermocellum (1e-21) | GO:0016052 : carbohydrate catabolic process |
ND = no biological data available
3.4 Analysis by quantitative real time RT-PCR of representative clones identified by SSH cloning strategy in snails exposed to normal miracidia
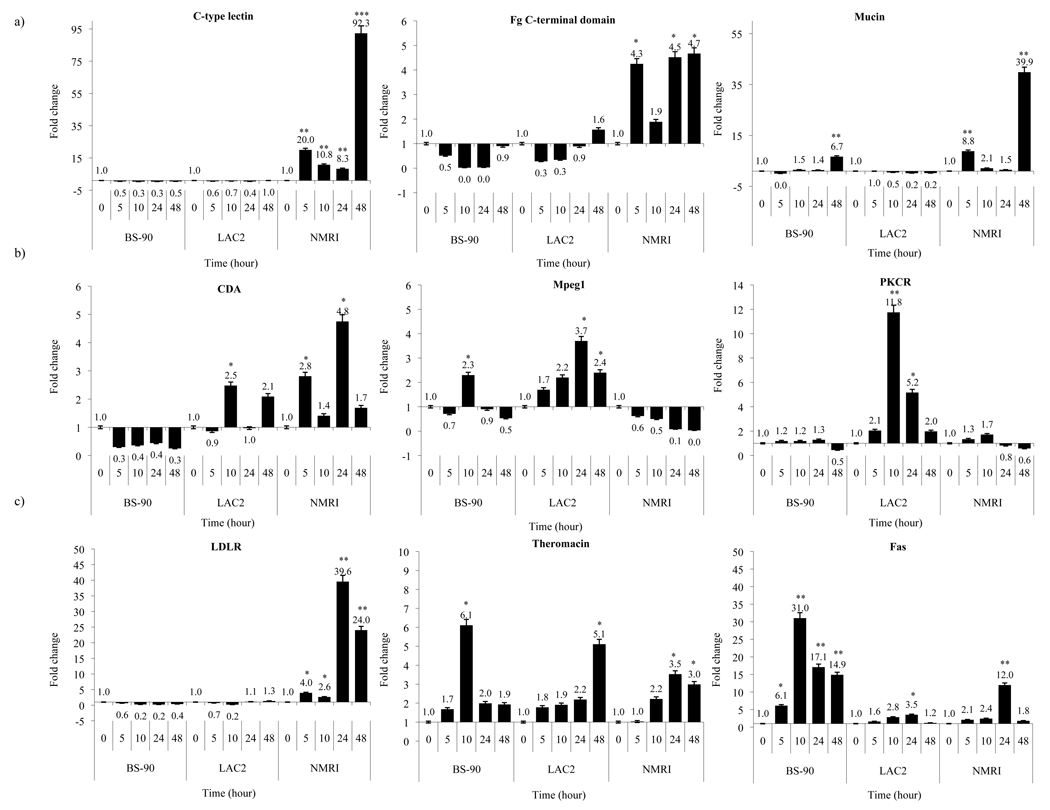
To determine changes in gene- expression between 5 to 48 h post- exposure in resistant (BS-90), non-susceptible (LAC2) and susceptible (NMRI) snails, real time Q-RT-PCR using primer sets shown in Table 3 was performed. Out of the 21 transcripts that we analyzed (Table 3), 12 transcripts, including ferritin, Hsp 70, and cathepsin B (11, 17) were modulated (up/and down regulated) after parasite exposure, and the remaining showed random expression in all the snails 5–24 h post exposure (data not shown). In Figure 2, results using this assay to measure fold- difference (induction) for transcripts encoding, the 3 shared transcripts (Fig. 2a), C- type lectin, Fg C- terminal domain, and mucin and representative resistant-specific transcripts, CDA, Mpeg1 and PKCR (Fig. 2b) are shown. In Figure 2c, fold change in gene expression of three representative susceptible- specific transcripts related to LDLR, theromacin, and Fas in all the snails pre- (0) and post - (5, 10, 24 and 48 h) exposure to normal miracidia are also presented. Results with cDNA templates prepared using individual snails showed that for the shared transcripts, C- type lectin, Fg C- terminal domain and mucin, all were induced early in the susceptible but not in resistant or non- susceptible snails. Thus, significant induction of C- type lectin occurred early (20 fold change at 5 h) and remained dramatically up-regulated (92.3 fold) at 48 h post exposure in the susceptible snail. Also in this snail stock, the early (5 h) up- regulation of transcripts related to Fg C- terminal domain and mucin (4.3 and 8.8 fold change, respectively) remained up- regulated (4.7 and 39.9 fold change, respectively) at 48 h post- exposure. For the representative resistant transcripts (Fig. 2b), CDA, Mpeg1, and PKCR, while no induction was detected in the BS-90 resistant snail for the transcript related to CDA, low induction (2.5 fold) was detected at 10 h post exposure in the non- susceptible LAC2 snail and also in the susceptible NMRI snail. For the Mpeg1 early (5 and 10 h) but low induction was detected only in the resistant (BS-90; 2.3 fold) and non- susceptible (LAC2; 1.7 fold) stocks. For the resistant specific transcript related to PKCR, significant up-regulation was triggered by infection in the non-susceptible LAC2 snail while levels in the resistant (BS-90) and NMRI susceptible snail remained unchanged at all time points (5 to 48 h) examined post-exposure. In the LAC2 non- susceptible snail, the level of the PKCR transcript showed a 2.1 fold change early (5 h) after exposure, increasing (11.8 fold) at 10 h post exposure and gradually declining after 24 h (5.2 fold) and 48 h (2.0 fold) post exposure. For the susceptible specific transcripts (Fig. 2c), LDLR, theromacin and Fas, early (5 h; 4.0 fold) and late (24 and 48 h post- exposure) up- regulation (39.6 and 24.0 fold, respectively) of LDLR was detected in the NMRI but not in the BS-90 and LAC2 snails. While the transcript related to theromacin increased early in all the snails, significant up- regulation of the transcript related to Fas was observed upon early exposure to miracidia especially in the resistant snail. Thus, in BS-90 upon early post exposure (from 5 to 10 h) there was a rapid increase from 6.1 fold change to a 31.0 fold increase of this transcript. In comparison to the other two snails (non- susceptible LAC2 and susceptible NMRI), early parasite exposure produced no significant up- regulation of the Fas transcript but an increase was detected, especially in the susceptible NMRI snail, 24 h post exposure (12.0 fold). Unlike the resistant snail, however, up- regulation of this transcript in the NMRI susceptible snail was not sustained and the level declined at 48 h post- exposure.
Figure 2.
Fold difference of gene expression after normal miracidia exposure in BS-90, LAC and NMRI stocks of B. glabrata evaluated by real time Q-RT-PCR for 0, 5, 10, 24 and 48 h post- exposure. a) Fold differences in transcript expressions of shared transcripts (C-type lectin, FgC- terminal domain and mucin, b) resistant specific transcripts (CDA, Mpeg1 and PKCR) and c) susceptible specific transcripts (LDLR, theromacin and Fas) were calculated by comparative delta-delta Ct method using the housekeeping transcript encoding myoglobin to normalize transcript levels. The significance of gene expression between unexposed and exposed groups was calculated by Student’s t-test. P-values of < 0.05 and < 0.01 are indicated by * and ** at the top of each error bar, respectively.
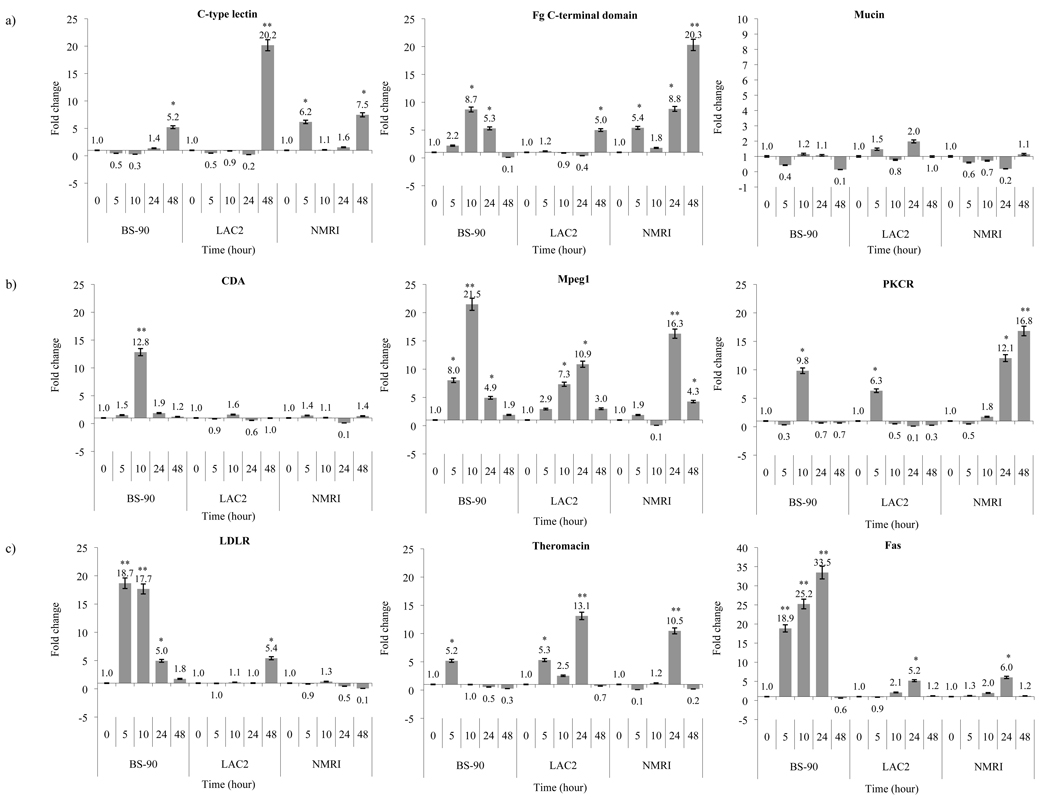
3.5 Analysis by quantitative real time RT- PCR of representative clones identified by SSH cloning strategy in snails exposed to attenuated miracidia
To determine whether changes observed in the expression of representative SSH- transcripts are due to the infection process rather than to any molecular event(s) associated with the intra-dermal penetration of the parasite into the snail, RNA samples prepared from snails exposed to attenuated miracidia were utilized for Q-RT -PCR. Results shown in Figure 3 indicate differences in induction of the 9 transcripts in resistant (BS-90), non- susceptible (LAC2) and susceptible (NMRI) snails pre -(0) and post-exposure (5, 10, 24, 48 h) to attenuated miracidia. Thus, using primer sets for the shared transcripts (Fig. 3a) matching C- type lectin, Fg C- terminal domain, and mucin showed that upon exposure to attenuated miracidia, there was no early (5 to 10 h) induction of the C- type lectin related transcript in either the resistant or non- susceptible snail. Although attenuated miracidia failed to trigger the early induction of this gene in these snails (resistant and non- susceptible) there was an increase in expression of this transcript in both snails (5.2 fold in BS-90 and 20.2 fold in LAC2) after late exposure (48 h) to the attenuated parasites. In contrast, up- regulation of C- type lectin related transcript (6.2 fold) occurred early (5 h) as well as late (7.5 fold at 48 h post- exposure) in the susceptible snail following exposure to attenuated miracidia. However, induction of this transcript (C- type lectin) was lower than levels detected within the same time period after exposure of the susceptible NMRI snail to normal miracidia (Fig. 2a). For the gene related to Fg C- terminal domain, however, significant higher induction of this transcript was observed in all the snails, especially following longer periods of exposure (from 24 to 48 h) to attenuated miracidia with some differences in expression in the resistant (BS-90) and non-susceptible LAC2 snails. In the case of the shared transcript related to mucin, results indicated that attenuated miracidia failed to trigger the induction of this gene in the susceptible snail contrary to the significant induction observed when these snails were exposed to normal miracidia (Fig. 2a).
Figure 3.
Fold difference of gene expression after irradiated miracidia exposure in BS-90, LAC and NMRI stocks of B. glabrata evaluated by quantitative real time PCR for 0, 5, 10, 24 and 48 h post- exposure. Fold differences in gene expressions of a) shared transcripts (C-type lectin, FgC- terminal domain and mucin; b) resistant specific transcripts (CDA, Mpeg1 and PKCR and c) susceptible specific transcripts (LDLR, theromacin and Fas) were calculated by comparative delta-delta Ct method using the housekeeping transcript encoding myoglobin to normalize transcript levels. The significance of gene expression between unexposed and exposed groups was calculated by Student’s t-test. P-values of < 0.05 and < 0.01 are indicated by * and ** at the top of each error bar, respectively.
Examination of the fold change of resistant specific transcripts, CDA, Mpeg1, and PKCR in the resistant, non- susceptible and susceptible snails following exposure to attenuated miracidia, showed (Fig. 3b) that the transcript encoding CDA while not up-regulated in LAC2 (non-susceptible) and NMRI (susceptible) was, however, induced (12.8 fold) in the BS-90 (resistant) snail after 10hr exposure. The transcripts related to Mpeg1 and PKCR were also up- regulated upon exposure to attenuated parasites (in all the snails) with even higher levels of induction than when these snails were exposed for the same time periods to normal miracidia (Fig. 2b).
Interestingly for the transcript related to PKCR, the up- regulation observed following the exposure of the susceptible snail to attenuated miracidia was substantially more significant (16.8 fold at 48 h post- exposure) than induction levels observed within the same time period post- exposure to normal miracidia (Fig. 2c). The increase in these transcripts following exposure to attenuated but not to normal miracidia may indicate that their specific involvement in molecular events associated with the parasite penetrating process of the snail e.g wound-healing and not to the early phase of parasite development per se.
Examination of the kinetics of early versus late induction of the representative transcripts categorized as susceptible specific (LDLR, theromacin, Fas) in the resistant, non- susceptible and susceptible snails upon exposure to irradiated miracidia by RT-Q-PCR is shown in Figure 3c. In the case of the transcript matching the LDLR, early induction (18.7 fold change at 5 h and 17.7 fold change at 10 post-exposure) of this transcript was observed in the resistant snail in response to the irradiated parasite. The up-regulation of this transcript (LDLR) while not observed in LAC2 snails after exposure to normal parasites (Fig. 2c) was, however, seen in these snails following 48 hr exposure (5.4 fold) to attenuated miracidia (Fig. 3c). Also in contrast to the up- regulation of the LDLR related transcript seen early in the susceptible snail when exposed to normal parasites (Fig. 2c) no similar induction of this transcript occurred when these snails were exposed instead for the same time periods to attenuated miracidia (Fig. 3c). For the transcripts related to theromacin and Fas, however, the induction pattern remained relatively unchanged in the snails when exposed either to normal (Fig. 2c) or attenuated miracidia; indicting that molecular mechanisms related to the penetration process may also elicit expression of these genes in the snail.
Discussion
In this study we have demonstrated the efficacy of using the SSH- cloning strategy to identify transcripts that are differentially up- regulated between resistant (BS-90), non- susceptible (LAC2), and susceptible (NMRI) juvenile B. glabrata snails upon early exposure to S. mansoni. As other differential gene expression studies (some also using the same SSH cloning approach) have shown, several of the enriched ESTs isolated from the SSH libraries we constructed failed to show similarity to nucleotide sequences currently deposited in GenBank and trace reads of B. glabrata genomic DNA. Until the genome of B. glabrata is sequenced and annotated, a project that is still underway [40], it will remain difficult to deduce with any degree of certainty the identity and putative function of these induced but yet ‘unknown’ transcripts in relation to the snail/ schistosome interaction.
A considerable number of the SSH- EST sequences (Tables 1 and Fig. 1), alignment analyzed by BlastX, did show significant hits to sequences in GenBank. These enriched sequences were classified as shared, resistant specific (present only in the resistant library), or susceptible specific (identified only in the susceptible library). Shared transcripts, most likely comprising high- copy abundant transcripts insufficiently subtracted under the hybridization conditions employed, consisted of genes involved in diverse molecular functions pertaining to biosynthetic/ catabolic/ metabolic processes, cellular component assembly, growth/ development, organelle organization, signal transduction, homeostatic process, immune regulation/ negative regulation biological process and response to stimulus. Although transcripts belonging to different functional groups were detected within both resistant- and susceptible- specific categories, significantly 24% of the genes involved in immune defense were identified among the resistant SSH- ESTs, but absent in susceptible SSH-ESTs. In the latter category, however, a significantly higher number of transcripts related to morphogenesis/ development/ proliferation (10%), negative regulation of biological process (8%) and response to stress/ other stimulus (12%) were instead identified, indicating that upon infection of susceptible snails, the parasite may manipulate the snail to enable its successful development and proliferation.
To confirm that the subtractive cloning approach had indeed met with the objective of enriching for transcripts that underlie either parasite- snail resistance or susceptibility, we undertook both qualitative and quantitative RT- PCR. Selected representative transcripts selected (nine SSH- ESTs) from the above categories (shared, resistant snail specific, and susceptible snail specific) were used for validation. Thus, from semi quantitative RT-PCR analysis (data not shown) with gene expression levels normalized against the basal transcript level of the housekeeping gene, myoglobin, transcripts showing significant homology to genes encoding C- type lectin, Fg C terminal domain and mucin were identified as being dominant immediate response genes in all the three snail stocks (transcript levels 5.8 to 16.5- normalized against myoglobin expression), resistant (BS-90), non- susceptible (LAC2) and susceptible (NMRI). With transcripts categorized as resistant specific, those similar to CDA, Mpeg1, and PKCR occurred at higher levels in the resistant (BS-90) and non- susceptible (LAC2) snails than in the susceptible snail. Likewise, expression levels of SSH- ESTs identified as susceptible- specific (LDLR, theromacin and Fas) were found to be higher in the susceptible (NMRI) than in either the resistant (BS-90) or non-susceptible (LAC2) snail (data not shown).
From quantitative real-time Q-RT-PCR (Fig. 2), we studied the transcript induction at pre and post- exposure (5 to 48 h) to normal miracidia. Thus, in resistant (BS-90) and non- susceptible (LAC2) snails, genes showing similarity to PKCR, Mpeg1, Fas and theromacin were all induced early (5 to 10 h) after parasite exposure. Likewise, in the susceptible snail (NMRI), transcripts encoding LDLR, mucin, C- type lectin and Fg C terminal domain were induced early and remained dramatically up- regulated even after 48 h post exposure.
In contrast, when all three snail stocks (resistant, non- susceptible and susceptible) were instead exposed to attenuated miracidia (Fig. 3) some of these transcripts, specifically, C- type lectin, mucin, and LDLR, were no longer up- regulated, while transcripts, such as Mpeg1, PKCR and CDA, theromacin, Fg C- terminal domain, and Fas remained intensely up- regulated following exposure to the attenuated parasite; an indication that these transcripts, in particular, may also be induced by molecular events associated specifically with the parasite penetration process into the snail-host, such as wound -repair. Based on these results it is possible that the B. glabrata homologue of theromacin, a cysteine-rich anti-microbial peptide [41] may be triggered in response to both challenges, infection and injury, as may the expression of genes for Fg C domain and Fas. Both these transcripts may function in adhesion/ inflammatory responses towards the schistosome pathogen and in wound repair. Resistant- specific transcripts; homologues for PKCR and Mpeg1 that remained up- regulated even in response to attenuated miracidia may, likewise, play dual roles in mechanisms related to parasite destruction and healing. From BLASTX analysis, the sequence of the B. glabrata SSH- EST encoding Mpeg-1 was found to be homologous to the abalone perforin- like protein that has been described as a regulatory protein involved in innate defence in this mollusk [42].
From induction responses observed in the susceptible snail exposed to either attenuated or normal parasites, we can conclude that for this stock, however, the SSH-ESTs similar to LDLR, C- type lectin, and mucin, are triggered only by normal, but not attenuated miracidia. These results indicate that the parasite and not molecular events related to the parasite intradermal penetration process provides the stimulus for specific induction of these three genes in this snail.
Lately, it has been shown that schistosome mucins may function as important molecular determinants in compatible/ incompatible outcomes of the parasite/ snail host interaction [27]. Thus, it may not be entirely surprising to observe that mucin is also highly induced (during early and late infection, Fig. 2a) by the parasite in the susceptible/ compatible (NMRI) but not in resistant/ incompatible (BS-90 and LAC2) snails. Preliminary results from Northern blot hybridization using the mucin related SSH- EST as probe have shown the existence of multiple and variable sized transcripts between the resistant and susceptible snail stocks (data not shown). Follow up studies will focus on sequencing these diverse mucin transcripts from resistant and susceptible snails in an attempt to determine if the expression of different snail mucin isoforms correlates with the new model proposed for the involvement of this gene in defining or undermining snail- schistosome relationships [43].
Lectins have been shown to play a pivotal role in ‘self’ / ‘non- self’ recognition in B. glabrata/ trematode interactions. For example, the transcripts encoding the fibrinogen related lectin (FREPs) containing immunoglobulin- like domains was shown to be up- regulated in the snail in response to trematode infection [22–26]. Since mollusks lack antibodies it is believed that lectins as major carbohydrate binding proteins, are important for the recognition and binding of invading pathogens. It is possible, that despite their known role in both vertebrate and invertebrate innate defense [5, 7, 44] the significant up- regulation of the C- type lectin homologue (calcium dependent carbohydrate binding protein) by the parasite, in susceptible but not resistant and non- susceptible snails, may point to a role whereby the over expression of this gene (specifically in the susceptible snail) may effectively prevent the binding of hemocytes to the parasite thus circumventing their destruction in the snail host. In a previous study [41], over- expression of the B. glabrata homologue of the lectin, selectin/ dematospodin, in echinostome susceptible but not resistant snails was described as also possibly reflecting an immune evasion tactic preventing parasite recognition/ binding in the snail. C- lectins have also been shown to function in cell adhesion and agglutination and are highly expressed in the hepatopancreas and haemocytes of other mollusks in response to microbial infection [41]. Therefore, the induction of this gene following exposure (48 h post- exposure, Fig. 3) of resistant and non- susceptible snails to attenuated miracidia could point to a role in wound healing following the parasite’s penetration into the snail involving C- lectin. Efforts are currently underway to express the B. glabrata C- type lectin orthologue to determine the tissue specificity of expression, and the temporal induction of protein in these snail stocks after exposure to either normal or irradiated parasites.
The strong up- regulation of the transcript related to LDLR (Fig. 2c) in response to infection (by normal parasite) in susceptible but not resistant and non- susceptible snails may be required to boost the uptake of host low density lipoprotein (LDL) that may be required for the development and survival of the parasite in the snail host. The immune evasion of schistosomes by masking via the acquisition of host LDL has been well documented [45–47]. Although schistosomes possess their own receptor for binding this important ligand [48, 49], involvement of a snail- host receptor in lipid homeostasis (cholesterol) during infection remains to be studied.
It will be interesting to further investigate the significance of the up- regulation of all these transcripts, especially those that were induced in the snail stocks (resistant and susceptible) in response to normal but not attenuated schistosomes by gene- silencing technologies, such as siRNA. From these ‘knock down’ studies we will be in a better position to assess whether blocking individual gene expression can indeed provide us with candidate gene targets in the snail that can be used in a realistic approach towards the development of alternative novel intervention tools to reduce schistosomiasis transmission.
In summary, using the SSH- cDNA cloning strategy, several enriched transcripts from libraries constructed from juvenile resistant, non- susceptible and susceptible snails shortly after exposure to normal miracidia have been isolated. Results also showed differences in gene expression between the snail strains in response to infection indicating differences in gene regulation between the snails, even between resistant (BS-90) and non-susceptible (LAC 2) snails. Further validation of the differential regulation of representative SSH- ESTs in response to either normal or attenuated parasites has revealed for the first time that the induction of transcripts triggered during the parasite penetration process into the soft body parts of the snail may be important in assessing changes in the gene expression profiles between resistant and susceptible snails after exposure to S. mansoni.
Acknowledgements
We would like to thank Drs. Nithya Raghavan and Fred Lewis for their helpful suggestions and critique of the manuscript, Dr. Peter FitzGerald for help with some of the bioinformatics, Pat Caspar for irradiating the miracidia, and Frances Barnes for technical assistance. This work was funded by a grant from NIH-NIAID R01-AI063480
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Note: Nucleotide sequence data reported in this paper have been submitted to GenBank with the following Accession nos.: GH716011- GH716292 (BS-90 specific ESTs), GH716293-GH716709 (LAC specific ESTs) and GH716710-GH717690 (NMRI specific ESTs).
References
- 1.Chitsulo L, Loverde P, Engels D. Schistosomiasis. Nat Rev Microbiol. 2004;2:12–13. doi: 10.1038/nrmicro801. [DOI] [PubMed] [Google Scholar]
- 2.Gryseels B, Polman K, Clerinx J, Kestens L. Human schistosomiasis. Lancet. 2006;368:1106–1118. doi: 10.1016/S0140-6736(06)69440-3. [DOI] [PubMed] [Google Scholar]
- 3.Dongbao Y, Ross A, Musheng X, Yuesheng L, Yan C. Highlights on the World Bank Loan Schistosomiasis Control Program in China (1991–1998): a special focus on Human Province. Southeast Asian J Trop Med Public Health. 1999;30:657–663. [PubMed] [Google Scholar]
- 4.Sumanth S, D'Souza PE, Jagannath MS. A study of nasal and visceral schistosomosis in cattle slaughtered at an abattoir in Bangalore, South India. Rev Sci Tech. 2004;23:937–942. doi: 10.20506/rst.23.3.1537. [DOI] [PubMed] [Google Scholar]
- 5.Yoshino TP, Dinguirard N, Kunert J, Hokke CH. Molecular and functional characterization of a tandem-repeat galectin from the freshwater snail Biomphalaria glabrata, intermediate host of the human blood fluke Schistosoma mansoni. Gene. 2008;411:46–58. doi: 10.1016/j.gene.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coppin JF, Lefebvre C, Caby S, Cocquerelle C, Vicogne J, Coustau C, et al. Gene expression changes in Schistosoma mansoni sporocysts induced by Biomphalaria glabrata embryonic cells. Parasitol Res. 2003;89:113–119. doi: 10.1007/s00436-002-0643-2. [DOI] [PubMed] [Google Scholar]
- 7.Zhang SM, Zeng Y, Loker ES. Characterization of immune genes from the schistosome host snail Biomphalaria glabrata that encode peptidoglycan recognition proteins and gram-negative bacteria binding protein. Immunogenetics. 2007;59:883–898. doi: 10.1007/s00251-007-0245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lockyer AE, Noble LR, Rollinson D, Jones CS. Schistosoma mansoni: resistant specific infection-induced gene expression in Biomphalaria glabrata identified by fluorescent-based differential display. Exp Parasitol. 2004;107:97–104. doi: 10.1016/j.exppara.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 9.Lockyer AE, Spinks JN, Walker AJ, Kane RA, Noble LR, Rollinson D, et al. Biomphalaria glabrata transcriptome: identification of cell-signalling, transcriptional control and immune-related genes from open reading frame expressed sequence tags (ORESTES) Dev Comp Immunol. 2007;31:763–782. doi: 10.1016/j.dci.2006.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lockyer AE, Spinks J, Noble LR, Rollinson D, Jones CS. Identification of genes involved in interactions between Biomphalaria glabrata and Schistosoma mansoni by suppression subtractive hybridization. Mol Biochem Parasitol. 2007;151:18–27. doi: 10.1016/j.molbiopara.2006.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Myers J, Ittiprasert W, Raghavan N, Miller A, Knight M. Differences in cysteine protease activity in Schistosoma mansoni-resistant and -susceptible Biomphalaria glabrata and characterization of the hepatopancreas cathepsin B Full-length cDNA. J Parasitol. 2008;94:659–668. doi: 10.1645/GE-1410R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bender RC, Broderick EJ, Goodall CP, Bayne CJ. Respiratory burst of Biomphalaria glabrata hemocytes: Schistosoma mansoni-resistant snails produce more extracellular H2O2 than susceptible snails. J Parasitol. 2005;91:275–279. doi: 10.1645/GE-415R. [DOI] [PubMed] [Google Scholar]
- 13.Humphries JE, Yoshino TP. Regulation of hydrogen peroxide release in circulating hemocytes of the planorbid snail Biomphalaria glabrata. Dev Comp Immunol. 2008;32:554–562. doi: 10.1016/j.dci.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitta G, Galinier R, Tisseyre P, Allienne JF, Girerd-Chambaz Y, et al. Gene discovery and expression analysis of immune-relevant genes from Biomphalaria glabrata hemocytes. Dev Comp Immunol. 2005;29:393–407. doi: 10.1016/j.dci.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 15.Vergote D, Bouchut A, Sautiere PE, Roger E, Galinier R, Rognon A, et al. Characterisation of proteins differentially present in the plasma of Biomphalaria glabrata susceptible or resistant to Echinostoma caproni. Int J Parasitol. 2005;35:215–224. doi: 10.1016/j.ijpara.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Guillou F, Roger E, Mone Y, Rognon A, Grunau C, Theron A, et al. Excretory-secretory proteome of larval Schistosoma mansoni and Echinostoma caproni, two parasites of Biomphalaria glabrata. Mol Biochem Parasitol. 2007;155:45–56. doi: 10.1016/j.molbiopara.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 17.Ittiprasert W, Nene R, Miller A, Raghavan N, Lewis F, Hodgson J, Knight M. Schistosoma mansoni infection of juvenile Biomphalaria glabrata induces a differential stress response between resistant and susceptible snails. Exp Parasitol. 2009;123:203–211. doi: 10.1016/j.exppara.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loker ES, Bayne CJ, Buckley PM, Kruse KT. Ultrastructure of encapsulation of Schistosoma mansoni mother sporocysts by hemocytes of juveniles of the 10-R2 strain of Biomphalaria glabrata. J Parasitol. 1982;68:84–94. [PubMed] [Google Scholar]
- 19.Miller AN, Raghavan N, FitzGerald PC, Lewis FA, Knight M. Differential gene expression in haemocytes of the snail Biomphalaria glabrata: effects of Schistosoma mansoni infection. Int J Parasitol. 2001;31:687–696. doi: 10.1016/s0020-7519(01)00133-3. [DOI] [PubMed] [Google Scholar]
- 20.Raghavan N, Miller AN, Gardner M, FitzGerald PC, Kerlavage AR, Johnston DA, et al. Comparative gene analysis of Biomphalaria glabrata hemocytes pre- and post-exposure to miracidia of Schistosoma mansoni. Mol Biochem Parasitol. 2003;126:181–191. doi: 10.1016/s0166-6851(02)00272-4. [DOI] [PubMed] [Google Scholar]
- 21.Nowak TS, Woodards AC, Jung Y, Adema CM, Loker ES. Identification of transcripts generated during the response of resistant Biomphalaria glabrata to Schistosoma mansoni infection using suppression subtractive hybridization. J Parasitol. 2004;90:1034–1040. doi: 10.1645/GE-193R1. [DOI] [PubMed] [Google Scholar]
- 22.Adema CM, Hertel LA, Miller RD, Loker ES. A family of fibrinogen-related proteins that precipitates parasite-derived molecules is produced by an invertebrate after infection. Proc Natl Acad Sci USA. 1997;94:8691–8696. doi: 10.1073/pnas.94.16.8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang SM, Loker ES. Representation of an immune responsive gene family encoding fibrinogen-related proteins in the freshwater mollusc Biomphalaria glabrata, an intermediate host for Schistosoma mansoni. Gene. 2004;341:255–266. doi: 10.1016/j.gene.2004.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang SM, Zeng Y, Loker ES. Expression profiling and binding properties of fibrinogen-related proteins (FREPs), plasma proteins from the schistosome snail host Biomphalaria glabrata. Innate Immun. 2008;14:175–189. doi: 10.1177/1753425908093800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang SM, Adema CM, Kepler TB, Loker ES. Diversification of Ig superfamily genes in an invertebrate. Science. 2004;305:251–254. doi: 10.1126/science.1088069. [DOI] [PubMed] [Google Scholar]
- 26.Hertel LA, Adema CM, Loker ES. Differential expression of FREP genes in two strains of Biomphalaria glabrata following exposure to the digenetic trematodes Schistosoma mansoni and Echinostoma paraensei. Dev Comp Immunol. 2005;29:295–303. doi: 10.1016/j.dci.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Roger E, Mitta G, Mone Y, Bouchut A, Rognon A, Grunau C, et al. Molecular determinants of compatibility polymorphism in the Biomphalaria glabrata/Schistosoma mansoni model: new candidates identified by a global comparative proteomics approach. Mol Biochem Parasitol. 2008;157:205–216. doi: 10.1016/j.molbiopara.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Roger E, Grunau C, Pierce RJ, Hirai H, Gourbal B, Galinier R, et al. Controlled Chaos of Polymorphic Mucins in a Metazoan Parasite (Schistosoma mansoni) Interacting with Its Invertebrate Host (Biomphalaria glabrata) PLoS Negl Trop Dis. 2008;2:e330. doi: 10.1371/journal.pntd.0000330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards CS. Genetic factors in susceptibility of Biomphalaria glabrata for different strains of Schistosoma mansoni. Parasitology. 1975;70:231–241. doi: 10.1017/s0031182000049696. [DOI] [PubMed] [Google Scholar]
- 30.Richards CS, Minchella DJ. Transient non-susceptibility to Schistosoma mansoni associated with atrial amoebocytic accumulations in the snail host Biomphalaria glabrata. Parasitology. 1987;95:499–505. doi: 10.1017/s0031182000057929. [DOI] [PubMed] [Google Scholar]
- 31.Niemann GM, Lewis FA. Schistosoma mansoni: influence of Biomphalaria glabrata size on susceptibility to infection and resultant cercarial production. Exp Parasitol. 1990;70:286–292. doi: 10.1016/0014-4894(90)90110-x. [DOI] [PubMed] [Google Scholar]
- 32.Lie KJ, Jeong KH, Heyneman D. Acquired resistance in snails. Induction of resistance to Schistosoma mansoni in Biomphalaria glabrata. Int J Parasitol. 1983;13:301–304. doi: 10.1016/0020-7519(83)90041-3. [DOI] [PubMed] [Google Scholar]
- 33.Azevedo CM, Borges CC, Andrade ZA. Changes induced in Biomphalaria glabrata (Say, 1818) following trials for artificial stimulation of its internal defense system. Mem Inst Oswaldo Cruz. 2006;101:199–203. doi: 10.1590/s0074-02762006000900031. [DOI] [PubMed] [Google Scholar]
- 34.Cooper LA, Richards CS, Lewis FA, Minchella DJ. Schistosoma mansoni: relationship between low fecundity and reduced susceptibility to parasite infection in the snail Biomphalaria glabrata. Exp Parasitol. 1994;79:21–28. doi: 10.1006/expr.1994.1055. [DOI] [PubMed] [Google Scholar]
- 35.Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci USA. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Zhao Y, Zhang LY, Liu CZ, Zhou XH. Full-length of human papillomavirus type 18L1 gene optimized using the plant prefered condons and synthesized by overlapping PCR. Nan Fang Yi Ke Da Xue Xue Bao. 2009;29:387–392. [PubMed] [Google Scholar]
- 40.Raghavan N, Knight M. The snail (Biomphalaria glabrata) genome project. Trends Parasitol. 2006;22:148–151. doi: 10.1016/j.pt.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 41.Tasiemski A, Vandenbulcke F, Mitta G, Lemoine J, Lefebvre C, Sautiere PE, et al. Molecular characterization of two novel antibacterial peptides inducible upon bacterial challenge in an annelid, the leech Theromyzon tessulatum. J Biol Chem. 2004;279:30973–30982. doi: 10.1074/jbc.M312156200. [DOI] [PubMed] [Google Scholar]
- 42.Mah SA, Moy GW, Swanson WJ, Vacquier VD. A perforin-like protein from a marine mollusk. Biochem Biophys Res Commun. 2004;316:468–475. doi: 10.1016/j.bbrc.2004.02.073. [DOI] [PubMed] [Google Scholar]
- 43.Theron A, Coustau C. Are Biomphalaria snails resistant to Schistosoma mansoni? J Helminthol. 2005;79:187–191. doi: 10.1079/joh2005299. [DOI] [PubMed] [Google Scholar]
- 44.Loker ES, Adema CM, Zhang SM, Kepler TB. Invertebrate immune systems--not homogeneous, not simple, not well understood. Immunol Rev. 2004;198:10–24. doi: 10.1111/j.0105-2896.2004.0117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bennett MW, Caulfield JP. Specific binding of human low-density lipoprotein to the surface of schistosomula of Schistosoma mansoni and ingestion by the parasite. Am J Pathol. 1991;138:1173–1182. [PMC free article] [PubMed] [Google Scholar]
- 46.Chiang CP, Caulfield JP. Human lipoprotein binding to schistosomula of Schistosoma mansoni. Displacement by polyanions, parasite antigen masking, and persistence in young larvae. Am J Pathol. 1989;135:1015–1024. [PMC free article] [PubMed] [Google Scholar]
- 47.Chiang CP, Caulfield JP. The binding of human low-density lipoproteins to the surface of schistosomula of Schistosoma mansoni is inhibited by polyanions and reduces the binding of anti-schistosomal antibodies. Am J Pathol. 1989;134:1007–1018. [PMC free article] [PubMed] [Google Scholar]
- 48.Rumjanek FD, Campos EG, Afonso LC. Evidence for the occurrence of LDL receptors in extracts of schistosomula of Schistosoma mansoni. Mol Biochem Parasitol. 1988;28:145–152. doi: 10.1016/0166-6851(88)90062-x. [DOI] [PubMed] [Google Scholar]
- 49.Fan J, Gan X, Yang W, Shen L, McManus DP, Brindley PJ. A Schistosoma japonicum very low-density lipoprotein-binding protein. Int J Biochem Cell Biol. 2003;35:1436–1451. doi: 10.1016/s1357-2725(03)00105-5. [DOI] [PubMed] [Google Scholar]