Abstract
Kinase activity is known as the key biochemical property of MAPKs. Here, we report that ERK1/2 also utilizes its noncatalytic function to mediate certain signal transductions. Sustained activation of the Raf/MEK/ERK pathway induces growth arrest, accompanied by changes in cell cycle regulators (decreased retinoblastoma phosphorylation, E2F1 down-regulation, and/or p21CIP1 up-regulation) and cell type-specific changes in morphology and expression of c-Myc or RET in the human tumor lines LNCaP, U251, and TT. Ablation of ERK1/2 by RNA interference abrogated all these effects. However, active site-disabled ERK mutants (ERK1-K71R, ERK2-K52R, and ERK2-D147A), which competitively inhibit activation of endogenous ERK1/2, could not block Raf/MEK-induced growth arrest as well as changes in the cell cycle regulators, although they effectively blocked phosphorylation of the ERK1/2 catalytic activity readouts, p90RSK and ELK1, as well as the cell type-specific changes. Because this indicated a potential noncatalytic ERK1/2 function, we generated stable lines of the tumor cells in which both ERK1 and ERK2 were significantly knocked down, and we further investigated the possibility using rat-derived kinase-deficient ERK mutants (ERK2-K52R and ERK2-T183A/Y185F) that were not targeted by human small hairpin RNA. Indeed, ERK2-K52R selectively restored Raf-induced growth inhibitory signaling in ERK1/2-depleted cells, as manifested by regained cellular ability to undergo growth arrest and to control the cell cycle regulators without affecting c-Myc and morphology. However, ERK2-T183A/Y185F was less effective, indicating the requirement of TEY site phosphorylation. Our study suggests that functions of ERK1/2 other than its “canonical” kinase activity are also involved in the pathway-mediated growth arrest signaling.
INTRODUCTION
ERK12 and its homologue ERK2, the MAPK components of the Raf/MEK/ERK cascade of Ras signaling, are ubiquitously expressed serine/threonine kinases with more than 160 substrates identified to date (1). ERK1/2 interacts with a wide variety of proteins (2, 3). Upon phosphorylation by MEK1/2, the only known activator of ERK1/2, ERK1/2 phosphorylates transcription factors, other kinases, phosphatases, cytoskeletal proteins, scaffolds, receptors, and signaling components that mediate diverse cellular processes. Although kinase activity of ERK1/2 is central in activation or inactivation of these ERK targets, it was also reported that ERK, in an in vitro reaction, can mediate noncatalytic activation of DNA topoisomerase IIα, suggesting that ERK1/2 also has noncatalytic function (4). Nonetheless, the possibility that ERK1/2 has functions other than kinase has not yet been clearly addressed in cells.
Many studies have shown that ERK1/2 signaling is pivotal in controlling cell survival and cell cycle progression (5). Constitutive activation of the MAPK cascade is also a central signature of many cancers with dysregulated Ras/Raf signaling (6, 7). Paradoxically, sustained activation of the Ras/Raf pathway induces growth arrest in primary cultured normal cells and in vivo, suggesting that cells possess anti-oncogenic defense mechanisms against aberrant activation of the pathway (8–13). Interestingly, Ras/Raf activation also elicits growth arrest in certain malignant tumor cell lines, mainly derived from medullary thyroid carcinoma, small cell lung carcinoma, pheochromocytoma, glioma, and prostate carcinoma (14–24). These tumor cell lines exhibit cell cycle arrest in G0/G1 or G2/M phases and differentiation in response to sustained activation of the Raf/MEK/ERK pathway. Because these Ras/Raf-responsive tumor cell lines are generally derived from tumor types in which mutation of Ras/Raf or elevated signaling of the pathway is rarely detected, it is considered that the pathway does not provide growth advantage to these tumor types and that they may retain intact innate tumor-suppressive mechanisms that respond to aberrant Ras/Raf activation. Elucidation of molecular mechanisms underlying the growth arrest barrier may not only provide insight into the steps involved in Ras/Raf tumorigenesis but also lead to potential strategies to suppress tumor growth.
In different cell types, the Ras/Raf/MEK/ERK pathway mediates growth arrest by controlling the key cell cycle regulatory or tumor-suppressive proteins, including Rb, E2F, cyclin-dependent kinase inhibitors, or p53 (11–13, 21, 25–29). This apparently straightforward mechanism is complicated by cell type-dependent participation of various intermediate signaling pathways, including p38 MAPK/PRAK, Wnt/glycogen synthase kinase 3/β-catenin, secretion of soluble factors, and modulators of cellular redox balance (30–35). We also have shown that the Raf/MEK/ERK pathway mediates growth arrest utilizing leukemia inhibitory factor, the JAK/STAT pathway, or IFI16 in a subset of tumor cell lines (22–24, 36). Although ERK1/2, as the focal point of the Raf/MEK/ERK pathway, would be expected to play a pivotal role in regulating these diverse growth arrest signaling networks and use of the MEK1/2-specific inhibitors, U0126 and PD98059, strongly support this notion (12, 16, 22, 37), the necessity of ERK1/2 has not been directly addressed. Study of ERK signaling is hampered because many cell types are sensitive to the absence of ERK1/2. Apart from the lethal effects of ERK1/2 gene deletion (38, 39), decreases in ERK1/2 activity, either through expression of kinase-deficient ERK mutants (40, 41) or gene knockdown (42–44), significantly suppressed cell proliferation in all cell types examined thus far. Accordingly, our knowledge of mechanisms underlying ERK signaling in the context of growth arrest is still limited.
In this study, we hypothesized that the Ras/Raf-responsive tumor lines may provide an advantage to study the role of ERK1/2 in the pathway-mediated growth arrest by serving as a model that is less sensitive to ERK1/2 depletion. Using lentiviral RNA interference systems designed for ERK1- and ERK2-specific knockdown, we demonstrate that ERK1 and ERK2 have redundant roles in mediating Raf/MEK-induced growth arrest in the human prostate carcinoma line LNCaP. Furthermore, using LNCaP, the human glioma line U251, and the human medullary thyroid carcinoma line TT, we generate cell line models in which both ERK1 and ERK2 are stably knocked down to the level sufficient to maintain cell survival and to suppress Raf/MEK-induced growth arrest. In these models, we asked whether functions of ERK1/2 other than kinase activity are also involved in its growth inhibitory signaling. Using catalytically inactive ERK mutants, we demonstrate that noncatalytic function of ERK1/2 is also utilized in mediating the growth arrest signaling.
EXPERIMENTAL PROCEDURES
Cell Culture, Generation of Stable Lines
The human prostate carcinoma line LNCaP (ATCC) and the human medullary thyroid carcinoma line TT (ATCC) were maintained in RPMI 1640 medium (Invitrogen) supplemented with 10 or 16% fetal bovine serum, respectively. The human glioma line U251 (ATCC) and the primary normal human diploid fibroblast IMR90 cells were maintained in minimum Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum. Culture conditions for these cell lines were also described previously (17, 21, 22). The ΔRaf-1:ER-expressing lines LNCaPRaf and U251Raf were generated by stably transducing LNCaP and U251 with the lentivirus produced from the pHAGE vector containing the activatable ΔRaf-1:ER construct and selecting against puromycin resistance. ΔRaf-1:ER is the CR3 catalytic domain of Raf-1 fused to the hormone binding domain of the human estrogen receptor and was activated with 1 μm 4-hydroxytamoxifen (Sigma) as described previously (45). TTRaf was described previously (22). The stable ERK1, ERK2, and ERK1/2 double knockdown cell lines were generated by stably transducing cell lines with lentivirus produced from pLL3.7-shERK1 (human) and pLL3.7-shERK2 (human) and selecting for puromycin resistance or for GFP expression.
Cell Proliferation Assay
For cell growth curves, cells were seeded in 24-well plates (Corning Glass) at a density of 104 cells per well. Cell proliferation was measured by the colorimetric 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT) assay, as described previously (24). Briefly, cells in 24-well plates were treated with 40 μl of 5 mg/ml MTT (Sigma) in phenol red-free RPMI 1640 medium containing 10% fetal bovine serum for 3 h at 37 °C. The medium was then replaced with 600 μl of DMSO and shaken for 15 min prior to measuring absorbance at 540 nm. A540 was measured every 2 days. Cell proliferation was also measured by counting cells every 2 days using a hemocytometer.
Cell Cycle Analysis
Cells were washed with ice-cold 0.2% bovine serum albumin in phosphate-buffered saline, resuspended in 250 mm sucrose, 40 mm citrate buffer (pH 7.6) containing 0.5% DMSO. Nuclei were prepared, stained with propidium iodide (46), and analyzed by LSR flow cytometer (BD Biosciences) with a gate that selects single nuclei within a normal size range. The cell cycle parameters from 10,000 gated nuclei were determined by CellQuest software.
Viral Infection
The lentiviral expression vector pHAGE and the lentiviral shRNA expression vector pLL3.7 (ATCC) were used as described previously (47, 48). Briefly, for viral production, pHAGE or pLL3.7 was co-transfected with packaging vectors into 293T cells, and the resulting supernatant was collected after 48 h. Viral titers were determined by infecting HEK293 or the recipient cell lines with serially diluted viral supernatants and scoring cells expressing GFP at 48 h post-infection. Cells for experiments were infected with lentivirus mixed with Polybrene (Sigma) at 4–8 μg/ml and switched into fresh culture medium on the following day before further treatment.
Plasmids and Recombinant Lentiviruses
pHAGE-GFP-ERK1-K71R was generated by subcloning the kinase-deficient ERK1-K71R (49) into the NotI/BamHI site of the pHAGE vector. pHAGE-GFP-ERK2wt and pHAGE-GFP-ERK2-K52R were generated by subcloning rat wild type ERK2 and the kinase-deficient ERK2-K52R genes (49) into the XhoI/XbaI site of pHAGE, respectively. To generate ERK2-D147A and ERK2-T183A/Y185F, the wild type ERK2 in pBluescript SK(−) was mutagenized using the QuickChange II site-directed mutagenesis kit (Stratagene, Cedar Creek, TX) and the primers TAATGTTCTGCACCGTGCCCTCAAGCCTTCCAAC and GTTGGAAGGCTTGAGGGCACGGTGCAGAACATTA (for D147A), and CATACAGGGTTCTTGGCAGAGTTTGTAGCCACGCGTTGG and CCAACGCGTGGCTACAAACTCTGCCAAGAACCCTGTATG (for T183A/Y185F), respectively. The resulting mutant genes were then subcloned into the XhoI/XbaI site of pHAGE. To generate pHAGE-puro-Raf:ER, the HindIII/ClaI fragment of the pLNCX-ΔRaf-1:ER vector (45) was ligated into the XhoI site of the pHAGE-puro vector, containing a puromycin resistance gene. To generate virus containing constitutively active MEK1 (MEK1CA) or MEK2 (MEK2CA), MEK1-R4F (ΔN3/S218E/S222D) and MEK2-KW71 (ΔN4/S222D/S226D) in pCEP4 (50, 51) were subcloned into the PmeI site of the pHAGE vector, respectively.
Small Hairpin RNA (shRNA)-mediated Knockdown of ERK1 and ERK2
To construct ERK1 and ERK2 knockdown systems specific to human or rat, we screened and selected individual siRNA oligomers from the SMART PoolTM reagent (Dharmacon, Lafayette, CO) based on their knockdown efficacy and specificity. Sequences of selected oligomers were used to design lentiviral shRNA systems targeting ERK1 or ERK2, which were constructed in the HpaI/XhoI site of the pLL3.7 vector (ATCC). pLL3.7-shERK1 (human) expressed the targeting sequence GACCTGAATTGTATCATC. pLL3.7-shERK2 (human) expressed the targeting sequence CCAAAGCTCTGGACTTATT. pLL3.7-shERK2 (rat) expressed the targeting sequence CCAAAGCTCTGGATTTACT, which has a difference in the two bases underlined in comparison with the human counterpart. Virus was then generated from these vectors as described above. Successful and specific knockdown of ERK1 and ERK2 was confirmed by Western blot analysis.
Immunoblot Analysis
Cells harvested at various times were lysed in 62.5 mm Tris (pH 6.8), 2% SDS mixed with the protease inhibitor mixture (Sigma) that contains 4-(2-aminoethyl) benzenesulfonyl fluoride, pepstatin A, E-64, bestatin, leupeptin, and aprotinin and briefly sonicated before determining the protein concentration using the BCA reagent (Pierce). 50 μg of protein was resolved by SDS-PAGE, transferred to a polyvinylidene difluoride membrane filter (Bio-Rad), and stained with Fast Green reagent (Fisher). Membrane filters were then blocked in 0.1 m Tris (pH 7.5), 0.9% NaCl, 0.05% Tween 20 with 5% nonfat dry milk and incubated with appropriate antibodies. Antibodies were diluted as follows: MEK1/2, 1:2,500; phospho-MEK1/2 (Ser-217/221), 1:2,500; ERK1/2, 1:2,500; phospho-ERK1/2 (Thr-202/Tyr-204), 1:2,500; p90RSK, 1:2,500; phospho-p90RSK (Thr-359/Ser-363), 1:2,500; ELK1, 1:2,000; phospho-ELK1 (Ser-383), 1:2,000; phospho-Rb (Ser-780), 1:1,000; GAPDH, 1:5,000 (Cell Signaling); E2F1, 1:1,000; c-Myc, 1:1,000; poly(ADP-ribose) polymerase, 1:1,000 (Thermo Fisher Scientific, Waltham, MA); p21CIP1, 1:1,000; RET, 1:1,000 (Santa Cruz Biotechnology, Santa Cruz, CA); Rb, 1:1,000 (BD Biosciences). For analysis of nuclear extracts, we extracted nuclear fractions using the nuclear extraction kit (Pierce) according to the manufacturer's instruction. The Supersignal West Pico and Femto chemiluminescence kits (Pierce) were used for visualization of the signal. For densitometry, immunoblots were scanned and analyzed using LabWorksTM (UVP BioImaging Systems, Upland, CA).
RESULTS
Raf Induces, via MEK1/2 Activation, Growth Arrest Accompanied by Changes in Cell Morphology and Expression of Cell Proliferation Regulators
Sustained activation of the Raf/MEK/ERK pathway induces growth inhibitory signaling, characterized by cell cycle arrest in G0/G1 or G2/M phases and morphological changes, in certain malignant cancer cell types, including the human prostate carcinoma line LNCaP, the human glioma line U251, and the human medullary thyroid carcinoma line TT (17, 21, 22). Using these tumor cell lines as models, we attempted to determine the requirement of ERK1 and ERK2 for the pathway-mediated growth arrest and to investigate underlying mechanisms of ERK1/2 signaling. For a specific control of the Raf/MEK/ERK pathway activation in these cells, we used the ΔRaf-1:ER construct that is regulated by the estrogen analogue 4-hydroxytamoxifen (45). ΔRaf-1:ER can induce ERK1/2 activity similar to the levels detected in human cancer cell lines displaying deregulated ERK1/2 signaling (supplemental data 1), and it has been important in the studies of the mechanism of growth arrest induced by oncogenically altered Raf/MEK/ERK signals (12, 16–19, 21, 22, 52).
When LNCaP cells, stably infected with ΔRaf-1:ER (LNCaPRaf), were exposed to 4-hydroxytamoxifen, cells exhibited morphological changes and decreased cell proliferation rates (Fig. 1A, top two panels, and B; supplemental data 2, A and B), which were correlated with decreased S phase and increased G2/M phase cell populations (supplemental data 2C), and altered levels of cell cycle regulators, including decreased phospho-Rb and E2F1, and increased p21CIP1 (Fig. 1C, 1st two lanes; supplemental data 2D). The cyclin-dependent kinase inhibitor p21CIP1 can mediate growth arrest and senescence by inhibiting cyclin-dependent kinases (53), whereas E2F1 is a critical transcription factor involved in S phase cell cycle progression, which is sequestered by Rb and is released upon phosphorylation of Rb (54). In addition to these previously reported changes (17), Raf activation in LNCaP cells led to down-regulation of the pleiotropic proto-oncogene c-Myc (Fig. 1C; supplemental data 2D), which has recently been identified as a critical component required to overcome Ras/Raf-mediated senescence-like growth arrest in melanoma cells (55). In that study (55), c-Myc up-regulation was correlated with the tumor stages that have overcome N-Ras/B-Raf-induced senescence, whereas c-Myc knockdown was sufficient to restore senescence responses in N-Ras/B-Raf-mutated melanoma cells. Raf activation also induced E2F1 down-regulation and p21CIP1 up-regulation, but not down-regulation of phospho-Rb and c-Myc, in U251 cells (supplemental data 2E), whereas it induced down-regulation of E2F1 and the RET receptor tyrosine kinase in TT cells (see Fig. 4D for TT). RET oncogene is necessary for cell survival of medullary thyroid carcinoma, and its down-regulation was shown to be associated with Raf-induced growth arrest in TT cells (18, 22). All of these ΔRaf-1:ER-induced changes were specific to Raf activation, because 4-hydroxytamoxifen alone did not induce any similar changes (supplemental data 2), as shown previously (12, 16–19, 21, 22, 52). In this study, given the potential of these cell cycle regulators (Rb, E2F1, and p21CIP1), c-Myc and RET, to influence cell proliferation, these proteins are used as surrogate markers to evaluate the mechanisms of Raf/MEK/ERK growth arrest signaling in the tumor cell lines.
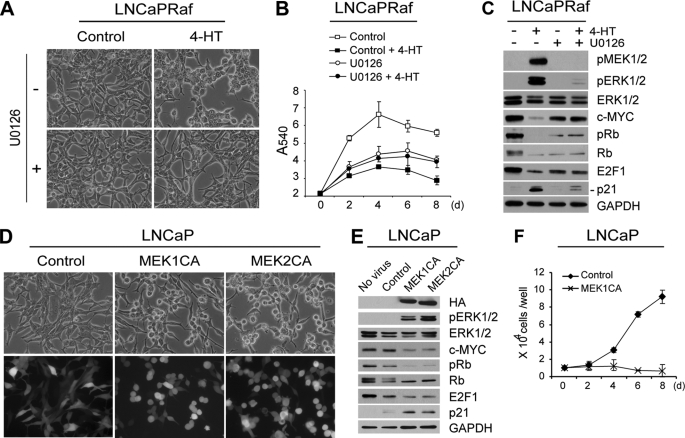
FIGURE 1.
MEK1/2 is essential and sufficient to mediate Raf-induced growth inhibitory signaling. A–C, LNCaPRaf cells harboring the hormone-activatable Raf:ER were treated with 1 μm 4-HT in the presence or absence of 10 μm U0126 (MEK1/2 inhibitor) and examined for morphological changes at day 2 (A), cell proliferation for 8 days by MTT assay (B), and expression of phosphorylated MEK1/2 (pMEK1/2), phosphorylated ERK1/2 (pERK1/2), ERK1/2, c-Myc, phosphorylated Rb (pRb), Rb, E2F1, and p21CIP1 (lower band) by Western blot analysis at day 2 (C). The downshift of Rb bands indicates down-regulated Rb phosphorylation. GAPDH was detected to validate equal protein loading. Data (means ± S.E.) are from a representative experiment performed in triplicate. p value is <0.05 for U0126 + 4-HT compared with control + 4-HT (Student's t test). 4-Hydroxytamoxifen alone had no effect on morphology, cell proliferation, and expression of these proteins (supplemental data 2). D–F, LNCaP cells, infected with lentivirus containing constitutively active MEK1 (MEK1CA) or MEK2 (MEK2CA), were observed for morphological changes at day 2 post-infection (D), expression of the indicated proteins by Western blot analysis at day 2 (E), and cell proliferation for 8 days by cell counting. The empty pHAGE lentivirus was used as control. Similar infection ratio was verified by GFP expression (D, lower panels). HA indicates expression of MEK1CA and MEK2CA. Cell counts (means ± S.E.) are from a representative experiment performed in triplicate.
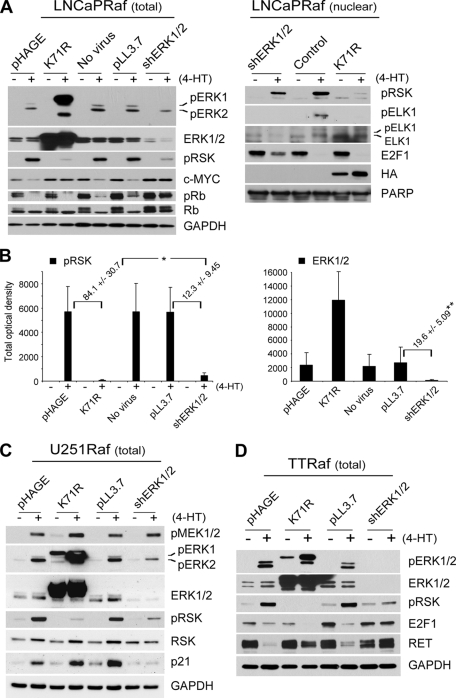
FIGURE 4.
Inability of ERK1-K71R in blocking Raf-induced growth arrest is not due to insufficient depletion of ERK1/2 kinase activity in different cell lines. A and B, LNCaPRaf cells, infected with the virus containing kinase-deficient ERK1 gene (K71R) or co-infected with the shERK1 and shERK2 viruses, were treated with 4-hydroxytamoxifen for 2 days. Total cell lysates (total) and nuclear extracts (nuclear) of harvested cells were examined for expression of the indicated proteins by Western blot analysis. ELK1 is a nuclear substrate of ERK1/2, indicating the catalytic activity of ERK1/2 in the nucleus. Upshift of ELK1 bands also indicates its phosphorylation. GAPDH and poly(ADP-ribose) polymerase (PARP) were used to validate equal protein loading of total cell lysates and nuclear extracts, respectively. B, Western blotting results of pRSK and ERK1/2 in total cell lysates were analyzed by densitometry to calculate the fold changes in pRSK and ERK1/2 levels affected by the two experimental conditions, RNA interference and overexpression of kinase-deficient ERK. ERK1/2 levels are the sum of ERK1 and ERK2 levels. Data (means ± S.E.) are from four independent experiments. *, p value is <0.05 for ERK1-K71R effects compared with shERK1/2 effects. **, p value is <0.001 for shERK1/2 compared with pLL3.7 (Student's t test). C and D, U251Raf and TTRaf cells, infected with the virus containing kinase-deficient ERK1 gene (K71R) or co-infected with the shERK1 and shERK2 viruses, were treated with 4-hydroxytamoxifen for 2 days. Total cell lysates of harvested cells were examined for expression of the indicated proteins by Western blot analysis. In this figure, c-Myc, Rb phosphorylation, E2F1, p21CIP1, RET, and p90RSK contrast the biological effects caused by the expression of kinase-deficient ERK1 and the ERK1/2 knockdown.
All of the Raf-induced changes in LNCaP cells, including morphology, growth arrest, and expression of c-Myc and the cell cycle regulatory proteins, were abrogated by U0126, although the MEK1/2 inhibitor also affected basal cell growth and basal levels of these proteins (Fig. 1, A–C), indicating that MEK1/2 is required for Raf-induced growth arrest signaling as well as for maintaining basal cell growth. On the other hand, expression of the constitutively active MEK1 or MEK2, which contain the phosphomimetic mutations (S218E/S222D in MEK1 and S222D/S226D in MEK2) and deletion of an N-terminal α-helix (ΔN3 in MEK1 and ΔN4 in MEK2) (50), was sufficient to induce “Raf activation”-like changes in morphology and expression of the cell cycle regulators and c-Myc (Fig. 1, D and E). The capability of the active MEK to mediate growth arrest signaling was further manifested when overexpression of the constitutively active MEK1 suppressed proliferation of LNCaP by inducing a similar pattern of cell cycle arrest as Raf activation (Fig. 1F; supplemental data 3). These data indicate that MEK1/2 is necessary and sufficient to mediate Raf-induced growth inhibitory signaling and also validate the use of the surrogate markers to indicate Raf/MEK signaling.
Depletion of Both ERK1 and ERK2 Is Required to Block Raf-induced Growth Arrest Signaling
ERK1 and ERK2 are the only known substrates of MEK1/2 (56). To investigate whether ERK1/2 is essential for Raf/MEK-induced growth inhibitory signaling, we examined the effect of ERK1/2 ablation using the pLL3.7 lentiviral systems that express shRNA targeting ERK1 or ERK2.
In most cell types, ERK1/2 knockdown is growth-suppressive (42–44). This effect would prevent our assessment of the role of ERK1 and ERK2 in Raf/MEK-mediated growth suppression. Indeed, when both ERK1 and ERK2 were knocked down in the normal human diploid fibroblast IMR90, which is a model of Ras/Raf-induced growth arrest (11–13, 33, 34), cell proliferation was significantly suppressed, hindering further manipulation of the cells (supplemental data 4). However, the growth of LNCaP, TT, and U251 cells was not significantly affected by ERK1/2 knockdown, and therefore we could determine the effect of ERK1/2 depletion on Raf-mediated growth arrest in these tumor cells.
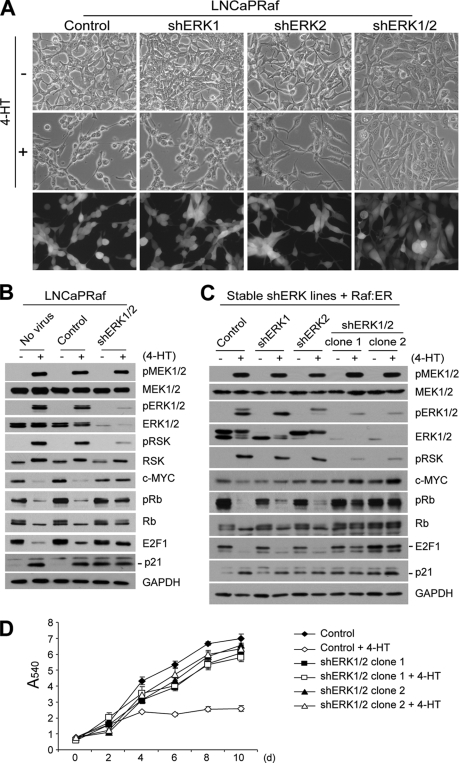
When both ERK1 and ERK2 were knocked down, LNCaPRaf cells no longer displayed the typical morphology changes in response to Raf activation (Fig. 2A). The double knockdown of ERK1 and ERK2 also rescued cells from Raf-mediated growth arrest, as demonstrated by the cells transiently infected with the two shRNA viruses (supplemental data 5) as well as the two independently generated ERK1/2 knockdown stable clones (Fig. 2D and Table 1). We were able to generate stably infected LNCaP cells in which ERK1 and ERK2 are individually or doubly knocked down, because LNCaP cells, transiently infected with the shRNA viruses, could still proliferate (supplemental data 5). Depletion of both ERK1 and ERK2, at the levels achieved in our study (about 20-fold), substantially decreased the levels of Raf-induced phosphorylation of ERK1/2 and the ERK substrate, ribosomal S6 kinase (p90RSK), indicating a significant reduction in ERK activity (Fig. 2B for transient infection; Fig. 2C for stable infection; ERK1/2 densitometry shown in Fig. 4B); p90RSK is a Ser/Thr kinase serving as a bona fide readout of in vivo ERK1/2 kinase activity (57). Basal levels of the upstream kinase MEK1/2, its activation by Raf, and total p90RSK levels were not affected by ERK1/2 depletion, indicating that the decreased ERK activity was a specific effect caused by ERK1/2 depletion. Depletion of both ERK1 and ERK2 at this level also did not affect basal levels of c-Myc, Rb, and E2F1, although it increased p21CIP1 basal levels (Fig. 2, B and C). Under this condition, all of the Raf-induced changes, including down- or up-regulation of c-Myc and the cell cycle regulatory proteins, were also significantly abrogated, indicating that ERK1/2 is necessary for the growth inhibitory signaling.
FIGURE 2.
Depletion of ERK1/2 by RNA interference blocks Raf-induced growth inhibitory signaling. A and B, LNCaPRaf cells, transiently infected with lentivirus containing shRNA targeting ERK1 (shERK1) or ERK2 (shERK2), or with both viruses (shERK1/2), were treated with 1 μm 4-HT for 2 days and examined for morphological changes (A) and expression of pMEK1/2, MEK1/2, pERK1/2, ERK1/2, phosphorylated p90RSK (pRSK), p90RSK (RSK), c-Myc, phosphorylated Rb (pRb), Rb, E2F1, and p21CIP1 by Western blot analysis (B). The empty pLL3.7 virus was used as control. Similar infection ratio was verified by GFP expression (A, bottom panels). C and D, cells of LNCaP stable clones, in which ERK1 and ERK2 are individually or doubly knocked down, were infected with the inducible Raf:ER virus and treated with 4-hydroxytamoxifen to examine expression of the indicated proteins by Western blot analysis at day 2 (C) or to monitor cell proliferation for 10 days by MTT assay (D). Data (means ± S.E.) are from a representative experiment performed in triplicate. p value is <0.005 for clone 1 and clone 2, respectively, when cell growth rates affected by Raf activation were compared with the control (Student's t test).
TABLE 1.
Effects of ERK1/2 knockdown or ERK1-K71R overexpression on Raf-mediated cell cycle arrest
LNCaPRaf cells, infected with pLL3.7 (control for shERK1/2), shERK1, and shERK2, pHAGE (control) or pHAGE-ERK1-K71R virus, were treated with 1 μm 4-HT for the days indicated. Data (mean ± S.E.) are from a representative experiment performed in triplicate. These data indicate that depletion of ERK1/2 could abrogate Raf-mediated cell cycle arrest, but overexpression of the kinase-deficient ERK1-K71R could not abrogate but rather augmented cell cycle arrest.
| % of cells in phase |
||||
|---|---|---|---|---|
| pLL3.7 |
shERK1/2 |
|||
| − | + | − | + (4-HT) | |
| Day 2 | ||||
| G0/G1 | 63.2 ± 0.31 | 65.9 ± 0.88 | 66.9 ± 0.02 | 63.7 ± 0.35 |
| S | 25.5 ± 0.19 | 14.2 ± 1.27 | 20.9 ± 0.31 | 22.0 ± 0.01 |
| G2/M | 11.3 ± 0.49 | 19.9 ± 0.39 | 12.3 ± 0.29 | 14.3 ± 0.17 |
| pHAGE |
ERK1-K71R |
|||
|---|---|---|---|---|
| − | + | − | + (4-HT) | |
| Day 2 | ||||
| G0/G1 | 72.6 ± 1.01 | 74.1 ± 1.03 | 72.6 ± 1.31 | 79.9 ± 1.00 |
| S | 18.7 ± 1.01 | 14.1 ± 1.48 | 18.6 ± 1.05 | 0.49 ± 0.01 |
| G2/M | 8.72 ± 1.07 | 11.8 ± 1.33 | 8.84 ± 1.03 | 19.7 ± 1.06 |
| Day 4 | ||||
| G0/G1 | 68.5 ± 1.01 | 77.3 ± 1.05 | 70.4 ± 1.01 | 80.7 ± 1.01 |
| S | 22.3 ± 1.01 | 11.6 ± 2.15 | 21.7 ± 1.02 | 0.51 ± 0.01 |
| G2/M | 9.18 ± 1.02 | 11.0 ± 1.51 | 7.93 ± 1.05 | 18.8 ± 1.02 |
Knockdown of ERK1 or ERK2 alone effectively depleted the cell of either ERK1 or ERK2, but it did not significantly affect Raf-induced phosphorylation of p90RSK and changes in morphology and the levels of c-Myc, Rb, E2F1, and p21CIP1 (Fig. 2, A and C). These data indicate that ERK1 and ERK2 are functionally redundant in the context of growth inhibitory signaling.
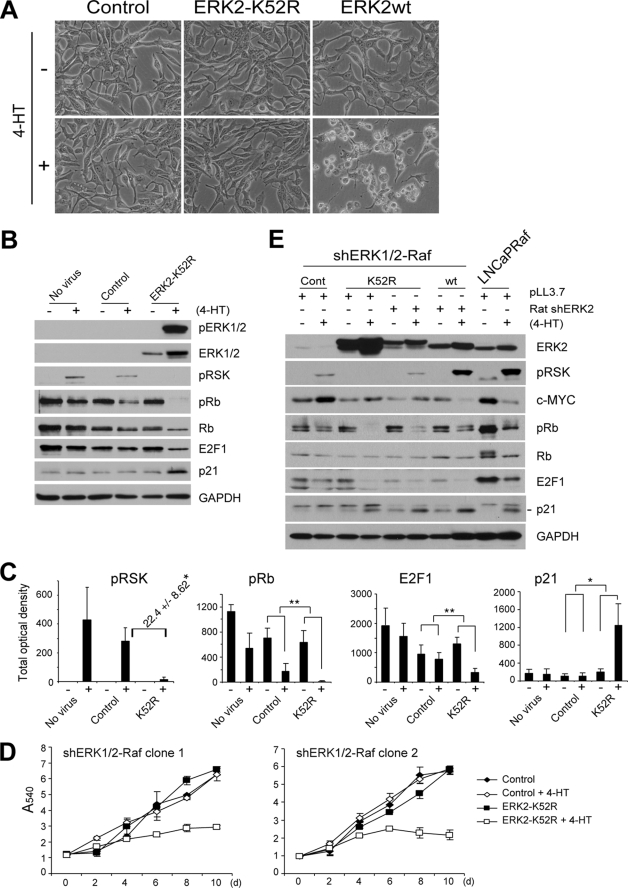
The necessity of ERK1/2 for Raf-induced growth arrest signaling was also observed in U251 and TT cells, and furthermore, ERK1/2 stably knocked down cells were also generated from these cell lines, and the cells derived from U251 were polyclonal (Fig. 4, C and D, for transient infection; Fig. 7, A and B, for stable infection). Introduction of wild type ERK2 into these ERK1/2 stable knockdown cells restored Raf-mediated growth arrest responses (Figs. 6 and 7) and the details are described below.
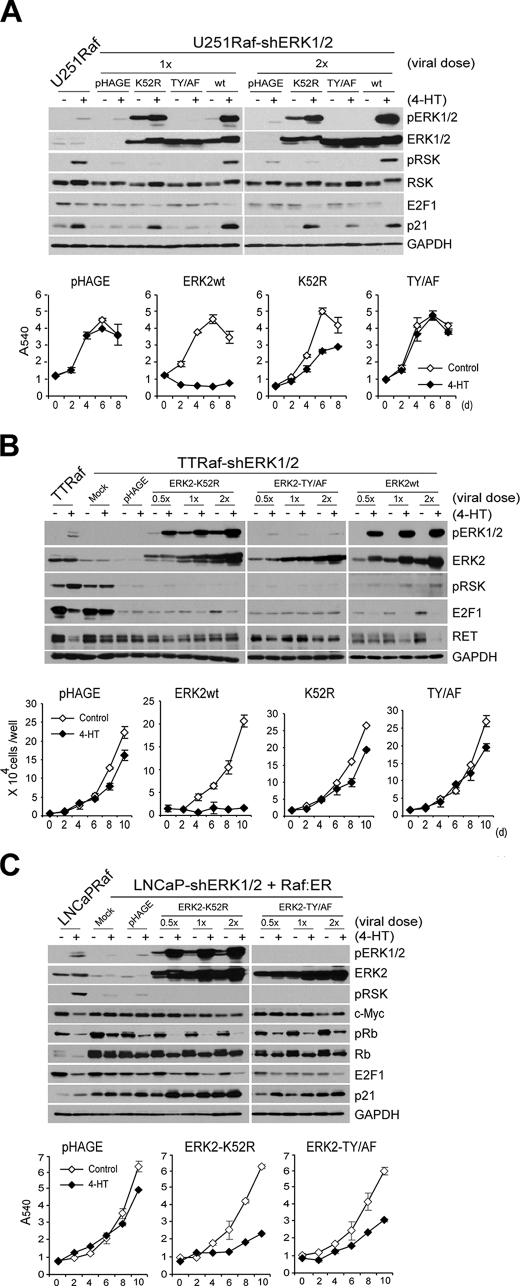
FIGURE 7.
Phosphorylation on TEY site is important, but not necessary, for noncatalytic function of ERK2. A, cells of U251Raf-shERK1/2, infected with the lentivirus containing the rat-derived ERK2 genes (K52R, T183A/Y185F (TY/AF), wild type (wt)) at two different doses, were treated with 1 μm 4-HT and examined for expression of the indicated proteins by Western blot analysis at day 2 and cell proliferation for 8 days by MTT assay. U251Raf is the parental cells for U251Raf-shERK1/2. The empty pHAGE virus was used as the control. Cells used for growth curve were from 1× viral dose-infected. Data (means ± S.E.) are from a representative experiment performed in triplicate. p value is <0.05 for ERK2-K52R effects compared with wild type ERK2 or ERK2-TY/AF (Student's t test). B, cells of TTRaf-shERK1/2, infected with the ERK2 lentivirus at three different doses, were treated with 4-hydroxytamoxifen and examined for expression of the indicated proteins by Western blot analysis at day 2 and cell proliferation for 10 days by cell counting. TTRaf is the parental cells for TTRaf-shERK1/2. Cells used for growth curve were from 2× viral dose-infected. Data (means ± S.E.) are from a representative experiment performed in triplicate. C, cells of the LNCaP-shERK1/2 stable clones, serially infected with the inducible Raf:ER virus and the lentivirus containing the rat-derived kinase-deficient ERK2 (K52R, T183A/Y185F), were treated with 1 μm 4-hydroxytamoxifen and examined for expression of the indicated proteins by Western blot analysis and cell proliferation for 10 days by MTT assay. Cells used for growth curve were from 1× viral dose-infected. Data (means ± S.E.) are from a representative experiment performed in triplicate.
FIGURE 6.
Kinase-deficient ERK2 selectively reconstitutes Raf-induced growth inhibitory signaling in ERK1 and ERK2 stably knocked down LNCaP cells. A–D, cells of the LNCaP-shERK1/2 stable clones, serially infected with the inducible Raf:ER virus and the lentivirus containing the rat-derived kinase-deficient ERK2 (ERK2-K52R), were treated with 1 μm 4-HT and examined for morphological changes at day 2 (A), expression of the indicated proteins by Western blot analysis and densitometry at day 2 (B and C), and cell proliferation for 10 days by MTT assay (D). The rat ERK2 genes (wild type (wt) and K52R) are not recognized by shERK2 (human). The empty pHAGE virus was used as the control. Morphology and Western blot results shown are from the stable clone 1. Similar results were obtained from the clone 2 (data not shown). C, densitometry data (means ± S.E.) are from three independent experiments. *, p value is <0.005 for ERK2-K52R effects compared with the control. **, p < 0.05 (Student's t test). D, MTT assay data (means ± S.E.) are from a representative experiment performed in triplicate. p value is <0.005 for clone 1 and p < 0.05 for clone 2 when cell growth rates affected by Raf activation were compared with the control (Student's t test). E, to adjust expression of the exogenous ERK2 genes close to physiologically more relevant levels, LNCaP-shERK1/2 stable cell lines were serially infected with the following: (i) the Raf:ER virus; (ii) virus containing ERK2-K52R (K52R) or ERK2wt (wt) gene; and (iii) virus expressing shRNA that specifically targets the exogenous ERK2 genes (rat shERK2). Cells were then treated with 4-HT for 2 days and examined for expression of the indicated proteins by Western blot analysis. LNCaPRaf cells were used as the control (Cont) for endogenous ERK2 expression levels.
Kinase-deficient ERK1 and ERK2 Mutants Cannot Block Raf-induced Growth Arrest Signaling although They Effectively Inhibit Activation of Endogenous ERK1/2
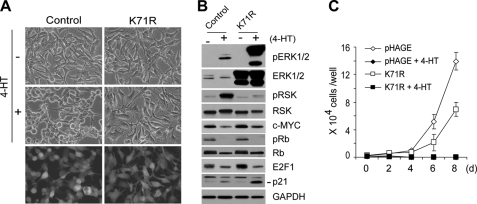
Exploiting the relatively low sensitivity of these tumor lines to ERK1/2 depletion, we next investigated whether noncatalytic functions of ERK1/2 might exist and be involved in Raf/MEK/ERK-induced growth arrest. For this, we used the kinase-deficient ERK mutant, ERK1-K71R, which has K71R replacement in its active site but the intact TEY site in its activation loop (58). Therefore, ERK1-K71R can be phosphorylated by MEK1/2 and undergo the activating conformational changes, although it lacks kinase activity. ERK1/2 mutants containing the Lys-Arg replacement have been used as competitive inhibitors of endogenous ERK1 and ERK2 activation (41, 49). Accordingly, we expected that expression of ERK1-K71R would establish an intracellular condition under which Raf activation increases phosphorylated ERK protein levels but not ERK kinase activity.
To effectively block ERK1/2 activity without causing significant cell stress, we used a lentivirus harboring the kinase-deficient ERK1 gene. More than 90% of LNCaPRaf cells were infected with the virus as determined by GFP expression (Fig. 3A). Expression of ERK1-K71R, by itself, did not affect cell morphology (Fig. 3A) and expression of p90RSK, c-Myc, and the cell cycle regulators (Fig. 3B), although it slightly retarded cell growth (Fig. 3C). Upon Raf activation, the overexpressed kinase-deficient ERK1 effectively competed for the active site of MEK1/2 (as indicated by predominant phosphorylation of the exogenous ERK) and inhibited activation of the endogenous ERK1/2 (as indicated by the significant decreases in phospho-p90RSK) (Fig. 3B). Under this condition, morphological changes and c-Myc down-regulation were effectively blocked (Fig. 3A), indicating that these changes are controlled by the catalytic activity of ERK1/2. However, surprisingly, ERK1-K71R could not inhibit the effects of Raf activation on Rb, E2F1, and p21CIP1 (Fig. 3B). The presence of ERK1-K71R rather augmented Raf-mediated p21CIP1 induction (Fig. 3B; this is consistently observed in Fig. 5, B and C). At the levels of physiological consequences, ERK1-K71R could not rescue cells from growth arrest (Fig. 3C) but, in fact, augmented cell cycle arrest (Table 1). Because depletion of ERK1/2 protein blocked Raf-mediated growth arrest, but inhibition of ERK1/2 kinase activity did not, this indicated that ERK1/2 may have noncatalytic function, which is required for Raf-induced growth arrest signaling.
FIGURE 3.
Kinase-deficient ERK1 inhibits Raf-induced morphological changes and c-Myc down-regulation but not growth arrest. LNCaPRaf cells, infected with the lentivirus containing kinase-deficient ERK1-K71R (K71R) or the empty pHAGE virus (control), were treated with 1 μm 4-HT and examined for morphological changes at day 2 (A), expression of pERK1/2, ERK1/2, pRSK, RSK, c-Myc, pRb, Rb, E2F1, and p21CIP1 by Western blot analysis at day 2 (B), and cell proliferation for 8 days (C). Similar infection ratio was verified by GFP expression (A, bottom panels). Cell counts (means ± S.E.) are from a representative experiment performed in triplicate.
FIGURE 5.
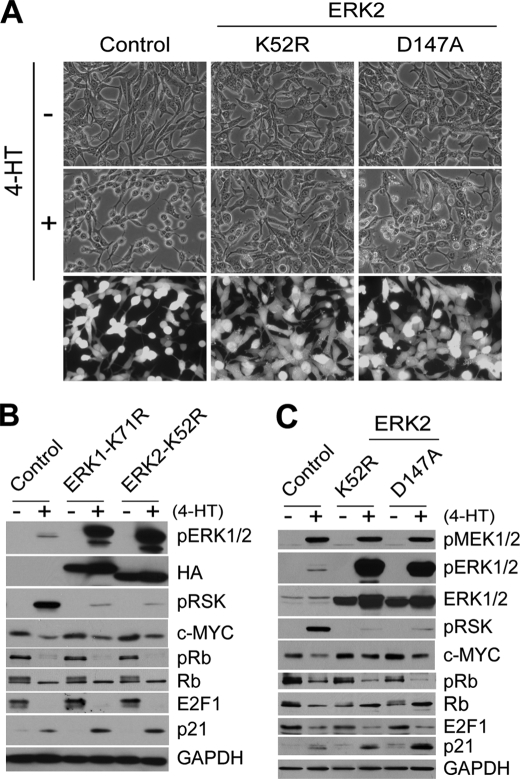
Kinase-deficient ERK2 mutants have similar inability as kinase-deficient ERK1 in blocking Raf-induced growth inhibitory signaling. LNCaPRaf cells, infected with lentivirus containing ERK1-K71R, ERK2-K52R (K52R), or another kinase-deficient ERK2 (D147A), were treated with 1 μm 4-HT for 2 days and examined for morphological changes (A) and expression of pMEK1/2, pERK1/2, ERK1/2, pRSK, c-Myc, Rb, E2F1, and p21CIP1 by Western blot analysis (B and C). The empty pHAGE virus was used as control. ERK1-K71R serves as the positive control for the comparison with ERK2-K52R. Similar infection ratio was verified by GFP expression (A, bottom panels). GAPDH was detected to validate equal protein loading.
This selective inability of kinase-deficient ERK1 to block Raf-induced growth arrest was not due to insufficient inhibition of endogenous ERK1/2 activation. When we compared the levels of ERK1/2 catalytic activity inhibited by the two approaches, RNA interference and overexpression of kinase-deficient ERK, we detected relatively lower levels of endogenous phospho-ERK2 in total cell lysates and phospho-p90RSK in total and nuclear extracts of cells expressing ERK1-K71R (Fig. 4, A and B). Phosphorylation of ELK1, a member of the ternary complex transcription factor subfamily that may serve as a readout of nuclear ERK1/2 activity (41), was inhibited at equivalent levels by both approaches (Fig. 4A). Nevertheless, only RNA interference could block Raf-induced down-regulation of Rb phosphorylation and E2F1, although these two approaches similarly blocked c-Myc down-regulation (Fig. 4A). This observation was not limited to LNCaP cells. In U251Raf cells, overexpression of kinase-deficient ERK-K71R also could not block Raf-mediated p21CIP1 induction, although ERK1-K71R expression was as effective as ERK1/2 depletion in blocking phosphorylation of p90RSK (Fig. 4C). Similarly in TTRaf cells, overexpression of ERK-K71R could not block Raf- mediated E2F1 down-regulation, although it blocked down-regulation of the RET receptor tyrosine kinase similarly as ERK1/2 depletion did (Fig. 4D).
These intriguing effects were not limited to ERK1-K71R but also were observed with kinase-deficient ERK2 mutants. We generated ERK2-K52R, which has the same Lys-Arg replacement in its active site, and another form of kinase-deficient mutant ERK2-D147A, which has an inactivating Asp-Ala replacement in its catalytic domain (Hanks subdomain VIb; Asp-147 acts as the catalytic base) (59). These kinase-deficient ERK2 mutants also showed similar selective inability in blocking Raf signaling in LNCaP cells; these mutants could not block the effects of Raf activation on Rb, E2F1, and p21CIP1, although they could inhibit morphological changes, c-Myc down-regulation, and phosphorylation of p90RSK (Fig. 5,A–C). These data indicated that different downstream events of Raf/MEK/ERK-mediated signaling are mediated via different ERK1/2 signaling mechanisms and that some parts of this signaling may be mediated by non-kinase function of ERK1/2. In particular, growth inhibition appears to require non-kinase functions of ERK1/2, although the morphological changes require ERK1/2 kinase activity.
Kinase-deficient ERK Can Selectively Restore Raf-induced Growth Arrest in ERK1/2-depleted Cells
To directly address the possibility that noncatalytic ERK function is involved in the pathway-mediated growth inhibitory signaling, we determined whether rat-derived ERK2-K52R (not recognized by shRNA targeting human ERK2) could selectively restore Raf-induced growth arrest signaling in the tumor cell lines in which ERK1 and ERK2 are stably knocked down.
Expression of ERK2-K52R or wild type ERK2 did not affect morphology of the ERK1/2-depleted LNCaP cells, and when Raf was activated, only wild type ERK2 restored the typical morphology changes (Fig. 6A), consistent with our earlier finding that the Raf-mediated morphological changes require ERK catalytic function (Figs. 3 and 5). Expression of ERK2-K52R inhibited the activation of the residual ERK1/2 and further depleted these cells of ERK catalytic activity (about 22-fold decrease), as indicated by phospho-p90RSK levels (Fig. 6, B and C); phospho-p90RSK was not detected in cells expressing ERK2-K52R even after prolonged development of the Western blot. Under this condition, the cells expressing ERK2-K52R exhibited down-regulated phospho-Rb and E2F1 levels and up-regulated p21CIP1 levels (Fig. 6B, the densitometry is shown in C), although their morphology did not change (Fig. 6A). Remarkably, these cells could also undergo growth arrest in response to Raf activation, as demonstrated in the two independently generated LNCaP clones (Fig. 6D), indicating that the kinase-deficient ERK2 has an ability to mediate Raf-induced growth arrest.
We also attempted to verify this observation under the condition that kinase-deficient ERK2 is expressed at physiologically relevant levels. For this, we controlled the expression levels of the exogenous rat ERK2 using an additional lentivirus encoding shRNA that specifically targets rat ERK2. Successfully adjusted expression levels of ERK2-K52R or wild type ERK2 in the ERK1/2 knockdown cells are shown in comparison with the endogenous ERK2 level in LNCaPRaf cells (Fig. 6E); the inhibition levels of p90RSK phosphorylation (indicating the ERK catalytic activity) was also changed accordingly. Under this condition, the cells expressing the kinase-deficient ERK2 still exhibited its capability to down-regulate phospho-Rb and E2F1 and to up-regulate p21CIP1 in response to Raf activation similarly to the cells expressing equivalent levels of wild type ERK2, although phospho-p90RSK levels, c-Myc down-regulation, and morphological changes in these two groups were clearly contrasted (Fig. 6E, data not shown for morphology). This was consistent with the results obtained before the control of protein expression levels, as shown in Fig. 6, A and B. However, under this modified condition, down-regulation of Rb phosphorylation was mild in cells expressing not only the kinase-deficient mutant but also wild type ERK2, and basal levels of the marker proteins were also affected (Fig. 6E), possibly indicating a technical difficulty in reconstituting a signal transduction pathway using multiple gene knockdown as well as expression systems. Restoration of Raf-mediated growth arrest signaling using kinase-deficient rat ERK2 mutants was also tested in ERK1/2-knocked down U251Raf and TTRaf cells and is described below and in Fig. 7.
Phosphorylation of ERK on Its Activation Loop Is Important, but Not Necessary, for Its Noncatalytic Function
Dual phosphorylation of the TEY site in the activation loop of ERK1/2 is essential for its activation conformational changes (60). We determined the significance of these phosphorylations for the role of noncatalytic ERK mutants using a rat ERK2 mutant in which the Thr and Tyr residues are switched with Ala and Phe, respectively (ERK2-TY/AF). In ERK1/2 doubly knocked down U251Raf cells, titrated expression of ERK2-K52R or ERK2-TY/AF further depleted ERK1/2 catalytic activity, as indicated by decreased p90RSK phosphorylation, whereas wild type ERK2 restored Raf-induced p90RSK phosphorylation (Fig. 7A). When Raf was activated, the cells expressing ERK2-K52R clearly exhibited p21CIP1 up-regulation and E2F1 down-regulation at similar levels to the cells expressing wild type ERK2 (Fig. 7A). However, cells expressing ERK2-TY/AF showed only mild changes, which was various depending on the dose of virus used (Fig. 7A). When cell proliferation rates were compared, ERK2-K52R expression restored Raf-induced growth arrest, although not as effectively as wild type ERK2, whereas ERK2-TY/AF could not (Fig. 7A). This clearly contrasts the difference among wild type ERK2, the active site mutant, and the activation loop mutant. In ERK1/2 knocked down TTRaf cells, ERK2-K52R partially restored Raf-mediated E2F1 down-regulation but not RET down-regulation, whereas ERK2-TY/AF could not restore anything (Fig. 7B). Nevertheless, Raf-mediated growth arrest was not restored by ERK2-K52R in TT cells (Fig. 7B), indicating that TT cells are less reliant on the noncatalytic ERK1/2 function. In the ERK1/2 knocked down LNCaPRaf cells, the difference between the active site mutant and the activation loop mutant was not as clearly contrasted as in U251 cells. Although slightly less efficient, ERK2-TY/AF could also restore the growth arrest signaling, as determined by the surrogate markers and proliferation rates (Fig. 7C). These data indicate that the activating phosphorylation of the TEY site is important for the non-kinase function of ERK1/2, although not necessary, and that different cell types have different levels of susceptibility to the growth arrest signaling promoted by the noncatalytic ERK mutants. Taken together, our findings strongly suggest that different levels of kinase activity as well as morality thresholds of ERK1/2 are involved in regulating growth arrest signaling of the Raf/MEK/ERK pathway and that ERK1/2 utilizes not only its “canonical” kinase activity but also its, as yet unidentified, noncatalytic function to mediate the signaling.
DISCUSSION
In this study, we present several lines of evidence demonstrating the following: (i) ERK1/2 is necessary for Raf/MEK-induced growth inhibitory signaling to occur; (ii) the growth arrest signaling involves different mechanisms of ERK1/2 action that contrast its catalytic activity and its, as yet unidentified, noncatalytic function; and (iii) the novel noncatalytic ERK1/2 function is also affected by its phosphorylation status. First, depletion of ERK1 and ERK2 using RNA interference abrogates Raf-induced growth inhibitory signaling in LNCaP, U251, and TT cells, which are characterized by growth arrest accompanied by morphological changes and up/down-regulation of several key regulators of cell proliferation. Second, as opposed to ERK1/2 depletion, expression of kinase-deficient ERK1 or ERK2 mutants does not block Raf-induced growth arrest, although it effectively depletes cells of ERK kinase activity and selectively blocks certain downstream incidents (e.g. morphological changes, c-Myc down-regulation in LNCaP, and RET down-regulation in TT cells). Third, introduction of kinase-deficient ERK2 into ERK1/2-depleted cells selectively restores Raf-induced growth arrest (e.g. selective restoration of Raf control on cell cycle regulators). Finally, expression of ERK2-K52R was more effective than expression of ERK2-TY/AF in restoring Raf-mediated growth arrest signaling.
Heretofore, participation of ERK1/2 in Raf-induced growth inhibitory signaling was mainly supported by two indirect pieces of evidence. First, the MEK1/2 inhibitors, U0126 and PD98059, abrogate the growth inhibitory signaling, as reported previously by us and others (12, 16, 22, 37). These inhibitors have high specificity to MEK1/2, which is expected for an inhibitor not competitive with ATP (61, 62), and were tested for various kinases (63); this makes the possibility unlikely that other kinases are also involved. Second, ERK1/2 is the only known substrate of MEK1/2 (56). Taken together, these findings strongly suggested the essential requirement of ERK1/2 for Raf-mediated growth arrest. Our study using RNA interference clearly demonstrates direct evidence that ERK1/2 is necessary for the growth inhibitory signaling to occur and, furthermore, that ERK1 and ERK2 have overlapping roles in that signaling context. ERK1 and ERK2 are highly homologous. Nevertheless, gene deletion studies in mice have shown distinct roles of ERK1 and ERK2 at different stages of development, including embryonic stem cell lineage commitment, T cell development, thymocyte maturation, and trophoblast development, with the characterization of ERK2 as being more important (38, 39, 64, 65). The significance of ERK2 over ERK1 for cell proliferation and survival has also been suggested in a study conducted in NIH3T3 cells using RNA interference (44). However, a more recent study suggests that ERK1 and ERK2 activities are indistinguishable and that the expression levels of ERK1 and ERK2 drive their biological differences in vitro and in vivo (43). Our data also indicate overlapping or interchangeable roles for ERK1 and ERK2 in Raf-mediated growth inhibition based on the following: (i) LNCaP expresses similar levels of ERK1 and ERK2; (ii) depletion of ERK1 or ERK2 similarly affects LNCaP responses to Raf activation; and (iii) ERK1 and ERK2 mutants display similar abilities to mediate growth arrest.
In this study, we generated tumor cell lines, in which ERK1 and ERK2 are stably knocked down. These cell lines are likely to provide a unique advantage in characterizing molecular mechanisms of ERK1/2 signaling in the context of the pathway-induced growth arrest, which might not be available in other cell types with higher sensitivity to ERK1/2 depletion. These tumor cells no longer responded to Raf-induced growth arrest signaling. Controlled expression of ERK2 restored Raf responsiveness of these cells. Nevertheless, RNA interference, by nature, cannot completely abrogate a gene expression. Therefore, it remains possible that the residual ERK1/2 present at low levels may still sustain cell survival and proliferation of these tumor lines. Likewise, it also remains possible that the residual ERK1/2 may have exerted a role in restoring the growth arrest signaling (discussed below).
In many studies, mainly in the context of cell survival and proliferation, kinase activity has been characterized as the key biochemical property required for ERK action. Indeed, inhibition of ERK1/2 activity using the kinase-deficient ERK2 mutants used in our study is sufficient to block Raf-induced CC139 cell proliferation or NIH3T3 cell transformation (40, 41). Interestingly, our study suggests that ERK1/2 signaling in the opposite biological context (growth arrest) requires not only its kinase activity but also its noncatalytic function. In addition, p90RSK and Elk1, which have been characterized as the key mediators of ERK signaling for cell survival and proliferation (66, 67), do not appear to be necessary for ERK-mediated growth arrest. Therefore, we suggest that the key mechanistic distinction between the Raf/MEK/ERK pathway-mediated opposing “proliferation” and “growth arrest” signaling is determined at the level of ERK. Noncatalytic ERK function was also contrasted by the different regulations of several important proteins that have the potential to influence cell proliferation (i.e. c-Myc, Rb, E2F1, and p21CIP1 in LNCaP cells; p21CIP1 and E2F1 in U251; and RET and E2F1 in TT cells). Because diverse intermediate pathways are mobilized to mediate the growth arrest signaling in different cell types (described in the Introduction) and it would involve different ERK signaling mechanisms according to our current findings, the different susceptibility to the novel ERK signaling detected in LNCaP, U251, and TT cells may indicate that different growth arrest-specific ERK targets are activated via different ERK signaling mechanisms in a cell type-specific manner to achieve the same goal (i.e. growth arrest) in response to aberrant pathway activation.
It will be necessary to understand the mechanisms by which the noncatalytic ERK1/2 mutants promoted the growth arrest signaling. Dual phosphorylation on the TEY site appears important for the action of the noncatalytic ERK mutants, indicating that MEK1/2 catalysis is also required to stimulate the noncatalytic ERK function. This is coherent with the result that MEK1/2 is essential and sufficient for growth arrest to occur. In support of this, compelling evidence was demonstrated in an in vitro reaction that kinase-deficient ERK2 (K52R) in “active” conformation could activate DNA topoisomerase IIα through a physical interaction (4). The phosphorylation-deficient ERK2-T183A/Y185F showed lower but some degree of capability to restore growth arrest signaling depending on its molarity and cell types. Extrapolating from this and the fact that biphosphodimers of ERK1/2 generate much higher activity than monophosphodimers and phosphomonomers (68), we speculate that a role of the noncatalytic mutants is to promote the activity of other proteins via protein-protein interactions that are augmented by ERK phosphorylation. Indeed, more recently, it was reported that ERK1/2 can act as a spatial regulator between other ERK1/2 and scaffolds to promote the formation of a signaling complex, which is important in determining ERK substrate specificity as well as subcellular localization (69). Similarly, in the ERK1/2-depleted cells, the catalytically inactive but conformationally active ERK proteins may have helped the residual catalytically active ERK1/2 to reach a specific intracellular compartment where the growth arrest-specific targets reside, although this possibility is diminished by the rescue experiments in which the residual ERK1/2 kinase activity was further depleted. It may also be speculative that ERK1/2 helped other MAPK signaling in a similar way as ERK1/2 can interact with a certain MAPK such as the p38α isoform, Mxi2 (70). This is an interesting possibility to test because p38 has recently been characterized as an important downstream mediator of Ras/Raf-induced senescence for the control of several cell cycle regulators, including p21CIP1 (30, 71). Finally, a growing number of evidence indicates that a variety of noncatalytic adaptor proteins are involved in MAPK signaling (2, 3). Therefore, it is also conceivable that ERK1/2 may form a unique signaling complex through protein-protein interactions to mediate noncatalytic signaling. An important aspect taken into consideration for future work will be to identify the proteins that specifically interact with ERK1/2 to mediate growth arrest and also to identify residues and/or motifs of ERK involved in the signaling.
Complicated mechanisms involving the magnitude of signaling intensity, spatio-temporal control, negative feedback regulation, or different scaffolds and regulators play key roles in determining physiological outputs of Raf/MEK/ERK signaling (2, 3, 72). The noncatalytic function of EKR1/2 that we have shown to be involved in the growth arrest signaling may be an important part of this complex signaling repertoire.
Supplementary Material
Acknowledgments
We thank Natalie Ahn and Melanie Cobb for MEK and ERK genes and Amy Hudson, Stephen Duncan, and Richard Mulligan for lentiviral vectors. We are also grateful to Barry Nelkin and Andrew Chan for critical reading of the manuscript.
This work was supported by FAMRI Young Investigator Award 062438, Department of Defense Prostate Cancer Research Program Grant W81XWH-07-01-0089, American Cancer Society IRG Grant 2204256, and the Wisconsin Breast Cancer Showhouse (to J. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental data 1–5.
- ERK
- extracellular signal-regulated kinase
- MTT
- 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide
- MAPK
- mitogen-activated protein kinase
- MEK
- MAPK kinase
- shRNA
- small hairpin RNA
- 4-HT
- 4-hydroxytamoxifen
- Rb
- retinoblastoma
- pRb
- phosphorylated Rb
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- GFP
- green fluorescent protein.
REFERENCES
- 1.Yoon S., Seger R. (2006) Growth Factors 24, 21–44 [DOI] [PubMed] [Google Scholar]
- 2.Kolch W. (2005) Nat. Rev. Mol. Cell Biol. 6, 827–837 [DOI] [PubMed] [Google Scholar]
- 3.Chuderland D., Seger R. (2005) Mol. Biotechnol. 29, 57–74 [DOI] [PubMed] [Google Scholar]
- 4.Shapiro P. S., Whalen A. M., Tolwinski N. S., Wilsbacher J., Froelich-Ammon S. J., Garcia M., Osheroff N., Ahn N. G. (1999) Mol. Cell Biol. 19, 3551–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambard J. C., Lefloch R., Pouysségur J., Lenormand P. (2007) Biochim. Biophys. Acta 1773, 1299–1310 [DOI] [PubMed] [Google Scholar]
- 6.McCubrey J. A., Steelman L. S., Chappell W. H., Abrams S. L., Wong E. W., Chang F., Lehmann B., Terrian D. M., Milella M., Tafuri A., Stivala F., Libra M., Basecke J., Evangelisti C., Martelli A. M., Franklin R. A. (2007) Biochim. Biophys. Acta 1773, 1263–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roberts P. J., Der C. J. (2007) Oncogene 26, 3291–3310 [DOI] [PubMed] [Google Scholar]
- 8.Michaloglou C., Vredeveld L. C., Soengas M. S., Denoyelle C., Kuilman T., van der Horst C. M., Majoor D. M., Shay J. W., Mooi W. J., Peeper D. S. (2005) Nature 436, 720–724 [DOI] [PubMed] [Google Scholar]
- 9.Collado M., Gil J., Efeyan A., Guerra C., Schuhmacher A. J., Barradas M., Benguría A., Zaballos A., Flores J. M., Barbacid M., Beach D., Serrano M. (2005) Nature 436, 642. [DOI] [PubMed] [Google Scholar]
- 10.Braig M., Lee S., Loddenkemper C., Rudolph C., Peters A. H., Schlegelberger B., Stein H., Dörken B., Jenuwein T., Schmitt C. A. (2005) Nature 436, 660–665 [DOI] [PubMed] [Google Scholar]
- 11.Serrano M., Lin A. W., McCurrach M. E., Beach D., Lowe S. W. (1997) Cell 88, 593–602 [DOI] [PubMed] [Google Scholar]
- 12.Zhu J., Woods D., McMahon M., Bishop J. M. (1998) Genes Dev. 12, 2997–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin A. W., Barradas M., Stone J. C., van Aelst L., Serrano M., Lowe S. W. (1998) Genes Dev. 12, 3008–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mabry M., Nakagawa T., Baylin S., Pettengill O., Sorenson G., Nelkin B. (1989) J. Clin. Invest. 84, 194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakagawa T., Mabry M., de Bustros A., Ihle J. N., Nelkin B. D., Baylin S. B. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 5923–5927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ravi R. K., Weber E., McMahon M., Williams J. R., Baylin S., Mal A., Harter M. L., Dillehay L. E., Claudio P. P., Giordano A., Nelkin B. D., Mabry M. (1998) J. Clin. Invest. 101, 153–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ravi R. K., McMahon M., Yangang Z., Williams J. R., Dillehay L. E., Nelkin B. D., Mabry M. (1999) J. Cell. Biochem. 72, 458–469 [DOI] [PubMed] [Google Scholar]
- 18.Carson E. B., McMahon M., Baylin S. B., Nelkin B. D. (1995) Cancer Res. 55, 2048–2052 [PubMed] [Google Scholar]
- 19.Ravi R. K., Thiagalingam A., Weber E., McMahon M., Nelkin B. D., Mabry M. (1999) Am. J. Respir. Cell Mol. Biol. 20, 543–549 [DOI] [PubMed] [Google Scholar]
- 20.Wood K. W., Qi H., D'Arcangelo G., Armstrong R. C., Roberts T. M., Halegoua S. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 5016–5020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fanton C. P., McMahon M., Pieper R. O. (2001) J. Biol. Chem. 276, 18871–18877 [DOI] [PubMed] [Google Scholar]
- 22.Park J. I., Strock C. J., Ball D. W., Nelkin B. D. (2003) Mol. Cell. Biol. 23, 543–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park J. I., Powers J. F., Tischler A. S., Strock C. J., Ball D. W., Nelkin B. D. (2005) Exp. Cell Res. 303, 79–88 [DOI] [PubMed] [Google Scholar]
- 24.Kim E. J., Park J. I., Nelkin B. D. (2005) J. Biol. Chem. 280, 4913–4920 [DOI] [PubMed] [Google Scholar]
- 25.Groth A., Weber J. D., Willumsen B. M., Sherr C. J., Roussel M. F. (2000) J. Biol. Chem. 275, 27473–27480 [DOI] [PubMed] [Google Scholar]
- 26.Malumbres M., Pérez De Castro I., Hernández M. I., Jiménez M., Corral T., Pellicer A. (2000) Mol. Cell Biol. 20, 2915–2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roper E., Weinberg W., Watt F. M., Land H. (2001) EMBO Rep. 2, 145–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olsen C. L., Gardie B., Yaswen P., Stampfer M. R. (2002) Oncogene 21, 6328–6339 [DOI] [PubMed] [Google Scholar]
- 29.Beauséjour C. M., Krtolica A., Galimi F., Narita M., Lowe S. W., Yaswen P., Campisi J. (2003) EMBO J. 22, 4212–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun P., Yoshizuka N., New L., Moser B. A., Li Y., Liao R., Xie C., Chen J., Deng Q., Yamout M., Dong M. Q., Frangou C. G., Yates J. R., 3rd, Wright P. E., Han J. (2007) Cell 128, 295–308 [DOI] [PubMed] [Google Scholar]
- 31.Ye X., Zerlanko B., Kennedy A., Banumathy G., Zhang R., Adams P. D. (2007) Mol. Cell 27, 183–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S., Fang X., Hall H., Yu S., Smith D., Lu Z., Fang D., Liu J., Stephens L. C., Woodgett J. R., Mills G. B. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5248–5253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wajapeyee N., Serra R. W., Zhu X., Mahalingam M., Green M. R. (2008) Cell 132, 363–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takahashi A., Ohtani N., Yamakoshi K., Iida S., Tahara H., Nakayama K., Nakayama K. I., Ide T., Saya H., Hara E. (2006) Nat. Cell Biol. 8, 1291–1297 [DOI] [PubMed] [Google Scholar]
- 35.Lee A. C., Fenster B. E., Ito H., Takeda K., Bae N. S., Hirai T., Yu Z. X., Ferrans V. J., Howard B. H., Finkel T. (1999) J. Biol. Chem. 274, 7936–7940 [DOI] [PubMed] [Google Scholar]
- 36.Park J. I., Strock C. J., Ball D. W., Nelkin B. D. (2005) Cytokine 29, 125–134 [DOI] [PubMed] [Google Scholar]
- 37.Carson-Walter E. B., Smith D. P., Ponder B. A., Baylin S. B., Nelkin B. D. (1998) Oncogene 17, 367–376 [DOI] [PubMed] [Google Scholar]
- 38.Pagès G., Guérin S., Grall D., Bonino F., Smith A., Anjuere F., Auberger P., Pouysségur J. (1999) Science 286, 1374–1377 [DOI] [PubMed] [Google Scholar]
- 39.Saba-El-Leil M. K., Vella F. D., Vernay B., Voisin L., Chen L., Labrecque N., Ang S. L., Meloche S. (2003) EMBO Rep. 4, 964–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pagès G., Lenormand P., L'Allemain G., Chambard J. C., Meloche S., Pouysségur J. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 8319–8323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kortenjann M., Thomae O., Shaw P. E. (1994) Mol. Cell Biol. 14, 4815–4824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bessard A., Fremin C., Ezan F., Fautrel A., Gailhouste L., Baffet G. (2008) Oncogene [DOI] [PubMed] [Google Scholar]
- 43.Lefloch R., Pouysségur J., Lenormand P. (2008) Mol. Cell Biol. 28, 511–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vantaggiato C., Formentini I., Bondanza A., Bonini C., Naldini L., Brambilla R. (2006) J. Biol. 5, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samuels M. L., Weber M. J., Bishop J. M., McMahon M. (1993) Mol. Cell. Biol. 13, 6241–6252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vindeløv L. L., Christensen I. J., Nissen N. I. (1983) Cytometry 3, 323–327 [DOI] [PubMed] [Google Scholar]
- 47.Mostoslavsky G., Fabian A. J., Rooney S., Alt F. W., Mulligan R. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16406–16411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rubinson D. A., Dillon C. P., Kwiatkowski A. V., Sievers C., Yang L., Kopinja J., Rooney D. L., Zhang M., Ihrig M. M., McManus M. T., Gertler F. B., Scott M. L., Van Parijs L. (2003) Nat. Genet. 33, 401–406 [DOI] [PubMed] [Google Scholar]
- 49.Frost J. A., Geppert T. D., Cobb M. H., Feramisco J. R. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 3844–3848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mansour S. J., Matten W. T., Hermann A. S., Candia J. M., Rong S., Fukasawa K., Vande Woude G. F., Ahn N. G. (1994) Science 265, 966–970 [DOI] [PubMed] [Google Scholar]
- 51.Lewis T. S., Hunt J. B., Aveline L. D., Jonscher K. R., Louie D. F., Yeh J. M., Nahreini T. S., Resing K. A., Ahn N. G. (2000) Mol. Cell 6, 1343–1354 [DOI] [PubMed] [Google Scholar]
- 52.Woods D., Parry D., Cherwinski H., Bosch E., Lees E., McMahon M. (1997) Mol. Cell Biol. 17, 5598–5611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abbas T., Dutta A. (2009) Nat. Rev. Cancer 9, 400–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.La Thangue N. B. (2003) Nat. Cell Biol. 5, 587–589 [DOI] [PubMed] [Google Scholar]
- 55.Zhuang D., Mannava S., Grachtchouk V., Tang W. H., Patil S., Wawrzyniak J. A., Berman A. E., Giordano T. J., Prochownik E. V., Soengas M. S., Nikiforov M. A. (2008) Oncogene 27, 6623–6634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shaul Y. D., Seger R. (2007) Biochim. Biophys. Acta 1773, 1213–1226 [DOI] [PubMed] [Google Scholar]
- 57.Lazar D. F., Wiese R. J., Brady M. J., Mastick C. C., Waters S. B., Yamauchi K., Pessin J. E., Cuatrecasas P., Saltiel A. R. (1995) J. Biol. Chem. 270, 20801–20807 [DOI] [PubMed] [Google Scholar]
- 58.Robbins D. J., Zhen E., Owaki H., Vanderbilt C. A., Ebert D., Geppert T. D., Cobb M. H. (1993) J. Biol. Chem. 268, 5097–5106 [PubMed] [Google Scholar]
- 59.Turjanski A. G., Hummer G., Gutkind J. S. (2009) J. Am. Chem. Soc. 131, 6141–6148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Seger R., Ahn N. G., Posada J., Munar E. S., Jensen A. M., Cooper J. A., Cobb M. H., Krebs E. G. (1992) J. Biol. Chem. 267, 14373–14381 [PubMed] [Google Scholar]
- 61.Duncia J. V., Santella J. B., 3rd, Higley C. A., Pitts W. J., Wityak J., Frietze W. E., Rankin F. W., Sun J. H., Earl R. A., Tabaka A. C., Teleha C. A., Blom K. F., Favata M. F., Manos E. J., Daulerio A. J., Stradley D. A., Horiuchi K., Copeland R. A., Scherle P. A., Trzaskos J. M., Magolda R. L., Trainor G. L., Wexler R. R., Hobbs F. W., Olson R. E. (1998) Bioorg. Med. Chem. Lett. 8, 2839–2844 [DOI] [PubMed] [Google Scholar]
- 62.Favata M. F., Horiuchi K. Y., Manos E. J., Daulerio A. J., Stradley D. A., Feeser W. S., Van Dyk D. E., Pitts W. J., Earl R. A., Hobbs F., Copeland R. A., Magolda R. L., Scherle P. A., Trzaskos J. M. (1998) J. Biol. Chem. 273, 18623–18632 [DOI] [PubMed] [Google Scholar]
- 63.Davies S. P., Reddy H., Caivano M., Cohen P. (2000) Biochem. J. 351, 95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Binétruy B., Heasley L., Bost F., Caron L., Aouadi M. (2007) Stem Cells 25, 1090–1095 [DOI] [PubMed] [Google Scholar]
- 65.Fischer A. M., Katayama C. D., Pagès G., Pouysségur J., Hedrick S. M. (2005) Immunity 23, 431–443 [DOI] [PubMed] [Google Scholar]
- 66.Anjum R., Blenis J. (2008) Nat. Rev. Mol. Cell Biol. 9, 747–758 [DOI] [PubMed] [Google Scholar]
- 67.Sharrocks A. D. (2002) Biochem. Soc. Trans. 30, 1–9 [DOI] [PubMed] [Google Scholar]
- 68.Philipova R., Whitaker M. (2005) J. Cell Sci. 118, 5767–5776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Casar B., Pinto A., Crespo P. (2008) Mol. Cell 31, 708–721 [DOI] [PubMed] [Google Scholar]
- 70.Sanz-Moreno V., Casar B., Crespo P. (2003) Mol. Cell Biol. 23, 3079–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Han J., Sun P. (2007) Trends Biochem. Sci. 32, 364–371 [DOI] [PubMed] [Google Scholar]
- 72.Ebisuya M., Kondoh K., Nishida E. (2005) J. Cell Sci. 118, 2997–3002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.