Abstract
Lewis lung carcinoma-derived high metastatic A11 cells constitutively overexpress hypoxia-inducible factor (HIF)-1α mRNA compared with low metastatic P29 cells. Because A11 cells exclusively possess a G13997A mutation in the mitochondrial NADH dehydrogenase subunit 6 (ND6) gene, we addressed here a causal relationship between the ND6 mutation and the activation of HIF-1α transcription, and we investigated the potential mechanism. Using trans-mitochondrial cybrids between A11 and P29 cells, we found that the ND6 mutation was directly involved in HIF-1α mRNA overexpression. Stimulation of HIF-1α transcription by the ND6 mutation was mediated by overproduction of reactive oxygen species (ROS) and subsequent activation of phosphatidylinositol 3-kinase (PI3K)-Akt and protein kinase C (PKC) signaling pathways. The up-regulation of HIF-1α transcription was abolished by mithramycin A, an Sp1 inhibitor, but luciferase reporter and chromatin immunoprecipitation assays indicated that Sp1 was necessary but not sufficient for HIF-1α mRNA overexpression in A11 cells. On the other hand, trichostatin A, a histone deacetylase (HDAC) inhibitor, markedly suppressed HIF-1α transcription in A11 cells. In accordance with this, HDAC activity was high in A11 cells but low in P29 cells and in A11 cells treated with the ROS scavenger ebselene, the PI3K inhibitor LY294002, and the PKC inhibitor Ro31-8220. These results suggest that the ROS-generating ND6 mutation increases HIF-1α transcription via the PI3K-Akt/PKC/HDAC pathway, leading to HIF-1α protein accumulation in hypoxic tumor cells.
INTRODUCTION
Somatic mutations in mitochondrial DNA (mtDNA) have been shown to accumulate in cancer cells and proposed to contribute to the progression of cancers of a variety of tissue origins. Mitochondria are the key regulators of the oxidative phosphorylation system that is composed of five complexes (I–V). Some somatic mtDNA mutations are envisioned as inhibiting the electron transport chain, resulting in a marked increase in mitochondrial reactive oxygen species (ROS)3 production (1). Actually, for example, a heteroplasmic frameshift mtDNA mutation in the NADH dehydrogenase subunit 5 (ND5) gene and a deletion mutant of cytochrome B (CYTB) gene promote ROS generation (2, 3). In addition, we have recently reported that a missense mutation in the ND6 gene causes the reduction of complex I activity, ROS overproduction, and increased metastatic potential of Lewis lung carcinoma cells (4).
Hypoxia is a common characteristic of locally advanced solid tumors. Hypoxic tumor cells activate many genes, including those related to cell survival, glycolysis, and angiogenesis, and invasion and metastasis to adapt to and escape from the microenvironment (5, 6). The oxygen-sensing mechanisms have been studied extensively and revealed hypoxia-inducible factors (HIFs) as the key regulatory transcription factors that are composed of HIF-α subunit and HIF-β/ARNT subunit. Under normoxic conditions, the α subunit (HIF-1α) is hydroxylated at Pro564 and Pro402 by specific Fe2+, oxoglutarate, and oxygen-dependent prolyl hydroxylases, recognized and ubiquitinated by an E3 ubiquitin ligase complex consisting of the tumor suppressor VHL (von Hippel-Lindau), elongin B and elongin C, and rapidly degraded through the ubiquitin-proteasome pathway, whereas the β subunit of HIF-1 (HIF-1β) is constitutively expressed. Under hypoxic conditions, HIF-1α protein is stabilized, allowing its nuclear translocation and dimerization with HIF-1β. In the nucleus, HIF binds to the hypoxia response element of hypoxia-inducible genes, including vascular endothelial growth factor (VEGF), and transactivates their transcription (5, 6).
Elevated HIF-1α protein levels are commonly observed in many tumor tissues and associated with increased angiogenesis, resistance to apoptosis and chemo- and radiotherapy, and poor patient prognosis (6, 7). Hypoxia generated by aberrant vasculature formation and high interstitial pressure is undoubtedly a major factor, but other factors such as activation of HIF-1α gene transcription may also play a role in up-regulation of HIF-1α protein in tumor tissues. Actually, we and others have reported the up-regulation of HIF-1α mRNA in some tumor types (8–10). Although the precise mechanism of HIF-1α gene activation is largely unknown, an increase in gene dosage is reported as one of the mechanisms of constitutive up-regulation of HIF-1α mRNA expression (9, 10).
ROS are the physiological mediators to stabilize and increase the transcriptional activity of HIF-1α protein. Incubation of cells with H2O2 or an oxidative stressor leads to the stabilization of HIF-1α protein and activation of HIF target genes under normoxic conditions (11). Conversely, treatment of cells with antioxidants such as N-acetylcysteine and glutathione attenuates HIF-1α protein accumulation and the expressions of HIF target genes in various cell types (11). HIF-1α protein levels increase under normoxia in response to growth factors, hormones, coagulation factors, cytokines, and vasoactive peptides, which also stimulate ROS generation (12, 13). Mitochondria-derived ROS produced by electron transport chain complex III are also reported to be able to stabilize HIF-1α protein under hypoxic conditions (14). Although the stabilization of HIF-1α protein by ROS has been highlighted, HIF-1α mRNA expression is also stimulated by ROS from NADPH oxidase (15).
So far, there are no reports of the involvement of mtDNA mutations in the activation of the HIF-1α gene. Given a high frequency of mtDNA mutation rate in tumor cells and ROS-mediated HIF-1α accumulation at both the protein and mRNA levels, we reasoned that mtDNA mutations could be a cause of HIF-1α transcriptional activation. In the present study, we addressed this issue and investigated the potential mechanism. We report here that certain ROS-generating mtDNA mutations can stimulate HIF-1α transcription via the phosphatidylinositol 3-kinase (PI3K)/protein kinase C (PKC)/histone deacetylase (HDAC) pathway.
EXPERIMENTAL PROCEDURES
Reagents
Actinomycin D was purchased from Sigma-Aldrich; and SB203580, LY294002, Ro31-8220, ebselene, pyrrolidine dithiocarbamate (PDTC), trichostatin A (TSA), mithramycin A, sulfasarazine, and curcumin were from Calbiochem. SP600125 was obtained from TOCRIS Cookson, Ellisville, MO, and PD98059 was from Cell Signaling Technology, Beverly, MA.
Antibodies
Monoclonal anti-HIF-1α antibody was obtained from Novus Biologicals, Littleton, CO. Polyclonal anti-p38 MAP kinase, anti-phospho-p38 MAP kinase (Thr180/Tyr182), anti-p44/42 MAP kinase, anti-phospho-p44/42 MAP kinase (Thr202/Tyr185), anti-SAPK/JNK, anti-phospho-SAPK/JNK (Thr183/Tyr185), anti-Akt, and anti-phospho-Akt (Ser473), were purchased from Cell Signaling Technology. Polyclonal anti-Sp1, anti-Sp3, anti-E2F-1 antibodies and normal rabbit IgG were obtained from Santa Cruz Biotechnology, monoclonal anti-β-actin antibody was from Sigma-Aldrich, and polyclonal anti-acetylhistone H4 antibody was from Upstate, Charlottesville, VA.
Cell Lines and Culture Conditions
Low metastatic P29 and P34 cells and high metastatic D6 and A11 cells, all of which were derived from Lewis lung carcinoma, were characterized previously (4, 8, 16). Trans-mitochondrial cybrids were established as described previously (4). P29mtA11 cybrids carry nuclear DNA from P29 cells and mtDNA from A11 cells, and A11mtP29 cybrids carry nuclear DNA from A11 cells and mtDNA from P29 cells. As controls, P29mtP29 cybrids, which have nuclear DNA from P29 cells and mtDNA from P29 cells, and A11mtA11 cybrids, which have nuclear DNA from A11 cells and mtDNA from A11 cells, were used. Lewis lung carcinoma cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with heat-inactivated (56 °C, 30 min) fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Trans-mitochondrial cybrids were maintained in DMEM supplemented with heat-inactivated fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin, 0.01% pyruvate, and 0.005% uridine. For hypoxic culture, they were incubated under hypoxic conditions (1% O2) in a NAPCO® automatic O2/CO2 incubator (Precision Scientific, Chicago, IL).
Sequencing of the ND6 Gene
Total DNAs extracted from P29, P34, D6, and A11 cells were used for amplification of the ND6 gene. The primers used for PCRs were as follows: the forward primer (n.p. 14,030 to 14,053, 5′-CAATTTCACAGCACCAAATCTCCA-3′) and the reverse primer (n.p. 14,759 to 14,779, 5′-TATTAGGGGGTTAGTTTTGCG-3′). All PCR amplifications were performed in a 50 μl of solution consisting of 1× PCR buffer, 0.2 mm dinucleotide triphosphates, 0.6 μm primers, 1 unit of ExTaq DNA polymerase (TaKaRa BIO, Shiga, Japan), and 10 ng of genomic DNA as a template. Reaction conditions were 94 °C for 1 min with cycle times of 30 s for denaturation at 94 °C, 30 s for annealing at 53 °C, and 1 min for extension at 72 °C for 30 cycles. The final extension was for 1 min. Amplified ND6 fragments were separated on 1% agarose gels and extracted and then directly sequenced using a Big Dye Terminator version 3.1 cycle sequencing kit (Applied Biosystems).
Measurement and Visualization of ROS Generation
ROS generation was detected with 2′,7′-dichlorofluorescin diacetate (DCFH-DA) (Molecular Probes, Eugene, OR). Briefly, the cells cultured in 35-mm-diameter glass-bottom culture dishes (MatTeck, Ashland, MA) were incubated with 10 μm DCFH-DA for 10 min at 37 °C in serum-free DMEM, washed twice with Dulbecco's phosphate-buffered saline (DPBS), and then immediately observed under a confocal laser microscope (Fluoview; Olympus, Tokyo) or analyzed with a FACScan flow cytometer (Beckton Dickinson). Mean fluorescence intensity was analyzed using CellQuest software (Becton Dickinson).
RNA Isolation and Northern Blotting
Total RNA was extracted with guanidinium thiocyanate. Total RNA (20 μg) was electrophoresed on 1% agarose gels containing formaldehyde and transferred onto nylon filters. Blots were hybridized with a 32P-labeled mouse HIF-1α cDNA probe or a mouse VEGF cDNA probe (8), which was prepared by the random primer method. Filters were finally washed at 50 °C in 30 mm NaCl, 3 mm sodium citrate, and 0.1% SDS.
SDS-PAGE and Western Blotting
Total cell lysates were prepared by directly solubilizing cells in SDS sample buffer. For analyzes of phosphorylated proteins, cells were lysed in 1% Nonidet P-40, 150 mm NaCl, 10% glycerol, 2 mm EDTA, 20 mm Tris-HCl (pH 8.0), 1 mm dithiothreitol, 1 mm Na3VO4, 1 mm phenylmethylsulfonyl fluoride (PMSF), and protease inhibitor mixture (Roche Applied Science). Nuclear extracts were prepared using a nuclear extraction kit (Active Motif, Carlsbad, CA) according to the manufacturer's protocol. Proteins were resolved by SDS-PAGE under reducing conditions. Protein concentration was determined by the method of Bradford using bovine serum albumin as a standard. The resolved proteins were transferred electrophoretically to nitrocellulose membrane. After incubating with 5% dry milk in TBS-T (150 mm NaCl, 50 mm Tris-HCl (pH 7.4), 0.05% Tween 20) for at least 1 h at room temperature, the membrane was incubated with polyclonal or monoclonal antibody for the appropriate time, washed extensively with TBS-T, and then incubated with horseradish peroxidase-conjugated goat anti-rabbit or anti-mouse IgG, respectively. Proteins were detected using ECL Western blotting detection reagents (Amersham Biosciences).
Luciferase Reporter Plasmid Construction
Murine HIF-1α promoter from nucleotide −1958 to +93 (the transcriptional start site was defined as +1) inserted in the KpnI/SacI site of a luciferase reporter plasmid pGL2-basic, hereafter termed pGL2-HIFpro(−1958/+93), was a generous gift of Dr. C. A. Bradfield, University of Wisconsin Medical School (17). Various truncated forms of the promoter were made by utilizing restriction enzyme recognition sites in the promoter and the vector (XbaI, KpnI/BbrPI, and SacII for making pGL2-HIFpro(−1422/+93), pGL2-HIFpro(−293/+93), and pGL2-HIFpro(−150/+93), respectively) or by PCR using the 5′ primers carrying the KpnI site at the 5′ end and the 3′ primer carrying SacI site at the 3′ end, 5′-GGGAGCTCCGGCTCGGGTTC-3′. The 5′ primers are: 5′-GAGGTACCTCAAGGTCTGTAGGTTGA-3′ for pGL2-HIFpro (−1048/+93), 5′-GAGGTACCATAGCAAAGAGCGGAGAC-3′ for pGL2-HIFpro(−668/+93), 5′-GAGGTACCTTTCCCTCCCCTCGCCGC-3′ for pGL2-HIFpro(−101/+93), and 5′-GAGGTACCCTTCAGCGCCTCAGTGCA-3′ for pGL2-HIFpro(−38/+93). The amplified PCR products were inserted into the KpnI/SacI site of a pGL2-basic vector. A pGL2-HIFpro(−150/+93)mutSp1 plasmid that harbors a mutation in the putative Sp1 binding site was generated by using the QuikChange Site-directed Mutagenesis kit (Stratagene, La Jolla, CA). The sense and the antisense primers used were 5′-CGCCCTTGCCCGAACCCTGCCGCTGC-3′ and 5′-GCAGCGGCAGGGTTCGGGCAAGGGCG-3′, respectively.
Luciferase Reporter Assay
Transient transfection of the luciferase reporter constructs harboring the HIF-1α promoter sequence was carried out using Lipofectamine Plus (Invitrogen). As a control for transfection efficiency, pRL-TK vector (Promega, Madison, WI) was cotransfected with test plasmids. pGL2-control vector (Promega) was used as a positive control. Luciferase activity in cell extracts was assayed 45 h after transfection according to the Dual-Luciferase reporter assay system protocols (Promega) using a luminometer (model TD-20/20; Turner Designs, Sunnyvale, CA).
Electrophoretic Mobility Shift Assay (EMSA)
The nuclear protein fractions for EMSA were prepared as described above. Consensus Sp1 (wtSp1, 5′-ATTCGATCGGGGCGGGGCGAGC-3′) and mutant Sp1 (mutSp1, 5′-ATTCGATCGTTCGGGGCGAGC-3′) double-stranded oligonucleotides were purchased from Santa Cruz Biotechnology. The HIF-1α gene promoter-specific double-stranded oligonucleotide probe (positions −72 to −48) (wtHIFpro-Sp1) or its mutant form (mutHIFpro-Sp1) was prepared by annealing the sense 5′-CGCCTCCGCCCTTGCCCGCCCCCTG-3′ and the antisense 5′-CAGGGGGCGGGCAAGGGCGGAGGCG-3′ oligonucleotides or the sense 5′-CGCCTCCGCCCTTGCCCGAACCCTG-3′ and the antisense 5′-CAGGGTTCGGGCAAGGGCGGAGGCG-3′, respectively. The probes were labeled using [γ-32P]ATP (Amersham Biosciences) and MEGALABELTM kit (TaKaRa BIO). Five micrograms of nuclear protein, 32P-labeled double-stranded probe (5000 cpm), 1 μg of poly(dI·dC), and 17 μl of binding buffer (20 mm Hepes (pH 7.9), 50 mm NaCl, 5% glycerol, 0.1 mm dithiothreitol) were mixed in a total volume of 20 μl. In competition assays, a 50-fold molar excess amount of unlabeled competitors was included in the reaction mixture. The mixture was incubated at room temperature for 30 min, then loaded on a 5% polyacrylamide gel in TGE buffer (50 mm Tris-HCl (pH 8.5), 380 mm glycine, 2 mm EDTA), and subjected to electrophoresis at 4 °C. The gel was dried and exposed to x-ray film at −70 °C. A supershift assay was performed using 10 μg of specific goat polyclonal anti-Sp1 or anti-Sp3 antibody.
Chromatin Immunoprecipitation Assay
Cells were fixed with 1% formaldehyde for 10 min at 37 °C, and the reaction was quenched by adding glycine to a final concentration of 125 mm. The cells were washed with DPBS containing 1 mm PMSF; centrifuged; swelled in 5 mm Hepes (pH 8.0), 85 mm KCl, 0.5% Nonidet P-40, 0.5 mm PMSF, 100 ng/ml leupeptin, 100 ng/ml aprotinin; incubated for 10 min on ice; and then lysed with a Dounce homogenizer. Nuclei were collected by centrifugation and resuspended in sonication buffer (1% SDS, 10 mm EDTA, 50 mm Tris-HCl (pH 8.0), 0.5 mm PMSF, 100 ng/ml leupeptin, 100 ng/ml aprotinin). The nuclei were sonicated on ice to an average length of 500 to 1000 bp and then centrifuged at 10,000 × g for 15 min at 4 °C. The chromatin solution was diluted 10-fold in chromatin immunoprecipitation dilution buffer (500 mm Tris-HCl (pH 8.0), 1670 mm NaCl, 11% Triton X-100, 1.1% sodium deoxycholate, 10 mm PMSF, 10 μg/ml leupeptin, 10 μg/ml aprotinin, 10 μg/ml pepstatin), precleared by the addition of protein A-Sepharose beads for 1 h at 4 °C. Precleared chromatin solution was incubated with 5 μg of anti-Sp1 antibody, anti-Sp3 antibody, or anti-acetylhistone H4 antibodies at 4 °C for 13 h. Normal rabbit IgG served as a control. Protein·DNA complexes were immunoprecipitated by protein A beads, and the cross-links were reversed by heating to 65 °C for 5 h. The DNA was recovered by phenol:chloroform extraction and precipitated by ethanol. Then, the association of Sp1, Sp3, and acetylated histone H4 with the Sp1/Sp3 recognition site in the HIF-1α promoter was examined by hot-start PCR using GoTaqDNA polymerase (Promega). The sense and the antisense primers used were 5′-ACCTCCTCCTGATTGGCTG-3′ (positions −258 to −239) and 5′-TCGCGTCCCCTCAGCCGA-3′ (positions −12 to +5), respectively. The PCR conditions were: 96 °C for 5 min, 30 cycles with 96 °C for 30 s, 57 °C for 30 s, and 72 °C for 30 s, and 72 °C for 5 min. Final PCR products were analyzed on 2% agarose gels with ethidium bromide staining.
CD31 Staining of Tumor Blood Vessels
P29mtP29, P29mtA11, A11mtA11, and A11mtP29 cells (1 × 106 cells) were inoculated subcutaneously into the abdominal flank of C57BL/6 mice. When an estimated tumor volume reached ∼2 cm3, subcutaneous tumors were surgically removed and immediately frozen in OCT compound. Cryostat sections (10-μm thickness) were made at every 100-μm distance, fixed with 4% paraformaldehyde, and then washed with DPBS. After blocking with 10% normal goat serum in DPBS, sections were incubated with rat anti-mouse CD31 antibody (1:100 dilutions) (BD Pharmingen) overnight at 4 °C. They were washed with DPBS and incubated with fluorescein isothiocyanate-conjugated goat anti-rat IgG. After extensive washing with DPBS, samples were counterstained with 1 μg/ml propidium iodide and observed under a confocal laser microscope (Fluoview). Images were captured and transferred to the ImageJ 1.34s software, and CD31-positive areas were analyzed. In this way, at least six randomly selected fields (0.2 cm2/field) were analyzed, and the percentage of CD31-positive area per field was calculated.
HDAC Activity Assay
Nuclear extracts were prepared using a nuclear extraction kit. HDAC activity was measured using a HDAC fluorometric assay/drug discovery kit (BioMol International, Plymouth Meeting, PA) according to the manufacturer's instruction. Briefly, 4.5-μg nuclear extracts were incubated with Fluor de Lys substrate buffer at 37 °C for 30 min followed by incubation with Fluor de Lys developer concentrate at 25 °C for 10 min. Fluorescence was measured with a multiwell plate reader (excitation at 355 nm and emission at 460 nm).
RESULTS
Activation of HIF-1α Gene Transcription in High Metastatic Cells
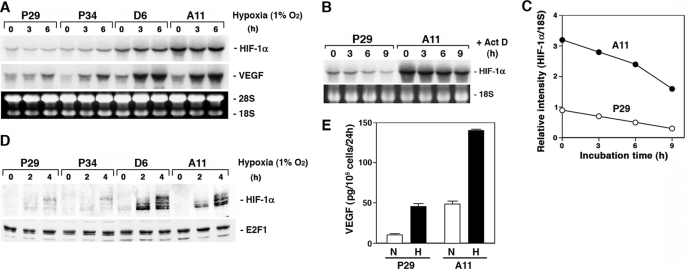
We compared the expression level of HIF-1α mRNA between the low metastatic (P29 and P34) and the high metastatic (D6 and A11) cells originated from Lewis lung carcinoma. The results showed that D6 and A11 cells expressed a larger amount of HIF-1α mRNA than P29 and P34 cells (Fig. 1A). Hypoxia did not affect the expression level of the mRNA. One of the possible mechanisms of HIF-1α mRNA up-regulation in D6 and A11 cells may be the difference in HIF-1α mRNA stability in the cells. To test this possibility, we cultured P29 and A11 cells in the presence of actinomycin D for up to 9 h and examined the mRNA level at each time point (Fig. 1B). The results showed that the half-life of HIF-1α mRNA in A11 cells was nearly equal to that in P29 cells (∼8 h) (Fig. 1C). Thus, the transcription of the HIF-1α gene was found to be more activated in A11 cells than in P29 cells.
FIGURE 1.
HIF-1α and VEGF expressions are higher in high metastatic D6 and A11 cells than in low metastatic P29 and P34 cells. A, P29, P34, D6, and A11 cells were cultured under hypoxic conditions (1% O2) for up to 6 h. Total RNA was extracted and subjected to Northern blot analysis. The blots were hybridized with either a 32P-labeled HIF-1α or VEGF cDNA probe. Ethidium bromide staining of the gel is also shown. B and C, P29 and A11 cells were incubated with 5 μg/ml actinomycin D (Act D) for up to 9 h. Total RNA was extracted and analyzed as above. Ethidium bromide staining of 18 S ribosomal RNA is also shown. After semiquantifying the intensities of bands, the levels of HIF-1α mRNA were normalized to those of 18 S ribosomal RNA. D, P29, P34, D6, and A11 cells were cultured under hypoxic conditions for up to 4 h. Nuclear extracts prepared from the cells were dissolved by SDS-PAGE. HIF-1α and E2F1, which served as a control, were detected by immunoblotting. E, P29 and A11 cells were cultured under normoxic (N) or hypoxic (H) conditions for 18 h. VEGF produced in the supernatants was measured by enzyme-linked immunosorbent assay. Bars, S.D.
Under normoxic conditions, HIF-1α protein was scarcely detected in both the low and the high metastatic cells. However, upon hypoxic exposure, HIF-1α protein level markedly increased in D6 and A11 cells compared with P29 and P34 cells (Fig. 1D). Accordingly, hypoxia enhanced the expression of VEGF in D6 and A11 cells more than in P29 and P34 cells at both the mRNA and protein levels (Fig. 1, A and E). Thus, the up-regulation of HIF-1α mRNA in D6 and A11 cells resulted in overexpression of HIF-1α under hypoxic conditions, leading to VEGF overexpression.
ND6 Mutation Activates HIF-1α Transcription
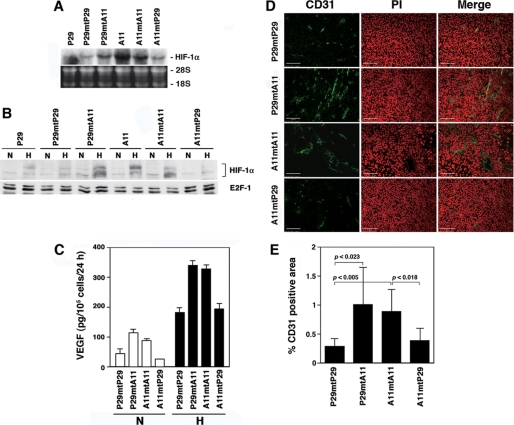
Sequencing of the ND6 gene revealed that D6 and A11 cells harbored a G13997A mutation, which changes evolutionally conserved proline 25 to leucine, whereas P29 and P34 cells did not (Fig. S1 and Table S1). To examine a causal relationship between the ND6 mutation and HIF-1α transcription, we examined HIF-1α mRNA levels in trans-mitochondrial cybrids, P29mtA11 and A11mP29 cells, which carry mtDNA from A11 and P29 cells and nuclear DNA from P29 and A11 cells, respectively. We used P29mtP29 and A11mtA11 cells as control cybrids. The results showed that the expression level of HIF-1α mRNA was higher in the cybrids with A11 mtDNA (P29mtA11 and A11mtA11) irrespective of whether their nuclear DNA is derived from P29 or A11 cells, than in the cybrids with mtDNA from P29 cells (P29mtP29 and A11mtP29) (Fig. 2A). Accordingly, HIF-1α protein and VEGF were highly induced in P29mtA11 and A11mtA11 cybrids under hypoxic conditions compared with A11mtP29 and P29mtP29 cybrids (Fig. 2, B and C). Furthermore, P29mtA11 and A11mtA11 cybrids showed enhanced angiogenesis in vivo (Fig. 2, D and E). These results indicate that the HIF-1α mRNA overexpression in A11 and D6 cells is attributed to the ND6 mutation.
FIGURE 2.
HIF-1α and VEGF expressions are high in the cybrids with mtDNA carrying the ND6 mutation. A, total RNA extracted from P29, A11, and the cybrids was subjected to Northern blot analysis. The blots were hybridized with a 32P-labeled HIF-1α cDNA. Ethidium bromide staining of the gel is also shown. B, P29 cells, A11 cells, and the cybrids were cultured under normoxic (N) or hypoxic (H) conditions for 4 h. Nuclear extracts prepared from the cells were dissolved by SDS-PAGE. HIF-1α and E2F1, which served as a control, were detected by immunoblotting. C, the cybrids were cultured under normoxic (N) or hypoxic (H) conditions for 18 h. VEGF produced in the supernatants was measured by enzyme-linked immunosorbent assay. Bars, S.D. D, cryostat sections prepared from subcutaneous tumors were stained with anti-CD31 antibody. Sections were counterstained with propidium iodide (PI). E, blood vessel density in subcutaneous tumors formed by the cybrids was assessed by staining with anti-CD31 antibody. The fluorescent images of at least six fields (0.2 cm2/field) were analyzed, and the percentage of CD31-positive area/field (columns) was calculated. Bars, S.D.
ROS Are Involved in HIF-1α Transcriptional Activation by the ND6 Mutation
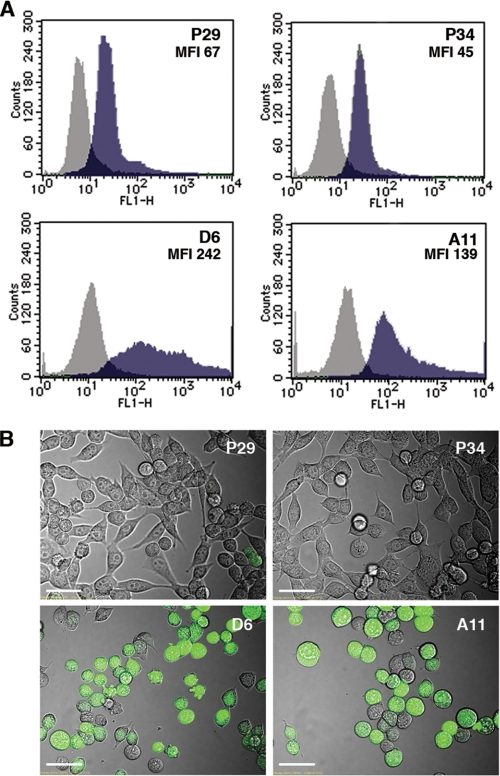
It is possible that mitochondrial ROS production caused by the ND6 mutation mediates the activation of HIF-1α transcription. To examine this possibility, we measured the intracellular ROS level. Fluorescence-activated cell sorter (FACS) analysis and confocal images showed that D6 and A11 cells produced a larger amount of ROS than P29 and P34 cells (Fig. 3). In addition, the cybrids with mtDNA from A11 cells (P29mtA11 and A11mtA11) overproduced ROS compared with the cybrids with mtDNA from P29 cells (P29mtP29 and A11mtP29) (Fig. S2). Thus, the ND6 mutation is correlated well with both ROS overproduction and HIF-1α mRNA up-regulation.
FIGURE 3.
ROS production is elevated in the cells with mtDNA carrying the ND6 mutation. A, P29, P34, D6, and A11 cells were incubated with 10 μm DCFH-DA for 10 min at 37 °C in serum-free DMEM and then immediately were analyzed with a FACScan flow cytometer. Mean fluorescence intensity (MFI) is also shown. B, the cells treated as above were observed under a confocal laser microscope.
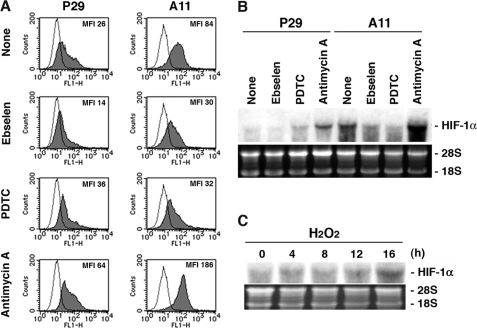
Next, to gain evidence of a causal relationship between ROS and HIF-1α mRNA expression, we examined the effects of general antioxidants ebselene and PDTC, and antimycin A, which inhibits electron transport pathway (18). FACS analysis showed that intracellular ROS level was low in ebselene- and in PDTC-treated cells whereas high in antimycin A-treated cells compared with untreated cells, showing more distinct changes in A11 cells than in P29 cells (Fig. 4A). Ebselene and PDTC effectively suppressed the expression of HIF-1α mRNA in A11 cells, whereas antimycin A increased the expression in both P29 and A11 cells (Fig. 4B). These results strongly suggest that the HIF-1α transcriptional activation is regulated by mitochondrial ROS. Supporting this, we found that exogenously added H2O2 stimulated the expression of HIF-1α mRNA in P29 cells (Fig. 4C).
FIGURE 4.
ROS production is correlated with HIF-1α mRNA expression. A, P29 and A11 cells were treated with solvent alone (dimethyl sulfoxide) (None), ebselen (20 μm), PDTC (20 μg/ml), and antimycin A (20 μg/ml) for 18 h. The cells were incubated with 10 μm DCFH-DA for 10 min at 37 °C in serum-free DMEM and then immediately were analyzed with a FACScan flow cytometer. Mean fluorescence intensity (MFI) is also shown. B, P29 and A11 cells were treated as above, and total extracted RNA was subjected to Northern blot analysis. The blots were hybridized with 32P-labeled HIF-1α cDNA. Ethidium bromide staining of the gel is also shown. C, P29 cells were treated with 25 μm H2O2 for up to 16 h. Total extracted RNA was analyzed as in B.
PI3K-Akt and PKC Signaling Pathways Are Involved in Mitochondrial ROS-mediated HIF-1α Overexpression
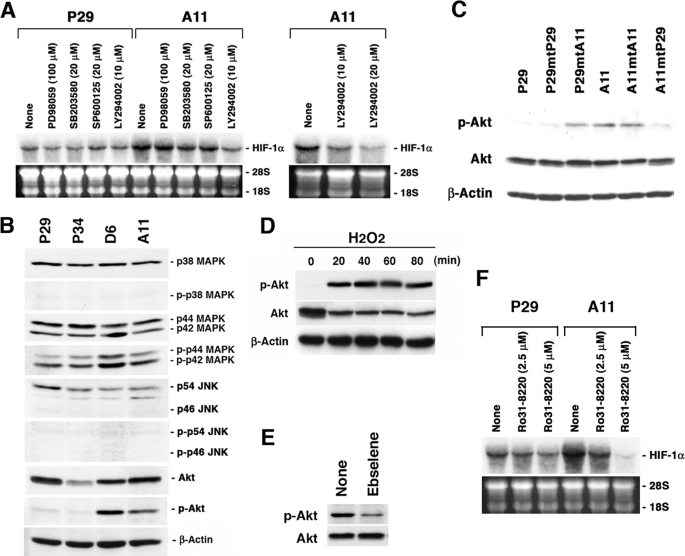
We next investigated signaling pathways of the mitochondrial ROS-mediated HIF-1α gene activation. For this, we treated P29 and A11 cells with PD98059, a MEK1 inhibitor, SB203580, a p38 MAP kinase inhibitor, SP600125, a JNK inhibitor, and LY294002, a PI3K inhibitor. As shown in Fig. 5A, LY294002 significantly inhibited HIF-1α mRNA expression in A11 cells in a dose-dependent manner, whereas PD98059, SB203580, and SP600125 did not, suggesting the involvement of the PI3K-Akt pathway in the mitochondrial ROS-mediated HIF-1α gene transcription. None of these inhibitors significantly suppressed HIF-1α mRNA expression in P29 cells, implying that the PI3K-Akt pathway does not contribute to the basal expression level of HIF-1α mRNA. In accordance with the data, Akt was highly activated, indicated by its protein phosphorylation, in D6 and A11 cells but not in P29 and P34 cells, and there was no consistent difference in the phosphorylation level of p38 MAP kinase, p44/42 MAP kinase, or JNK (Fig. 5B). Akt was also highly activated in P29mtA11 and A11mtA11 cybrids, but not in P29mtP29 and A11mtP29 cybrids (Fig. 5C). H2O2 strongly induced Akt activation in P29 cells in a time-dependent manner (Fig. 5D). Moreover, ebselene inhibited Akt phosphorylation in A11 cells (Fig. 5E). Thus, Akt phosphorylation is linked to the level of HIF-1α mRNA. In addition, Ro31-8220, a pan-specific PKC inhibitor, markedly inhibited HIF-1α mRNA expression in A11 cells, but only slightly in P29 cells, suggesting the involvement of PKC in the ROS-mediated HIF-1α mRNA expression (Fig. 5F). On the other hand, rapamycin, an mTOR inhibitor, sulfasarazine, an NF-κB inhibitor, and curcumin, an AP-1 inhibitor, did not significantly inhibit HIF-1α mRNA expression in A11 cells (Fig. S3). Collectively, these data indicate that the PI3K-Akt and PKC pathways are involved in the ROS-mediated HIF-1α transcriptional activation in A11 cells.
FIGURE 5.
PI3K-Akt and PKC pathways are involved in the ROS-mediated HIF-1α mRNA overexpression. A, P29 and A11 cells were treated with dimethyl sulfoxide, PD98059, SB203580, SP600125, and LY294002 at the indicated concentrations for 18 h. Total RNA was extracted and subjected to Northern blot analysis. The blots were hybridized with a 32P-labeled HIF-1α cDNA. Ethidium bromide staining of the gel is also shown. B, cell lysates prepared from P29, P34, D6, and A11 cells were dissolved by SDS-PAGE. Proteins and phosphorylated proteins and β-actin, which served as a loading control, were detected by immunoblotting. C, cell lysates prepared from P29, A11, and the cybrids were subjected to immunoblotting to detect Akt and phosphorylated Akt. β-Actin served as a loading control. D, P29 cells were treated with 25 μm H2O2 for up to 80 min. Cell lysates were prepared and subjected to immunoblotting as in C. E, A11 cells were treated with ebselene (20 μm) for 18 h. Cell lysates were prepared and subjected to immunoblotting as in C. F, P29 and A11 cells were treated with Ro31-8220 at the indicated concentrations for 18 h. Total RNA was analyzed as in A.
Sp1 Is Necessary but Not Sufficient for ROS-mediated HIF-1α Gene Activation
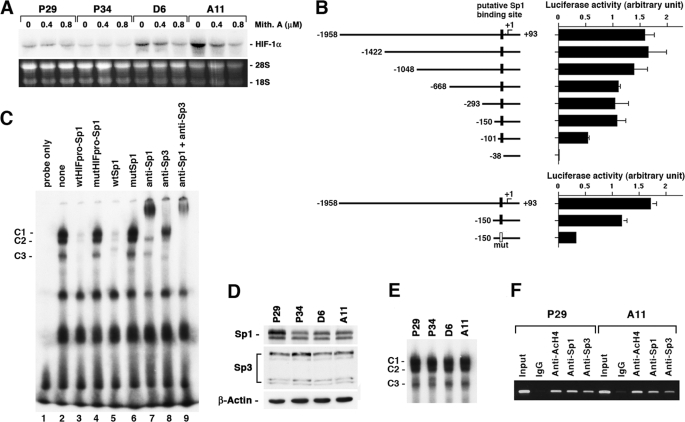
To gain further insight into the underlying mechanisms of HIF-1α gene activation in the high metastatic cell lines, we treated the cells with mithramycin A, an Sp1 inhibitor. The results showed that it significantly suppressed the expression of HIF-1α mRNA in D6 and A11 cells but not in P29 and P34 cells (Fig. 6A), suggesting the involvement of Sp1 in the ROS-mediated HIF-1α mRNA overexpression. To examine which region of the HIF-1α promoter is responsible for the activated transcription of the gene in A11 cells, we constructed luciferase reporter plasmids harboring the full-length (−1958/+93) and a series of truncated promoters (Fig. 6B). We transiently transfected them into A11 cells and examined their activities. The results showed that deletion of the region from −1958 to −101 effectively reduced the promoter activity compared with the full-length promoter, whereas deletion from −1958 to −150 did not significantly reduce the activity. Deletion of the region from −1958 to −38 abrogated the promoter activity, indicating that an important sequence for the promoter activity resides in the region from −149 to −38. Sequence analysis of this region using a computer software (TFSEARCH, Papia system) revealed a putative Sp1 binding sequence (−60/−51). Mutation of this sequence TGCCCGCCCC to TGCCCGAACC significantly reduced the promoter activity (Fig. 6B), demonstrating the importance of this sequence for the promoter activity.
FIGURE 6.
Sp1 is necessary but not sufficient for the ROS-mediated HIF-1α mRNA overexpression. A, P29, P34, D6, and A11 cells were treated with mithramycin A at the indicated concentrations for 24 h. Total RNA was extracted and subjected to Northern blot analysis. The blots were hybridized with a 32P-labeled HIF-1α cDNA. Ethidium bromide staining of the gel is also shown. B, pGL2-basic luciferase reporter plasmids harboring the full-length (−1958/+93) and a series of truncated HIF-1α promoters were transfected in A11 cells, and luciferase activity was assayed 45 h after transfection. Filled and open boxes indicate putative wild-type and mutated Sp1 binding sites, respectively. C, nuclear extracts prepared from A11 cells were subjected to EMSAs in which 32P-labeled oligonucleotides (HIFpro-Sp1) containing a putative Sp1 binding site was used as a probe. wtHIFpro-Sp1, mutHIFpro-Sp1, wtSp1, and mutSp1 indicate wild-type HIFpro-Sp1, mutated HIFpro-Sp1, consensus Sp1 and mutated Sp1 oligonucleotides, respectively. C1–C3 indicate specific Sp-dependent binding complexes. D, total cell lysates prepared from P29, P34, D6, and A11 cells were subjected to immunoblotting using anti-Sp1, anti-Sp3 and anti-β-actin antibodies. E, nuclear extracts prepared from P29, P34, D6, and A11 cells were subjected to EMSAs as in C. F, nuclear extracts prepared from P29 and A11 cells were subjected to chromatin immunoprecipitation assays in which anti-acetylated histone H4 (anti-AcH4), anti-Sp1, and anti-Sp3 antibodies were used. Normal rabbit IgG served as a control.
To obtain direct evidence that Sp family members bind to this putative Sp1 binding sequence, we carried out EMSAs using wtHIFpro-Sp1 (−72/−48) as a DNA probe. As shown in Fig. 6C, these assays revealed three constitutive binding complexes (C1–C3) (lane 2) that were almost entirely Sp-dependent, as shown by competition with excess wtHIFpro-Sp1 or Sp1 consensus oligonucleotides (wtSp1) (lanes 3 and 5), but not with their mutant form mutHIFpro-Sp1 or mutSp1 (lanes 4 and 6). Addition of antibodies directed against either Sp1 or Sp3 induced a supershift and/or a significant reduction of Sp1/Sp3-dependent binding activities (lanes 7 and 8). Simultaneous addition of both antibodies led to a nearly complete supershift (lane 9). These data indicate that Sp1 and Sp3 proteins actually bind to the region proximal to the transcription initiation site.
We then compared the expression levels of Sp1 and Sp3 between the high and the low metastatic cell lines. However, we could not detect any difference (Fig. 6D). Also, the DNA binding activity of Sp1/Sp3, as demonstrated by EMSA analysis, did not correlate with the HIF-1α transcriptional level (Fig. 6E). Moreover, chromatin immunoprecipitation assays revealed that there were no differences in the binding of Sp1 and Sp3 to the Sp1/Sp3 binding site and the level of histone H4 acetylation around the site between P29 and A11 cells (Fig. 6F). These results indicate that Sp1 is necessary but not sufficient for explaining the higher expression of HIF-1α mRNA in the high metastatic cell lines.
HDAC Activation Contributes to the ROS-mediated HIF-1α Transcriptional Activation
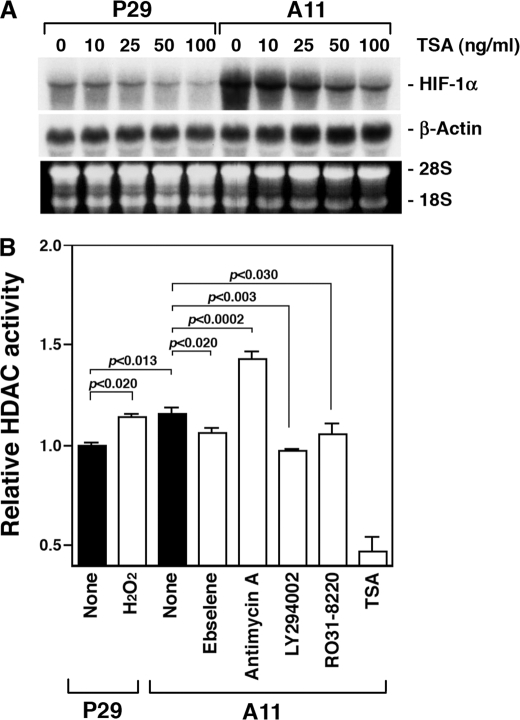
To investigate a possible mechanism of the HIF-1α transcriptional activation in the high metastatic cells further, we treated P29 and A11 cells with TSA, a nonselective HDAC inhibitor. The results showed that TSA dramatically suppressed HIF-1α mRNA expression in A11 cells but slightly in P29 cells (Fig. 7A), suggesting the relationship between HDAC activity and the ROS-mediated HIF-1α mRNA overexpression. Then, we measured HDAC activity in P29 cells, P29 cells treated with H2O2, A11 cells, and A11 cells treated with ebselene, antimycin A, LY294002, and Ro31-8220. The results in Fig. 7B show that HDAC activity was significantly higher in A11 cells than in P29 cells. It was high in H2O2-treated P29 cells and antimycin A-treated A11 cells, but low in A11 cells treated with ebselene, LY294002, and Ro31-8220 compared with the respective untreated cells. Thus, the HDAC activity is positively correlated with the HIF-1α mRNA up-regulation.
FIGURE 7.
HDAC activity is involved in the ROS-mediated HIF-1α transcription. A, P29 and A11 cells were treated with TSA at the indicated concentrations for 18 h. Total RNA was extracted and subjected to Northern blot analysis. The blots were hybridized with a 32P-labeled HIF-1α cDNA. Ethidium bromide staining of the gel is also shown. B, HDAC activity is shown in untreated P29 and P29 cells treated with 25 μm H2O2 for 16 h, and untreated A11 and A11 cells treated with ebselene (20 μm), antimycin A (20 μm), LY294002 (20 μm), Ro31-8220 (5 μm), and TSA (100 ng/ml) for 18 h.
DISCUSSION
The present study demonstrates that an ROS-generating mtDNA mutation in the ND6 gene leads to HIF-1α mRNA overexpression, resulting in marked up-regulation of HIF-1α protein and VEGF production levels under hypoxic conditions. This study also suggests the possibility for the first time that some of pathogenic mtDNA mutations can activate HIF-1α transcription.
mtDNA mutations are frequently observed in tumor cells and implicated to be a factor in the progression of tumors. mtDNA mutations in tumor cells include severe mutations such as insertion-deletion and chain termination mutations and mild missense mutations. The mutation in the ND6 gene found in A11 cells is a missense mutation that reduces complex I activity (4). This mutation was also found in the other high metastatic D6 cells but not in the low metastatic P29 or P34 cells. In both A11 and D6 cells, up-regulation of HIF-1α gene transcription was detected, suggesting a causal linkage between the ND6 mutation and HIF-1α transcription. In the present study, we used trans-mitochondrial cybrids to prove this linkage, and as expected, the cybrids carrying mtDNA from A11 cells overexpressed HIF-1α mRNA, despite the source of nuclear DNA.
Several lines of evidence supported that ROS caused by the ND6 mutation primarily mediates HIF-1α transcription. First, the cells carrying A11 mtDNA overproduced ROS. Second, ebselene and PDTC reduced the intracellular ROS level and concomitantly abolished HIF-1α transcription. Third, antimycin A that inhibits the function of complex III, thereby generating large quantities of superoxide radicals, increased the expression of HIF-1α mRNA in both P29 and A11 cells. Fourth, exogenous H2O2 enhanced the expression. ROS from NADPH oxidase are also mediators of HIF-1α mRNA induction in lipopolysaccharide-stimulated microglial cells and thrombin-stimulated pulmonary artery smooth muscle cells (15, 19). Furthermore, we showed that PI3K-Akt and PKC, but not ERK or JNK, regulate HIF-1α mRNA expression. Because both LY29004 and Ro31-8220 suppressed HIF-1α mRNA expression more effectively in A11 cells than in P29 cells, PI3K-Akt and PKC may engage in the ROS-mediated expression of HIF-1α mRNA. Consistent with these results, either PI3K or PKC or both are shown to regulate HIF-1α transcription in lipopolysaccharide-stimulated glial cells, BCR/ABL-expressing Ba/F3 hematopoietic cells, and angiotensin II-treated vascular smooth muscle cells (12, 15, 20). In contrast, ERK and JNK are reported to mediate lipopolysaccharide-stimulated HIF-1α mRNA induction in human monocytes/macrophages and hepatoma cells, respectively (21, 22). Further study is required to determine which PKC isoform is responsible for the ROS-mediated expression of HIF-1α mRNA using a molecular approach.
The HIF-1α gene promoter contains putative binding sites for several transcription factors, including Sp1, AP-1, and NF-κB (23–25). Treatment of A11 cells with mithramycin A resulted in a marked suppression of HIF-1α mRNA expression in A11 cells, whereas sulfasarazine and curcumin showed no effect, suggesting the importance of Sp1 for the promoter activity. Luciferase reporter assays also indicated that the Sp1 binding site is indispensable for the promoter activity. Unexpectedly, however, we could not find any difference in the level of Sp1 binding to and histone acetylation around the binding site between A11 and P29 cells. In contrast, Oh et al. (19) reported that lipopolysaccharide induces HIF-1α mRNA in an Sp1-dependent pathway. It is necessary to determine whether other regions of the promoter and transcription factors are involved in the overexpression of HIF-1α mRNA in A11 cells.
In the present study, we showed that TSA markedly repressed the expression of HIF-1α mRNA in A11 cells. Based on this observation, we found a correlation between HDAC activity and HIF-1α transcription; that is, HDAC activity was higher in A11 than in P29 cells. It was also higher in H2O2-treated P29 cells and antimycin A-treated A11 cells than in the respective control cells. Furthermore, HDAC activity in A11 cells was repressed by ebselene, LY294002, and Ro31-8220. Together, these data indicate that ROS lead to HDAC activation through PI3K and PKC pathways, thereby activating HIF-1α transcription. In general, histone acetylation enhances gene expression through the chromatin remodeling caused by histone modification (26). Therefore, it is not clear how HDAC inhibition can lead to the transcriptional repression of the HIF-1α gene. However, many genes such as proinflammatory genes, including tumor necrosis factor-α, interleukin-1β, interferon-γ, and inducible nitric-oxide synthase, are reported to be repressed by HDAC inhibitors (27–30). The repression of these proinflammatory genes has been suggested to be a result of inhibition of NF-κB activation and the acetylation of non-histone proteins (30). Because our data indicate little contribution of NF-κB in the ROS-mediated HIF-1α mRNA overexpression in A11 cells, acetylation of other non-histone proteins may be important. It should be noted that Noh et al. (31) have recently shown that TSA decreases mRNA of extracellular matrix components. They also show that HDAC2 plays an important role in the development of extracellular matrix accumulation and that ROS mediate transforming growth factor-β1-induced activation of HDAC2 (31). HDACs constitute a family of 18 enzymes (32). Therefore, it will be interesting to determine which HDAC is responsible for the ROS-mediated HIF-1α transcription in the cells carrying mtDNA with the ND6 mutation.
In conclusion, our findings show that the ROS-generating ND6 mutation causes HIF-1α transcription via PI3K-Akt/PKC/HDAC pathway. Because mtDNA mutations have been implicated to be a factor in cancer etiology and shown to be gradually accumulated in tumor cells, some of them, especially pathogenic somatic mutations, may contribute to malignant progression by causing the up-regulation of HIF-1α protein in tumors.
Supplementary Material
This work was supported in part by Grants-in-aid for Third Term Comprehensive Control Research for Cancer from the Ministry of Health, Labor, and Welfare (to K. T.) and by Grants-in-aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to N. K. and K. T.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and Table S1.
- ROS
- reactive oxygen species
- DCFH-DA
- 2′,7′-dichlorofluorescin diacetate
- DMEM
- Dulbecco's modified Eagle's medium
- DPBS
- Dulbecco's phosphate-buffered saline
- EMSA
- electrophoretic mobility shift assay
- ERK
- extracellular signal-regulated kinase
- FACS
- fluorescence-activated cell sorter
- HDAC
- histone deacetylase
- HIF-1α
- hypoxia-inducible factor-1α
- JNK
- c-Jun N-terminal kinase
- MAP
- mitogen-activated protein
- ND6
- NADH dehydrogenase subunit 6
- PDTC
- pyrrolidine dithiocarbamate
- PI3K
- phosphatidylinositol 3-kinase
- PKC
- protein kinase C
- PMSF
- phenylmethylsulfonyl fluoride
- TSA
- trichostatin A
- VEGF
- vascular endothelial growth factor.
REFERENCES
- 1.Brandon M., Baldi P., Wallace D. C. (2006) Oncogene 25, 4647–4662 [DOI] [PubMed] [Google Scholar]
- 2.Park J. S., Sharma L. K., Li H., Xiang R., Holstein D., Wu J., Lechleiter J., Naylor S. L., Deng J. J., Lu J., Bai Y. (2009) Hum. Mol. Genet. 18, 1578–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dasgupta S., Hoque M. O., Upadhyay S., Sidransky D. (2008) Cancer Res. 68, 700–706 [DOI] [PubMed] [Google Scholar]
- 4.Ishikawa K., Takenaga K., Akimoto M., Koshikawa N., Yamaguchi A., Imanishi H., Nakada K., Honma Y., Hayashi J. (2008) Science 320, 661–664 [DOI] [PubMed] [Google Scholar]
- 5.Semenza G. L. (2009) Semin. Cancer Biol. 19, 12–16 [DOI] [PubMed] [Google Scholar]
- 6.Semenza G. L. (2008) IUBMB Life 60, 591–597 [DOI] [PubMed] [Google Scholar]
- 7.Vaupel P., Mayer A. (2007) Cancer Metastasis Rev. 26, 225–239 [DOI] [PubMed] [Google Scholar]
- 8.Koshikawa N., Iyozumi A., Gassmann M., Takenaga K. (2003) Oncogene 22, 6717–6724 [DOI] [PubMed] [Google Scholar]
- 9.Secades P., Rodrigo J. P., Hermsen M., Alvarez C., Suarez C., Chiara M. D. (2009) Genes Chromosomes Cancer 48, 441–454 [DOI] [PubMed] [Google Scholar]
- 10.Saramäki O. R., Savinainen K. J., Nupponen N. N., Bratt O., Visakorpi T. (2001) Cancer Genet. Cytogenet. 128, 31–34 [DOI] [PubMed] [Google Scholar]
- 11.Chandel N. S., McClintock D. S., Feliciano C. E., Wood T. M., Melendez J. A., Rodriguez A. M., Schumacker P. T. (2000) J. Biol. Chem. 275, 25130–25138 [DOI] [PubMed] [Google Scholar]
- 12.Pagé E. L., Robitaille G. A., Pouysségur J., Richard D. E. (2002) J. Biol. Chem. 277, 48403–48409 [DOI] [PubMed] [Google Scholar]
- 13.Pouysségur J., Mechta-Grigoriou F. (2006) Biol. Chem. 387, 1337–1346 [DOI] [PubMed] [Google Scholar]
- 14.Klimova T., Chandel N. S. (2008) (2008) Cell Death Differ. 15, 660–666 [DOI] [PubMed] [Google Scholar]
- 15.Bonello S., Zähringer C., BelAiba R. S., Djordjevic T., Hess J., Michiels C., Kietzmann T., Görlach A. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 755–761 [DOI] [PubMed] [Google Scholar]
- 16.Koshikawa N., Maejima C., Miyazaki K., Nakagawara A., Takenaga K. (2006) Oncogene 25, 917–928 [DOI] [PubMed] [Google Scholar]
- 17.Luo G., Gu Y. Z., Jain S., Chan W. K., Carr K. M., Hogenesch J. B., Bradfield C. A. (1997) Gene Expr. 6, 287–299 [PMC free article] [PubMed] [Google Scholar]
- 18.Indo H. P., Davidson M., Yen H. C., Suenaga S., Tomita K., Nishii T., Higuchi M., Koga Y., Ozawa T., Majima H. J. (2007) Mitochondrion 7, 106–118 [DOI] [PubMed] [Google Scholar]
- 19.Oh Y. T., Lee J. Y., Yoon H., Lee E. H., Baik H. H., Kim S. S., Ha J., Yoon K. S., Choe W., Kang I. (2008) Neurosci. Lett. 431, 155–160 [DOI] [PubMed] [Google Scholar]
- 20.Mayerhofer M., Valent P., Sperr W. R., Griffin J. D., Sillaber C. (2002) Blood 100, 3767–3775 [DOI] [PubMed] [Google Scholar]
- 21.Frede S., Stockmann C., Freitag P., Fandrey J. (2006) Biochem. J. 396, 517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H. Y., Kim Y. H., Nam B. H., Kong H. J., Kim H. H., Kim Y. J., An W. G., Cheong J. (2007) Exp. Cell Res. 313, 1866–1876 [DOI] [PubMed] [Google Scholar]
- 23.Iyer N. V., Leung S. W., Semenza G. L. (1998) Genomics 52, 159–165 [DOI] [PubMed] [Google Scholar]
- 24.Minet E., Ernest I., Michel G., Roland I., Remacle J., Raes M., Michiels C. (1999) Biochem. Biophys. Res. Commun. 261, 534–540 [DOI] [PubMed] [Google Scholar]
- 25.Das C., Kundu T. K. (2005) IUBMB Life 57, 137–149 [DOI] [PubMed] [Google Scholar]
- 26.Adcock I. M. (2007) Br. J. Pharmacol. 150, 829–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joseph J., Mudduluru G., Antony S., Vashistha S., Ajitkumar P., Somasundaram K. (2004) Oncogene 23, 6304–6315 [DOI] [PubMed] [Google Scholar]
- 28.Leoni F., Zaliani A., Bertolini G., Porro G., Pagani P., Pozzi P., Donà G., Fossati G., Sozzani S., Azam T., Bufler P., Fantuzzi G., Goncharov I., Kim S. H., Pomerantz B. J., Reznikov L. L., Siegmund B., Dinarello C. A., Mascagni P. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 2995–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu Z., Zhang W., Kone B. C. (2002) J. Am. Soc. Nephrol. 13, 2009–2017 [DOI] [PubMed] [Google Scholar]
- 30.Quivy V., Van Lint C. (2004) Biochem. Pharmacol. 68, 1221–1229 [DOI] [PubMed] [Google Scholar]
- 31.Noh H., Oh E. Y., Seo J. Y., Yu M. R., Kim Y. O., Ha H., Lee H. B. (2009) Am. J. Physiol. Renal. Physiol. 297, 729–739 [Google Scholar]
- 32.Witt O., Deubzer H. E., Milde T., Oehme I. (2009) Cancer Lett. 277, 8–21 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.