Abstract
We have previously identified integrin αvβ3 and Fas as receptors for the streptococcal pyrogenic exotoxin B (SPE B), and G308S, a mutant of SPE B that binds to Fas only. In the current study we found that after binding to αvβ3, SPE B stimulated the tyrosine phosphorylation of JAK2 and STAT1. STAT1 tyrosine phosphorylation was inhibited by a JAK2 inhibitor, AG490, short interfering RNA (siRNA) silencing of JAK2, and anti-αVβ3 antibody. AG490 also decreased the binding of tyrosine-phosphorylated STAT1 to the procaspase 8 promoter, decreasing procaspase 8 expression, suggesting that SPE B up-regulates procaspase 8 expression via the JAK2/STAT1 pathway. Alternatively, both SPE B and G308S increased STAT1 phosphorylation at serine 727, which was inhibited by anti-Fas antibody, a p38 inhibitor, SB203580, and siRNA silencing of p38. In addition, SPE B and G308S increased binding of serine-phosphorylated STAT1 to the Bax promoter and Bax expression, which was decreased by SB203580. SPE B and G308S-stimulated Bax expression was also inhibited by anti-Fas antibody. These findings suggest that Fas mediate SPE B-induced Bax expression through p38. Silencing of JAK2 or p38 by siRNA blocked procaspase 8 expression, whereas only p38 siRNA decreased Bax expression. Furthermore, JAK2 inhibition and p38 inhibition reduced SPE B-induced apoptosis, but only p38 inhibition blocked G308S-induced apoptosis.
INTRODUCTION
Streptococcus pyogenes (group A streptococcus) causes a wide spectrum of infection, including pharyngitis, cellulitis, and severe invasive diseases, such as necrotizing fasciitis and streptococcal toxic shock syndrome (1–3). Streptococcal pyrogenic exotoxin B (SPE B)2 is secreted by all group A streptococcus and is an important factor in streptococcal infections. It is a cysteine protease synthesized as a 40-kDa zymogen that is cleaved to a 28-kDa active enzyme by autocatalysis or proteolysis (4–7).
Our recent study (8) showed that SPE B-induced apoptosis in human lung epithelial A549 cells is mediated through a receptor-like mechanism and mitochondrion-dependent pathway and that the protease activity of SPE B is required for the initiation of apoptotic signaling, most likely by exposing the binding site for SPE B. The time course analysis indicated that during apoptosis, molecules were activated in the following sequence: caspase 8, Bid, Bax, cytochrome c release, caspase 9, and caspase 3 (8). Further analysis indicated that transcription of procaspase 8 and Bax were stimulated by SPE B. In the present study we further characterize the signal pathways that lead to the expression of procaspase 8 and Bax.
Signal transducers and activators of transcription (STAT) protein family members are important for growth, development, proliferation, and cell death because they modulate the expression of target genes (9). Tyrosine phosphorylation provides the binding site for the SH2 domain of STAT protein to form homo- or heterodimers. Dimer formation results in the translocation of STAT proteins to the nucleus, where they bind to target genes and modulate transcription. The COOH-terminal transactivation domains of some STAT proteins contain a conserved serine residue that can be phosphorylated to serve as a coactivator to modulate the function of other transcription factors independent of STAT binding to DNA (9). As for cytokine-activated STATs, Janus kinases (JAKs) phosphorylate tyrosine residues, whereas mitogen-activated protein kinases (MAPKs) phosphorylate serine residues (10). STAT transcription factors regulate both apoptotic and anti-apoptotic signal pathways (11–16).
The three major MAPK pathways, extracellular signal-regulated kinase (ERK), c-Jun NH2-terminal kinase (JNK), and p38 MAPK, mediate phosphorylation on serine residues. ERKs are more important for the anti-apoptotic signal pathway, e.g. ERK inhibits Fas-induced tumor cell apoptosis (17), whereas JNKs and p38 MAPK are involved in the pro-apoptotic signal pathways, e.g. the JNK-dependent pathway mediates TNF-α-induced apoptosis (18), and p38α MAPK seems to sensitize cells to apoptosis by up-regulating Bax (19).
We have previously identified integrin αVβ3 and Fas as receptors for SPE B-induced apoptosis, mediated by RGD motif-dependent and -independent pathways, respectively (20). In the present study we further elucidate the roles of the STAT1 and MAPK pathways in the SPE B-induced apoptotic pathway. Our presented evidence indicates that (i) SPE B triggers the integrin αVβ3-mediated JAK2/STAT1 signal pathway to induce the expression of procaspase 8 and (ii) SPE B also binds to the Fas receptor to activate p38 MAPK that phosphorylates STAT1 at serine 727 and increases expression of Bax.
EXPERIMENTAL PROCEDURES
Preparation of Recombinant SPE B and Its Mutant G308S
The expression and purification of recombinant SPE B (rSPE B) have been previously described (5). The gene encoding ProSPE B was amplified using a PCR with six histidine tags and a BamH1 recognition site. The gene was then cloned into the BamH1 site of the pET21a vector (Novagen), which was then transformed into Escherichia coli BL21 pLys. A wild-type construct was used to produce a G308S mutant, a conversion of the RGD motif to RSD, using overlap extension PCR (21). An inoculum (250 μl) of stock culture was added to 250 ml of LB/AMP medium (Sigma) and allowed to grow to an optical density (590 nm) of 0.5–1.0. To induce rSPE B expression, 250 μl of isopropyl-β-d-thiogalactopyranoside (100 mg/ml, MDBio) was added to the culture medium. For rSPE B, cells were grown at 33 °C overnight, whereas for G308S, cells were grown at 37 °C overnight. After induction, cells were centrifuged at 4000 × g for 15 min, and the pellets were resuspended in 80 ml of buffer A (20 mm Tris and 200 mm NaCl (pH 7.0). The resuspended pellet was broken three times using a French press at 1500 kg/cm2 for 30 s. The entire content was centrifuged at 10,000 × g at 4 °C for 10 min to collect supernatant, which was then separated on a nickel-chelated column (Amersham Biosciences) and eluted by a 0–200 mm imidazole gradient. The collected fractions were concentrated using Amicon ultrafiltration with a 10-kDa cutoff membrane (Millipore Corp.) and dialyzed with phosphate-buffered saline (PBS). Purity was verified using SDS-PAGE. rSPE B and G308S were expressed as a 42-kDa zymogen and converted to a 28-kDa active enzyme during the course of purification. The purified rSPE B and G308S were passed through Detoxi-Gel (Pierce) to remove lipopolysaccharide contamination.
Cell Culture
A549 cells were grown in defined minimal essential medium (Sigma) supplemented with 10% heat-inactivated fetal calf serum (Invitrogen), 2 mm l-glutamate (Sigma), and 50 μg/ml gentamicin (Sigma) at 37 °C in a CO2 incubator. Cells (8 × 104/well) were seeded into a 24-well plate for 24 h and then washed with PBS and incubated in complete medium containing specified agents for the time indicated.
Immunoblotting
Cells (2 × 105/well) were seeded into a 6-well plate for 24 h, washed with cold PBS, and then incubated with the indicated agents for the indicated times. The cells were collected and boiled in sample buffer for 5 min. After SDS-PAGE, proteins were transferred to a polyvinylidene difluoride membrane (BioTrace) and blocked with PBS containing 5% lowfat milk at 4 °C overnight. Polyvinylidene difluoride membranes were probed with one of the following primary antibodies diluted in PBST (PBS containing 0.05% Tween 20) or TBST (Tris-buffered saline containing Tween 20): 1/100 dilution of anti-procaspase 8 (Cell Signaling), 1/200 dilution of anti-caspase 8 (Cell Signaling), 1/500 dilution of anti-Bax (Oncogene), 1/500 dilution of anti-JAK2 (Santa Cruz Biotechnology), 1/500 dilution of anti-STAT1 (Calbiochem), 1/500 dilution of anti-phosphotyrosine (Calbiochem), 1/100 dilution of phospho-specific (Thr180/Tyr182) anti-p38 MAPK (Santa Cruz Biotechnology), 1/500 dilution of anti-p38α MAPK (Santa Cruz Biotechnology), 1/200 dilution of anti-ERK1/2 (Calbiochem), 1/100 dilution of phospho-specific (Thr202/Tyr204) anti- ERK1/2 (Calbiochem), 1/100 dilution of anti-p-JNK (Santa Cruz Biotechnology), 1/500 dilution of anti-JNK (Santa Cruz Biotechnology), 1/100 dilution of phospho-specific (Tyr701) anti-STAT1 (Calbiochem), or 1/100 dilution of phospho-specific (Ser727) anti-STAT1 (Calbiochem). The membranes were washed 4 times with PBST or TBST and probed with 0.1 μg/ml of the corresponding horseradish peroxidase-conjugated secondary antibody. The protein bands were visualized using an enhanced chemiluminescence (ECL) kit (Amersham Bioscience). Protein loading was controlled using anti-β-actin (1/10,000) (Sigma) or anti-proliferating cell nuclear antigen antibody (1/1000) (Calbiochem). Each experiment was independently repeated three times. In the immunoprecipitation assay, the cell lysates were preincubated with 2.5 μg of anti-JAK2 antibody at 4 °C overnight and then with protein A-agarose at 4 °C for 3 h. Protein A beads were washed 4 times with cold PBS, resuspended in Laemmli buffer (25 μl; 0.0625 m Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, 5% 2-mercaptoethanol, and 0.001% bromphenol blue), and boiled. Proteins were detected using Western blotting with anti-phosphotyrosine antibody, as described above.
RT-PCR
Total RNA was extracted from A549 cells, and 5 μg of total RNA was subjected to an oligo(dT)-primed reverse transcriptase reaction. cDNA was synthesized by a PCR. The primers used for procaspase 8 amplification were 5′-AGGAAAGTTGACATCCTGAAAA-3′ and 5′-GGAGAGTCCGAGATTGTCATTA-3′, which recognized fragments of α1 (127 bp) and α2 (173 bp), respectively. The conditions for PCR amplification of procaspase 8 were 95 °C for 5 min, then 30 cycles at 95 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s, and a final elongation step at 72 °C for 10 min. The primers used for Bax amplification were 5′-ATCCAGGATCGAGCAGGGCG-3′ and 5′-ACTCGCTCAGCTTCTTGGTG-3′. The conditions for PCR amplification of Bax were 95 °C for 2 min, then 35 cycles at 95 °C for 30 s, 64 °C for 30 s, and 72 °C for 30 s and a final elongation step at 72 °C for 5 min. Before PCR amplification with each primer pair, an aliquot of the cDNA was amplified with β-actin primers to determine whether the equivalent amounts of RNA were prepared. All PCR products were separated on 2% (w/v) agarose gels.
Nuclear Extraction
Cells were scraped from the culture dish and washed with cold PBS. Pellets were resuspended in 500 μl of cell lysis buffer (10 mm Hepes-KOH (pH 7.9), 1.5 mm MgCl2, 0.5 mm dithiothreitol, 0.2 mm phenylmethylsulfonyl fluoride, and 10 mm KCl), incubated on ice for 15 min, and then vortexed for 15 s. Whole contents were centrifuged at 15,000 × g at room temperature for 10 s. STAT1 in the supernatants was detected by Western blotting. The pellet was resuspended in 100 μl of cold nuclear lysis buffer (20 mm Hepes-KOH (pH 7.9), 25% glycerol, 420 nm NaCl, 0.2 mm EDTA, 0.5 mm dithiothreitol, 0.2 mm phenylmethylsulfonyl fluoride, and 1.5 mm MgCl2) and incubated on ice for 20 min. The sample was then centrifuged at 15,000 × g for 10 s to remove the nuclear debris. Supernatants were used to detect STAT1 using Western blotting.
Short Interfering RNA (siRNA) Preparation
The expression of short hairpin RNA is described elsewhere (8). The pSUPER/enhanced green fluorescent protein vector was kindly provided by R. Agami (The Netherlands Cancer Institute, Amsterdam, The Netherlands). The pSUPER/enhanced green fluorescent protein vector was digested with BglII and HindIII, and the oligonucleotides were ligated into the vector. The oligonucleotides are: 5′-gatccccGTTCCTGAGCCTGGACTACttcaagagaGTAGTCCAGGCTCAGGAACtttttggaaa-3′ and 5′- agcttttccaaaaaGTTCCGAGCCTGGACTACtctcttgaaGTAGTCCAGGCTCAGGAACggg-3′; the lowercase letters at the 5′ and 3′ ends represent restriction enzyme cutting sites for construction, and the lowercase letters in the middle sections of the sequences represent the spacers for siRNA to form a loop structure (8). For transfection, cells (5 × 106) were washed with serum-free minimal essential medium and incubated in a medium containing 2.5 μg of DNA and 5 μl of Lipofectamine 2000. After 24 h of incubation, the medium was removed, and cells were maintained in minimal essential medium supplemented with 10% heat-inactivated fetal calf serum for an additional 24 h before the assay. The enhanced green fluorescent protein-positive cells were sorted by FACSAria (BD Biosciences) with a purity ranging from 72 to 76% in different experiments.
For the JAK2, p38, and Bax silencing assay, cells were transfected with control siRNA (Santa Cruz Biotechnology), JAK2 siRNA (Santa Cruz Biotechnology), p38 siRNA (Santa Cruz Biotechnology), or Bax siRNA (Cell signaling) according to the manufacturer's protocol. A549 cells at a density of 2 × 105/well were seeded into medium without antibiotics for 1 day before transfection. siRNA and siTransfection reagent were incubated separately with siTransfection medium at room temperature for 10 min, subsequently, both incubation media were combined and incubated at room temperature for another 30 min. The medium was then replaced with fresh medium without antibiotics, and the mixtures were added to the cells. The cells were incubated for 24 h. To verify the effectiveness of siRNAs, Western blotting was performed as described above (Supplement 1).
Analysis of Apoptosis
Cells treated with SPE B, or other agents were fixed at −20 °C with 70% ethanol. After being washed once with PBS, the cells were stained with propidium iodide (40 μg/ml in PBS) (Sigma) containing ribonuclease (100 μg/ml) (Sigma) at room temperature in the dark for 30 min. Apoptotic cells were quantified using FACScan with CELLQuest software (BD Biosciences) and presented as the percentage of hypodiploid cells.
Chromatin Immunoprecipitation (ChIP)
The interaction of STAT1 with DNA in A549 cells was evaluated by a ChIP assay (Upstate Biotechnology) according to the manufacturer's instructions. Cells (1 × 107) were washed with cold PBS and stimulated with new medium containing SPE B or G308S for 3 h. The cells were then treated with formaldehyde (1%) at room temperature for 60 min, washed 3 times with PBS, and centrifuged at 800 × g for 5 min. The nuclei were prepared in SDS lysis buffer, and the lysates were sonicated 5 times for 10 s, each at 30% power using a sonic dismembrator (model 300; Fisher) to shear genomic DNA. Anti-STAT1-specific antibody (Calbiochem) and polyclonal rabbit IgG antibodies (Pierce) were used for immunoprecipitation. Samples (5 μl each) were analyzed by 35 rounds of PCR amplification. The procaspase 8 promoter region was analyzed with primers of 5′-ACAGTGCCAGGAAGTGAGAAAC-3′ and 5′-CAGATGCTCCAGAAATGCTAAT-3′. These primers were used, respectively, as upper and lower primers for the region −188/+126. The Bax promoter region was analyzed using the primers 5′-TAATCCCAGCGCTTTGGAAG-3′ and 5′-TGCAGAGACCTGGATCTAGC-3′.
Statistical Analysis
Statistical analysis was done using Student's t test or one way analysis of variance and then Tukey's post test. Statistical significance was set at p < 0.05.
RESULTS
SPE B, but Not G308S, Up-regulates Procaspase 8 Expression
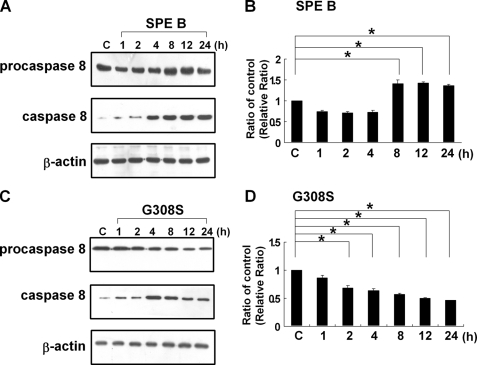
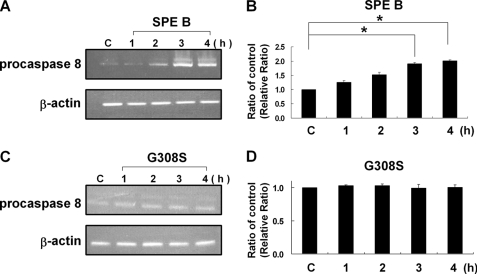
We previously found that caspase 8 was activated first during SPE B-induced apoptosis (8). To further delineate the activation process of caspase 8, a time course analysis of levels of procaspase 8 and caspase 8 in the presence of SPE B was conducted. As shown in Fig. 1, A and B, the level of caspase 8 increased in a time-dependent manner. As caspase 8 increased, the procaspase 8 level did not decrease concomitantly but, rather, increased slightly over time. A similar time course analysis was also performed in the presence of G308S, and different results were observed. The level of caspase 8 increased with time until 4 h post-treatment and then decreased slightly with time, whereas the level of procaspase 8 decreased with time (Fig. 1, C and D). Because activation of caspase 8 is through the cleavage of procaspase 8, an increase in caspase 8 is usually accompanied with a decrease in procaspase 8. The continuous increase of procaspase 8 in the presence of SPE B, but not in the presence of G308S, suggested that there must be a SPE B-specific pathway that up-regulates procaspase 8 production. The total RNA was then extracted, and the levels of procaspase 8 mRNA were analyzed by RT-PCR. Procaspase 8 mRNA expression increased with time in the presence of SPE B (Fig. 2A and B). In contrast, no differences in the expression of procaspase 8 mRNA were found in the presence of G308S (Fig. 2, C and D). The up-regulation of procaspase 8 by SPE B, but not G308S, indicated that the process was mediated through integrin αVβ3.
FIGURE 1.
The effect of SPE B and G308S on levels of procaspase 8 and caspase 8. A549 cells (2 × 105) were treated with SPE B (20 μg/ml) (A) and G308S (20 μg/ml) (C) for the indicated times or with PBS (control (C)) for 4 h. Total cell lysates were immunoblotted using anti-procaspase 8 and anti-caspase 8 antibodies. β-Actin was an internal control. A quantitative result of procaspase 8 versus β-actin is expressed as the relative ratio (B and D). The ratio of procaspase 8 to β-actin in the control sample is defined as 1, the ratios of each procaspase 8 to β-actin in other time points are compared with control, and their ratios are presented. Results are the mean ± S.E. of three independent experiments. *, p < 0.05.
FIGURE 2.
SPE B induced procaspase 8 gene expression. Cells (3 × 106) were treated with SPE B (20 μg/ml) (A) and G308S (20 μg/ml) (C) or PBS (control (C)) for the indicated times. Total RNA was prepared from A549 cells and subjected to RT-PCR analysis of procaspase 8 mRNA expression. The cDNA was amplified with specific oligos for procaspase 8 and β-actin. A quantitative result of procaspase 8 versus β-actin is expressed as the relative ratio in the same way as described in Fig. 1, B and D. Results are the mean ± S.E. of three independent experiments. *, p < 0.05.
SPE B Triggers Tyrosine Phosphorylation of JAK2 and STAT1 through Integrin αVβ3
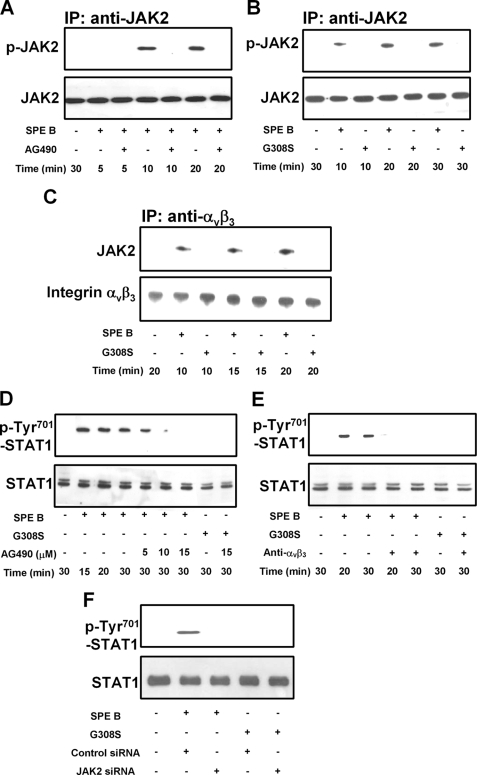
By modulating the expression of target genes, STAT transcription factors play important roles in apoptosis. It has been demonstrated that STAT1 up-regulates proapoptotic genes such as caspases and death receptors to promote apoptosis (12, 22). To determine whether STAT1 activation is a key event in SPE B-induced expression of procaspase 8, we evaluated the involvement of JAK2. When cells were incubated with SPE B for various times, tyrosine phosphorylation of JAK2 was detected 10 min after SPE B treatment, whereas no phosphorylation could be detected in cells pretreated with the JAK2 inhibitor, AG490 (Fig. 3A). However, when cells were incubated with G308S, the tyrosine phosphorylation of JAK2 was not detected (Fig. 3B).
FIGURE 3.
SPE B activates the JAK2/STAT1 signal pathway through αVβ3. A549 cells (2 × 105) were preincubated with AG490 (15 μm) for 1 h and then treated with SPE B (20 μg/ml) for the indicated times (A). Cells were also incubated with SPE B (20 μg/ml) or G308S (20 μg/ml) for the indicated times (B and C), and cell lysates were immunoprecipitated (IP) with anti-JAK2 antibody (A and B) or anti-αVβ3 antibody (C) and immunoblotted using anti-phosphotyrosine or anti-JAK2 antibody (A and B) and anti-JAK2 or anti-αVβ3 antibody (C). Cells were also preincubated with various concentrations of AG490 for 1 h and then treated with SPE B (20 μg/ml) or G308S (20 μg/ml) for the indicated times (D). Cells were treated with SPE B (20 μg/ml) or G308S (20 μg/ml) for the indicated times with or without anti-αVβ3 antibody (4 μg/ml) (E), cells (2 × 105) were also transfected with siRNA of JAK2 and incubated with SPE B or G308S (20 μg/ml) for 30 min (F), and cell lysates were immunoblotted using anti-phospho-Tyr-701 STAT1 or anti-STAT1 antibody.
Previously we demonstrated that there is a direct association between SPE B and integrin αVβ3. We then examined interactions between integrin αVβ3 and JAK2. As shown in Fig. 3C, JAK2 coprecipitated with αVβ3 in the presence of SPE B and not in the presence of G308S. The phosphorylation of STAT1 on tyrosine 701 was detected 15 min after SPE B treatment and was inhibited by AG490 in a dose-dependent manner (Fig. 3D). When cells were pretreated with anti-αVβ3 antibody, STAT1 phosphorylation of tyrosine 701 was inhibited (Fig. 3E). When cells were transfected with JAK2 siRNA, STAT1 phosphorylation of tyrosine 701 was also abrogated (Fig. 3F). These observations indicate that SPE B induces STAT1 tyrosine phosphorylation through integrin αVβ3 and JAK2 tyrosine phosphorylation.
SPE B Induces STAT1 Translocation to the Nucleus
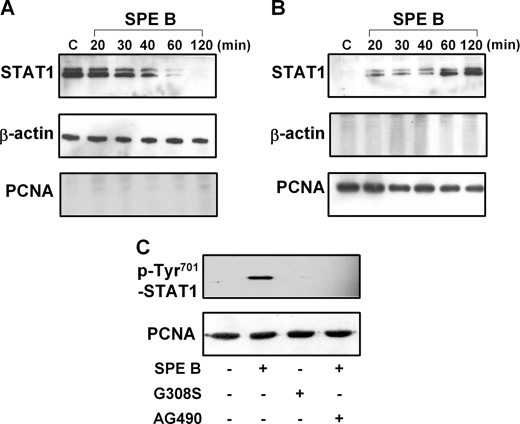
Because STAT1 is a transcription factor that functions in the nucleus, it is important to determine whether SPE B induces STAT1 translocation from the cytosol to the nucleus in A549 cells. Our results show that in the presence of SPE B, the level of STAT1 in the cytoplasmic extract decreased (Fig. 4A), whereas its level in the nucleus increased over time (Fig. 4B). STAT1 in the nuclear fraction was also confirmed with phospho-specific (Tyr701) anti-STAT1 antibody (Fig. 4C). Its formation and translocation was also inhibited by AG490 and not produced by G308S. We also investigated whether pathways in A549 cell induced by SPE B occur in other epithelial cell types. The results indicated that in HEp-2 cells, JAK2 and STAT1 were phosphorylated on tyrosine in response to SPE B treatment, and JAK2 acted upstream of STAT1 (Supplement 2, A and B).
FIGURE 4.
SPE B induces phosphorylated STAT1 translocation to the nucleus. Cells (2 × 105) were treated with SPE B (20 μg/ml) for the indicated times or PBS for 2 h. The cytoplasmic (A) and nuclear (B) extracts were immunoblotted using anti-STAT1 antibody. β-Actin and proliferating cell nuclear antigen (PCNA) were internal controls for cytoplasmic and nuclear extracts, respectively. Cells were also preincubated with AG490 (15 μm) for 1 h and then treated with SPE B or G308S (20 μg/ml) for 2 h. The phosphorylation of STAT1 on tyrosine 701 in nuclear extracts was determined (C).
STAT1 Phosphorylation at Serine 727 in SPE B-induced Apoptosis
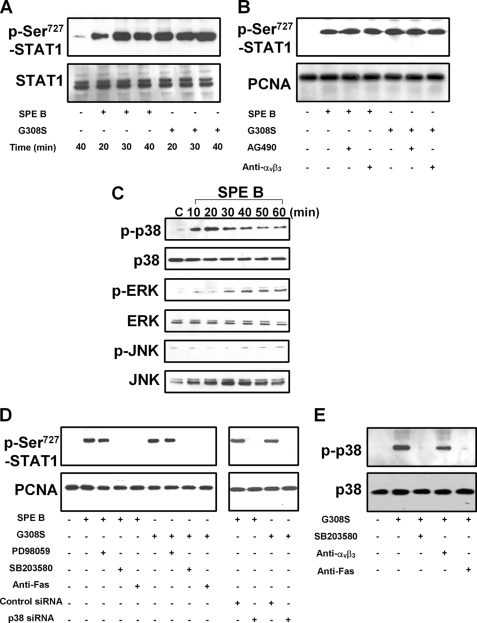
Because phosphorylation at the serine residue in the COOH terminus of the STAT protein is important to its transcriptional activity, we also evaluated this process in SPE B-induced apoptosis. As shown in Fig. 5A, levels of STAT1 phosphorylated at serine 727 were increased both in SPE B- and G308S-treated cells. Pretreatment of cells with anti-αVβ3 antibody or AG490 did not inhibit this phosphorylation (Fig. 5B). Our data suggest that an integrin-independent pathway mediates the phosphorylation of STAT1 at serine 727. The increased level of phosphoserine 727 in the nucleus also indicates that phosphorylation of serine 727 alone is enough for nuclear entry.
FIGURE 5.
SPE B increases nuclear phospho-Ser-727 STAT1 through Fas and p38 MAPK. Cells (2 × 105) were treated with SPE B or G308S (20 μg/ml) for the indicated times or PBS for 40 min. Cell lysates were immunoblotted using anti-phospho-Ser-727 STAT1 or anti-STAT1 antibody (A). Similar experiments were carried out to evaluate the effect of AG490 (15 μm), anti-αVβ3 antibody (4 μg/ml) (B), PD98059, SB203580 (10 μm), and p38 siRNA (D) on the level of phospho-Ser-727 STAT1 in the nucleus after SPE B or G308S (20 μg/ml) treatment for 40 min. The activation of MAPKs by SPE B was studied by incubating cells with SPE B (20 μg/ml) or PBS, and cell lysates were immunoblotted using antibodies against phosphorylated or total p38, ERK, and JNK (C). Cells were also treated with G308S (20 μg/ml) for 20 min with or without SB203580 (10 μm), anti-αVβ3 antibody (4 μg/ml), and anti-Fas antibody (0.5 μg/ml). Cell lysates were immunoblotted using anti-phospho-p38 or anti-p38 antibody (E).
The involvement of ERK, JNK, and p38 MAPK in the phosphorylation of STAT1 at serine 727 was also studied. We found that SPE B stimulated the phosphorylation of ERK and p38 MAPK in A549 cells (Fig. 5C). SPE B or G308S treatment increased the nuclear level of STAT1 phosphorylated at serine 727, whereas the p38 MAPK inhibitor SB203580, anti-Fas antibody, and p38 siRNA each inhibited serine phosphorylation of STAT1 induced either by SPE B or by G308S (Fig. 5D). The ERK inhibitor, PD98059, had no effect on serine 727 phosphorylation. Furthermore, the G308S-induced p38 phosphorylation decreased in the presence of SB203580 and anti-Fas antibody (Fig. 5E), suggesting that phosphorylation of STAT1 at serine 727 is mediated through the Fas and p38 MAPK pathways. Similarly, SPE B also activated p38 and phosphorylated p38-regulated STAT1 serine 727 phosphorylation in HEp-2 cells (Supplement 3, A and B).
SPE B-induced Bax Expression Is p38 MAPK-dependent
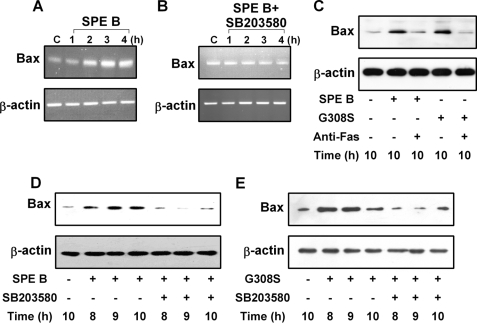
We next analyzed the transcriptional levels of several pro-apoptotic genes. We found no differences in mRNA levels of caspase 2, caspase 3, or caspase 9 either in the presence or in the absence of SPE B (data not shown). However, the mRNA level of Bax increased over time in SPE B-treated cells (Fig. 6A). In the presence of SB203580, this increase was eliminated (Fig. 6B). Several studies have reported that p38 MAPK acts as a mediator of apoptosis under different physiological conditions (23–26). It was also shown that p38 MAPK sensitized cells to apoptosis by increasing the levels of Bax and Fas (19). We found that SPE B and G308S both enhanced the generation of Bax protein and that the anti-Fas antibody inhibited this process (Fig. 6C). In addition, the increased levels of Bax induced by SPE B or G308S were inhibited by SB203580 (Fig. 6, D and E). These findings indicate that the Fas receptor and p38 MAPK are important mediators for the production of Bax by SPE B.
FIGURE 6.
Activated p38 induces Bax gene expression. Cells (3 × 106) were treated with SPE B (20 μg/ml) without (A) or with (B) SB203580 (10 μm) for the indicated times or PBS for 4 h. Total RNA was prepared and analyzed by RT-PCR for Bax mRNA expression. β-Actin was an internal control. Cells (2 × 105) were also treated with SPE B or G308S (20 μg/ml) with or without anti-Fas antibody (0.5 μg/ml) for 10 h. Cell lysates were immunoblotted using anti-Bax antibody. β-Actin was an internal control (C). For Bax production, cells (2 × 105) were treated with SPE B (D) or G308S (E) (20 μg/ml) with or without SB203580 (10 μm) for the indicated times or with PBS for 10 h. Cell lysates were immunoblotted using anti-Bax antibody.
Phosphorylated STAT1 Increases Expression of Procaspase 8 and Bax
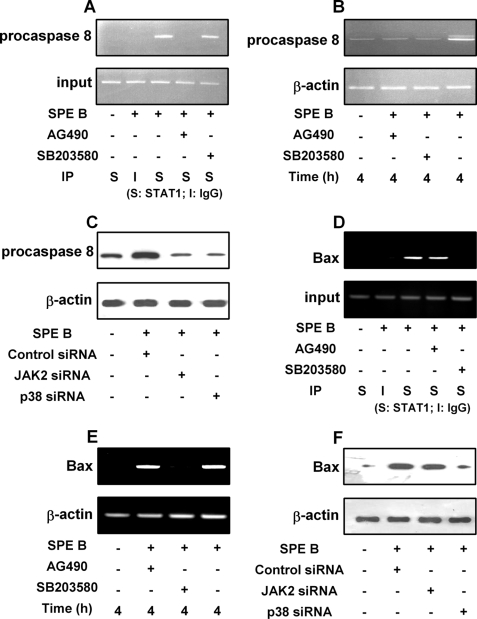
To investigate the molecular basis of SPE B-induced up-regulation of procaspase 8, we studied whether the expression of procaspase 8 mRNA depends on STAT1 transcriptional activity. Using the ChIP assay, we found that in the presence of SPE B, STAT1 was bound to procaspase 8 promoter and that this binding was eliminated by AG490 but not affected by SB203580 (Fig. 7A). This suggests that phosphorylation of STAT1 at tyrosine 701 was necessary for its binding to procaspase 8 promoter, whereas the phosphorylation of serine 727 was not. Furthermore, AG490, SB203580, and siRNA of JAK2 and p38 inhibited procaspase 8 mRNA transcription (Fig. 7, B and C), which indicated that the phosphorylation of tyrosine 701 and serine 727 of STAT1 are required for the expression of procaspase 8. In HEp-2 cells procaspase 8 promoter could recruit activated STAT1, and their binding was affected by tyrosine phosphorylation but not by serine phosphorylation (Supplement 2C). Both tyrosine and serine phosphorylation of STAT1 increased procaspase 8 expression (Supplement 2D).
FIGURE 7.
Phosphorylated STAT1 binds to procaspase 8 and Bax promoters after SPE B treatment. ChIP assay was used to detect the binding of STAT1 on the promoter of procaspase 8 or Bax. Cells (3 × 106) were treated with SPE B (20 μg/ml) with or without AG490 (15 μm) or SB203580 (10 μm) for 4 h. Control cells were treated with SPE B only. Chromatin was extracted and immunoprecipitated (IP) with antibodies specific to STAT1. Promoter of procaspase 8 (A) or Bax (D) was amplified with indicated primers by PCR. Input indicates the amount of genomic DNA used for immunoprecipitation. For the expression of mRNA, cells (3 × 106) were treated with SPE B (20 μg/ml) with or without AG490 (15 μm) or SB203580 (10 μm) for 4 h. Total RNA was prepared and analyzed by RT-PCR for procaspase 8 (B) or Bax (E) mRNA expression. Cells (2 × 105) were also transfected with siRNA of JAK2 or p38 and incubated with SPE B (20 μg/ml) for 10 h, and total cell lysates were immunoblotted using anti-procaspase 8 (C) or anti-Bax antibody (F).
The ChIP analysis of the Bax promoter showed that in the presence of SPE B, STAT1 was bound to the promoter and binding was abrogated by SB203580 but not by AG490 (Fig. 7D). SPE B increased the level of Bax mRNA, which was inhibited by SB203580 and by siRNA of p38 but not by AG490 or siRNA of JAK2 (Fig. 7, E and F). These results suggest that serine 727 phosphorylation of STAT1 via the p38 signaling pathway is important for STAT1 binding to the Bax promoter and that the expression of Bax mRNA is not mediated by the JAK2 pathway. Similarly, in HEp-2 cells, the serine-phosphorylated STAT1 bound to the Bax promoter in response to SPE B stimulation, and Bax expression was regulated by p38 (Supplement 3, C and D).
SPE B-induced Apoptotic Cell Death Is Associated with Procaspase 8 and Bax Expression
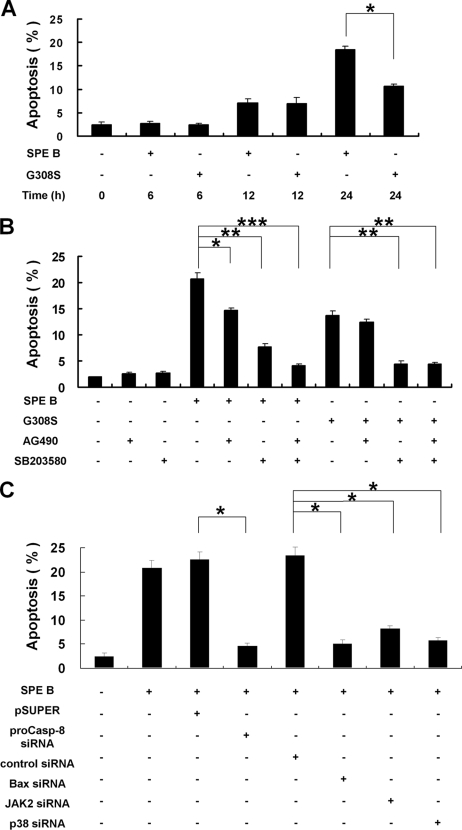
The findings above show that G308S increased only Bax expression, and SPE B increased both procaspase 8 and Bax expression. We then compared the contributions of these two pathways to the level of apoptosis. The results show that with short incubations i.e. 6 and 12 h, SPE B and G308S induced a similar level of apoptosis. However, after 24 h treatment, SPE B induced a higher level of apoptosis (Fig. 8A). In addition, in the presence of either AG490 or SB203580, SPE B-induced apoptosis was lower, and in the presence of both inhibitors the SPE B-induced apoptosis decreased even more, suggesting a possible synergistic or additive effect. For G308S-induced apoptosis, it was inhibited by SB203580 but not by AG490 (Fig. 8B). Cells transfected with procaspase 8, Bax, JAK2, or p38 siRNA could survive after SPE B treatment (Fig. 8C). Similar to A549 cells, inhibition of JAK2 or p38 activation reduced the SPE B-induced apoptosis in HEp-2 cells (Supplement 3E).
FIGURE 8.
SPE B-induced cell apoptosis is associated with procaspase 8 and Bax expression. Cells (8 × 104) were treated with SPE B or G308S (20 μg/ml) for the indicated times (A). Apoptosis was then determined using propidium iodide staining followed by flow cytometry analysis. Results are the mean ± S.E. of three experiments. *, p < 0.05. Cells (8 × 104) were also treated with SPE B or G308S (20 μg/ml) with or without AG490 (15 μm) and SB203580 (10 μm) pretreatment for 1 h (B). Apoptosis was determined after 24 h. Results are the mean ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001. After transfection with various siRNAs, cells (8 × 104) were treated with SPE B (20 μg/ml), and the level of apoptosis was measured after 24 h (C). Data are the mean ± S.E. of three experiments. *, p < 0.05.
DISCUSSION
We had demonstrated previously that in SPE B-induced apoptosis, caspase 8 is activated first followed by t-Bid and Bax activation, release of cytochrome c from mitochondria to cytosol, and finally, activation of caspase 9 and 3. Both extrinsic and intrinsic death pathways are involved in the process (8). We have also identified integrin αVβ3 and Fas as receptors for SPE B, which mediates RGD motif-dependent and -independent pathways (20). In the present study we have further identified two of the pathways mediated by integrin αVβ3 and by Fas for increasing the expression of procaspase 8 and Bax. Two recombinant proteins, SPE B and G308S, were used. G308S is a mutant of SPE B in which the glycine residue at 308 is converted to serine so that the RGD motif is replaced with RSD. To examine the structural difference between SPE B and G308S mutant, circular dichroism (CD) analyses were performed to determine their secondary structures. The results showed that SPE B and G308S had 23.8 and 23.3% α-helix, 53.5 and 52.6% β-structures, and 22.7 and 24.1% coil, respectively. Basically, there is no difference in their secondary structures. SPE B was used as the ligand for both RGD-dependent and -independent pathways, whereas G308S was used as the ligand for the RGD-independent pathway.
Our results show that the activation of JAK2 and STAT1 by tyrosine phosphorylation was stimulated by SPE B but not by G308S. This was inhibited by the anti-αVβ3 antibody, suggesting that the SPE B-induced JAK/STAT pathway in A549 cells was mediated through αVβ3. Although the molecular mechanism for activating integrin-mediated JAK is not clear, cytokine-mediated transphosphorylation of JAK has been well documented (27). Cytokine and receptor binding forms a complex, including JAK and its receptors, that induces the formation of phosphorylated tyrosine on receptor tail. Our coimmunoprecipitation experiments suggested that there is a direct association between integrin αVβ3 and JAK2. A similar mechanism to cytokine-mediated transphosphorylation of JAK may account for the activation of JAK by SPE B. The activated STAT1 translocated into the nucleus, bound to the promoter of procaspase 8, and increased the expression of procaspase 8 mRNA. The signal pathways leading to integrin-mediated gene transcription through the activation of JAK/STAT have been reported in various systems. Cells treated with growth factors or cytokines trigger the JAK/STAT signal pathway for protein expression (27, 28). In addition, integrin-mediated cell adhesion to the extracellular matrix also activates the JAK/STAT signal pathway. For example, the JAK2/STAT5 pathway via integrin-dependent adhesion of endothelial cells regulates c-fos gene expression (29).
The phosphorylation of STAT1 on serine is also important for its transcriptional activity (9). Our results show that STAT1 was also phosphorylated at serine 727 in the presence of SPE B and G308S. Interestingly, this phosphorylation was not inhibited by AG490 or anti-αVβ3 antibody but was inhibited by the inhibitor of p38. Furthermore, the activation of p38 was inhibited by anti-Fas antibody and not by anti-αVβ3 antibody. These results suggest that phosphorylation of STAT1 at serine 727 was stimulated by the Fas/p38 pathway.
In light of the role of STAT1 in apoptosis, recent studies (11, 30) have indicated that STAT1 up-regulates the expression of apoptotic genes, such as caspases, death ligand, and Bcl-xL. In addition to regulating the constitutive expression of caspases by unphosphorylated STAT1, STAT1 activation by phosphorylation may also lead to caspase induction (11). Our findings show that SPE B is responsible for inducing phosphorylation of STAT1 at tyrosine 701 and serine 727, which play different roles in the modulation of procaspase 8 expression. Phosphorylation of STAT1 on tyrosine 701 is critical for its binding to the procaspase 8 promoter, whereas phosphorylation of tyrosine 701 and serine 727 are required for the increased transcriptional activity.
Phosphorylation of serine in STAT1, the binding of phosphoserine 727 STAT1 to the Bax promoter, and the expression of Bax mRNA and protein were all inhibited by p38 inhibitor SB203580. In addition, the expression of Bax was inhibited by the anti-Fas antibody. These results suggest that the SPE B-induced Bax was regulated by the Fas/p38 pathway. The p38-regulated Bax expression is not well understood. However, it has been reported that STAT1 coactivates p53 to increase the induction of p53-responsive genes (31). p53 has a positive role in Bax gene expression, and p38 MAPK is required for the activation of p53 in UV-induced apoptosis (32, 33). The p53-mediated signaling events have been reported in several accounts of toxin-induced apoptosis, which involves the p38-dependent activation of p53 (34, 35). Our preliminary results also show that SPE B can trigger the Fas-mediated p38/p53 signal pathway (data not shown).
SPE B binds to integrin αVβ3 and Fas, and G308S binds only to Fas due to the lack of an RGD motif. The level of SPE B-induced apoptosis (15–20% of cells) was higher than G308S-induced apoptosis (∼10% of cells) after 24 h of incubation. The level of SPE B-induced apoptosis was lower in the presence of AG490 and SB203580; however, G308S-induced apoptosis was sensitive only to the p38 inhibitor. These results are consistent with the hypothesis that the mechanism through which SPE B induces apoptosis is a pathway involving increased levels of procaspase 8 and Bax, whereas G308S-induced apoptosis enhanced only Bax expression. Modulating the expression of apoptotic proteins to increase the sensitivity to apoptosis by various agents has been demonstrated in different cell types. For example, interferon-γ treatment of human adenocarcinoma cells contributes to the higher sensitivity of cell apoptosis by inducing the expression of apoptosis-signaling receptors such as Fas, TNFR1, the interleukin-1β-converting enzyme (Ice) family, and Bak (36). Interferon-γ acts on human erythroid progenitor cells to induce the expression of Fas and caspases 1, 3, and 8, which promote cell death (37). Stachybotrys chartarum toxin induced apoptosis in murine alveolar macrophages involved the gene expression of caspase-related genes (35). Although some other streptococcal virulence factors have been reported to be involved in apoptosis, their mode of action is less clear (38, 39). It is not known how they may interact with the pathways presented in this study. This issue remains under further investigation.
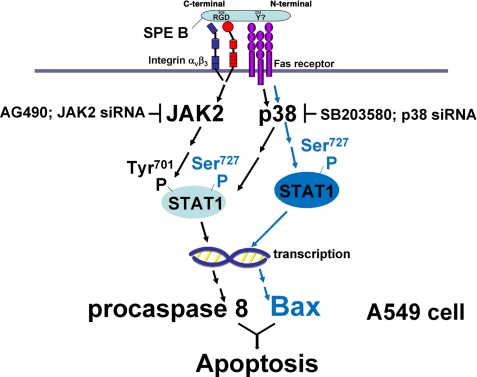
In summary (Fig. 9), our findings in the presented studies indicate that SPE B activates distinct pathways to up-regulate the expression of procaspase 8 and Bax by binding to different receptors via different molecular motifs. Binding of SPE B to αVβ3 activates the JAK2/STAT1 pathway, and binding of SPE B to Fas activates the p38 pathway. Complementary up-regulation of procaspase 8, an initiator of the extrinsic death pathway, and Bax, an important component of the intrinsic pathway, sensitizes cells to a higher level of apoptosis.
FIGURE 9.
JAK2/STAT1 and p38 signal pathways regulate SPE B-induced apoptosis in A549 cells. SPE B binds to integrin αVβ3 and Fas. Signals from both receptors activate the JAK2 and p38 pathways to phosphorylate tyrosine 701 and serine 727 of STAT1, respectively. Activated STAT1 translocates to the nucleus to activate the transcription of procaspase 8. Phospho-Ser-727 STAT1 is also a co-transactivator for the transcription of Bax.
Epithelial cell apoptosis is always observed in lung injury and inflammation (40). However, the pathophysiologic role of group A streptococcus in epithelial cell apoptosis is not well understood. A previous study indicated that apoptosis in response to bacterial infection may function to clear infected and damaged epithelial cells and restore cell growth regulation and epithelial integrity (41). Other reports indicated that the surfaces of mucosal linings are mainly composed of epithelial cells and serve as a first line of defense. A large number of apoptotic epithelial cells provide invading bacteria sufficient time to invade the deeper mucosal layers and spread.
It was demonstrated in our previous study that SPE B entered into the cell via receptor-mediated endocytosis and was degraded in the lysosome within 60 min (42), and the activation of caspase 8 to initiate apoptosis occurred at 90 min (8). Thus, apoptosis seems to be part of a host defense mechanism to remove invading bacteria.
Supplementary Material
Acknowledgment
We thank Jonathan Courtenay and Jason Kim for editorial assistance.
This work was supported by National Health Research Institute Grant NHRI-EX 96-9027SP and the National Science Council Grant NSC96-2745-B-320-003-URD, Taiwan.

The on-line version of this article (available at http://www.jbc.org) contains Supplements 1–3.
- SPE B
- streptococcal pyrogenic exotoxin B
- rSPE B
- recombinant SPE B
- siRNA
- short interfering RNA
- STAT
- signal transducers and activators of transcription
- JAK
- Janus kinase
- ERK
- extracellular signal-regulated kinase
- JNK
- c-Jun NH2-terminal kinase
- RT
- reverse transcription
- MAPK
- mitogen-activated protein kinase
- PBS
- phosphate-buffered saline
- ChIP
- chromatin immunoprecipitation.
REFERENCES
- 1.Musser J. M., Hauser A. R., Kim M. H., Schlievert P. M., Nelson K., Selander R. K. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 2668–2672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schlievert P. M., Assimacopoulos A. P., Cleary P. P. (1996) J. Lab. Clin. Med. 127, 13–22 [DOI] [PubMed] [Google Scholar]
- 3.Stevens D. L., Tanner M. H., Winship J., Swarts R., Ries K. M., Schlievert P. M., Kaplan E. (1989) N. Engl. J. Med. 321, 1–7 [DOI] [PubMed] [Google Scholar]
- 4.Chaussee M. S., Gerlach D., Yu C. E., Ferretti J. J. (1993) Infect. Immun. 61, 3719–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C. Y., Luo S. C., Kuo C. F., Lin Y. S., Wu J. J., Lin M. T., Liu C. C., Jeng W. Y., Chuang W. J. (2003) J. Biol. Chem. 278, 17336–17343 [DOI] [PubMed] [Google Scholar]
- 6.Hauser A. R., Schlievert P. M. (1990) J. Bacteriol. 172, 4536–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuo C. F., Wu J. J., Lin K. Y., Tsai P. J., Lee S. C., Jin Y. T., Lei H. Y., Lin Y. S. (1998) Infect. Immun. 66, 3931–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsai W. H., Chang C. W., Chuang W. J., Lin Y. S., Wu J. J., Liu C. C., Chang W. T., Lin M. T. (2004) Infect. Immun. 72, 7055–7062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Levy D. E., Darnell J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3, 651–662 [DOI] [PubMed] [Google Scholar]
- 10.Ihle J. N. (2001) Curr. Opin. Cell Biol. 13, 211–217 [DOI] [PubMed] [Google Scholar]
- 11.Battle T. E., Frank D. A. (2002) Curr. Mol. Med. 2, 381–392 [DOI] [PubMed] [Google Scholar]
- 12.Huang Y. Q., Li J. J., Karpatkin S. (2000) J. Biol. Chem. 275, 6462–6468 [DOI] [PubMed] [Google Scholar]
- 13.Minami M., Inoue M., Wei S., Takeda K., Matsumoto M., Kishimoto T., Akira S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 3963–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephanou A., Brar B. K., Scarabelli T. M., Jonassen A. K., Yellon D. M., Marber M. S., Knight R. A., Latchman D. S. (2000) J. Biol. Chem. 275, 10002–10008 [DOI] [PubMed] [Google Scholar]
- 15.Meister N., Shalaby T., von Bueren A. O., Rivera P., Patti R., Oehler C., Pruschy M., Grotzer M. A. (2007) Eur. J. Cancer 43, 1833–1841 [DOI] [PubMed] [Google Scholar]
- 16.Refaeli Y., Van Parijs L., Alexander S. I., Abbas A. K. (2002) J. Exp. Med. 196, 999–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmström T. H., Tran S. E., Johnson V. L., Ahn N. G., Chow S. C., Eriksson J. E. (1999) Mol. Cell. Biol. 19, 5991–6002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng Y., Ren X., Yang L., Lin Y., Wu X. (2003) Cell 115, 61–70 [DOI] [PubMed] [Google Scholar]
- 19.Porras A., Zuluaga S., Black E., Valladares A., Alvarez A.M., Ambrosino C., Benito M., Nebreda A. R. (2004) Mol. Biol. Cell 15, 922–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai W. H., Chang C. W., Lin Y. S., Chuang W. J., Wu J. J., Liu C. C., Tsai P. J., Lin M. T. (2008) Infect. Immun. 76, 1349–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aiyar A., Xiang Y., Leis J. (1996) Methods Mol. Biol. 57, 177–191 [DOI] [PubMed] [Google Scholar]
- 22.Kumar A., Commane M., Flickinger T. W., Horvath C. M., Stark G. R. (1997) Science 278, 1630–1632 [DOI] [PubMed] [Google Scholar]
- 23.De Zutter G. S., Davis R. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6168–6173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sarkar D., Su Z. Z., Lebedeva I. V., Sauane M., Gopalkrishnan R. V., Valerie K., Dent P., Fisher P. B. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10054–10059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y., Huang S., Sah V. P., Ross J., Jr., Brown J. H., Han J., Chien K. R. (1998) J. Biol. Chem. 273, 2161–2168 [DOI] [PubMed] [Google Scholar]
- 26.Zhuang S., Demirs J. T., Kochevar I. E. (2000) J. Biol. Chem. 275, 25939–25948 [DOI] [PubMed] [Google Scholar]
- 27.Schindler C., Darnell J. E., Jr. (1995) Annu. Rev. Biochem. 64, 621–651 [DOI] [PubMed] [Google Scholar]
- 28.Ihle J. N., Kerr I. M. (1995) Trends Genet. 11, 69–74 [DOI] [PubMed] [Google Scholar]
- 29.Brizzi M. F., Defilippi P., Rosso A., Venturino M., Garbarino G., Miyajima A., Silengo L., Tarone G., Pegoraro L. (1999) Mol. Biol. Cell 10, 3463–3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fu X. Y. (1999) Cell Death Differ. 6, 1201–1208 [DOI] [PubMed] [Google Scholar]
- 31.Townsend P. A., Scarabelli T. M., Davidson S. M., Knight R. A., Latchman D. S., Stephanou A. (2004) J. Biol. Chem. 279, 5811–5820 [DOI] [PubMed] [Google Scholar]
- 32.Bulavin D. V., Saito S., Hollander M. C., Sakaguchi K., Anderson C. W., Appella E., Fornace A. J., Jr. (1999) EMBO J. 18, 6845–6854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thornborrow E. C., Manfredi J. J. (2001) J. Biol. Chem. 276, 15598–15608 [DOI] [PubMed] [Google Scholar]
- 34.Kim H., Kokkotou E., Na X., Rhee S. H., Moyer M. P., Pothoulakis C., Lamont J. T. (2005) Gastroenterology 129, 1875–1888 [DOI] [PubMed] [Google Scholar]
- 35.Wang H., Yadav J. S. (2007) Apoptosis 12, 535–548 [DOI] [PubMed] [Google Scholar]
- 36.Ossina N. K., Cannas A., Powers V. C., Fitzpatrick P. A., Knight J. D., Gilbert J. R., Shekhtman E. M., Tomei L. D., Umansky S. R., Kiefer M. C. (1997) J. Biol. Chem. 272, 16351–16357 [DOI] [PubMed] [Google Scholar]
- 37.Dai C., Krantz S. B. (1999) Blood 93, 3309–3316 [PubMed] [Google Scholar]
- 38.Cywes Bentley C., Hakansson A., Christianson J., Wessels M. R. (2005) Cell. Microbiol. 7, 945–955 [DOI] [PubMed] [Google Scholar]
- 39.Timmer A. M., Timmer J. C., Pence M. A., Hsu L. C., Ghochani M., Frey T. G., Karin M., Salvesen G. S., Nizet V. (2009) J. Biol. Chem. 284, 862–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Medan D., Wang L., Yang X., Dokka S., Castranova V., Rojanasakul Y. (2002) J. Cell. Physiol. 191, 320–326 [DOI] [PubMed] [Google Scholar]
- 41.Kim J. M., Eckmann L., Savidge T. C., Lowe D. C., Witthöft T., Kagnoff M. F. (1998) J. Clin. Invest. 102, 1815–1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang C. W., Tsai W. H., Chuang W. J., Lin Y. S., Wu J. J., Liu C. C., Tsai P. J., Lin M. T. (2007) J. Biomed. Sci. 14, 419–427 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.