Abstract
Periplasmic binding proteins (PBPs) constitute a protein superfamily that binds a wide variety of ligands. In prokaryotes, PBPs function as receptors for ATP-binding cassette or tripartite ATP-independent transporters and chemotaxis systems. In many instances, PBPs bind their cognate ligands with exquisite specificity, distinguishing, for example, between sugar epimers or structurally similar anions. By contrast, oligopeptide-binding proteins bind their ligands through interactions with the peptide backbone but do not distinguish between different side chains. The extremophile Thermotoga maritima possesses a remarkable array of carbohydrate-processing metabolic systems, including the hydrolysis of cellulosic polymers. Here, we present the crystal structure of a T. maritima cellobiose-binding protein (tm0031) that is homologous to oligopeptide-binding proteins. T. maritima cellobiose-binding protein binds a variety of lengths of β(1→4)-linked glucose oligomers, ranging from two rings (cellobiose) to five (cellopentaose). The structure reveals that binding is semi-specific. The disaccharide at the nonreducing end binds specifically; the other rings are located in a large solvent-filled groove, where the reducing end makes several contacts with the protein, thereby imposing an upper limit of the oligosaccharides that are recognized. Semi-specific recognition, in which a molecular class rather than individual species is selected, provides an efficient solution for the uptake of complex mixtures.
INTRODUCTION
Periplasmic binding proteins (PBPs)2 are soluble ligand-binding components of ATP-binding cassette (1) or tripartite ATP-independent (2) transporters and chemotaxis systems (3). Members of the PBP superfamily mediate uptake of many primary metabolites in bacteria such as amino acids (4), carbohydrates (5), ions (6), and polyamines (7). Ligands are subsequently transported across the membrane by accessory proteins that couple transport to ATP hydrolysis (1) or H+/M+ motive force (2).
PBPs form a structural superfamily that is classified into three groups according to the ordering of β-strands in the core of the two domains that characterize this protein fold: group I/ribose-binding protein fold (8), group II/maltose-binding protein fold (9), and group III/vitamin B12-binding protein fold (10, 11). Each of the domains forms a three-layered α/β/α-fold, divided by a two- or three-β-strand hinge. The ligand-binding site is situated between the two domains. The interactions of these binding surfaces are contributed largely by amino acid side chains located in the loops connecting the alternating β-strands and α-helices of each domain.
PBPs undergo a hinge-bending motion upon addition of ligand, allowing the two interfaces, which are solvent-exposed in the absence of ligand, to completely envelop the ligand in the closed form, thus mimicking the desolvated environment of a protein core (8, 12–14). The resulting binding sites typically form extensive specific interactions with their cognate ligands, enabling highly specific discrimination between anomeric or epimeric carbohydrates (15), differently sized carbohydrates (15, 16), or chemically similar anions (17). By contrast, group II di/oligopeptide-binding proteins (OpBPs), which bind peptides that range from two to nine amino acids (18, 19), are semi-specific and show little discrimination between side chains of bound peptides (20). In these proteins, recognition is mediated primarily through hydrogen bonds to the peptide main chain atoms, whereas the side chains are placed in nonspecific pockets that accommodate both polar and non-polar amino acid side chains through interactions with differentially ordered water molecules (19–21). Any of the 20 amino acids are bound by OpBPs with little sequence-dependent variations in dissociation constants (Kd) (19).
The hyperthermophilic bacterium Thermotoga maritima MSB8 is an extensively studied model for extremophiles and a potential source of many enzymes and metabolic pathways of biotechnological interest (22). Its genomic sequence reveals a remarkable array of diverse carbohydrate metabolic pathways (23, 24). Among these are systems for the uptake and processing of a variety of β-linked oligosaccharides. Metabolite transport is encoded by a large number of ATP-binding cassette transporters and related proteins (23). It has been difficult to assign function reliably to the PBP components of these because of the distant sequence relationships between T. maritima and biochemically characterized model organisms. A combination of in vitro binding studies (25) and transcriptional expression profiling of cultures grown in the presence of various carbohydrate substrates (26, 27) has shown that 5 of 12 PBPs that are homologous to OpBPs bind not oligopeptides but various carbohydrates. The x-ray crystal structure of one of these, the β-1,4-mannobiose-binding protein (Protein Data Bank code 1VR5), was recently solved in the open conformation in the absence of its cognate ligand (structure determined and deposited by the Joint Center for Structural Genomics (28) but otherwise undocumented). As expected, based on amino acid similarity, the overall structure of 1VR5 is similar to known OpBPs. In the absence of a ligand, the mode of ligand binding in this protein remains unknown. These observations raise the question of how the OpBP fold has adapted to recognize sugars and what, if any, specificity is encoded.
To answer these questions, we undertook structural studies of tm0031, which was shown previously to bind the β-linked disaccharides cellobiose and laminaribiose (25). In additional ligand-binding studies, we established that this cellobiose-binding protein (tmCBP) binds not only disaccharides but also oligosaccharides up to five sugar monomers in length. X-ray crystallographic structural analysis revealed that only the first two sugar rings are recognized specifically though hydrogen bonds and van der Waals interactions with the protein. Additional rings are placed into a large solvent-filled cavity in the interior of the binding pocket, where there is little specific recognition of the ligand polar and non-polar groups, in a manner similar to the recognition of peptides in the OpBP fold (19). Comparison of superimposed apo-1VR5 with ligand-bound tmCBP suggests that the former possesses similar adaptations to bind its cognate ligand(s). Consequently, we postulate that this binding pocket imposes limited specificity between oligosaccharides other than through specific recognition of the first two sugar rings and the sterics of the cavity that imposes length constraints. This mode of sugar binding has not been observed in other carbohydrate-binding PBPs and represents a novel mechanism of carbohydrate recognition in this protein superfamily.
EXPERIMENTAL PROCEDURES
Overexpression and Purification
The tmCBP plasmid was a gift from the laboratory of K. Noll. Protein was expressed and purified as described (25).
Circular Dichroism
CD measurements were carried out on a JASCO CD spectrophotometer. Thermal denaturations were determined by measuring the CD signal at 222 nm (1-cm path length) as a function of temperature using 1.0 μm protein (10 mm Tris-HCl (pH 7.8) and 150 mm NaCl). In the absence of the chemical denaturant guanidinium chloride (GdmCl), tmCBP is too stable to exhibit temperature-induced denaturation. To determine the apparent thermal transition midpoint (Tm(app)) in the absence of GdmCl, a series of thermal melts in the presence of decreasing amounts of GdmCl was used to extrapolate to 0 m GdmCl (16). Protein samples were incubated for 15 min prior to collecting data. Each measurement included a 3-s averaging time for data collection and a 60-s equilibration period at each temperature. Data were fit to a two-state model to determine the Tm(app) values (29).
Crystallization and Data Collection
Crystals of tmCBP were grown in the presence of 1 mm ligand. The cellopentaose complex was crystallized by hanging drop vapor diffusion in drops containing 2 μl of the protein solution mixed with 2 μl of 0.1 m sodium cacodylate (pH 6.3–6.5), 18–24% (w/v) polyethylene glycol 4000, and 0.2 m magnesium acetate. The cellobiose complex was crystallized by hanging drop vapor diffusion in drops containing 2 μl of the protein solution mixed with 2 μl of 0.1 m MES (pH 6.5), 0.2 m ammonium sulfate, 30% (w/v) polyethylene glycol monomethyl ether 5000. Diffraction-quality crystals typically grew within 1 week at 17 °C. Crystals grown in the presence of cellohexaose resulted in a cellopentaose complex (data not shown) due to the low purity of the oligosaccharides (∼95% pure) and the presence of contaminating cellopentaose. The crystals of the cellobiose and cellopentaose complexes diffract to 1.50 Å resolution and belong to the P41212 space group (a/b = 107.3 and c = 118.2 Å) and the P21 space group (a = 62.1, b = 101.5, and c = 108.3 Å; β = 94.1°), respectively (see Table 1). Crystals were transferred stepwise to the precipitant solution containing 10% ethylene glycol for cryoprotection, mounted in a nylon loop, and flash-frozen in liquid nitrogen. All data were collected at 100 K at the Southeast Regional Collaborative Access Team 22-ID beam line at the Advanced Photon Source. Diffraction data were scaled and integrated using XDS (30).
TABLE 1.
Data collection and refinement statistics
APS/SER-CAT, Advanced Photon Source/Southeast Regional Collaborative Access Team; r.m.s.d., root mean square deviation.
| Ligand |
||
|---|---|---|
| Cellobiose | Cellopentaose | |
| Data collection | ||
| Resolution range (Å) | 50–1.50 | 50–1.50 |
| Data source | APS/SER-CAT | APS/SER-CAT |
| Unique reflections | 109,675 | 205,603 |
| Mean I/σ(I)a | 18.8 (2.9) | 15.7 (4.0) |
| Rsym (%)a | 8.1 (47.5) | 5.0 (32.9) |
| Completeness (%)a | 99.9 (99.8) | 96.3 (83.9) |
| Unit cell dimensions | a/b = 107.3, c = 117.2 Å | a = 62.1, b = 101.5, c = 108.3 Å; β = 94.1° |
| Space group | P41212 | P21 |
| Refinement | ||
| Non-hydrogen atoms in refinement | ||
| Protein | 4812 | 9775 |
| Water | 500 | 887 |
| Ligand | 23 | 112 |
| Rcryst/Rfree (%)b | 19.2/21.6 | 20.4/22.5 |
| r.m.s.d. from ideal | ||
| Bond lengths (Å) | 1.12 | 1.07 |
| Bond angles | 0.007° | 0.005° |
| B-factors (Å2) | ||
| Main chain | 15.8 | 21.2 |
| Side chain | 16.8 | 24.3 |
| Ligand | 12.5 | 20.8 |
| Water | 26.1 | 30.7 |
| Ramachandran plot | ||
| Allowed (%) | 100 | 100 |
| Favored (%) | 97.7 | 97.8 |
a Numbers in parentheses represent values in the highest resolution shell.
b Rfree is the R-factor based on 5% of data excluded from refinement.
Structure Determination Methods, Model Building, and Refinement
The structure of the cellobiose complex was determined to a resolution of 1.50 Å by molecular replacement using the N- and C-terminal domains of the β-1,4-mannobiose-binding protein (Protein Data Bank code 1VR5) separately as search models in the program PHASER (31); the individual domains rather than the entire molecule were used to allow for changes in the interdomain hinge-bending angles. One protein molecule bound with cellobiose in the binding pocket was found in the asymmetric unit and was refined to Rcryst and Rfree values of 19.2 and 21.6%, respectively (see Table 1). The final model for the cellobiose complex includes two intact monomers, two cellobiose molecules, and 500 water molecules. The structure of the cellopentaose complex was determined to a resolution of 1.5 Å by molecular replacement using the cellobiose-bound form as a search model in the program PHASER (31). Two molecules bound with cellopentaose in the binding pocket were found in the asymmetric unit and were refined to Rcryst and Rfree values of 20.4 and 22.5%, respectively (see Table 1). The final model for the cellopentaose complex includes two intact monomers, two cellopentaose molecules, and 887 water molecules. The cellobiose and cellopentaose complexes are in nearly identical conformations, with a root mean square deviation of 0.21 Å for alignment of all atoms in both crystal forms (excluding water molecules and ligand). Manual model building was carried out in the programs O and COOT and refined using REFMAC5 (32–34). The models exhibit good stereochemistry as determined by PROCHECK and MolProbity; final data collection and refinement statistics are listed in Table 1 (35, 36). No residues are Ramachandran outliers. The Protein Data Bank coordinates and structure factors of the cellobiose and cellopentaose complexes have been deposited in the Protein Data Bank under codes 2O7I and 3I5O, respectively.
RESULTS
Thermal Stability and Ligand Binding
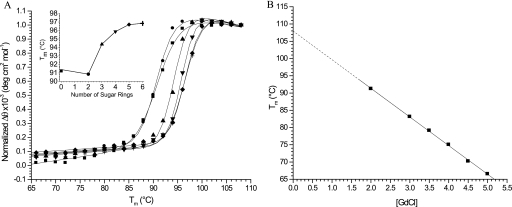
The stability of tmCBP was determined by thermal denaturation using CD. In the absence of the chemical denaturant GdmCl, no significant change in the CD signal (up to 100 °C) could be observed as a function of temperature (data not shown), and all subsequent measurements were carried out in the presence of GdmCl (Fig. 1). Melting curves were found to fit a two-state model (29, 37); a Tm(app) of 108 °C in the absence of GdmCl was determined by linear extrapolation of a series of melting point determinations carried out at different concentrations of GdmCl (Fig. 1) (15).
FIGURE 1.
Thermal denaturation of tmCBP determined by CD. A, thermal denaturation of tmCBP in 2 m GdmCl in the absence (■) and presence of 1 mm cellobiose (●), cellotriose (▲), cellotetraose (▼), cellopentaose (♦), and cellohexaose (+). Solid lines in A were fit to a two-state model that accounts for the native and denatured base-line slopes (37). Inset, Tm(app) values as a function of the number of sugar rings. deg, degrees. B, Tm(app) values in the absence of denaturant obtained by extrapolation of a series of thermal melting curves determined at different concentrations of GdmCl (GdCl). Solid lines represent linear fits to the observations.
Ligand-mediated shifts in Tm(app) values in the presence of 2 m GdmCl were used to assess the binding of various cellulose hydrolysates ranging from two-sugar rings (cellobiose) to six-sugar rings (cellohexaose). Protein stability increased nonlinearly with oligosaccharide length (Fig. 1), clearly delineating a minimal recognition unit and a maximal length where no more contribution is made to protein stability. The disaccharide cellobiose bound relatively weakly, as evidenced by a small shift in Tm(app) and a change in cooperativity; a significant increase was observed with the trisaccharide cellotriose; beyond five sugar units, there was no significant increase in stability. These results indicate that the first three sugar rings make the greatest contribution to the free energy of ligand binding, although the binding pocket binds five optimally but not necessarily maximally.
Overall Three-dimensional Structure
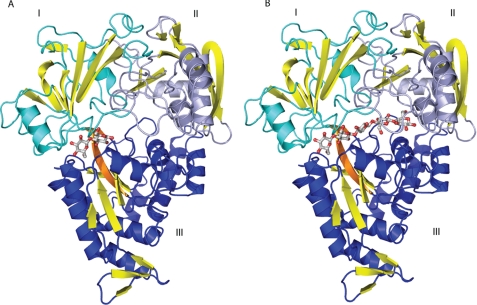
The protein was co-crystallized with cellobiose and cellopentaose. In these two complexes, the tmCBP structure adopts the closed conformation and has the α/β-fold that is characteristic of OpBPs and shares the highest structural homology with Salmonella typhimurium OpBP (Protein Data Bank code 1JEV; Cα root mean square deviation of 2.8 Å) (38, 39) even though the amino acid sequence similarity between them is low (18% identity). Alignment of the individual domains of the 1VR5 search model revealed Cα root mean square deviations of 1.3 and 1.1 Å for the N- and C-terminal domains, respectively. Like S. typhimurium OpBP, tmCBP is a three-domain protein with the carbohydrate-binding site located in a deep groove at the domain interface (Fig. 2). The topology of β-strands in domains I and III belongs to the group II PBP subfamily.
FIGURE 2.
Overall structure of tmCBP. A, cellobiose complex; B, cellopentaose complex. Domain I (blue), domain II (cyan), and domain III are indicated. Yellow, β-strands; orange, β-strands in the hinge. Bound sugars are shown in ball and stick representation: red, oxygens; gray, carbons.
Oligosaccharide-binding Pocket
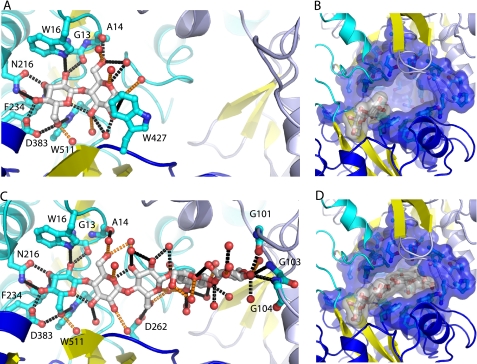
The cellopentaose complex defines a ligand-binding site that spans the entire interface formed by domains I–III (Fig. 2). The minimal cellobiose-binding site is found at one extreme of this groove, at the interface between domains I and III (Fig. 2). As is observed in other highly specific carbohydrate-binding PBPs, the two sugar rings are bound by an extensive network of hydrogen bonds that largely satisfy the dual potential of the hydroxyls (i.e. both the proton donor and the oxygen acceptor). A total of 19 hydrogen bonds are made with the cellobiose. Of these, 15 hydrogen bonds are formed by a network of polar interactions with protein side chain, main chain, and specifically bound water molecules mediating recognition of the hydroxyls in the two sugars (Fig. 3). Two polar amino acids (Asn216 and Asp383) contribute 4 of the 10 hydrogen bonds, three are made by the protein main chain (Ala14 oxygen, Gly13 oxygen, and Phe234 nitrogen), and three are made with aromatic binding pocket residues through either polar or aromatic hydrogen bonds. The remaining hydrogen-bonding potential is satisfied by water molecules (Fig. 3). The oxygen from the hemiacetal of the second sugar ring (ring B) forms a hydrogen bond with two water molecules, one of which also interacts with the protein; the hemiacetal oxygen from the first ring (ring A) forms an intramolecular hydrogen bond with the C3 hydroxyl of the second ring (ring B) (Fig. 3). Three additional tryptophan residues (Trp381, Trp384, and Trp536) form extensive van der Waals contacts with both rings of the cellobiose.
FIGURE 3.
Sugar-binding pocket and cavity. A and B, cellobiose complex; C and D, cellopentaose complex. Hydrogen-bonding interactions are indicated with black dashed lines drawn between the ligand and amino acid side chains and waters (red spheres) that do no interact with the protein. Hydrogen bonds between the ligand and water molecules that interact with the protein are indicated with orange dashed lines.
Adjacent to this minimal disaccharide-binding site is a large cavity that is shielded from bulk solvent but completely filled with well defined water molecules (Figs. 3 and 4). This region lacks the aromatic and polar amino acids lining the disaccharide-binding site. In the cellopentaose-bound form, the extra three rings are placed into this cavity (Figs. 3 and 4). However, unlike the cellobiose-bound form, there is little specific recognition of the sugar rings (Fig. 3). These additional rings have the potential to form a total of 26 hydrogen bonds through their hydroxyls and hemiacetal oxygens, but only four hydrogen bonds are made with the protein. One is formed with the C6 hydroxyl of the third ring (ring C) and the carboxylate of Asp262; three other hydrogen bonds are formed by the main chain of three glycine residues (Gly101, Gly103, and Gly104), at the tip of the cavity (the cap), with the hydroxyls of the fifth ring (ring E). Ordered water molecules satisfy the remaining hydrogen-bonding potential of the three sugar rings (Fig. 3).
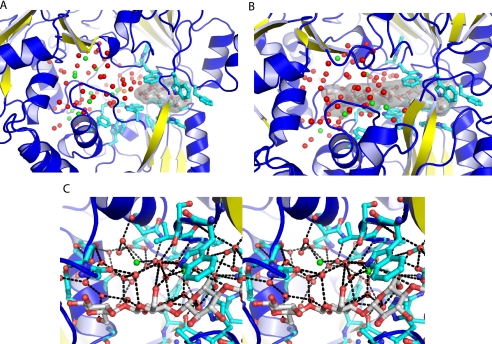
FIGURE 4.
Solvent structure in the ligand-binding cavity. A, the disaccharide binds specifically at one end of the binding site groove in a sited defined by a cluster of aromatic residues (the hook). The adjacent cavity is filed with 80 well defined water molecules. B, the nonreducing end of the cellopentaose also binds to the hook. The three other rings are placed into the adjacent cavity, which contains 75 well defined water molecules. C, a stereo diagram of the water structure in the cellopentaose complex is shown. Hydrogen-bonding interactions are indicated with black dashed lines. Water molecules are colored according to their solvation layer: red, primary solvation shell; green, secondary solvation shell; orange, tertiary solvation shell.
Ligand-binding Cavity Water Structure
80 water molecules fill the cavity of the cellobiose complex, of which 66 form a primary solvation shell that directly contacts the protein or carbohydrate polar groups (Fig. 4). An additional 13 form a secondary solvation shell that is bound only by water molecules in the primary solvation shell. The remaining one ordered water molecule is hydrogen-bonded to the secondary shell (tertiary solvation shell). 75 water molecules are retained in the cavity of the cellopentaose complex, of which 67 are in the primary solvation shell and eight in secondary solvation shell (Fig. 4). No ordered tertiary shell water molecules are found in the cellopentaose complex, with the single tertiary shell water molecule being displaced by the extra rings of the carbohydrate. Many of the water molecules in the primary solvation shell form an approximately tetrahedral hydrogen-bonding network, thereby constructing an ice-like structure within the cavity (Fig. 4). The secondary solvation shell forms between one to four hydrogen bonds, with the remaining hydrogen-bonding potential being satisfied by interactions with bulk water.
The positions of the ordered water molecules change little upon ligand binding. Binding of the pentasaccharide displaces 1 of the 13 water molecules in the secondary shell and the single tertiary shell water molecule. Only five primary shell water molecules are displaced upon cellopentaose binding. In several cases, the hydroxyls of the incoming sugar fully replace the hydrogen-bonding geometry of the lost water molecule.
Comparison of tmCBP and 1VR5 Ligand-binding Sites
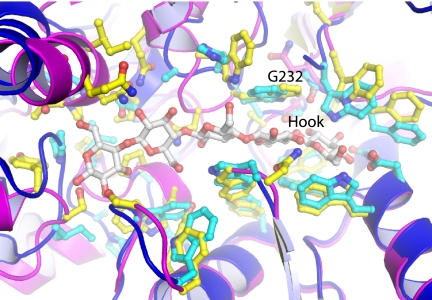
Superposition of the individual domains of the 1VR5 search model suggests that the β-1,4-mannobiose ligand binds in a similar manner as the tmCBP ligand (Fig. 5) because of similarities in the surfaces that line the ligand-binding pockets. Glucose and mannose, which compose the monomers of the oligosaccharides bound by 1VR5 and tmCBP, differ in their hydroxyl at C2. The 12 amino acids and the backbone conformation in the region of the binding pocket that recognizes the first sugar rings (the “hook”) are highly conserved between the two proteins (Fig. 5). The seven amino acids involved in hydrogen bonding with the ligand are conserved in all but one position, which in tmCBP recognizes the C3 and C4 hydroxyls of the ligand. The remaining five amino acids differ in only two positions: Leu233, which forms van der Waals interactions with the O3 hydroxyl, is replaced with phenylalanine in 1VR5, and Gly232, which forms van der Waals interactions with the O2 hydroxyl, is replaced with asparagine in 1VR5. The latter change is accompanied by a backbone movement in 1VR5, which prevents a steric clash that would occur with the axial C2 hydroxyl of mannose.
FIGURE 5.
Comparison of the binding sites of tmCBP and the apoprotein of the β-1,4-mannobiose-binding protein (Protein Data Bank code 1VR5). The N- and C-terminal domains of 1VR5 (cyan with side chains shown in ball and stick representation: red, oxygens; yellow, carbons) were superimposed individually onto the cellopentaose complex of tmCBP (blue with side chains shown in ball and stick representation: red, oxygens; yellow, carbons).
The remainder of the ligand-binding cavity, which in tmCBP is relatively nonspecific, differs significantly between the aligned structures: the loops and helices contain substitutions and insertions/deletions, and the backbone is found in different conformations (Fig. 5). No definitive conclusions can be drawn about 1VR5 in the absence of a cognate ligand, but it appears that this region also forms a cavity that is larger than the molecular envelope of an individual oligosaccharide chain, suggesting semi-specific recognition.
DISCUSSION
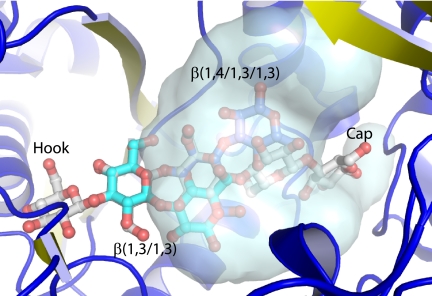
We have demonstrated that the T. maritima tm0031 open reading frame encodes a periplasmic polymeric carbohydrate-binding protein that binds β(1→4)-linked glucose molecules that range in size from two to five sugar rings. The mode of oligosaccharide recognition is unusual in that this binding protein is specific for only part of the ligand, whereas other sugar-binding proteins in the PBP superfamily typically are exquisitely specific. Instead, tmCBP selects a molecular class rather than a single species. Based on comparison with the apo structure of the β-1,4-mannobiose-binding protein, it appears that its ligands may bind in a similar manner, although the boundaries of the cavity, which determine sugar length and branch selectivity, are not well defined in the absence of a cognate ligand. Oligosaccharides are selected by specific recognition of the disaccharide at the nonreducing end (the hook), whereas the rest of the oligosaccharide with mixed linkages is placed into a solvent-filled cavity. The reducing end of cellopentaose forms a few specific hydrogen bonds. This “capping” interaction might impose an upper limit on the length of an oligosaccharide that is recognized by tmCBP and subsequently transported across the membrane. The nonselective cavity is filled with a remarkable arrangement of ordered water molecules, divided into a semipermanent layer that forms a tetrahedral ice-like hydrogen-bonding network and a more loosely bonded layer that is partially displaced by the bound oligosaccharide. It is therefore possible that different branches and linkages can be accommodated within the steric limitation of the cavity, enabling pre-concentration of a wider array of plant-based (40), marine algae-based (41), and fungus-based (42) β-carbohydrates. For instance, the pentameric β(1→4)-sugar does not occupy the entire volume of the cavity, occupying only a fraction of one region; other linkages of oligosaccharides can be positioned in the remaining cavity space (Fig. 6).
FIGURE 6.
Carbohydrate selectivity of the ligand-binding cavity. Different linkages of carbohydrates (blue, β(1→4/1→3/1→3); cyan, β(1→3/1→3); gray, cellopentaose) beyond the hook can be modeled to occupy different regions of the cavity adjacent to the cellobiose-binding site (gray surface representation).
T. maritima possesses an extensive saccharolytic metabolic system based on extracellular cellulases that hydrolyze polysaccharides to shorter chains, which are then transported across the membrane for phosphorylation and further cleavage (22). In this scheme, tmCBP acts as a molecular sieve, with the hook selecting only a subset of β-oligosaccharides and the “cap” imposing a size limit oligosaccharides that can be transported and function as a substrate for the intracellular β-glucose hydrolases. Such semi-specific recognition systems present an efficient solution for the processing of complex mixtures down to common metabolic feedstocks.
Acknowledgments
We thank G. Shirman for protein expression and purification and the laboratory of K. Noll for the tm0031 plasmid. Data were collected at the Southeast Regional Collaborative Access Team 22-ID and Structural Biology Center 19-ID beam lines at the Advanced Photon Source, Argonne National Laboratory.
This work was supported, in whole or in part, by National Institutes of Health Pioneer Award 5 DP1 OD000122-02 and Homeland Security Advanced Research Projects Agency Grant W81XWH-05-C-0161 (to H. W. H.). Use of the Advanced Photon Source was supported by the United States Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract W-31-109-Eng-38.
The atomic coordinates and structure factors (codes 2O7I and 3I5O) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PBP
- periplasmic binding protein
- OpBP
- oligopeptide-binding protein
- tmCBP
- T. maritima cellobiose-binding protein
- GdmCl
- guanidinium chloride
- MES
- 4-morpholineethanesulfonic acid.
REFERENCES
- 1.Boos W., Shuman H. (1998) Microbiol. Mol. Biol. Rev. 62, 204–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kelly D. J., Thomas G. H. (2001) FEMS Microbiol. Rev. 25, 405–424 [DOI] [PubMed] [Google Scholar]
- 3.Aksamit R. R., Koshland D. E., Jr. (1974) Biochemistry 13, 4473–4478 [DOI] [PubMed] [Google Scholar]
- 4.Hsiao C. D., Sun Y. J., Rose J., Wang B. C. (1996) J. Mol. Biol. 262, 225–242 [DOI] [PubMed] [Google Scholar]
- 5.Duan X., Quiocho F. A. (2002) Biochemistry 41, 706–712 [DOI] [PubMed] [Google Scholar]
- 6.Shouldice S. R., Dougan D. R., Williams P. A., Skene R. J., Snell G., Scheibe D., Kirby S., Hosfield D. J., McRee D. E., Schryvers A. B., Tari L. W. (2003) J. Biol. Chem. 278, 41093–41098 [DOI] [PubMed] [Google Scholar]
- 7.Sugiyama S., Vassylyev D. G., Matsushima M., Kashiwagi K., Igarashi K., Morikawa K. (1996) J. Biol. Chem. 271, 9519–9525 [DOI] [PubMed] [Google Scholar]
- 8.Björkman A. J., Mowbray S. L. (1998) J. Mol. Biol. 279, 651–664 [DOI] [PubMed] [Google Scholar]
- 9.Quiocho F. A., Spurlino J. C., Rodseth L. E. (1997) Structure 5, 997–1015 [DOI] [PubMed] [Google Scholar]
- 10.Karpowich N. K., Huang H. H., Smith P. C., Hunt J. F. (2003) J. Biol. Chem. 278, 8429–8434 [DOI] [PubMed] [Google Scholar]
- 11.Fukami-Kobayashi K., Tateno Y., Nishikawa K. (1999) J. Mol. Biol. 286, 279–290 [DOI] [PubMed] [Google Scholar]
- 12.Sharff A. J., Rodseth L. E., Spurlino J. C., Quiocho F. A. (1992) Biochemistry 31, 10657–10663 [DOI] [PubMed] [Google Scholar]
- 13.Cuneo M. J., Beese L. S., Hellinga H. W. (2008) BMC Struct. Biol. 8, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marvin J. S., Hellinga H. W. (2001) Nat. Struct. Biol. 8, 795–798 [DOI] [PubMed] [Google Scholar]
- 15.Cuneo M. J., Changela A., Warren J. J., Beese L. S., Hellinga H. W. (2006) J. Mol. Biol. 362, 259–270 [DOI] [PubMed] [Google Scholar]
- 16.Cuneo M. J., Changela A., Beese L. S., Hellinga H. W. (2009) J. Mol. Biol. 389, 157–166 [DOI] [PubMed] [Google Scholar]
- 17.Bruns C. M., Nowalk A. J., Arvai A. S., McTigue M. A., Vaughan K. G., Mietzner T. A., McRee D. E. (1997) Nat. Struct. Biol. 4, 919–924 [DOI] [PubMed] [Google Scholar]
- 18.Levdikov V. M., Blagova E. V., Brannigan J. A., Wright L., Vagin A. A., Wilkinson A. J. (2005) J. Mol. Biol. 345, 879–892 [DOI] [PubMed] [Google Scholar]
- 19.Sleigh S. H., Seavers P. R., Wilkinson A. J., Ladbury J. E., Tame J. R. (1999) J. Mol. Biol. 291, 393–415 [DOI] [PubMed] [Google Scholar]
- 20.Tame J. R., Murshudov G. N., Dodson E. J., Neil T. K., Dodson G. G., Higgins C. F., Wilkinson A. J. (1994) Science 264, 1578–1581 [DOI] [PubMed] [Google Scholar]
- 21.Tame J. R., Dodson E. J., Murshudov G., Higgins C. F., Wilkinson A. J. (1995) Structure 3, 1395–1406 [DOI] [PubMed] [Google Scholar]
- 22.Conners S. B., Mongodin E. F., Johnson M. R., Montero C. I., Nelson K. E., Kelly R. M. (2006) FEMS Microbiol. Rev. 30, 872–905 [DOI] [PubMed] [Google Scholar]
- 23.Nelson K. E., Clayton R. A., Gill S. R., Gwinn M. L., Dodson R. J., Haft D. H., Hickey E. K., Peterson J. D., Nelson W. C., Ketchum K. A., McDonald L., Utterback T. R., Malek J. A., Linher K. D., Garrett M. M., Stewart A. M., Cotton M. D., Pratt M. S., Phillips C. A., Richardson D., Heidelberg J., Sutton G. G., Fleischmann R. D., Eisen J. A., White O., Salzberg S. L., Smith H. O., Venter J. C., Fraser C. M. (1999) Nature 399, 323–329 [DOI] [PubMed] [Google Scholar]
- 24.Vanfossen A. L., Lewis D. L., Nichols J. D., Kelly R. M. (2008) Ann. N.Y. Acad. Sci. 1125, 322–337 [DOI] [PubMed] [Google Scholar]
- 25.Nanavati D. M., Thirangoon K., Noll K. M. (2006) Appl. Environ. Microbiol. 72, 1336–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conners S. B., Montero C. I., Comfort D. A., Shockley K. R., Johnson M. R., Chhabra S. R., Kelly R. M. (2005) J. Bacteriol. 187, 7267–7282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chhabra S. R., Shockley K. R., Conners S. B., Scott K. L., Wolfinger R. D., Kelly R. M. (2003) J. Biol. Chem. 278, 7540–7552 [DOI] [PubMed] [Google Scholar]
- 28.Blow N. (2008) Nat. Methods 5, 203–207 [DOI] [PubMed] [Google Scholar]
- 29.Schellman J. A. (1987) Annu. Rev. Biophys. Biophys. Chem. 16, 115–137 [DOI] [PubMed] [Google Scholar]
- 30.Kabsch W. (1993) J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 31.Collaborative Computational Project (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 32.Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard (1991) Acta Crystallogr. Sect. A 47, 110–119 [DOI] [PubMed] [Google Scholar]
- 33.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 35.Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 36.Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) Nucleic Acids Res. 32, W615–W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cohen D. S., Pielak G. J. (1994) Protein Sci. 3, 1253–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Holm L., Sander C. (1993) J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 39.Tame J. R., Sleigh S. H., Wilkinson A. J., Ladbury J. E. (1996) Nat. Struct. Biol. 3, 998–1001 [DOI] [PubMed] [Google Scholar]
- 40.Buliga G. S., Brant D. A., Fincher G. B. (1986) Carbohydr. Res. 157, 139–156 [DOI] [PubMed] [Google Scholar]
- 41.Alderkamp A. C., van Rijssel M., Bolhuis H. (2007) FEMS Microbiol. Ecol. 59, 108–117 [DOI] [PubMed] [Google Scholar]
- 42.Kondoh O., Tachibana Y., Ohya Y., Arisawa M., Watanabe T. (1997) J. Bacteriol. 179, 7734–7741 [DOI] [PMC free article] [PubMed] [Google Scholar]