Abstract
Mounting evidence underscores the importance of protein-protein interactions in the functional regulation of drug-metabolizing P450s, but few studies have been conducted in membrane environments, and none have examined P450s catalyzing sex steroid synthesis. Here we report specific protein-protein interactions for full-length, human, wild type steroidogenic cytochrome P450 (P450, CYP) enzymes: 17α-hydroxylase/17,20-lyase (P450c17, CYP17) and aromatase (P450arom, CYP19), as well as their electron donor NADPH-cytochrome P450 oxidoreductase (CPR). Fluorescence resonance energy transfer (FRET)3 in live cells, coupled with quartz crystal microbalance (QCM), and atomic force microscopy (AFM) studies on phosphatidyl choline ± cholesterol (mammalian) biomimetic membranes were used to investigate steroidogenic P450 interactions. The FRET results in living cells demonstrated that both P450c17 and P450arom homodimerize but do not heterodimerize, although they each heterodimerize with CPR. The lack of heteroassociation between P450c17 and P450arom was confirmed by QCM, wherein neither enzyme bound a membrane saturated with the other. In contrast, the CPR bound readily to either P450c17- or P450arom-saturated surfaces. Interestingly, N-terminally modified P450arom was stably incorporated and gave similar results to the wild type, although saturation was achieved with much less protein, suggesting that the putative transmembrane domain is not required for membrane association but for orientation. In fact, all of the proteins were remarkably stable in the membrane, such that high resolution AFM images were obtained, further supporting the formation of P450c17, P450arom, and CPR homodimers and oligomers in lipid bilayers. This unique combination of in vivo and in vitro studies has provided strong evidence for homodimerization and perhaps some higher order interactions for both P450c17 and P450arom.
INTRODUCTION
Sex steroids are essential for normal male and female development and fertility, and because testosterone is a substrate for estradiol synthesis, biopotent androgen and estrogen production are interdependent (1). Androgens and estrogens are synthesized by the microsomal enzymes 17α-hydroxylase/17,20-lyase (P450c17) and aromatase cytochrome P450 (P450arom), respectively. Balanced P450c17 and P450arom expression is necessary to maintain reproductive function and general health just as ablation of sex steroid production is an important therapeutic goal in treating cancers of the prostate and breast. Steroidogenesis is regulated and characterized by a remarkable level of organization that partitions different enzymes into specific tissues and cells as well as subcellular compartments. This is especially true of expression of P450c17 and P450arom, both of which are partitioned in ovarian follicles (2) and among fetal tissues in pregnancy (3). How, or if, these enzymes are organized within the endoplasmic reticulum is unknown. Evidence indicates that some microsomal P450s heterodimerize with one another, influencing catalytic activity (4), but, with a few notable exceptions (5, 6), most of these observations are derived from studies of reconstituted systems using solubilized components. Much less is understood of the organization of P450s or other steroidogenic enzymes and redox partner proteins in lipid bilayers that have rarely been studied in living cells (5, 6). P450c17 and P450arom are envisioned to interact with their redox partner protein, NADPH-cytochrome P450 oxido-reductase (CPR) as monomers at discrete docking sites (7–9), as for all microsomal P450s. Unlike most of the drug-metabolizing P450s, product synthesis catalyzed by P450c17 and P450arom involves sequential oxygenation reactions that could well be facilitated in higher order functional complexes. For instance, intermediates in androgen synthesis are not always detectable (10), but still it is not clear how these more polar steroids are effectively trapped in the enzyme through sequential oxidative cycles. Consequently, much remains to be understood about the basic biochemistry underpinning the synthesis of sex steroids.
The phenomenon of cooperativity in drug-metabolizing P450s involves simultaneous binding of multiple ligands that influence overall catalytic activity (11, 12), as well as direct P450-P450 interactions (4, 13). P450s catalyzing sex steroid synthesis, with much limited substrate specificities are unlikely to be able to bind or exchange multiple ligands in the way drug-metabolizing P450s can. The crystal structure of human P450arom indicates that it has one of the smallest active sites of any P450 so characterized to date (14). However, the possibility of molecular interactions among those P450s that form the backbone of the steroidogenic cascade has not previously been explored. In this study we investigate for the first time the biomolecular interactions between P450c17, P450arom, and CPR in membranes using a unique combination of cell-based and biophysical methods. Previous studies using similar biophysical approaches to investigate drug-metabolizing P450 enzymes utilized methods that immobilize the proteins, restricting conformational freedom (15, 16). The results can be difficult to correlate directly with in vivo studies (17). In the current studies, fluorescent probes fused with the recombinant P450 enzymes enabled the identification of intracellular localization and protein-protein interactions using FRET. Complementary biophysical data were obtained using a QCM to monitor the assembly of recombinant proteins on supported biomimetic membranes in real time. The amount of protein binding to the membrane and the relative affinity of the proteins for the membrane could be assessed directly. Finally, the structural organization of the P450c17, P450arom, and CPR in the membrane bilayers was probed using AFM in solution. This novel combinatorial approach provides complementary information about interaction among membrane-bound proteins involved in sex steroid synthesis, both in living cells and biomimetic membranes. Protein homo- and heterodimerization was defined, and the first topographical view of the molecular steroidogenic landscape was generated on a nanometer scale as enzyme complexes assembled in lipid membranes.
EXPERIMENTAL PROCEDURES
Materials
Tissue culture materials were purchased from Invitrogen, Fisher, and Sigma. CFP (eCFP-N1) and YFP (eYFP-N1 and eYFP-C1) vectors were purchased from Clontech (Mountain View, CA).
FRET Fusion Constructs
All of the cDNAs used in FRET analysis encoded full-length, wild type enzymes, and fusions were constructed so as to avoid any modification of the putative transmembrane binding domains. The cDNAs encoding human P450arom (provided by Dr. E. Simpson, Prince Henry's Institute of Medical Research, Clayton, Australia) and human P450c17 cDNA (provided by Dr. R. Auchus, Department of Internal Medicine, University of Texas Southwestern Medical Center, Dallas, TX) were engineered to generate proteins fused with eYFP and eCFP at the C terminus. The CPR-eYFP fusion construct was donated generously by Dr. Byron Kemper (University of Illinois), and its successful use in FRET experiments has been described previously (5).
Transfection of HEK293 and NCI-H295A Cells
HyClone (Logan, UT) media and fetal bovine serum were used for transfection procedures. HEK293 cells (American Type Culture Collection, Manassas, VA) were cultured on Primaria cultureware (BD Biosciences, San Jose, CA) in Dulbecco's modified Eagle's medium with 5% fetal bovine serum, 10 mm Hepes (pH 7.4), 100 units/ml penicillin, and 100 μg/ml streptomycin. After 24 h, the cells were transfected (4 μg/35 mm well) with plasmid fusion constructs using Lipofectamine 2000 (Invitrogen) as suggested by the manufacturer. For FRET experiments, the transfected cells were plated after 18 h into Lab-Tek chambered cover glass (1.0 borosilicate cover glass; Nunc, Rochester, NY) at 40% confluence and cultured for 24 h at 37 °C.
FRET Analysis
FRET was evaluated from the enhanced acceptor emission (18), which removed contamination of fluorescent signals from spectral bleed-through and cross-talk and quantified FRET efficiency across a wide wavelength range (see Fig. 1, A–C). HEK293 cells were transfected with P450c17-eCFP and P450arom-eYFP plasmids and replated into Lab-Tek chambered cover glass 24 h later. Fluorescence and spectral images (from multiple cells and multiple experiments) were captured on a computer-controlled, inverted Olympus IX-81 epifluorescence microscope (Olympus USA, Center Valley, PA) fitted with an Acton SpectraPro215 spectrograph (Roper Scientific, Tucson, AZ). A mercury light source was used for excitation in conjunction with filter cubes, each containing an excitation filter and dichroic mirror (CFP, excitation filter D436/20, dichroic 455DCLP; YFP, excitation filter HQ500/20, dichroic Q515LP). Two spectral images of each individual cell, expressing either the CFP construct or the YFP construct or co-expressing both constructs, were recorded using CFP and YFP excitation cubes, respectively. Fluorescence emission spectra were constructed from spectral images after background subtraction and analyzed using MetaMorph (Molecular Devices Corp., Sunnyvale, CA) in conjunction with Microsoft Excel. FRET ratios were calculated from these spectra and from eYFP and eCFP plasmids expressed and used as spectral standards, as described in detail elsewhere (18). Using this method, ratios significantly greater than 1.0 represent FRET between CFP and YFP fluorphores. FRET ratios of cells from experiments investigating interactions between various combinations of fusion constructs (P450c17-eCFP, P450c17-eYFP, P450arom-eCFP, and P450arom-eYFP) were analyzed by nested one-way analysis of variance SAS 9.1 (The SAS Institute, Cary, NC).
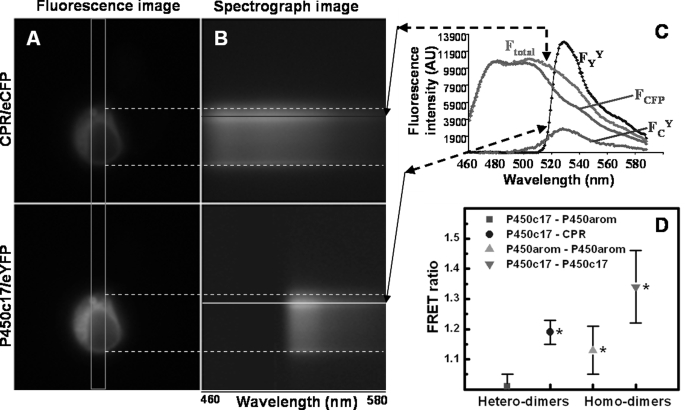
FIGURE 1.
Fluorescence and spectral images, along with experimental and standard emission spectra, of a HEK293 cell co-transfected with and co-expressing CPR-eCFP and P450c17-eYFP fusion constructs. A, pseudo-colored fluorescence images detailing the perinuclear localization of both fusion proteins. B, spectrograph images of both fusion proteins, taken through the slit indicated by the orange rectangle in A. The x axis of the spectrograph images corresponds to wavelength; the y axis corresponds to cell position. Note the presence of the nuclear ghost in the spectrograph images. C, experimental and standard emission spectra necessary for calculation of the FRET ratio of HEK293 cells transfected with P450c17-eYFP and CPR-eCFP fusion constructs. Emission spectra of a given cell region (see colored lines) obtained from excitation at 436 nm (CFP; Ftotal) and 500 nm (YFP; FYY). FCFP is the emission spectrum from cells transfected only with CPR-eCFP, scaled to the spectrum Ftotal. FCY is extracted by subtraction (Ftotal − FCFP) and includes two emission components: one from direct eYFP excitation at 436 nm (FCdirect) and the other from FRET (FCFRET). D, FRET ratios of P450c17, P450arom, and CPR-eCFP/eYFP fusion constructs with hetero- and homodimer interactions in HEK293 cells. FRET was not detected between P450c17-eYFP and P450arom-eCFP (red square; p > 0.5). Interactions were detected for all other combinations of fusion constructs with FRET ratios significantly greater than 1 (*, p < 0.05) and greater than between P450c17-eYFP and P450arom-eCFP.
Purified Protein Sources
Recombinant P450c17, P450arom, and CPR were expressed from plasmids; the proteins were purified, and the concentrations were determined as previously described (19–21). Stable overexpression of P450c17 and P450arom from Escherichia coli utilized constructs with modification of the N terminus to maximize yield. Modifications to the human P450c17 sequence involved substitutions within the first 10–12 N-terminal amino acids to optimize codon usage (22). Human P450arom was also N-terminally modified for stability of expression and purification from E. coli, as previously reported (21), but required replacing the first 46 residues with 11 amino acids from the N terminus of P450 2C11. Because in this case, changes modify sequence involving the putative membrane-spanning region, additional full-length, wild type human P450arom was also expressed in, and purified from, insect cells infected with recombinant baculovirus as described and reported earlier (23). Experiments conducted on biomimetic membranes for the quartz crystal microbalance and atomic force microscope utilized both of these unmodified (wild type) and modified human P450arom enzymes.
Quartz Crystal Microbalance
The membranes were created via liposome deposition onto 3-mercaptopropionic acid modified gold QCM sensor chips as described previously (24–26). A typical QCM experiment consisted of two to three steps. The first step involved formation of the 1,2-ditetradecanoyl-sn-glycero-3-phosphocholine (DMPC) lipid layer on the chip or alternatively the DMPC/cholesterol (20%) membrane. Next, the protein sample was introduced into the chamber, and the changes in frequency and dissipation (Δf and ΔD) were recorded for the third, fifth, seventh, and ninth overtone. For a subset of the experiments, as a third step a second protein was introduced onto the membrane already containing the first protein. The proteins were introduced as PBS (0.02 m potassium phosphate, 0.1 m NaCl, pH 6.9) solutions at varying concentration and at a flow rate of 50 μl·min·1. Each QCM experiment was repeated three to six times with high reproducibility, and the results presented in the figures were arbitrarily chosen as representative examples. Protein concentrations that were needed for saturation of the membrane surface were calculated based on a study recently published on Azurin adsorption on mecaptopropionic acid-modified gold QCM chips (25). The range of concentrations of protein, as well as variable flow rates were tested to optimize binding of protein to the membrane. For a comprehensive description of principles of QCM operation and for a review of its use to study biological materials, please refer to Refs. 15 and 27–34.
Experimental data were analyzed with OriginPro 7.5 (OriginLab) and QTools software (QSense). The estimation of the mass of protein (ng·cm−2) and the amount (pmol·cm−2) per unit area was calculated using Sauerbrey derived equation (Δm = −C/n·(Δfn), where C = 17.7 ng·cm−2 Hz−1 at 5 MHz quartz chip, and n is the overtone number), which assumes a tightly coupled, low dissipating, and uniform layer on the surface of the chip. The mercaptopropionic acid self-assembled monolayer promotes creation of multiple bilayers (two or three) on the chip surface that were found to behave as a viscoelastic body rather than a rigid single lipid bilayer (26). In such cases, we used the Voigt model available within the QTools software, which makes use of the differential signal output from the multiple frequency overtones measured when the viscoelastic material is coupling to the chip surface. The addition of protein to these layers reduced their viscoelastic character, and the classical Sauerbrey approach was found to be valid for the purpose of estimating the mass.
Atomic Force Microscopy
AFM imaging was performed with either a Nanoscope IV atomic force microscope equipped with a MultiMode head (Veeco Instruments) or a Ntegra scanning probe laboratory (NT-MDT, Zelenograd, Russia), operating in intermittent contact (or tapping) mode in aqueous solution using MikroMasch NSC36 probes. Images of 512 × 512 pixels with a scan size of 3 × 3, 1 × 1, 0.5 × 0.5 μm2 were acquired at scan rates of 1–2 Hz. AFM images were processed to correct plane tilt, arcing, and line fluctuations. For the presented images, a further 3 × 3 Gaussian noise filtering and three-dimensional rendering was also applied using WsXM freeware (version 4, build no. 11.6) (35).
In a typical experiment, the liposomes were deposited onto freshly cleaved mica using 20 μl of 0.5 mm DMPC in PBS buffer for 2 min, and then the sample was gently rinsed with deionized water to remove excess membrane; finally 20 μl of PBS buffer was applied prior to mounting the sample onto the liquid cell of the AFM. The lipid was imaged to establish the presence of a bilayer prior to the introduction of the protein samples. All of the AFM imaging was performed at room temperature.
The protein sample was injected into the liquid cell and incubated for 2–5 min before resuming imaging. The samples included pure CPR, 2 μl of 0.22 pmol·μl−1; P450arom, 2 μl of 0.01 pmol·μl−1; or P450c17, which was preincubated with 4 μl of 10 μm pregnenolone for 15 min at room temperature prior to the addition of 4 μl of 0.565 pmol·μl−1 of the P450c17 + pregnenolone mixture.
RESULTS
In Vivo Homodimer Formation in Steroidogenic Cells Revealed by FRET
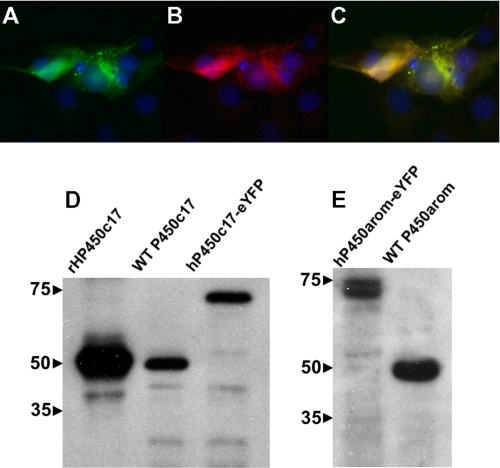
The results of a typical FRET experiment are depicted in Fig. 1 (A–C), and the FRET ratio data for all studies are summarized (Fig. 1D). FRET ratios significantly greater than 1.0 show definitive evidence of protein-protein interactions and were observed in experiments investigating homodimerization between P450c17-eCFP and P450c17-eYFP (1.34 ± 0.12, p < 0.05), as well as between P450arom-eCFP and P450arom-eYFP (1.13 ± 0.08, p < 0.05). As expected, heterodimerization was observed between P450c17-eYFP and CPR-eCFP (1.19 ± 0.04, p < 0.05), with values in a similar range to those of the P450 homodimers. In contrast, there was no evidence of heterodimerization between P450c17 and P450arom in HEK293 cells, which yielded a FRET ratio of 1.01 ± 0.04. Correct subcellular localization of the P450c17-eYFP fusion protein expressed in COS1 cells was verified by direct eYFP fluorescence (Fig. 2A) and indirect immunofluorescence for P450c17 (Fig. 2B) to be concentrated around the nucleus (Fig. 2C) in a pattern consistent with microsomal expression. The P450arom-eCFP, P450arom-eYFP, and CPR-eYFP fusion proteins showed an identical intracellular pattern of expression, confirming that all enzymes were expressed within the endoplasmic reticulum. This was verified further by enzyme activities and immunoblot analyses performed on isolated proteins from transfected cells. Western immunoblots conducted on enriched microsomal protein were consistent with the successful expression of the fusion proteins showing the expected increase in molecular size (Fig. 2, D and E; 25-kDa shift) in the microsomal fraction. Finally, detection of homodimerization of P450c17 by FRET was confirmed using a dominant negative approach, overexpressing nonfusion enzyme with an adenoviral vector expressing full-length wild type (wt) P450c17 (generously provided by Dr. Denis Magoffin, Department of Obstetrics and Gynecology, UCLA). HEK293 cells were transiently transfected with the P450c17-eYFP and P450c17-eCFP fusion constructs and then infected with wt-P450c17 expressed in adenovirus. P450c17-eCFP/P450c17-eYFP homodimer formation was disrupted by expression of wt-P450c17 with a complete loss of detectable interaction (FRET ratio reduced to 0.62 ± 0.06).
FIGURE 2.
Expression of P450c17-eYFP fusion proteins in transiently transfected COS1 cells. Expression of P450c17-eCFP detected by direct fluorescence (A; green) and by primary antisera against P450c17 (B; red, Alexa594-conjugated secondary antisera). A merged image (C) shows co-localization of P450c17 expression with direct fluorescence of eYFP. The nuclei are stained with Hoechst 33342 (blue). D and E, immunodetection of fluorescence fusion proteins in enriched microsomal fraction (20 μg/lane) of HEK293 cells by Western analysis. Expression of P450c17 and P450arom were detected by antisera directed against the respective human proteins. A shift in molecular mass of ∼25 kDa was observed with both fusion proteins as compared with wild type proteins. The molecular size markers (kDa) are shown at the left.
In Vitro Protein-Lipid and Protein-Protein Interactions Revealed by QCM
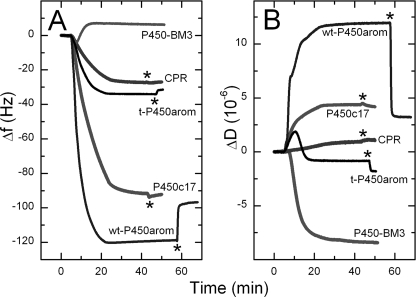
The interaction of P450c17, P450arom (truncated and full-length), and CPR with biomimetic lipid bilayers was explored in real time using QCM. Fig. 3 shows examples of typical changes in frequency (Δf; Fig. 3A) and dissipation (ΔD; Fig. 3B) because CPR, P450c17, and P450arom interact with DMPC membranes at concentrations of protein found to be adequate for saturation coverage. The initial base line (∼5 min) corresponded to the previously deposited lipid bilayer in PBS; thus the decrease in frequency observed upon the introduction of CPR, P450c17, and P450arom represents the mass of protein, and associated water of hydration, binding to the membrane (25). Similar rates of incorporation and mass deposited were seen with the full-length P450c17 and P450arom enzymes; much slower incorporation and less mass was deposited when the truncated P450arom and CPR were introduced into the membrane (Fig. 3A). These results were consistent and highly reproducible. Our estimates of surface saturation are 28 pmol·cm−2 for P450c17 and 15 pmol·cm−2 for P450arom. Protein incorporation within the membrane was also stable, and only a small amount of protein was removed with PBS washes at flow rates (300 μl/min) that were 6-fold higher than those utilized for deposition. This was equally true for the full-length and truncated forms of P450arom.
FIGURE 3.
Typical QCM, Δf-t, and ΔD-t profiles for deposition of CPR, P450c17 wt, and truncated (t-) P450arom on pure DMPC membrane. The lack of affinity of bacterial P450 BM3 toward DMPC is also shown. The protein concentrations used were 20 nm t-P450arom, 20 nm wt-P450arom, 21.5 nm P450c17, 44 nm CPR, and 33 nm P450 BM3. 1 ml of protein solution in PBS was introduced onto the membrane surface at 50 μl·min−1, at t = ∼5 min; flow was stopped ∼15 min after solution entered the chambers; a final PBS rinse (*) was used to rinse nonspecifically bound protein from the membrane. Note: a decrease in frequency (Δf) is equivalent to the mass increase on the membrane-modified chip surface caused by protein binding.
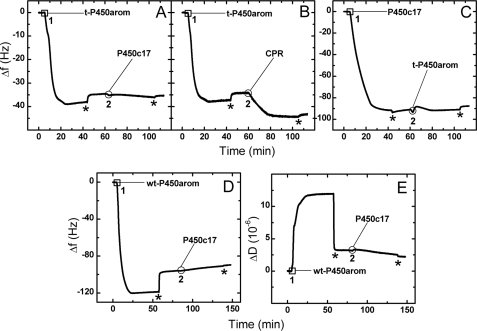
Initial experiments on the QCM exploring interactions between P450c17, P450arom, and CPR utilized the truncated P450arom. Evidence for heterodimerization was obtained for some protein pairs but not others, supporting the FRET data relative to the lack of heterodimer formation between the P450c17 and the P450arom. Specifically, there was no binding of P450c17 into DMPC membranes already saturated with the N-terminally truncated P450arom (Fig. 4A), even though incorporation was observed for CPR (Fig. 4B), a much larger protein. The same result was obtained when the membrane was first saturated with truncated P450arom, followed by P450c17 (Fig. 4C), and then CPR (data not shown), even though much less of the truncated P450arom than P450c17 was required to saturate the bilayer (Fig. 3A). Substituting the full-length enzyme for the truncated form yielded the same result, no binding of P450c17 in a membrane already saturated with P450arom (Fig. 4D). Experiments with DMPC/cholesterol membranes also gave results consistent with DMPC (data not shown), and the formation and characteristics of the membrane itself was verified as described elsewhere (26). Finally, as an additional control, the soluble, non-membrane-anchoring, bacterial enzyme P450 BM3 was introduced into the QCM cell in an identical fashion. In contrast to CPR, P450c17, and P450arom, there was no evidence that this protein was incorporated into the membrane, and instead, P450 BM3 caused a mild disruption of the lipid surface (increase in Δf ∼2 Hz, indicative of a loss of lipid; Fig. 3A). In summary, these data established the biomimetic environment so generated to be a robust platform on which to investigate enzyme protein complex assembly.
FIGURE 4.
QCM Δf-t profiles that show deposition of t-P450arom (1 in A and B), followed by deposition of CPR, despite surface saturation with t-P450arom (2 in B), but absence of binding of P450c17 under the same conditions (2 in A). Similarly, membrane saturated with P450c17 (1 in C) shows no binding of t-P450arom to this surface (2 in C). Similarly, no binding of P450c17 to wt-P450arom saturated membrane is observed (2 in D). The ΔD-t profile (E) for the wt-P450arom-P450c17 experiment (D) confirms that no exchange of proteins from the previously membrane bound protein with the incoming protein is observed. The asterisk denotes the final PBS rinse used to rinse nonspecifically bound protein from the membrane. The protein concentrations used were 20 nm t-P450arom, 20 nm wt-P450arom, 21.5 nm P450c17, and 20 nm CPR, and the flow rate was 50 μl·min−1.
Morphology of Proteins Incorporated into Lipid Membrane Studied by AFM
Although the QCM provided quantitative bulk data for the protein binding to the lipid, AFM was employed to visually assess the state of aggregation of these proteins. Each individual protein was imaged successfully using intermittent contact mode as previously described (24). Typically, the proteins appeared to be mobile in the DMPC membrane bilayer, seen as congregating proteins pushed by the AFM tip from side to side while being imaged. The addition of cholesterol to DMPC greatly reduced this distortion, allowing single protein entities to be imaged. Although images obtained on both lipid surfaces revealed similar protein morphologies and dimensions, far better quality data were obtained with cholesterol-containing membranes.
AFM images of P450c17 (Fig. 5A) showed the protein populating the lipid-modified mica surface. The images were optimal when the P450c17 protein was preincubated with the substrate, pregnenolone. The height image was initially collected as 3 × 3 μm2. The 1 × 1 μm2 off-line zoom showed mostly regular like-sized features over most of the surface with a few larger protein aggregates present. An example of the cross-sectional analysis of some of these features is shown in Fig. 5A where the encircled row of proteins exhibited remarkably uniform dimensions of ∼3.5 nm in height and appeared approximately spherical.
FIGURE 5.
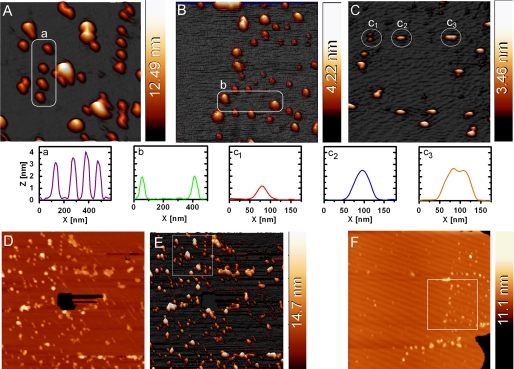
The three-dimensional AFM images show well defined morphologies of P450c17 (A), wt-P450arom (B), and CPR (C) proteins on a typical lipid surface (DMPC/cholesterol). Imaging was undertaken in liquid PBS buffer in tapping mode, and the P450 sample were preincubated with substrate prior to deposition (see text). The 1 × 1 μm2 image (A) is an off-line zoom from the 3 × 3-μm2 images (see D and E), whereas the 1 × 1 μm2 images in B and C are the actual scan region. The region highlighted in B is subsequent to the 3 × 3-μm2 image shown in F. The white circled areas contain proteins with regular sizes, and the cross-section analyses are displayed below the images. Panels D (two-dimensional) and E (three-dimensional) show the original AFM image (3 × 3 μm2) of P450c17 from which the area used for A is shown as a square region in E. To confirm that the protein was on a membrane layer, a hole was created through the membrane using the AFM tip (black hole in the center of D), with the appropriate height for a lipid bilayer membrane. Similarly, F shows the original (3 × 3 μm2) AFM image of P450arom from which B was obtained. The presence of the edge of the membrane patch (bottom right-hand corner of the image) once again confirmed the proteins lay on the membrane. The height scale for all the panels is shown as a color bar at the right-hand side of each AFM image.
An example of the corresponding AFM image collected for wild type P450arom on lipid bilayers is shown in Fig. 5B from a 1 × 1 μm2 image. As for P450c17, when P450arom was preincubated with androstenedione prior to introduction to the lipid, regular-sized, globular proteins were stably imaged in the DMPC/cholesterol bilayer. The most commonly observed features (circled in Fig. 5B) were ∼2 nm in height, and similar profiles were obtained for both the full-length and N-terminally modified enzymes.
It is significant that AFM imaging of the CPR protein (Fig. 5C) showed evidence of aggregation, with possible monomers (panel c1), dimers (panel c2), and perhaps a dimer of dimers (panel c3) visible on the lipid surface. The putative monomers all displayed a consistent height of 1–1.2 nm. Interestingly, multimeric forms (putative dimer and dimer of dimers; Fig. 5C, panels c2 and c3) were almost twice as tall as a putative monomer (Fig. 5C, panel c2) consistent with a substantial change in orientation in the lipid to achieve dimerization. The original AFM images from which Fig. 5 (A and B) were obtained are provided in Fig. 5 (D–F), and they also demonstrate the presence of the membrane as a 4–5-nm bilayer onto which the protein bound.
DISCUSSION
These studies provide the first evidence to indicate that key enzymes required for sex steroid synthesis, namely P450c17 and P450arom, are adapted in such a way as to organize themselves into specific complexes involving homodimers, but not heterodimers, while heterodimerizing effectively with their common redox partner, CPR, in the microsomal compartment. Our data, generated using a novel combination of biophysical techniques to probe protein-protein interactions in membranes, provide a unique perspective on the intracellular and intermolecular organization of sex steroid synthesis by enzymes in membranes. The specificity of homodimerization of steroid synthesizing P450s and perhaps higher order hetero-oligomeric complexes with CPR in vivo, as well as in vitro, suggests that this order serves to help regulate the rate of androgen and estrogen synthesis and might reasonably present novel targets for disruption and thereby pharmacological and therapeutic control.
Homodimerization of P450 Enzymes in Live Cells
Although proteins can form nonspecific aggregates at high expression levels (6), the lack of heterodimerization between P450c17 and P450arom together with disruption of P450c17-FP homodimerization by wt-P450c17 argues strongly that the homodimer (and/or homo-oligomer) complexes detected here for each of P450c17 and P450arom result from specific interactions. The lack of heterodimerization between P450c17 and P450arom may also be biologically relevant and relate to their differential expression into separate cellular or tissue compartments when estrogen synthesis is high, as in the preovulatory follicle, two-cell model (36), or human pregnancies, fetal adreno-placental unit (37). In each case, androgen synthesis is partitioned to one cell or tissue, whereas estrogen synthesis from those androgens occurs in another. Similarly, in the bovine placenta, P450c17 is expressed in uninucleate trophoblast, and P450arom is expressed in binucleate trophoblast cells (38). This suggests that high levels of estrogen synthesis may be best balanced physiologically by compartmentalization of these enzyme systems, which over time lost the potential to heterodimerize as other P450s have been shown to do. Compartmentalization may limit estrogen synthesis, which is in some cases orders of magnitude lower than the rate of androgen synthesis in gonadal tissues (20, 39); too much estrogen may be as damaging as too little. Studies such as those described herein will help to define what constitutes functionally competent complexes in steroid-secreting cells and their biochemical and physiological relevance.
The potential importance of homodimerization among the steroid hydroxylases has not been considered to our knowledge. Yet studies utilizing rotational diffusion and reconstituted systems have established various oligomeric relationships between hepatic P450s and CPR (4, 40, 41) with evidence for P450 hexamers in native membranes (42). Heterodimers were detectable by BiFC, although proteins tagged in this way exhibit intrinsic binding affinity that can generate artificial co-assembly (5, 6). These arrangements are hypothesized to modulate the activities of a number of microsomal P450s (4). Hetero- and homodimer complexes may actually facilitate the sequential metabolism of substrate by some P450s, such as the steroid hydroxylases, just as aggregation may disrupt the function of others (4, 5). Because both P450c17 and P450arom carry out multiple, distinct enzymatic activities (1, 43), formation of P450c17 and P450arom homo-oligomers may be functionally important in allowing for the transfer of metabolic intermediates from P450 to P450 in established complexes. This would be consistent with the study of recombinant P450c17 by Yamazaki et al. (44), who concluded that a significant percentage of the product of the first oxygenation reaction did not dissociate before the second oxygenation took place. In this way, oligomerization of P450c17 homodimers with CPR might facilitate the sequential substrate metabolism (17α-hydroxylation and subsequently 17,20-lyase) necessary for androgen and estrogen synthesis. The significantly lower FRET ratio associated with P450arom homodimerization may well reflect the existence of higher order homo-oligomers of this P450 than of P450c17, consistent with the additional oxygenation step required for estrogen compared with androgen formation. Conversely, altering the level of these associations might selectively modulate 17α-hydroxylase and 17,20-lyase activities, providing a novel strategy for development of selective inhibitors of adrenal androgen synthesis that do not also alter cortisol production.
Protein-Lipid and Protein-Protein Interactions Studied by QCM
The Δf-t traces for each protein in Fig. 3 suggest differences in protein affinity for the lipid surface. The soluble enzyme P450 BM3 showed no such absorptive propensity. The specific binding of CPR to the P450c17- or P450arom-saturated membrane surface was a result that was anticipated, because CPR is the natural redox partner for both P450s in vivo. Thus tight binding is observed in Fig. 4B at point 2. In contrast, the lack of binding of the P450c17 onto a lipid saturated with P450arom and vice versa (Fig. 4, A and C, point 2) was remarkable, because proteins are generally regarded as prone to adhesion. These data support the FRET results discussed above and demonstrate that heterodimerization between these two P450s does not occur either in vivo or in biomimetic membranes. Entirely consonant results were obtained with full-length (unmodified) human P450arom (Fig. 4D) and with the N-terminally modified enzyme. This suggests further that N-terminal domains are not likely involved in at least heterodimeric interactions of these P450s. Although a lack of change in the mass of the saturated surfaces does not preclude the possibility of exchange with bound proteins, the lack of change in dissipation (ΔD-t) (Fig. 4E) shows no change in the properties of the membrane and hence argues strongly against it. Taken together with FRET results, these observations indicate that there is no heterodimerization between P450c17 and P450arom.
The data presented also support a role for N-terminal residues including the putative transmembrane domain of P450arom in organization within lipid. The truncated P450arom still bound firmly to the bilayer, consistent with the involvement of other domains in membrane anchoring, as postulated by Ghosh et al. (14) from their crystal structure. However, protein incorporation was slower, less protein was able to bind at saturation, and the complexes imaged by AFM had lower vertical heights, all of which suggest that the N-terminal residues influence how P450arom is oriented in the bilayer. The QCM technique has been used previously to examine some P450 proteins, including P4501B1 (45), P450cam, and P4502B4 (15). In the latter study (15), P450s were adsorbed onto poly-ion films that required drying prior to mass measurements, whereas our results relate to interactions between the proteins and the membrane under hydrated conditions. Even so, our estimates of surface saturation (28 and 15 pmol·cm−2 for P450c17 and P450arom, respectively) are in close accord with those for P450 2B4 (12–25 pmol·cm−2). Together with the lower FRET ratios of P450arom compared with P450c17 homodimers that were seen in living cells, these independent but complementary data sets may correlate with the tendency of these P450s to oligomerize.
Morphology of Protein Homo-oligomers
AFM images of mammalian drug-metabolizing P450s dried on mica (46) and in lipid nanodiscs (47) (and references therein) have been reported, as have peptides and other proteins (27–31). Our high resolution images of steroidogenic P450c17 and P450arom (Fig. 5) provide confirmation that proteins were imbedded in the lipid surface and give further insight into their molecular organization. The lateral dimensions for the proteins measured by AFM in lipid bilayer environment were all larger than expected because of a combination of effects such as tip broadening and protein mobility in the membrane; nevertheless, the regular shape of globular molecules and their aggregates was apparent. Height measurement of the proteins provided a more reliable estimate of the size characteristics for each protein (as indicated by the cross-sectional dimension shown in Fig. 5), and comparison among the enzymes investigated was of primary interest.
Several previous studies have reported images of P450 2B4 and CPR on graphite- (46, 48), phospholipid- (49–51), and poly-ion-coated (15) surfaces. Images reported herein extend those observations to two different P450s in addition to CPR in lipid layers, allowing comparisons between enzymes and providing evidence further supporting FRET and QCM-generated data. Specifically, the images consistent with homodimerization and homo-oligomerization were obtained for P450 and CPR enzymes. Putative individual CPR monomers could be directly compared with dimers and dimer of dimers morphologies (Fig. 5C, panels c1–c3, respectively). The cross-sectional height of the CPR monomer at 1–1.2 nm is envisaged as consistent with a ∼75-kDa protein partially embedded into the membrane. Larger CPR structures were higher, with ∼2–2.2 nm proposed to be a dimer and a dimer of dimers, respectively. Phenomenologically, the lipophilic patch of the CPR monomer could well be the binding region of the CPR dimer resulting in a striking change in the orientation of the dimer in the lipid bilayer and consequent height increase. These dimensions are in good agreement with results obtained by Bayburt et al. (49), who imaged CPR protruding from lipid nanodiscs using both contact (height = 1.5 nm) and tapping (height = 2.0 nm) modes, although their later reports indicated 6–7-nm height (52). Interestingly, higher order aggregation states were much more common. Comparison with the x-ray crystal structure for the truncated CPR indicates a bowl shape for the monomers with ∼5 × 6 × 7-nm dimensions (53). Although there are challenges for a direct comparison of the x-ray derived dimensions with the AFM data, the CPR appears to be embedded deeply in the lipid bilayer in an “iceberg” fashion.
AFM images of P450c17 (Fig. 5A) show a remarkable similarity to those obtained by Bayburt and Sligar (50) for the microsomal P450 2B4. The computational model for the full-length P450c17 (8) provides an estimate for the protein dimensions, predicting a size of ∼4 × 6 × 6 nm. It is difficult to conclude that the protein structures observed by AFM are dimeric; however, the cross-sections of the row of proteins in Fig. 5A (panel a) display quite large widths-at-half-height, which argues against the monomer form as a candidate and favors a dimer or an oligomer of the P450c17, consistent with the demonstrated homodimerization of P450c17 by FRET analysis. Sampling heights for P450arom structures were less consistent with dimers of P450arom and are not supportive of monomers. Therefore, the majority of visible features could represent oligomers of this protein of uniform size and shape, suggesting nonrandom interaction among P450arom monomers. Our AFM results clearly show that the proteins examined exist in varying forms of aggregation rather than in the monomeric form when interacting with the lipid bilayer in vitro, supporting FRET results of homodimerization among the P450 enzymes in vivo.
Conclusion
We have demonstrated using a combination of biophysical approaches including FRET, a novel application of QCM coupled with AFM that the steroidogenic P450s (P450c17 and P450arom) bind to biomimetic phospholipid membranes as homodimers and perhaps homo-oligomers. Heteromeric associations between the two P450s did not occur, but each associated with their common electron donor, CPR. These findings of the specific associations were supported by FRET analysis in live cells expressing P450s and CPR in vivo. Furthermore, the complementarity of the in vitro QCM data with the in vivo FRET data illustrates the potential value of this novel use of QCM in elucidating the structure-function relationships of membrane-bound enzymes and potentially other proteins. These in vitro techniques show that the steroidogenic cytochrome P450s are more likely to function in vivo as dimers or multimers. The possible roles and functions of these homo- and heteroassociations among P450s have still to be investigated, but their specificity suggests that they are important for biological and biochemical function.
Acknowledgments
We express sincere gratitude to Christina Takanishi for assistance with cell culture and transfection procedures, as well as training with the spectral FRET technique. We acknowledge Prof. Ray Rodgers for enlightening discussions, Dr. Helene Vacher for immunofluorescent localization experiments, Prof. Walt Miller for generously sharing H295A cells, and Prof. Luet Wong for providing the P450 BM3 sample.
This work was supported by funds from the Australian Research Council (to L. L. M. and A. M.), the National Health & Medical Research Council (to L. L. M.), and National Institutes of Health Grant HD-048797 (to A. J. C.).
- FRET
- fluorescence resonance energy transfer
- QCM
- quartz crystal microbalance
- AFM
- atomic force microscopy
- CFP
- cyan fluorescent protein
- YFP
- yellow fluorescent protein
- DMPC
- 1,2-ditetradecanoyl-sn-glycero-3-phosphocholine
- PBS
- phosphate-buffered saline
- wt
- wild type
- t
- truncated
- CPR
- NADPH-cytrochrome P450 oxido-reductase.
REFERENCES
- 1.Conley A., Hinshelwood M. (2001) Reproduction 121, 685–695 [DOI] [PubMed] [Google Scholar]
- 2.Hillier S. G., Whitelaw P. F., Smyth C. D. (1994) Mol. Cell. Endocrinol. 100, 51–54 [DOI] [PubMed] [Google Scholar]
- 3.Conley A. J., Mason J. I. (1990) Baillieres Clin. Endo. Metab. 4, 249–272 [DOI] [PubMed] [Google Scholar]
- 4.Backes W. L., Kelley R. W. (2003) Pharmacol. Ther. 98, 221–233 [DOI] [PubMed] [Google Scholar]
- 5.Szczesna-Skorupa E., Mallah B., Kemper B. (2003) J. Biol. Chem. 278, 31269–31276 [DOI] [PubMed] [Google Scholar]
- 6.Ozalp C., Szczesna-Skorupa E., Kemper B. (2005) Drug Metab. Dispos. 33, 1382–1390 [DOI] [PubMed] [Google Scholar]
- 7.Graham-Lorence S., Amarneh B., White R. E., Peterson J. A., Simpson E. R. (1995) Protein Sci. 4, 1065–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auchus R. J., Miller W. L. (1999) Mol. Endocrinol. 13, 1169–1182 [DOI] [PubMed] [Google Scholar]
- 9.Hlavica P., Schulze J., Lewis D. F. V. (2003) J. Inorg. Biochem. 96, 279–297 [DOI] [PubMed] [Google Scholar]
- 10.Engel L. L. (1973) in Handbook of Physiology Endocrinology II (Greep R. O., Astwood E. B. eds) pp. 467–483, American Physiological Society, Washington, D.C. [Google Scholar]
- 11.Guengerich F. P. (2003) in 12th North American ISSX Meeting, Marcel Dekker Inc., Providence, RI [Google Scholar]
- 12.Denisov I. G., Frank D. J., Sligar S. G. (2009) Pharmacol. Ther. 124, 151–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davydov D. R., Halpert J. R. (2008) Expert Opin. Drug Metab. Toxicol. 4, 1523–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh D., Griswold J., Erman M., Pangborn W. (2009) Nature 457, 219–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schenkman J. B., Jansson I., Lvov Y., Rusling J. F., Boussaad S., Tao N. J. (2001) Arch Biochem. Biophys. 385, 78–87 [DOI] [PubMed] [Google Scholar]
- 16.Estavillo C., Lu Z., Jansson I., Schenkman J. B., Rusling J. F. (2003) Biophys. Chem. 104, 291–296 [DOI] [PubMed] [Google Scholar]
- 17.Johnson D. L., Martin L. L. (2005) J. Am. Chem. Soc. 127, 2018–2019 [DOI] [PubMed] [Google Scholar]
- 18.Takanishi C. L., Bykova E. A., Cheng W., Zheng J. (2006) Brain Res. 1091, 132–139 [DOI] [PubMed] [Google Scholar]
- 19.Johnson D. L., Conley A. J., Martin L. L. (2006) J. Mol. Endocrinol. 36, 349–359 [DOI] [PubMed] [Google Scholar]
- 20.Corbin C. J., Moran F. M., Vidal J. D., Ford J. J., Wise T., Mapes S. M., Njar V. C., Brodie A. M., Conley A. J. (2003) Biol. Reprod. 69, 390–397 [DOI] [PubMed] [Google Scholar]
- 21.Kagawa N., Hori H., Waterman M. R., Yoshioka S. (2004) Steroids 69, 235–243 [DOI] [PubMed] [Google Scholar]
- 22.Barnes H. J., Arlotto M. P., Waterman M. R. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 5597–5601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Corbin C. J., Mapes S. M., Lee Y. M., Conley A. J. (2003) Mol. Cell. Endocrinol. 206, 147–157 [DOI] [PubMed] [Google Scholar]
- 24.Mechler A., Praporski S., Atmuri K., Boland M., Separovic F., Martin L. L. (2007) Biophys. J. 93, 3907–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fleming B. D., Praporski S., Bond A. M., Martin L. L. (2008) Langmuir 24, 323–327 [DOI] [PubMed] [Google Scholar]
- 26.Mechler A., Praporski S., Piantavigna S., Heaton S. M., Hall K. N., Aguilar M. I., Martin L. L. (2009) Biomaterials 30, 682–689 [DOI] [PubMed] [Google Scholar]
- 27.Asakura N., Kamachi T., Okura I. (2003) Anal. Biochem. 314, 153–157 [DOI] [PubMed] [Google Scholar]
- 28.Caruso F., Furlong D. N., Kingshott P. (1997) J. Colloid Interface Sci. 186, 129–140 [DOI] [PubMed] [Google Scholar]
- 29.Fredriksson C., Kihlman S., Rodahl M., Kasemo B. (1998) Langmuir 14, 248–251 [Google Scholar]
- 30.Glasmästar K., Larsson C., Höök F., Kasemo B. (2002) J. Colloid Interface Sci. 246, 40–47 [DOI] [PubMed] [Google Scholar]
- 31.Höök F., Kasemo B., Nylander T., Fant C., Sott K., Elwing H. (2001) Anal. Chem. 73, 5796–5804 [DOI] [PubMed] [Google Scholar]
- 32.Höök F., Rodahl M., Brzezinski P., Kasemo B. (1998) J. Colloid Interface Sci. 208, 63–67 [DOI] [PubMed] [Google Scholar]
- 33.Kaufman E. D., Belyea J., Johnson M. C., Nicholson Z. M., Ricks J. L., Shah P. K., Bayless M., Pettersson T., Feldotö Z., Blomberg E., Claesson P., Franzen S. (2007) Langmuir 23, 6053–6062 [DOI] [PubMed] [Google Scholar]
- 34.Muguruma H., Kase Y., Murata N., Matsumura K. (2006) J. Phys. Chem. B 110, 26033–26039 [DOI] [PubMed] [Google Scholar]
- 35.Horcas I., Fernandez R., Gomez-Rodriguez J. M., Colchero J., Gomez-Herrero J., Baro A. M. (2007) Rev. Sci. Instrum. 78, 13705–13708 [DOI] [PubMed] [Google Scholar]
- 36.Tamura T., Kitawaki J., Yamamoto T., Osawa Y., Kominami S., Takemori S., Okada H. (1992) J. Endocrinol. 135, 589–595 [DOI] [PubMed] [Google Scholar]
- 37.Diczfalusy E. (1984) J. Steroid Biochem. Mol. Biol. 20, 945–953 [DOI] [PubMed] [Google Scholar]
- 38.Schuler G., Ozalp G. R., Hoffmann B., Harada N., Browne P., Conley A. J. (2006) Reproduction 131, 669–679 [DOI] [PubMed] [Google Scholar]
- 39.Moran F. M., Ford J. J., Corbin C. J., Mapes S. M., Njar V. C., Brodie A. M., Conley A. J. (2002) Endocrinology 143, 3361–3369 [DOI] [PubMed] [Google Scholar]
- 40.Kawato S., Gut J., Cherry R. J., Winterhalter K. H., Richter C. (1982) J. Biol. Chem. 257, 7023–7029 [PubMed] [Google Scholar]
- 41.Gut J., Richter C., Cherry R. J., Winterhalter K. H., Kawato S. (1983) J. Biol. Chem. 258, 8588–8594 [PubMed] [Google Scholar]
- 42.Myasoedova K. N., Berndt P. (1990) FEBS Lett. 275, 235–238 [DOI] [PubMed] [Google Scholar]
- 43.Nakajin S., Hall P. F. (1981) J. Steroid Biochem. Mol. Biol. 14, 1249–1252 [DOI] [PubMed] [Google Scholar]
- 44.Yamazaki T., Ohno T., Sakaki T., Akiyoshi-Shibata M., Yabusaki Y., Imai T., Kominami S. (1998) Biochemistry 37, 2800–2806 [DOI] [PubMed] [Google Scholar]
- 45.Shen Z., Yan H., Parl F. F., Mernaugh R. L., Zeng X. (2007) Anal Chem 79, 1283–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kiselyova O. I., Yaminsky I. V., Ivanov Y. D., Kanaeva I. P., Kuznetsov V. Y., Archakov A. I. (1999) Arch Biochem. Biophys. 371, 1–7 [DOI] [PubMed] [Google Scholar]
- 47.Nath A., Atkins W. M., Sligar S. G. (2007) Biochemistry 46, 2059–2069 [DOI] [PubMed] [Google Scholar]
- 48.Kuznetsov V. Y., Ivanov Y. D., Archakov A. I. (2004) Proteomics 4, 2390–2396 [DOI] [PubMed] [Google Scholar]
- 49.Bayburt T. H., Carlson J. W., Sligar S. G. (1998) J. Struct. Biol. 123, 37–44 [DOI] [PubMed] [Google Scholar]
- 50.Bayburt T. H., Sligar S. G. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 6725–6730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Civjan N. R., Bayburt T. H., Schuler M. A., Sligar S. G. (2003) BioTechniques 35, 556–563 [DOI] [PubMed] [Google Scholar]
- 52.Bayburt T. H., Carlson J. W., Sligar S. G. (2000) Langmuir 16, 5993–5997 [Google Scholar]
- 53.Wang M., Roberts D. L., Paschke R., Shea T. M., Masters B. S., Kim J. J. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 8411–8416 [DOI] [PMC free article] [PubMed] [Google Scholar]