Abstract
Electrophilic fatty acid derivatives, including nitrolinoleic acid and nitro-oleic acid (OA-NO2), can mediate anti-inflammatory and pro-survival signaling reactions. The transcription factor Nrf2, activated by electrophilic fatty acids, suppresses redox-sensitive pro-inflammatory gene expression and protects against vascular endothelial oxidative injury. It was therefore postulated that activation of Nrf2 by OA-NO2 accounts in part for its anti-inflammatory actions, motivating the characterization of Nrf2-dependent and -independent effects of OA-NO2 on gene expression using genome-wide transcriptional profiling. Control and Nrf2-small interfering RNA-transfected human endothelial cells were treated with vehicle, oleic acid, or OA-NO2, and differential gene expression profiles were determined. Although OA-NO2 significantly induced the expression of Nrf2-dependent genes, including heme oxygenase-1 and glutamate-cysteine ligase modifier subunit, the majority of OA-NO2-regulated genes were regulated by Nrf2-independent pathways. Moreover, gene set enrichment analysis revealed that the heat shock response is the major pathway activated by OA-NO2, with robust induction of a number of heat shock genes regulated by the heat shock transcription factor. Inasmuch as the heat shock response mediates anti-inflammatory and cytoprotective actions, this mechanism is proposed to contribute to the protective cell signaling functions of nitro-fatty acids and other electrophilic fatty acid derivatives.
INTRODUCTION
Nitration products of unsaturated fatty acids are formed via NO-dependent oxidative reactions (1) in inflammation (2, 3). Nitroalkene derivatives of linoleic acid (nitrolinoleic acid, LNO2)3 and oleic acid (nitro-oleic acid, OA-NO2) mediate pluripotent cell signaling actions, many of which are attributed to their electrophilic nature. Nitroalkenes undergo covalent, thiol-reversible reactions with cysteine and histidine residues in proteins such as glyceraldehyde-3-phosphate dehydrogenase (4) as well as with low molecular weight thiols (5, 6). Post-translational modification of signaling proteins by nitroalkenes mediates a component of observed anti-inflammatory actions. For example, LNO2 and OA-NO2 inhibit nuclear factor-κB (NF-κB) signaling via inhibiting p65 DNA binding and suppression of pro-inflammatory target gene expression in monocytes and human endothelial cells (7). Inhibition of p65 also contributes to the protective actions of OA-NO2 in focal cardiac ischemia and reperfusion (3). NF-κB inhibition is a property shared by other electrophilic lipid oxidation products, including 15-deoxy-Δ12,14-prostaglandin J2 (8). Moreover, nitroalkene fatty acid derivatives induce the expression of heme oxygenase-1 (HO-1, HMOX1), an important cytoprotective and anti-inflammatory gene, via a peroxisome proliferator-activated receptor γ-independent mechanism in human endothelial cells (9).
In addition to inhibiting inflammatory signaling, electrophilic lipid oxidation products such as 15-deoxy-Δ12,14-prostaglandin J2 and 4-hydroxynonenal, as well as the nitroalkene LNO2, activate nuclear factor E2-related factor 2 (Nrf2), a member of the cap ‘n’ collar family of basic region-leucine zipper transcription factors and a mediator of antioxidant and phase II detoxifying enzyme expression (10, 11). Upon electrophile exposure, Nrf2 protein stabilizes, translocates to the nucleus, heterodimerizes with other basic region-leucine zipper transcription factors (preferentially small Maf proteins), and binds to the antioxidant-response element (ARE), a common regulatory element found in the promoter regions of antioxidant and detoxication enzymes (12). Activity of Nrf2 is tightly regulated by Keap1 (Kelch-like ECH-associated protein 1), which binds to Nrf2 and directs it to Cullin-3-dependent ubiquitination and proteosomal degradation under homeostatic conditions. Upon exposure to electrophiles, reactive Keap1 thiols are adducted, dissociating Nrf2 from ubiquitin E3 ligase complex and facilitating nuclear accumulation and downstream effects on gene transcription (12).
Nrf2 regulation of antioxidant and phase II enzymes underscores a broader impact on the regulation of inflammatory processes. Nrf2-deficient mice display more pronounced inflammation in cigarette smoke-induced emphysema than wild type mice (13) and an increased susceptibility to severe airway inflammation and asthma (14). Moreover, Nrf2 is a critical regulator of innate immune response in sepsis (15). Interestingly, 15-deoxy-Δ12,14-prostaglandin J2 attenuates acute inflammation in a carrageenan-induced pleurisy model in wild type, but not Nrf2-deficient, mice (16). From a cardiovascular perspective, vascular endothelial cell overexpression of Nrf2 inhibits tumor necrosis factor-α-induced expression of the proinflammatory mediators VCAM-1 (vascular cell adhesion molecule-1) and MCP-1 (monocyte chemotactic protein-1) (17). Nrf2 is also activated by atheroprotective laminar flow (18–21). Finally, in vivo gene transfer of Nrf2 reduces inflammation in the balloon-denuded rabbit artery (22).
In addition to the ARE-regulated stress response, cells have other integrated stress-response pathways triggered by endogenous stimuli or environmental stresses. The heat shock response (HSR) is an ordered response to a wide array of acute and chronic stress conditions. HSR is regulated at the transcriptional level by heat shock factors (HSF). Upon activation, HSF undergoes multistep processing involving post-translational modifications, nuclear enrichment, trimerization, and binding to heat shock elements resulting in transcription of a large family of heat shock genes. Many of the proteins encoded by heat shock genes function as chaperones, proteases, or other proteins essential for protection of the cell against proteotoxic stress (23).
The notion that both nitroalkene fatty acid derivatives and Nrf2 display anti-inflammatory effects motivated the hypothesis that the immune modulatory actions of nitro-fatty acids are largely attributable to Nrf2. We thus determined the gene expression profile of human endothelial cells exposed to OA-NO2, and we evaluated Nrf2-dependent and -independent effects by genome-wide transcriptional profiling of cells transfected with Nrf2-specific or control siRNA. The results reveal that Nrf2 plays only a minor role in the gene regulation mediated by OA-NO2, with the heat shock response being the major activated pathway.
EXPERIMENTAL PROCEDURES
Reagents
OA-NO2 was prepared as described previously (24). The synthetic nitration product of OA-NO2 used in the study was an equimolar distribution of 9- and 10-nitro-octadeca-9-enoic acid (supplemental Fig. S1).
Cell Culture
Human umbilical vein endothelial cells (HUVECs) were isolated from umbilical cords obtained from the maternity ward of the Kuopio University Hospital by the approval of the Kuopio University Hospital Ethics Committee. HUVECs were cultivated as described previously (10).
siRNA
siRNA oligonucleotides targeting Nrf2, HSF1, and a nonspecific RNA control were obtained from Invitrogen. Four specific siRNA sequences targeting Nrf2 and two HSF1 siRNAs were tested prior to experimentation. The sequences of the most efficient siRNAs were as follows: Nrf2, 5′-UGA CAG AAG UU GAC AAU UA-3′, and HSF1, 5′-GCT CAT TCA GTT CCT GAT CTT-3′. For the experiments, HUVECs were seeded on 6-well plates or 10-cm plates at the density of 150,000 or 900,000 cells/well, respectively, and allowed to adhere for 24 h. Cells were transfected with 50 or 100 nm siRNA oligonucleotides using Oligofectamine (Invitrogen) for 24, 48, or 72 h. Cells were treated with OA-NO2 or OA for up to 48 h for quantitative real time PCR, Western blot, or Affymetrix analyses.
RNA Isolation and Quantitative Real Time PCR
HUVECs were collected and RNA extracted with TRI Reagent® (Sigma). For the cDNA synthesis, 1 μg of total RNA was used using random hexamer primers (Promega) and Moloney-murine leukemia virus reverse transcriptase (Finnzymes). The relative expression levels of Nrf2 (NFE2L2), HO-1 (HMOX1), GCLM, NQO1, HSPA1A, DNAJA4, HSPB8, HSPA6, and HSF1 mRNA in HUVECs were measured according to the manufacturer's protocol with quantitative real time PCR (ABI PRISM® 7700 sequence detector, Applied Biosystems) using specific assays-on-demand (Applied Biosystems) target mixes. The expression levels were normalized to β2-microglobulin expression and presented as fold change in the expression versus control.
Western Blotting
Cells were lysed, and total protein concentration was measured with BCA assay (Pierce). Nuclear and cytoplasmic fractions were isolated with NE-PER cytoplasmic and nuclear extraction reagents (Pierce). For electrophoresis, 10 μg of protein was used. The proteins were electrophoresed on a Tris/glycine SDS-polyacrylamide gel and transferred to nitrocellulose membrane. The primary antibodies used for detection were rabbit polyclonal anti-HO-1 (Assay Designs), anti-GCLM (10) (a gift from Dr. Terrance Kavanagh, University of Washington, Seattle), mouse monoclonal anti-NQO1 (clones A180 and B771, gifts from Dr. David Ross, University of Colorado, Denver), rabbit polyclonal anti-Nrf2 (Santa Cruz Biotechnology), rabbit polyclonal anti-lamin B1 (Abcam), anti-Hsp70 (Assay Designs), and anti-HSF1 (Cell Signaling) Blots were visualized using horseradish peroxidase-conjugated secondary antibodies and ECL Plus detection system (GE Healthcare) and detected with Typhoon 9400 (GE Healthcare) scanner. Protein expression was quantified with ImageQuant TL 7.0 software (GE Healthcare).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was done as described previously (25). For immunoprecipitation against HSF1, the anti-HSF1 antibody (Cell Signaling) was used. The forward and reverse primers used for PCR amplification were as follows: HSP70 forward, 5′-CCA TGG AGA CCA ACA CC CT-3′, and reverse, 5′-CCC TGG GCT TTT ATA AGT CG-3′; β-actin forward, 5′-AAC TCT CCC TCC TCC TCT TCC TC-3′, and reverse, 5′-GAG CCA TAA AAG GCA ACT TTC GG-3′.
Statistical Analysis
Statistical analysis was performed with GraphPad Prism, and the data were analyzed by one-way ANOVA with Bonferroni's post hoc comparisons. Data are expressed as means ± S.E., and differences were considered significant at p < 0.05.
Microarray Analysis
For the microarray experiment, HUVECs pooled from three separate donors were transfected with 50 nm Nrf2-siRNA or a nonspecific control siRNA using Oligofectamine (Invitrogen). After 24 h, cells were treated with methanol (vehicle), 3 μm OA, or 3 μm OA-NO2 for 8 h. Triplicate samples were used. Total RNA was extracted with TRI Reagent® (Sigma) according to the manufacturer's instructions. The amounts and purity of total RNAs were measured with NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE). The probes for gene expression analysis were made according to the Affymetrix protocol. Briefly, total RNA was reverse-transcribed to double strand cDNA. The double strand cDNA was purified and transcribed in vitro to biotin-labeled cRNA. Twenty micrograms of purified cRNA were fragmented, and the fragmentation was checked on agarose gel electrophoresis. The cRNA was hybridized to Human Genome U133 Plus 2.0 GeneChips (Affymetrix, Santa Clara, CA). The chips were stained, washed (Affymetrix Fluidics Station 400), and scanned (Affymetrix GeneChip Scanner 3000) according to the manufacturer's protocol.
Microarray Data Handling and Statistics
Microarray data were extracted from the scanned images by using MAS 5.1 software (Affymetrix). GeneChip operating software (Affymetrix) was used to generate CEL files. Quantile inter-array data normalization was carried out using CARMAweb (26). Primary statistical comparisons were done by using a Bayesian one-way ANOVA with post hoc Tukey HSD (R/Bioconductor) (27), and p values were corrected for multiple testing by the Benjamini/Hochberg false discovery rate method (28). Criteria for significance are as follows: false discovery rate (Benjamini/Hochberg) <5%; post hoc p value <0.05; and >1.5-fold intensity differences between different treatment groups. Gene set enrichment analysis software (GSEA version 2.0) (29, 30) was used for analysis of enrichment of transcription factor-binding sites (TRANSFAC 7.4) in promoters of differentially expressed genes.
RESULTS
OA-NO2 Induces HO-1, GCLM, and NQO1 mRNA and Protein Expression in Human Endothelial Cells
LNO2 induces HO-1, a well characterized Nrf2 target gene, in endothelial cells (9). To determine whether the more stable mono-unsaturated OA-NO2 also induces HO-1 as well as other ARE-regulated genes, including the glutamate-cysteine ligase modifier subunit (GCLM) and NAD(P)H:quinone oxidoreductase-1 (NQO1), HUVECs were treated with 0–5 μm OA-NO2 for 8 h, and target gene expression was determined via quantitative real time PCR. OA-NO2 induced the mRNA expression of all three genes in a dose-dependent manner. The induction of GCLM and NQO1 by 3 μm OA-NO2 was not statistically significant (Fig. 1A). However, in another set of experiments, in which the time-dependent changes in gene expression was assessed, 3 μm OA-NO2 significantly induced all three genes after 8 h, with the induction of NQO1 occurring later and sustaining longer than the other two genes (Fig. 1B). The discrepancy is likely due to donor-specific differences in gene expression obtained from a single donor. OA-NO2 also induced HO-1, GCLM, and NQO1 protein in a dose-dependent manner (Fig. 1C). The maximum induction was observed at 12 h for HO-1 and at 24 h for GCLM and NQO1 (Fig. 1D).
FIGURE 1.
OA-NO2 induces HO-1, GCLM, and NQO1 expression. HUVECs were treated with 0–5 μm OA-NO2 for 8 h (A) or 16 h (C) or with 3 μm OA-NO2 for 0–48 h (B and D). The expression of HO-1, GCLM, and NQO1 was analyzed with quantitative real time PCR (A and B) and Western blot (C and D). Values are expressed as mean ± S.E., (n = 3). *, p < 0.05 versus control. Western blots are representative of three independent experiments.
Induction of HO-1, GCLM, and NQO1 by OA-NO2 Is Nrf2-dependent
Having established the optimal time and concentration for the induction of HO-1, GCLM, and NQO1 by OA-NO2, the role of Nrf2 in the regulation of these genes was examined using siRNA to inhibit Nrf2 expression. Optimal siRNA concentration and transfection times were established, and it was observed that transfection with 50 nm Nrf2-specific siRNA for 24 h resulted in 85% silencing of Nrf2 mRNA expression and 89% silencing at the protein level under basal conditions, with no further suppression at higher siRNA concentrations or longer transfection times. (Fig. 2, A and B, and results not shown). Moreover, control or Nrf2-specific siRNA did not cause an interferon response, assessed by 2′,5′-oligoadenylate synthetase 2 (OAS) and interferon-induced protein with tetratricopeptide repeats 1 (IFIT1) mRNA expression (results not shown). Both OA-NO2-inducible mRNA and protein expression of HO-1, GCLM, and NQO1 were significantly inhibited in cells transfected with Nrf2-siRNA (Fig. 2, A and C). To further validate that OA-NO2 activates ARE-dependent genes, the NQO1 ARE luciferase reporter vector and a vector in which the core ARE was mutated (25) were transfected into HUVECs, and luciferase activity was assessed. OA-NO2, but not OA, activated the ARE of the NQO1 gene (supplemental Fig. S2A). Luciferase expression was abolished when the core ARE was mutated (supplemental S2A). Chromatin immunoprecipitation supported the binding of Nrf2 to the NQO1 and HO-1 promoters. Treatment with OA-NO2 increased Nrf2 binding to the NQO1 promoter area 477 bp upstream of the transcription start site-containing canonical ARE site (31) as well as the distal enhancer region of HO-1 gene that contains multiple antioxidant-response-like elements (supplemental Fig. S2B) (32).
FIGURE 2.
Inhibition of Nrf2 blocks OA-NO2-induced HO-1, GCLM, and NQO-1 expression. A, HUVECs were transfected with 50 nm control or Nrf2 siRNA. 24 h after transfection, cells were treated with 3 μm OA-NO2 for 8 h. The expression of Nrf2, HO-1, GCLM, and NQO1 was analyzed with quantitative real time PCR. ns, not significant. B, HUVECs were transfected with 50 nm control or Nrf2 siRNA and 24 h after transfection treated with 3 μm OA-NO2. Cytoplasmic and nuclear extracts were isolated, and Nrf2 expression was analyzed by Western blot analysis. Lamin B1 was used as control for nuclear extracts. C, HUVECs were transfected with 50 nm control or Nrf2 siRNA. 24 h after transfection, cells were treated with 3 μm OA-NO2 for 16 h. The expression of HO-1, GCLM, and NQO1 was analyzed with Western blot. Values are expressed as mean ± S.E., n = 3. *, p < 0.05 versus control. Western blots are representative of three independent experiments.
Microarray Transcriptome Analyses
Primary Statistics
To gain insight into the effect of OA-NO2 on the global gene expression profile in human endothelial cells and to more incisively define the role of Nrf2 in mediating these effects, we employed genome-wide transcriptional profiling of cells transfected with either control or Nrf2-specific siRNA and then exposed to vehicle, 3 μm OA, or OA-NO2 for 8 h. Under all conditions, no differences in cell injury or morphology were detected (data not shown). Initial transcriptome analyses comparing OA-NO2-treated cells with OA-treated and untreated cells revealed that mRNA expression levels were altered by OA-NO2 treatment with a high degree of significance (supplemental Table S1). Of these genes, 363 genes were up-regulated and 103 were down-regulated by OA-NO2. Hierarchical clustering analysis of the mRNA expression (Fig. 3A) illustrated that OA-NO2 specifically altered the expression of these genes, whereas OA had no effect. Of significance, within a sub-cluster of 50 genes with an elevated mRNA expression after OA-NO2 treatment (Fig. 3B and supplemental Table S1), eight of the genes with the greatest induction were heat shock proteins.
FIGURE 3.
Hierarchical clustering of ANOVA signature genes. A, clustering of 363 ANOVA signature genes with significant intensity differences between OA-NO2 (3 μm)-treated (n = 6) and OA (3 μm)-treated cells (n = 6). B, clustering of the top 50 genes with highest clustering ranks. The bar graph shows the fold intensity differences between OA-NO2 and OA treatments, expressed as mean ± S.D. Asterisks represent heat shock-related genes.
Role of Nrf2 in Mediating OA-NO2-dependent Gene Expression
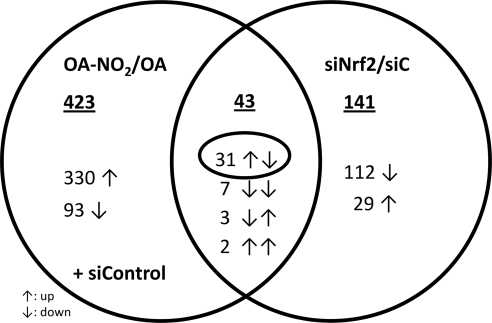
To assess the role of Nrf2 in gene expression changes evoked by OA-NO2, differential gene expression of OA-NO2-induced versus siNrf2-silenced HUVECs was compared (Fig. 4). In all treatment groups combined, silencing of Nrf2 by siRNA resulted in 78% down-regulation of Nrf2 mRNA (supplemental Fig. S3). Of the two datasets, only 43 genes were overlapping, 31 of which were up-regulated by OA-NO2 and repressed by siNrf2 (Fig. 4 and supplemental Table S1). This gene subset contained several previously characterized Nrf2 target genes, including ferritin heavy chain 1 (FTH1), sequestosome 1 (SQSTM1), and glutathione reductase (GSR), but the overall low number of genes indicates that other Nrf2-independent pathways play a significant role in mediating OA-NO2-dependent gene transcription.
FIGURE 4.
Venn diagram of ANOVA signature genes. Genes with significant intensity differences either with 3 μm OA-NO2 treatment or Nrf2-specific siRNA were compared in a Venn diagram. The underlined numbers indicate the number of genes regulated by each treatment alone or in combination. ↑, up-regulation; ↓, down-regulation.
Transcription Factor-binding Site Analyses
To define other transcriptional pathways important for OA-NO2-dependent gene regulation, transcriptomes were analyzed by GSEA to assess whether certain transcription factor-binding sites are over-represented within the promoter regions of the genes elevated by OA-NO2 in comparison with OA-treated and -untreated cells, respectively. In addition to Nrf2 and highly homologous MafG-binding sites, GSEA revealed binding sites for cell stress-related transcription factors, specifically HSF1, HSF2, and AP1 (activator protein 1), which were highly represented among OA-NO2-induced genes (supplemental Table S2). In this analysis, 50% of enriched transcription factor-binding sites were HSF-binding sites, indicating that the HSR pathway is the most pronounced response pathway in endothelial cells exposed to OA-NO2. Detailed analysis revealed a large number of HSF1/2 target genes within the OA-NO2-induced transcriptome, including such well characterized HSF target genes as HSPA6 (heat shock 70-kDa protein 6, also known as HSP70B′), HSPA1A (heat shock 70-kDa protein 1A, also known as HSP72, HSPA1, HSP70I, HSPA1B, HSP70-1, and HSP70-1A), and DNJA4 (DnaJ (Hsp40) homolog, subfamily A, member 4) (Fig. 5 and supplemental Table S3). Of note, with the exception of HO-1, which has previously been shown to be regulated by HSF1 on exposure to cadmium (33), silencing of Nrf2 had no impact on basal or inducible expression of HSF1 and HSF2 target genes (supplemental Fig. S4, A and B), indicating that the OA-NO2-mediated HSR is independent of Nrf2.
FIGURE 5.
OA-NO2 induces HSF target genes. Upon activation, HSF undergoes a multistep process involving post-translational modifications, nuclear enrichment, trimerization, and binding to heat shock elements resulting in transcription of a large family of heat shock genes. The table shows HSF-related, significantly regulated genes in the microarray after 3 μm OA-NO2 treatment. Genes were defined as HSF1 and -2 targets when an HSF1/2-binding site was present in the promoter region or an altered mRNA or protein expression level has been reported in response to HSF1/2 activation. The numbers at the right sides of the gene symbols indicate the fold differences (FD) between the relative mRNA expression of OA-NO2- and OA-treated cells.
Validation of Microarray Data
Quantitative real time PCR was utilized to select for and validate HSP genes showing the greatest induction by OA-NO2. Incubation with 3 μm OA-NO2 induced HSPA1A, DNAJA4, HSPB8, and HSPA6 mRNA in a time-dependent manner, with maximum induction occurring at 4–8 h (Fig. 6A). Inasmuch as HSPA1A (inducible HSP70) is the HSP most often used as an indicator of HSR, HSP70 expression was also evaluated at the protein level. OA-NO2 increased HSP70 protein in a concentration- and time-dependent manner (Fig. 6, B and C). Moreover, OA-NO2 treatment of control or HSF1 siRNA-transfected HUVECs revealed that HSF1-siRNA decreased HSF1 mRNA by 77% and HSF1 protein by 76%, quantified by densitometry (Fig. 7, A and B). The silencing of HSF1 caused a reduction of HSP70 expression at both mRNA (77%) and protein (64%) levels, affirming HSF-dependent regulation (Fig. 7, A and B). Heat shock was used as a positive control in these experiments. Similarly to OA-NO2, heat shock also induced HSP70 mRNA and protein significantly, and the expressions were reduced in HSF1 siRNA-transfected cells by 85 and 81%, respectively.
FIGURE 6.
OA-NO2 induces heat shock response. A, HUVECs were treated with 3 μm OA-NO2 for 0–24 h, and the expression of HSPA1A, DNAJA4, HSPB8, and HSPA6 was analyzed with quantitative real time PCR. Values are expressed as mean ± S.E., n = 3. *, p < 0.05 versus control. B and C, HUVECs were treated with 0–5 μm OA-NO2 for 8 h (B) or with 3 μm OA-NO2 for 0–24 h (C). The expression of HSP70 was analyzed with Western blot.
FIGURE 7.
OA-NO2 induces HSP70 expression via HSF1. A and B, HUVECs were transfected with 100 nm control or HSF1 siRNA. 72 h after transfection, cells were treated with 3 μm OA-NO2 for 4 h (A) or 8 h (B) or with heat shock for 30 min. The expression levels of HSF1 and HSP70 (HSPA1A) were analyzed with quantitative real time PCR (A) or Western blot (B). Values are expressed as mean ± S.E., n = 3. *, p < 0.05 versus control. Western blots are representative of three independent experiments. C, HUVECs were treated with 5 μm OA-NO2 for 4 h. The binding of HSF1 to the promoter region of HSP70 was determined with chromatin immunoprecipitation using β-actin as a control. D, HUVECs were transfected with 50 nm control or Nrf2 siRNA. 72 h after transfection, cells were treated with OA-NO2 for 4 h. The binding of HSF1 to the promoter region of HSP70 was determined with chromatin immunoprecipitation using β-actin as a control.
Although HO-1 has a functional heat shock element in its promoter (33) and is included in the GSEA HSF dataset (Fig. 5), the suppression of HSF1 had no impact on HO-1 mRNA and protein expression (supplemental Fig. S5). This indicates that HSF1 does not regulate residual Nrf2-independent induction of HO-1 by OA-NO2 (Fig. 2). The OA-NO2 activation of HSF1 was also confirmed by the chromatin immunoprecipitation analysis, which demonstrated increased binding of HSF1 to the proximal HSP70 promoter (34) on exposure to OA-NO2 (Fig. 7C). Furthermore, Nrf2 silencing did not affect binding of HSF1 to the HSP70 gene promoter (Fig. 7D), strengthening the notion that HSF1 activation is Nrf2-independent.
DISCUSSION
Unsaturated fatty acids of membranes and lipoproteins undergo both autocatalytic and enzymatically catalyzed peroxidation reactions yielding electrophilic derivatives that include α,β-unsaturated ketones, aldehydes, and nitroalkenes. Nitroalkenes are notable because the vinyl nitro group confers a kinetically rapid and reversible electrophilic reactivity that induces the post-translational modification of susceptible proteins, principally via S-alkylation of thiols (35). Current data support that cell survival and anti-inflammatory responses, rather than dysmetabolic and genotoxic events, are predominantly evoked by low concentrations of electrophiles (36). Here, genome-wide transcriptional profiling was utilized to identify specific gene regulatory pathways that mediate vascular endothelial responses to nitroalkenes. Suppression of Nrf2 translation supported that although phase II antioxidant responses are induced by exposure to OA-NO2, there was also a robust activation of HSR-related gene expression. The expression of HSR proteins are induced by a variety of stimuli, including heat, exposure to chemicals and heavy metals, and oxidative inflammatory mediators. Various electrophiles have been shown to be inducers of HSR (37, 38). HSR is orchestrated by HSF transcription factors, of which HSF1 is the most important in mediating responses to cell stress (39). HSF activation and ensuing induction of HSP chaperones not only affords protection against the adverse actions of misfolded proteins but also contributes to the regulation of inflammatory processes (23).
Fatty acid nitroalkene derivatives mediate adaptive and anti-inflammatory signaling via mechanisms frequently centered on post-translational protein modification of susceptible signaling mediators by electrophiles (40). This includes inhibition of NF-κB-dependent signaling (7) that in turn leads to protection against ischemic injury (3), activation of peroxisome proliferator-activated receptor γ-dependent gene expression (41), and inhibition of inflammatory cell function (7, 42). Also, lipopolysaccharide-induced STAT1 (signal transducer and activator of transcription 1) phosphorylation and the STAT1-dependent transcription are inhibited, thereby suppressing expression of the pro-inflammatory target genes inducible nitric-oxide synthase and MCP-1 (43). Induction of HSP expression adds to these operative mechanisms, as HSF1-dependent HSP70 expression attenuates NF- κB-dependent gene expression and signaling via HSP70 interaction with the IκB kinase (44). In addition, HSP27 (HSPB1), significantly induced by OA-NO2, similarly inhibits NF-κB activity (45). Notably, both heat shock and HSF-activating agents, as well as OA-NO2, protect kidneys from ischemia-reperfusion injury (46–48), supporting the notion that HSF-mediated anti-inflammatory actions of OA-NO2 may play a role in the protection against ischemia-reperfusion injury.
In addition to inhibiting inflammation, the HSR provides additional benefit to vascular endothelium. Endothelial cell injury is a critical component of atherosclerosis, thrombosis, and neointimal proliferation (49). HSPs display diverse cytoprotective functions, with heat shock pre-conditioning protecting against the cytotoxic effects of hydrogen peroxide in human endothelial cells (23, 50). Although the soluble plasma HSP60 may induce pro-atherogenic effects, intracellular HSPs are typically athero-protective (51). HSF1 also contributes to the statin-induced regulation of expression of the thrombin inhibitor thrombomodulin and the net antithrombotic effects of the HSR (52).
The detailed mechanism whereby OA-NO2 stimulates the HSR remains a matter of investigation, with HSF activation viewed to be an initial signaling event (39). On activation, HSFs trimerize and bind to heat shock elements, consisting of repetitive nGAAn sequences in target genes such as HSPs. Although stress-induced phosphorylation and other post-translational modifications of HSF contribute to activation via trimerization (39, 53), other mechanisms may also be operative. HSF1 contains redox-sensitive cysteine residues that are apparently oxidized by hydrogen peroxide to reversibly assemble into a homotrimer (54). It is therefore conceivable that electrophiles such as OA-NO2 can adduct and activate HSF1-dependent gene expression via adduction of these residues. Also, electrophilic lipid peroxidation products such as 4-hydroxynonenal bind to many heat shock proteins (e.g. HSP70 and HSP90) (55), with HSP90 cysteine 572 being exceptionally sensitive to thiol modification (56). Inasmuch as OA-NO2 is known to target cysteine residues in proteins (4), OA-NO2 may well adduct these chaperones and other redox-sensitive proteins involved in protein folding and turnover in an analogous manner, thereby altering HSP function and inducing compensatory activation of the HSR pathway. It is notable that interactions of HSF1 with HSP70 and HSP90 maintains it in an inactive state (57) and that modification of these co-chaperones can abolish their repressive functions. In support of the concept that post-translational modification of HSP70 can facilitate HSF1 activation by electrophilic lipids, the interaction of HSP70 with HSF1 is inhibited by 4-hydroxynonenal (37).
A critical insight from this study is the observation that the heat shock response is not dependent on Nrf2. Conversely, silencing of HSF1, important for HO-1 induction by cadmium exposure (33), had no impact on HO-1 expression in this study. The residual Nrf2-independent induction of HO-1 may involve a number of potential regulators, as the transcriptional regulation of HO-1 is complex (58). Upon exposure to oxidized phospholipids, both Nrf2 and the cAMP-responsive element-binding protein mediate HO-1 induction in human endothelial cells (25, 59). Genome-wide transcriptional profiling of HSF1 target genes using HSF1-null mouse fibroblasts did not show an induction of known ARE target genes (60), further supporting that these two stress responses are distinct and separate. However, the molecular attributes important for biological responses may be similar for both pathways. For example, the actions of the HSF1 activator celastrol, a quinone methide triterpene-based traditional Chinese medicinal herb, can be blocked by an added excess of thiol (61). The effects of celastrol that are blocked by thiols not only include triggering HSR but also inhibition of glucocorticoid receptor activity and ARE activation (61).
In summary, the exemplary nitroalkene fatty acid derivative OA-NO2 is a potent inducer of the HSR, adding a new dimension to the pleiotropic anti-inflammatory signaling mechanisms manifested by these products of oxidative inflammatory reactions (2, 40, 62). The small molecule activation of the HSR is an emerging therapeutic modality for a number of human diseases affecting protein folding and conformation, as well as inflammatory conditions such as ischemia-reperfusion injury (23). Electrophilic fatty acid nitroalkenes, in this context, thus represent a new class of potent HSR inducers. It is noteworthy that the effective concentrations of OA-NO2 sufficient to evoke HSR are comparable with the most efficacious HSR activators currently available, with the exception of some HSP90 inhibitors (23), suggesting that this biological activity is achievable in vivo.
Supplementary Material
Acknowledgments
We thank Lea Sistonen for valuable discussions and suggestions. B. A. F. acknowledges financial interests in Complexa, Inc.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL58115 and R01 HL64937 (to B. A. F.). This work was also supported by the Finnish Cultural Foundation (to E. K. and A. L. L.), the Academy of Finland, the Finnish Foundation of Cardiovascular Research, the Sigrid Juselius Foundation (to A. L. L.), American Diabetes Association Grant 7-08-JF-52, and American Heart Association Grant 0665418U (to F. J. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5, Tables S1–S3, and additional references.
- LNO2
- nitrolinoleic acid
- OA-NO2
- nitro-oleic acid
- siRNA
- small interfering RNA
- HO-1
- heme oxygenase-1
- HUVEC
- human umbilical vein endothelial cell
- ANOVA
- analysis of variance
- OA
- oleic acid
- ARE
- antioxidant-response element
- HSF
- heat shock factor
- HSR
- heat shock response
- GSEA
- Gene Set Enrichment Analysis
- GLCM
- glutamate-cysteine ligase modifier.
REFERENCES
- 1.Rubbo H., Radi R., Trujillo M., Telleri R., Kalyanaraman B., Barnes S., Kirk M., Freeman B. A. (1994) J. Biol. Chem. 269, 26066–26075 [PubMed] [Google Scholar]
- 2.Nadtochiy S. M., Baker P. R., Freeman B. A., Brookes P. S. (2008) Cardiovasc. Res. 82, 333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rudolph V., Rudolph T. K., Schopfer F. J., Bonacci G., Woodcock S. R., Cole M. P., Baker P. R., Ramani R., Freeman B. A. (2009) Cardiovasc. Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Batthyany C., Schopfer F. J., Baker P. R., Durán R., Baker L. M., Huang Y., Cerveñansky C., Branchaud B. P., Freeman B. A. (2006) J. Biol. Chem. 281, 20450–20463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker L. M., Baker P. R., Golin-Bisello F., Schopfer F. J., Fink M., Woodcock S. R., Branchaud B. P., Radi R., Freeman B. A. (2007) J. Biol. Chem. 282, 31085–31093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schopfer F. J., Batthyany C., Baker P. R., Bonacci G., Cole M. P., Rudolph V., Groeger A., Rudolph T. K., Nadtochiy S., Brookes P. S., Freeman B. A. (2009) Free Radic. Biol. Med. 46, 1250–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cui T., Schopfer F. J., Zhang J., Chen K., Ichikawa T., Baker P. R., Batthyany C., Chacko B. K., Feng X., Patel R. P., Agarwal A., Freeman B. A., Chen Y. E. (2006) J. Biol. Chem. 281, 35686–35698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Straus D. S., Pascual G., Li M., Welch J. S., Ricote M., Hsiang C. H., Sengchanthalangsy L. L., Ghosh G., Glass C. K. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 4844–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright M. M., Schopfer F. J., Baker P. R., Vidyasagar V., Powell P., Chumley P., Iles K. E., Freeman B. A., Agarwal A. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4299–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levonen A. L., Landar A., Ramachandran A., Ceaser E. K., Dickinson D. A., Zanoni G., Morrow J. D., Darley-Usmar V. M. (2004) Biochem. J. 378, 373–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Villacorta L., Zhang J., Garcia-Barrio M. T., Chen X. L., Freeman B. A., Chen Y. E., Cui T. (2007) Am. J. Physiol. Heart Circ. Physiol. 293, H770–H776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kensler T. W., Wakabayashi N., Biswal S. (2007) Annu. Rev. Pharmacol. Toxicol. 47, 89–116 [DOI] [PubMed] [Google Scholar]
- 13.Rangasamy T., Guo J., Mitzner W. A., Roman J., Singh A., Fryer A. D., Yamamoto M., Kensler T. W., Tuder R. M., Georas S. N., Biswal S. (2005) J. Exp. Med. 202, 47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rangasamy T., Cho C. Y., Thimmulappa R. K., Zhen L., Srisuma S. S., Kensler T. W., Yamamoto M., Petrache I., Tuder R. M., Biswal S. (2004) J. Clin. Invest. 114, 1248–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thimmulappa R. K., Lee H., Rangasamy T., Reddy S. P., Yamamoto M., Kensler T. W., Biswal S. (2006) J. Clin. Invest. 116, 984–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh K., Mochizuki M., Ishii Y., Ishii T., Shibata T., Kawamoto Y., Kelly V., Sekizawa K., Uchida K., Yamamoto M. (2004) Mol. Cell. Biol. 24, 36–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X. L., Dodd G., Thomas S., Zhang X., Wasserman M. A., Rovin B. H., Kunsch C. (2006) Am. J. Physiol. Heart Circ. Physiol. 290, H1862–H1870 [DOI] [PubMed] [Google Scholar]
- 18.Chen X. L., Varner S. E., Rao A. S., Grey J. Y., Thomas S., Cook C. K., Wasserman M. A., Medford R. M., Jaiswal A. K., Kunsch C. (2003) J. Biol. Chem. 278, 703–711 [DOI] [PubMed] [Google Scholar]
- 19.Dai G., Vaughn S., Zhang Y., Wang E. T., Garcia-Cardena G., Gimbrone M. A., Jr. (2007) Circ. Res. 101, 723–733 [DOI] [PubMed] [Google Scholar]
- 20.Fledderus J. O., Boon R. A., Volger O. L., Hurttila H., Ylä-Herttuala S., Pannekoek H., Levonen A. L., Horrevoets A. J. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 1339–1346 [DOI] [PubMed] [Google Scholar]
- 21.Hosoya T., Maruyama A., Kang M. I., Kawatani Y., Shibata T., Uchida K., Warabi E., Noguchi N., Itoh K., Yamamoto M. (2005) J. Biol. Chem. 280, 27244–27250 [DOI] [PubMed] [Google Scholar]
- 22.Levonen A. L., Inkala M., Heikura T., Jauhiainen S., Jyrkkänen H. K., Kansanen E., Määttä K., Romppanen E., Turunen P., Rutanen J., Ylä-Herttuala S. (2007) Arterioscler. Thromb. Vasc. Biol. 27, 741–747 [DOI] [PubMed] [Google Scholar]
- 23.Westerheide S. D., Morimoto R. I. (2005) J. Biol. Chem. 280, 33097–33100 [DOI] [PubMed] [Google Scholar]
- 24.Baker P. R., Lin Y., Schopfer F. J., Woodcock S. R., Groeger A. L., Batthyany C., Sweeney S., Long M. H., Iles K. E., Baker L. M., Branchaud B. P., Chen Y. E., Freeman B. A. (2005) J. Biol. Chem. 280, 42464–42475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jyrkkänen H. K., Kansanen E., Inkala M., Kivelä A. M., Hurttila H., Heinonen S. E., Goldsteins G., Jauhiainen S., Tiainen S., Makkonen H., Oskolkova O., Afonyushkin T., Koistinaho J., Yamamoto M., Bochkov V. N., Ylä-Herttuala S., Levonen A. L. (2008) Circ. Res. 103, e1–e9 [DOI] [PubMed] [Google Scholar]
- 26.Rainer J., Sanchez-Cabo F., Stocker G., Sturn A., Trajanoski Z. (2006) Nucleic Acids Res. 34, W498–W503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Long A. D., Mangalam H. J., Chan B. Y., Tolleri L., Hatfield G. W., Baldi P. (2001) J. Biol. Chem. 276, 19937–19944 [DOI] [PubMed] [Google Scholar]
- 28.Reiner A., Yekutieli D., Benjamini Y. (2003) Bioinformatics 19, 368–375 [DOI] [PubMed] [Google Scholar]
- 29.Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C. (2003) Nat. Genet. 34, 267–273 [DOI] [PubMed] [Google Scholar]
- 30.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaiswal A. K. (2000) Free Radic. Biol. Med. 29, 254–262 [DOI] [PubMed] [Google Scholar]
- 32.Banning A., Deubel S., Kluth D., Zhou Z., Brigelius-Flohé R. (2005) Mol. Cell. Biol. 25, 4914–4923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koizumi S., Gong P., Suzuki K., Murata M. (2007) J. Biol. Chem. 282, 8715–8723 [DOI] [PubMed] [Google Scholar]
- 34.Ostling P., Björk J. K., Roos-Mattjus P., Mezger V., Sistonen L. (2007) J. Biol. Chem. 282, 7077–7086 [DOI] [PubMed] [Google Scholar]
- 35.Rudolph V., Schopfer F. J., Khoo N. K., Rudolph T. K., Cole M. P., Woodcock S. R., Bonacci G., Groeger A. L., Golin-Bisello F., Chen C. S., Baker P. R., Freeman B. A. (2009) J. Biol. Chem. 284, 1461–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liebler D. C. (2008) Chem. Res. Toxicol. 21, 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jacobs A. T., Marnett L. J. (2007) J. Biol. Chem. 282, 33412–33420 [DOI] [PubMed] [Google Scholar]
- 38.Morimoto R. I., Santoro M. G. (1998) Nat. Biotechnol. 16, 833–838 [DOI] [PubMed] [Google Scholar]
- 39.Anckar J., Sistonen L. (2007) Adv. Exp. Med. Biol. 594, 78–88 [DOI] [PubMed] [Google Scholar]
- 40.Freeman B. A., Baker P. R., Schopfer F. J., Woodcock S. R., Napolitano A., d'Ischia M. (2008) J. Biol. Chem. 283, 15515–15519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schopfer F. J., Lin Y., Baker P. R., Cui T., Garcia-Barrio M., Zhang J., Chen K., Chen Y. E., Freeman B. A. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2340–2345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Coles B., Bloodsworth A., Clark S. R., Lewis M. J., Cross A. R., Freeman B. A., O'Donnell V. B. (2002) Circ. Res. 91, 375–381 [DOI] [PubMed] [Google Scholar]
- 43.Ichikawa T., Zhang J., Chen K., Liu Y., Schopfer F. J., Baker P. R., Freeman B. A., Chen Y. E., Cui T. (2008) Endocrinology 149, 4086–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ran R., Lu A., Zhang L., Tang Y., Zhu H., Xu H., Feng Y., Han C., Zhou G., Rigby A. C., Sharp F. R. (2004) Genes Dev. 18, 1466–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park K. J., Gaynor R. B., Kwak Y. T. (2003) J. Biol. Chem. 278, 35272–35278 [DOI] [PubMed] [Google Scholar]
- 46.Harrison E. M., Sharpe E., Bellamy C. O., McNally S. J., Devey L., Garden O. J., Ross J. A., Wigmore S. J. (2008) Am. J. Physiol. Renal Physiol. 295, F397–F405 [DOI] [PubMed] [Google Scholar]
- 47.Jo S. K., Ko G. J., Boo C. S., Cho W. Y., Kim H. K. (2006) J. Am. Soc. Nephrol. 17, 3082–3092 [DOI] [PubMed] [Google Scholar]
- 48.Liu H., Jia Z., Soodvilai S., Guan G., Wang M. H., Dong Z., Symons J. D., Yang T. (2008) Am. J. Physiol. Renal Physiol 295, F942–F949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rössig L., Dimmeler S., Zeiher A. M. (2001) Basic Res. Cardiol. 96, 11–22 [DOI] [PubMed] [Google Scholar]
- 50.Gill R. R., Poh A. C., Camp P. C., Allen J. M., Delano M. T., Jacobson F. L., Hunsaker A., Colson Y. L. (2008) AJR Am. J. Roentgenol. 191, 1046–1056 [DOI] [PubMed] [Google Scholar]
- 51.Xu Q. (2002) Arterioscler. Thromb. Vasc. Biol. 22, 1547–1559 [DOI] [PubMed] [Google Scholar]
- 52.Fu Q., Wang J., Boerma M., Berbée M., Qiu X., Fink L. M., Hauer-Jensen M. (2008) Circ. Res. 103, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Westerheide S. D., Anckar J., Stevens S. M., Jr., Sistonen L., Morimoto R. I. (2009) Science 323, 1063–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ahn S. G., Thiele D. J. (2003) Genes Dev. 17, 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vila A., Tallman K. A., Jacobs A. T., Liebler D. C., Porter N. A., Marnett L. J. (2008) Chem. Res. Toxicol. 21, 432–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carbone D. L., Doorn J. A., Kiebler Z., Ickes B. R., Petersen D. R. (2005) J. Pharmacol. Exp. Ther. 315, 8–15 [DOI] [PubMed] [Google Scholar]
- 57.Morimoto R. I. (1998) Genes Dev. 12, 3788–3796 [DOI] [PubMed] [Google Scholar]
- 58.Ryter S. W., Alam J., Choi A. M. (2006) Physiol. Rev. 86, 583–650 [DOI] [PubMed] [Google Scholar]
- 59.Krönke G., Bochkov V. N., Huber J., Gruber F., Blüml S., Fürnkranz A., Kadl A., Binder B. R., Leitinger N. (2003) J. Biol. Chem. 278, 51006–51014 [DOI] [PubMed] [Google Scholar]
- 60.Trinklein N. D., Murray J. I., Hartman S. J., Botstein D., Myers R. M. (2004) Mol. Biol. Cell 15, 1254–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trott A., West J. D., Klaić L., Westerheide S. D., Silverman R. B., Morimoto R. I., Morano K. A. (2008) Mol. Biol. Cell 19, 1104–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ferreira A. M., Ferrari M. I., Trostchansky A., Batthyany C., Souza J. M., Alvarez M. N., López G. V., Baker P. R., Schopfer F. J., O'Donnell V., Freeman B. A., Rubbo H. (2009) Biochem. J. 417, 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.