Abstract
Recent evidence suggests that signaling by the proinflammatory cytokine interleukin-1β (IL-1β) is dependent on reactive oxygen species derived from NADPH oxidase. Redox signaling in response to IL-1β is known to require endocytosis of its cognate receptor (IL-1R1) following ligand binding and the formation of redox-active signaling endosomes that contain Nox2 (also called redoxosomes). The consequent generation of reactive oxygen species by redoxosomes is responsible for the downstream recruitment of IL-1R1 effectors (IRAK, TRAF6, and IκB kinase kinases) and ultimately for activation of the transcription factor NFκB. Despite this knowledge of the signaling events that occur downstream of redoxosome formation, an understanding of the mechanisms that coordinate the genesis of redoxosomes following IL-1β stimulation has been lacking. Here, we demonstrate that lipid rafts play an important role in this process. We show that Nox2 and IL-1R1 localize to plasma membrane lipid rafts in the unstimulated state and that IL-1β signals caveolin-1-dependent endocytosis of both proteins into the redoxosome. We also show that inhibiting lipid raft-mediated endocytosis prevents NFκB activation. Finally, we demonstrate that Vav1, a Rac1 guanine exchange factor and activator of Nox2, is recruited to lipid rafts following IL-1β stimulation and that it is required for NFκB activation. Our results fill in an important mechanistic gap in the understanding of early IL-1R1 and Nox2 signaling events that control NFκB activation, a redox-dependent process important in inflammation.
INTRODUCTION
IL-1β2 is a potent proinflammatory cytokine that controls inflammation in response to a diverse collection of health problems, including ischemia/reperfusion injury, viral infection, bacterial infection, autoimmune diseases such as diabetes, allergies, trauma, and chemical exposure (1–8). A primary role for signaling by IL-1β in these inflammatory responses is the activation of NFκB, a transcription factor that regulates a large number of immune molecules, apoptotic factors, anti-apoptotic factors, and other transcription factors (9). The importance of IL-1β and NFκB in inflammation was highlighted by a clinical study on mortality in septic patients (10). A spectrum of cytokines and transcription factors was examined in this study, and two were identified as significant prognostic indictors of patient outcome. One prognostic indicator was NFκB activity, with this transcription factor twice as active in non-survivors relative to survivors. The second prognostic indicator kinase was the ratio between IL-1β and its competitive antagonist IL-1ra, with survivors having a 50% higher IL-1ra/IL-1β ratio than non-survivors.
Because of the clinical importance of IL-1β, elucidating the signaling events involved in IL-1β-mediated NFκB activation is of great significance. Among the early events that control IL-1β signaling is the induction of IL-1R1 dimerization following ligand binding (11, 12). This event initiates binding of MyD88 to the TIR (Toll/IL-1R1) domains within the cytoplasmic tail of IL-1R1 (13). Subsequently, multiple receptor/ligand pairs are endocytosed into a specialized signaling endosome (redoxosome) that contains the transmembrane protein Nox2 (NADPH oxidase 2) (14, 15). The recruitment of Nox2 into the redoxosome and the subsequent production of superoxide require Rac1, a co-activator of Nox2 (14, 15). Superoxide produced by Nox2 has been proposed either to dismutate spontaneously, which enables it to cross the endosomal membrane (16), or to be transported to the outside of the endosome via anion channels (17), where it rapidly dismutates into hydrogen peroxide. The generation of hydrogen peroxide by redoxosomes is required for the downstream recruitment of the IL-1R1 effectors TRAF6, IRAK1, and other mitogen-activated protein kinases that lead to the phosphorylation of IκB kinase (14, 15). IκB kinase activation triggers in turn the release of NFκB from IκB, allowing NFκB to move into the nucleus to activate downstream target genes transcriptionally.
Members of the Nox family of proteins have been associated with a number of signaling pathways, including those stimulated by IL-1β, tumor necrosis factor-α, angiotensin II, insulin, and platelet-derived growth factor (14, 18–20). Both Nox1 and Nox2 have been localized to lipid rafts (21–23), which are specialized membrane domains that are enriched in cholesterol and glycosphingolipids (24). Lipid rafts are ∼50–150 nm in size and have been assigned an important role in signaling by many different pathways. Taken together, the association of lipid rafts with Nox2, the role of lipid rafts as signaling platforms, and the involvement of lipid rafts with endocytosis suggest that redox signaling through IL-1β might involve lipid rafts. However, to date, the role of lipid rafts in IL-1β redox signaling has not been investigated.
Here, we report on a study that was designed to examine the role of lipid rafts in IL-1β-mediated redoxosome signaling. We demonstrate that inhibiting lipid raft formation reduces IL-1β-mediated NFκB activation and also that, in resting cells, both IL-1R1 and Nox2 are present in lipid rafts at the plasma membrane. Furthermore, we show that, following IL-1β stimulation, effectors of the IL-1R1 pathway are recruited into lipid rafts and that Nox2 and IL-1R1 are co-endocytosed into lipid raft/EEA1-positive early endosomes via a mechanism that requires caveolin-1. These findings support the importance of lipid rafts in transducing redoxosome signals by IL-1R1. Finally, we provide evidence that Vav1, a Rac GEF that has been associated with lipid rafts, is required for IL-1β-mediated NFκB activation.
EXPERIMENTAL PROCEDURES
Expression Vectors
For the lipid raft inhibitor experiments assaying NFκB activity, we infected MCF-7 cells with an NFκB luciferase reporter (Ad.NFκB.luc) using recombinant adenovirus as described previously (25). Cells were used for experiments 48 h post-infection. The expression plasmids for FLAG-tagged IL-1R1, Nox2, GFP, wild-type Vav1 (wtVav1), dominant-negative Vav1 (dnVav1), NFκB luciferase reporter (NFκB.luc), and GFP-caveolin-1 have been characterized previously (26–30). Transfection of the MCF-7 cells was performed by electroporation. Briefly, cells were grown to 80% confluency on 150-mm plates. Cells were detached from the plates via trypsinization, pelleted, washed once with Opti-MEM medium, and then resuspended at a concentration of 15 × 106 cells/800 μl of Opti-MEM medium. The cells were mixed with the experimental plasmid (20 μg/15 × 106 cells for the FLAG-IL-1R1 plasmid; 30 μg/15 × 106 cells for the Nox2, GFP, wtVav1, and dnVav1 plasmids; and 5 μg/15 × 106 cells for the NFκB.luc plasmid), and the mixture was placed in a Bio-Rad 0.4-cm gap cuvette. Electroporation was performed on a BTX ECM 830 electro-square porator with the following settings: voltage, 225 V; pulse length, 5.0 ms; number of pulses, two; and interval between pulses, 1.0 s. Following electroporation, the cells were allowed to sit undisturbed for 10 min, and then the contents of the cuvette were plated on a 150-mm cell culture dish with 20 ml of medium. The medium was changed at 24 h post-transfection, and the cells were used for experiments at 48–72 h post-transfection.
NFκB Activity Studies
For the lipid raft inhibition studies, MCF-7 cells were plated at 40% confluency in black 24-well cell culture plates. MCF-7 cells were transduced with Ad.NFκB.luc virus (1000 particles/cell) 48 h prior to treatment with nystatin (20 or 50 μg/ml), filipin (1 or 3 μg/ml), or vehicle (for mock treatment controls). After a 1-h incubation period, luciferin (15 μg/well in sterile PBS) was added to each well; the cells were incubated for 5 min at 37 °C; and the base line of NFκB luciferase activity was assessed using a biophotonic in vivo imaging system (IVIS, Xenogen). After base-line readings were obtained, cells were stimulated with IL-1β (5 ng/ml) or vehicle (PBS) at 37 °C. Subsequent readings of the NFκB luciferase reporter were taken at the indicated time points, with fresh luciferin added before each reading. For all readings, the IVIS system was set to take a 1-min luminescence exposure (with the field view setting to E, the focal plane set to 0.3 cm, binning set to medium, and the f-stop set to f1). Analysis of the results was performed using the Living Image software provided by Xenogen. For the wtVav1 and dnVav1 studies, MCF-7 cells were cotransfected with the NFκB.luc plasmid and the wtVav1, dnVav1, or GFP plasmid and then plated on black 24-well plates. After 48 h, a base-line reading of the NFκB luciferase reporter was taken, and then cells were stimulated with IL-1β (5 ng/ml) or vehicle (PBS) at 37 °C. Reporter activity was monitored at 4 h, with IVIS settings the same as in the lipid raft inhibitor studies.
Lipid Raft Isolation Studies
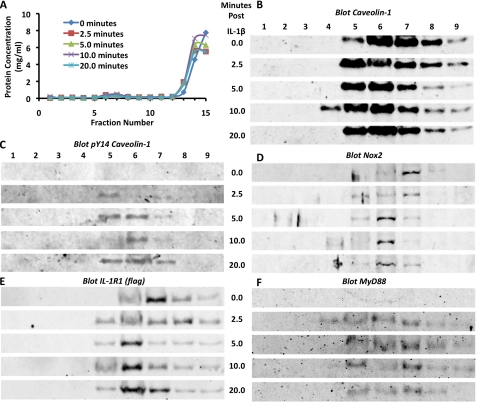
Lipid rafts were isolated using a previously described sodium carbonate method (31–33). Briefly, cells transfected with FLAG-IL-1R1 and Nox2 expression plasmids were stimulated with IL-1β for varying periods of time and then immediately placed on ice. The cells were scraped off the plate, washed with cold PBS, and then pelleted. The cell pellets were mixed with sodium carbonate buffer (250 mm sodium carbonate (pH 11.0), 2 mm EDTA, 1 mm NaF, 1 mm orthovanadate, and one Roche Complete protease inhibitor tablet/50 ml) and homogenized in a Duall cell homogenizer, followed by three 20-s pulses with a VirSonic 600 sonicator. The homogenate was mixed with 80% sucrose solution prepared in 25 mm MES, 0.15 m NaCl, 2 mm EDTA, 1 mm NaF, 1 mm orthovanadate, and one Roche Complete protease inhibitor table/50 ml to a final concentration of 40% sucrose. A three-step discontinuous gradient was generated, with the 40% sucrose sample on the bottom and 30 and 5% sucrose step gradients (both containing the MES buffer and 150 mm sodium carbonate) on the top. The gradient was spun in a Beckman SW 41 rotor at 200,000 × g for 18 h. Following centrifugation, gradient fractions were collected, and their protein concentrations were determined using the Bradford assay. A small protein peak corresponding to a visible, light-refracting band (typically in fractions 5–7) represented the fractions enriched for lipid rafts (31).
Immunoblotting
The lipid raft samples were concentrated by diluting each fraction and spinning in a fixed-angle rotor for 2 h at 100,000 × g. Proteins were separated on a 10% SDS-polyacrylamide gel and transferred to a nitrocellulose membrane. After transfer, the membrane was blocked overnight in 0.1% casein with 1 mm sodium fluoride and 1 mm orthovanadate. Western blotting was performed using standard protocols and casein buffer containing 0.2% Tween 20. The following antibodies were used for blotting: anti-caveolin-1 and anti-phospho-Tyr14 caveolin-1 (BD Transduction Laboratories); anti-Nox2, anti-MyD88, and anti-Rac1 (Millipore); anti-Vav1 (R&D Systems); and anti-FLAG (Sigma). Appropriate secondary antibodies with infrared tags were purchased from LI-COR Biosciences. Imaging of the infrared signal was performed on a LI-COR Odyssey scanner.
Immunofluorescence
Cells were transfected with FLAG-IL-1R1, Nox2, and GFP-caveolin-1 expression plasmids as indicated and grown on MatTek microscopy plates. 48 h post-transfection, the plates were placed at 4 °C for 15 min to inhibit all endocytosis. A portion of the cell medium was then removed from each well and mixed with the experimental labels at 4 °C (Alexa Fluor 488-labeled cholera toxin (0.1 μg/ml; Invitrogen), anti-Nox2 antibody (1:50 dilution; Medical & Biological Laboratories), and/or anti-FLAG antibody (1:300 dilution)) prior to being placed back onto the MCF-7 cells. The cells were then incubated for 1 h at 4 °C and washed three times with cold minimum essential medium, followed by mock stimulation or IL-1β (5 ng/ml) stimulation in conditioned medium. The cells were shifted to 37 °C for 20 min, washed once with cold PBS, fixed in 4% paraformaldehyde for 15 min, washed three times with filtered PBS, and blocked in blocking buffer (5% donkey serum, 0.1% Triton X-100, and 0.5% bovine serum albumin) for 1 h. For experiments investigating EEA1, anti-EEA1 antibody (1:150 dilution; BD Transduction Laboratories) in blocking buffer minus the donkey serum was incubated with the cells for 1 h, followed by three PBS washes. Cells were incubated with Alexa 488- and 546-labeled secondary antibodies (Invitrogen) at a 1:300 dilution in blocking buffer minus the donkey serum for 1 h. The cells were then washed three times with filtered PBS, and VECTASHIELD with 4′,6-diamidino-2-phenylindole was applied to the cells. Imaging was performed using a 63× objective on a spinning disc confocal microscope.
Image Analysis
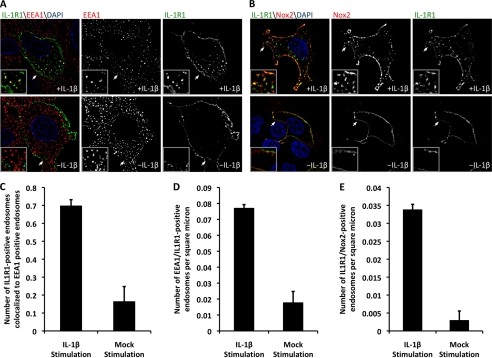
Image analysis was performed using MetaMorph software. A set of script programs was generated so that all images for a given fluorescent marker were treated exactly the same. Images were processed using two-dimensional deconvolution, and a threshold was applied to the images. MetaMorph was directed to count the number of intracellular endosomes (as defined by a size exclusion filter of 5–100 pixels) stained for each marker, as well as the number of endosomes in which pairs of markers co-localized. MetaMorph was also set to record the intracellular area over which the endosome count was performed. Quantification of marker co-localization was based either on the number of endosomal co-localization events divided by the total number of EEA1-positive endosomes in a given cell or on the number of endosomal co-localization events divided by the intracellular area for a given cell. Both methods gave similar results (see Fig. 3, C and D). A second method for normalization of the cellular area was needed when no definitive endosomal marker was used in the co-localization analysis (e.g. IL-1R1 and Nox2). Results were generated from experiments carried out in triplicate, and at least 24 cells were analyzed for each condition.
FIGURE 3.
Nox2 enters the redoxosome from the plasma membrane. MCF-7 cells were transfected with FLAG-IL-1R1 and Nox2 expression plasmids and used for experiments at 48 h post-transfection. A, for visualization of the movement of IL-1R1 into the early endosome, plasma membrane-bound IL-1R1 was labeled at 4 °C using anti-FLAG antibody. The cells were then pulsed at 37 °C with or without IL-1β stimulation for 20 min and fixed for immunostaining of FLAG and EEA1. Cells were treated with 4′,6-diamidino-2-phenylindole (DAPI) and visualized by confocal microscopy. The arrows mark the lower left corner of the region represented in each enlarged inset. B, for visualization of the movement of Nox2 and IL-1R1 into the endosomal compartment, plasma membrane-bound Nox2 and FLAG-IL-1R1 were prebound with antibodies to extracellular epitopes at 4 °C. The cells were then pulsed for 20 min at 37 °C in the presence or absence of IL-1β. Cells were fixed and immunostained with secondary antibodies. The arrows mark the lower left corner of the region represented in each enlarged inset. C, MetaMorph quantification was performed to evaluate IL-1β-induced changes in the co-localization of IL-1R1 and EEA1. The program was directed to count the total number of co-localized endosomes and the total number of EEA1-positive endosomes. The bar graph indicates the fraction of EEA1-positive endosomes that contain IL-1R1. D, as a second method to assess IL-1β-induced endosomal co-localization of EEA1 and IL-1R1, the total number of co-localized endosomes was normalized to the intracellular area of each cell imaged. E, shown is IL-1β-induced co-localization of Nox2 and IL-1R1 in the endosome as assessed using the morphometric approach described for D. For all image analyses, a minimum of 24 cells were imaged per condition from a total of three independent experiments. Values represent the mean ± S.D. (n = 3). C–E demonstrate a statistical difference between mock- and IL-1β-stimulated conditions as assessed by Student's t test (p < 0.005).
RESULTS
Disruption of Lipid Rafts Inhibits IL-1β-mediated NFκB Activation
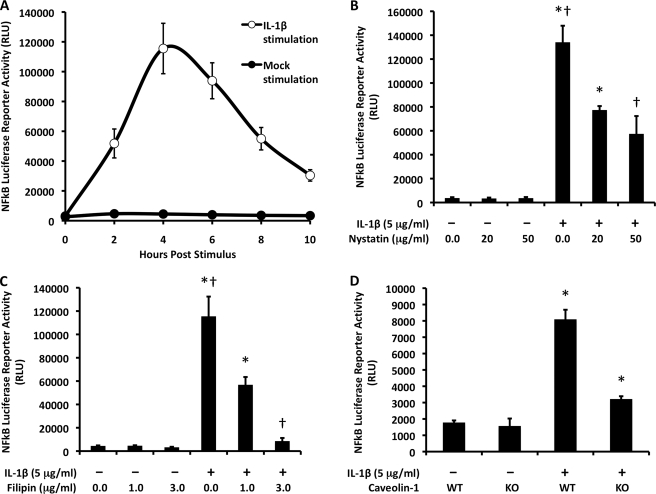
In this study, the mammary epithelial MCF-7 cell line was used. This cell line endogenously expresses Nox2 and the IL-1 receptor and has a robust Nox2-mediated induction of NFκB following IL-1β stimulation (14). To index changes in NFκB activity, we used an NFκB-responsive luciferase reporter in conjunction with biophotonic imaging over a period of 10 h following IL-1β stimulation. In this system, IL-1β induction of the NFκB luciferase reporter was found to peak at 4 h post-IL-1β stimulation (Fig. 1A).
FIGURE 1.
Lipid rafts facilitate IL-1β-mediated NFκB activation. A, MCF-7 cells were infected with a recombinant adenovirus expressing an NFκB luciferase reporter 48 h prior to IL-1β stimulation. Luciferase expression, an index of NFκB activation, was assessed as relative light units (RLU) at different time points post-stimulation by biophotonic imaging using an IVIS system. Values indicate the mean ± S.D. (n = 4). B and C, MCF-7 cells were infected as described for A but then preincubated with the lipid raft inhibitor nystatin (B) or filipin (C) for 1 h prior to IL-1β stimulation. NFκB activity was then assessed by luciferase assay at 4 h post-stimulation. Values indicate the mean ± S.D. (n = 4). D, caveolin-1 knock-out (KO) or wild-type (WT) mouse embryonic fibroblasts expressing an NFκB luciferase reporter were evaluated for NFκB induction at 2 h following IL-1β stimulation. Values indicate the mean ± S.D. (n = 3). Marked comparisons (* and †) are statistically different as assessed by Student's t test (p < 0.005).
Nox2 has been reported to reside within the lipid rafts of phagocytic cells (22, 23). Because Nox2 was shown previously to be required for IL-1β-dependent NFκB activation in MCF-7 cells (14), we hypothesized that disrupting lipid rafts would interfere with NFκB activation following IL-1β stimulation. To test this hypothesis, we treated MCF-7 cells expressing the NFκB luciferase reporter with nystatin or filipin. Nystatin disrupts lipid rafts by binding to cholesterol within the plasma membrane without extracting it and has been used widely to demonstrate the involvement of lipid rafts in biological processes (34). Similarly, filipin interacts with cholesterol at the plasma membrane and sequesters it into large complexes (35). Both agents inhibit lipid raft-mediated endocytosis without interfering with clathrin-mediated endocytosis (36). As hypothesized, nystatin treatment of MCF-7 cells reduced NFκB activation in response to IL-1β; the effect was dose-dependent, with a reduction of 40–60% by 4 h post-stimulation (Fig. 1B). In cells treated with nystatin in the absence of IL-1β, by contrast, base-line levels of NFκB were not altered (Fig. 1B). Similarly, filipin treatment of MCF-7 cells significantly reduced NFκB activation by IL-1β in a dose-dependent fashion; in this case, NFκB activation was almost completely abolished at the highest dose of 3 μg/ml (Fig. 1C).
The ability of nystatin and filipin to inhibit IL-1β-mediated NFκB activation suggested that this signaling pathway is dependent on lipid rafts. Caveolin-1 is a critical functional component of a subset of lipid rafts, in which it facilitates invagination and endocytosis from the plasma membrane (37). Therefore, we next sought to determine whether IL-1β-induced lipid raft signaling is caveolin-1-dependent. To this end, we utilized caveolin-1 knock-out and wild-type mouse embryonic fibroblasts (American Type Culture Collection) expressing the NFκB reporter. The results demonstrated that NFκB activation was significantly reduced in caveolin-1 knock-out cells compared with wild-type control cells (Fig. 1D). These findings suggest that IL-1β signals through the subset of lipid rafts that utilize caveolin-1.
IL-1R1 and Nox2 Are Constitutive Components of Lipid Rafts
To better understand the relationship between lipid rafts and IL-1R1 signaling, we sought to determine whether IL-1R1 is actively recruited to lipid rafts in a ligand-dependent fashion. This would be analogous to signaling by tumor necrosis factor-α receptor 1, which actively moves to lipid rafts following ligand stimulation (21). By contrast, the epidermal growth factor receptor is already present in lipid rafts prior to ligand binding (38). Because the IL-1β and tumor necrosis factor-α receptors share some early effectors such as Nox2 and Rac1 (14, 20), we favored the hypothesis that IL-1R1 enters lipid rafts after ligand stimulation. To test this hypothesis, we characterized the abundance of IL-1R1 in isolated lipid rafts prior to and following IL-1β stimulation.
As expected, total caveolin-1 was enriched in isolated lipid raft fractions (fractions 5–7), and its abundance in these fractions remained largely unchanged following IL-1β stimulation (Fig. 2B). In support of the involvement of caveolin-1 in IL-1β-mediated NFκB activation (Fig. 1D), the phosphorylated active form of caveolin-1 was enriched in the lipid raft fractions following IL-1β stimulation (Fig. 2C). Thus, IL-1R1 appears to signal through caveolae. Nox2 has also been shown to reside in caveolae and was indeed also constitutively present in the lipid raft fractions of MCF-7 cells (Fig. 2D). Contrary to the prediction of our original hypothesis, IL-1R1 was constitutively present in lipid raft factions regardless of IL-1β stimulation (Fig. 2E). However, consistent with the fact that MyD88 docking on IL-1R1 requires ligand stimulation, MyD88 was present in lipid rafts only after IL-1β stimulation (Fig. 2F). These findings suggest that the activation of IL-1R1 and its dependence on lipid rafts are more similar to the epidermal growth factor receptor (compared with the tumor necrosis factor-α receptor 1) signaling pathway (38).
FIGURE 2.
IL-1R1 signaling initiates from lipid rafts. MCF-7 cells were treated with IL-1β for varying periods of time (0, 2.5, 5, 10, and 20 min). Lipid rafts were then isolated from the cells using a sodium carbonate density gradient method. Following centrifugation, the gradients were collected into 15 fractions. A, shown are the results from Bradford protein quantification of the fractions from an experimental series of lipid raft isolations. Lipid rafts were typically contained within the small protein peak found in fractions 5–7. B–F, Western blotting was performed to examine the signaling components in the isolated lipid raft-containing fractions. The antigen detected in each blot is indicated at the top of each panel.
Nox2 Enters the Redoxosome with IL-1R1 from the Cell Surface
The biochemical localization of Nox2 and IL-1R1 to lipid rafts, together with the observation that IL-1β stimulates phosphorylation of caveolin-1, suggested that Nox2 and IL-1R1 might enter redoxosomes together from cell-surface caveolae. Alternatively, it remained a formal possibility that Nox2 from an intracellular source is recruited to the redoxosome independently by vesicle fusion following IL-1R1 endocytosis. To differentiate between these two possibilities, we sought to co-localize IL-1R1 and Nox2 in cells prior to and following IL-1β stimulation. Given that redoxosomes form within the early endosome (14), we first sought to establish a localization protocol capable of tracking ligand-dependent movement of FLAG-tagged IL-1R1 into the early endosome. Because the FLAG tag on IL-1R1 localizes to an extracellular region, it was possible to use anti-FLAG antibodies to label only IL-1R1 at the cell surface and to evaluate the endocytic fate of IL-1R1 following IL-1β stimulation. As predicted from previous studies evaluating the genesis of IL-1β-dependent redoxosomes (14, 16), we observed a significant increase in the percentage of IL-1R1 that co-localized with the early endosomal marker EEA1 by 20 min after cell stimulation with IL-1β (Fig. 3A). By contrast, in the absence of stimulation, the majority of IL-1R1 resided at the plasma membrane (Fig. 3A). Quantitative analysis using two independent morphometric methods demonstrated that IL-1β stimulation led to an ∼4-fold increase in the level of IL-1R1 in the early endosome (Fig. 3, C and D).
Using this approach, we next sought to visualize Nox2 co-localization with IL-1R1 using a monoclonal antibody that recognizes an extracellular domain of Nox2. In unstimulated cells, Nox2 was almost exclusively at the plasma membrane, and its localization significantly overlapped with that of IL-1R1 (Fig. 3B). Following IL-1β stimulation, the localization of both Nox2 and IL-1R1 within the endosomal compartment increased, and there was significant overlap in their intracellular staining pattern (Fig. 3B). Quantitative morphometric analysis demonstrated that IL-1β stimulation led to an ∼12-fold increase in IL-1R1 and Nox2 co-localization to the endosomal compartment (Fig. 3E). Given that Nox2 and IL-1R1 co-localized at the plasma membrane prior to IL-1β stimulation and also in the endosomal compartment following IL-1β stimulation, these findings support the hypothesis that these two components are co-endocytosed from the plasma membrane into newly formed redoxosomes.
IL-1R1-containing Redoxosomes Form through Endocytosis of Caveolin-1-positive Lipid Rafts
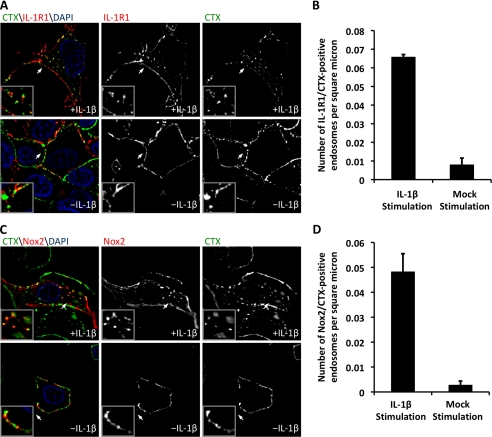
To investigate whether lipid rafts coordinate the movement of Nox2 and IL-1R1 from the plasma membrane into redoxosomes following IL-1β stimulation, we performed localization studies with the cholera toxin B subunit (CTX), which specifically binds to the lipid raft component glycosphingolipid-1 (39, 40). We hypothesized that Nox2 and IL-1R1 undergo lipid raft-mediated endocytosis following IL-1β stimulation, as suggested by the ability of nystatin and filipin to inhibit NFκB induction by IL-1β (Fig. 1) and by the biochemical fractionation of Nox2 and IL-1R1 with lipid rafts (Fig. 2). An alternative possibility was that lipid rafts act as a staging point for IL-1β signaling prior to the endocytic removal of IL-1R1/Nox2 from lipid rafts. The lipid raft isolation experiments could not adequately differentiate between these two hypotheses. Thus, we used CTX to label lipid rafts while simultaneously using selective antibodies to label IL-1R1 or Nox2 on the plasma membrane. We observed a significant (8-fold) increase in CTX/IL-1R1-positive endosomes following IL-1β stimulation compared with control mock-stimulated cells (Fig. 4, A and B). Similarly, Nox2/CTX-positive endosomes also significantly increased (15-fold) in the presence of IL-1β (Fig. 4, C and D). Furthermore, Nox2 and IL-1R1 co-localized to lipid rafts at the plasma membrane in both unstimulated and IL-1β-stimulated cells. These findings support the hypothesis that lipid rafts coordinate ligand-dependent internalization of Nox2 and IL-1R1 into the redoxosome. Furthermore, they demonstrate that lipid rafts remain in the newly formed redoxosome at least 20 min post-stimulation.
FIGURE 4.
IL-1R1 and Nox2 co-localize to lipid rafts both at the plasma membrane and within the redoxosome. MCF-7 cells were transfected with FLAG-IL-1R1 and Nox2 expression plasmids and used for experiments at 48 h post-transfection. A, for an evaluation of IL-1R1 localization to lipid rafts prior to and following stimulation with IL-1β, IL-1R1 and lipid rafts were labeled with anti-FLAG antibody and Alexa Fluor 488-labeled CTX at 4 °C. The cells were then pulsed at 37 °C with or without IL-1β stimulation for 20 min and fixed prior to immunostaining for FLAG. DAPI, 4′,6-diamidino-2-phenylindole. B, the number of IL-1R1- and CTX-positive endosomes per intracellular area was quantified. C, experiments similar to those described for A were performed using an antibody that recognizes an extracellular epitope in Nox2 and Alexa Fluor 488-labeled CTX. The panels show representative examples of Nox2 localization with CTX-positive lipid rafts in the presence and absence of IL-1β stimulation. The arrows mark the lower left corner of the region represented in each enlarged inset. D, the number of Nox2- and CTX-positive endosomes per intracellular area was quantified. For each analysis, a minimum of 24 cells were imaged per condition from a total of three independent experiments. Values represent the mean ± S.D. (n = 3). B and D demonstrate a statistical difference between mock- and IL-1β-stimulated cells as assessed by Student's t test (p < 0.005).
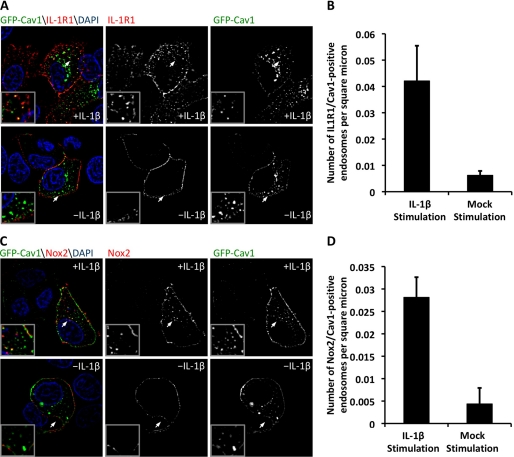
We next sought to confirm our hypothesis that the endocytosis of IL-1R1 is caveolin-1-dependent. To this end, we expressed GFP-tagged caveolin-1 in MCF-7 cells and again utilized selective labeling of IL-1R1 or Nox2 on the plasma membrane. These experiments revealed a 6.8-fold increase in IL-1R1/caveolin-1-positive endosomes in the IL-1β-stimulated cells relative to mock-stimulated cells (Fig. 5, A and B). Similarly, we observed a 6.4-fold increase in Nox2/caveolin-1-positive endosomes in IL-1β-stimulated cells relative to control cells (Fig. 5, C and D). Taken together with the observed increase in caveolin-1 phosphorylation following IL-1β stimulation and the decrease in IL-1β-mediated NFκB activation in caveolin-1 knock-out cells, these findings suggest that redoxosome formation and signal propagation in the IL-1R1 pathway are caveolin-1-dependent.
FIGURE 5.
IL-1R1 and Nox2 undergo caveolin-1-mediated endocytosis following IL-1β stimulation. A, to evaluate IL-1R1 localization with caveolin-1 (Cav1)-positive endosomes, MCF-7 cells were transfected with IL-1R1 and GFP-caveolin-1 expression plasmids. At 48 h post-transfection, IL-1R1 was labeled at the plasma membrane by incubation with anti-FLAG antibody at 4 °C. The cells were then pulsed at 37 °C with or without IL-1β stimulation for 20 min and fixed for immunostaining of FLAG. DAPI, 4′,6-diamidino-2-phenylindole. B, the number of IL-1R1- and caveolin-1-positive endosomes per intracellular area was quantified. C, MCF-7 cells were transfected with Nox2 and GFP-caveolin-1 expression plasmids. At 48 h post-transfection, plasma membrane-localized Nox2 was labeled with a monoclonal antibody at 4 °C. The cells were then pulsed at 37 °C with or without IL-1β stimulation for 20 min and fixed for immunostaining of Nox2. The arrows mark the lower left corner of the region in each enlarged inset. D, shown is the MetaMorph quantification of the fraction of Nox2- and caveolin-1-positive endosomes. For all image analysis studies, a minimum of 24 cells were imaged per condition from a total of three independent experiments. Values represent the mean ± S.D. (n = 3). B and D demonstrate a statistical difference between mock- and IL-1β-stimulated conditions as assessed by Student's t test (p < 0.01 (B) and p < 0.005 (D)).
Vav1 Is Actively Recruited to Rac1-containing Lipid Rafts following IL-1β Stimulation to Facilitate NFκB Activation
Activation of the Nox2 complex is a tightly regulated multistep event. Superoxide production by Nox2 is stimulated following recruitment of the cofactors p47phox, p67phox, and Rac1 (41). The dependence of Nox activation on a Rac1 GEF (which converts GDP-Rac1 to an active GTP-Rac1 form) has been demonstrated in phagocytes as well as other Nox signaling pathways (42–46). We hypothesized that, of the many Rac1 GEFs, Vav1 would act as the Rac1 GEF of the IL-1β redox signaling pathway. This hypothesis was based on three facts. First, Vav1 has been strongly associated with Nox2 activation (42). Second, Vav1 has been associated with lipid rafts and contains a caveolin-1-binding domain (47, 48). Finally, Vav1 acts as a Rac1 GEF in the highly related Toll-like receptor 4 pathway (49, 50).
To test our hypothesis, we isolated lipid rafts and evaluated the abundance of Rac1 and Vav1 before and after IL-1β stimulation. These experiments revealed that Rac1 was present in lipid rafts prior to and following IL-1β stimulation (Fig. 6A), suggesting that it is constitutively associated with lipid rafts. In contrast, Vav1 was recruited to lipid rafts in an IL-1β-dependent manner (Fig. 6B); it was present by 2.5 min post-stimulation and remained strong at 5 min post-stimulation but declined by 20 min post-stimulation, when Nox activity is maximal (14). These findings are consistent with Vav1 acting early and transiently in the process of redoxosome formation.
FIGURE 6.
Vav1 is actively recruited to Rac1-containing lipid rafts following IL-1β stimulation and facilitates NFκB activation. A and B, lipid rafts were isolated from MCF-7 cells at various times post-IL-1β stimulation, and the abundance of Rac1 and Vav1 within the lipid raft fractions was evaluated by Western blotting. C, MCF-7 cells were cotransfected with dnVav1, wtVav1, or GFP expression plasmids together with an NFκB luciferase reporter plasmid. At 48 h post-transfection, cells were stimulated with IL-1β, and luciferase activity was measured at 4 h post-stimulation. Data indicate the mean ± S.D. (n = 6). Marked comparisons (* and †) are statistically significant as assessed by Student's t test (p < 0.005). RLU, relative light units.
To verify the dependence of IL-1β signaling on Vav1, we tested the influence of wtVav1 and dnVav1 on NFκB activation. MCF-7 cells were transiently cotransfected with an NFκB luciferase reporter in combination with one of the Vav1 expression constructs or a GFP control plasmid. The results from this analysis demonstrated that dnVav1 expression significantly inhibited transcriptional activation of an NFκB reporter following IL-1β stimulation relative to that in wtVav1- or GFP-transfected cells. Cumulatively, these findings demonstrate that Vav1 is actively recruited to lipid rafts following IL-1β stimulation, where it facilitates NFκB activation, likely through its regulatory role in Rac1.
DISCUSSION
A growing number of receptors have been recognized to utilize Nox proteins and their reactive oxygen species products to mediate intracellular signaling. The compartmentalization of these receptors (to localized domains within the cell that produce the second messenger reactive oxygen species) limits cellular redox stress while also facilitating the redox changes required to propagate signal transduction. In the context of IL-1β signaling, these events are controlled at the level of the redoxosome (15). Despite our increasing knowledge of redoxosome signaling components used in the IL-1β pathway, little is known about the early molecular events that control redoxosome biogenesis.
It is well established that IL-1R1 moves from the plasma membrane into endocytic vesicles following ligand binding (14, 16, 51). Previous work by our laboratory has demonstrated that dynamin-dependent endocytosis is necessary for IL-1β-dependent Nox activation in the endosomal compartment and for redox activation of IL-1R1 and NFκB (14). Dynamin facilitates both clathrin-dependent and certain types of lipid raft-mediated endocytosis by promoting vesicle scission events from the plasma membrane (37). We therefore focused our efforts on differentiating between these two basic types of endocytosis. Our findings suggest that IL-1β stimulation leads to the caveolin-1-dependent lipid raft-mediated endocytosis of IL-1R1 and that this process is required for NFκB activation. Given that nystatin and filipin are known to disrupt lipid rafts but not to interfere with clathrin-dependent endocytosis, we interpreted our findings and previous work on the dynamin dependence of IL-1R1 signals (14) to mean that dynamin/caveolin-1-dependent lipid raft-mediated endocytosis is critical for the formation of redoxosomes and that clathrin is not involved in this process.
In support of the importance of caveolin-1-dependent lipid raft-mediated signaling in the IL-1β pathway, we found that IL-1β induces caveolin-1 phosphorylation at Tyr14 in isolated lipid rafts. This modification is known to be required in the endocytosis of ligands such as epidermal growth factor and to destabilize the interaction of caveolin-1 with the caveolar membrane. This destabilization allows for the recruitment of dynamin and the subsequent scission of caveolae from the plasma membrane (52). Our data are consistent with the notion that IL-1R1 likewise stimulates endocytosis by such a mechanism.
Using biochemical and immunofluorescence techniques to localize Nox2 and IL-1R1 to lipid rafts, we have demonstrated that both of these molecules are permanent constituents of lipid rafts at the plasma membrane in MCF-7 cells. Furthermore, IL-1β stimulates the co-endocytosis of Nox2 and IL-1R1 from plasma membrane lipid rafts into the early endosome. Interestingly, lipid rafts appear to maintain their integrity within the early endosome and may be required for sustained Nox2 activation following endocytosis because several Nox2 activators (p47phox and p67phox) are recruited to lipid rafts during Nox activation (23, 53). Hence, the lipid rafts may serve as a controlled microdomain for the synthesis and degradation of lipid moieties involved in redox signaling through Nox2. For example, p47phox stabilizes the Nox2 complex by binding specifically to the phospholipid phosphoinositol 3,4-bisphosphate during Nox activation.
Rac1 was shown previously to be required for the recruitment of Nox2 into the endosomal compartment following IL-1β stimulation (14). In this context, Rac1 associates with both IL-1R1 and Nox2 and is thought to tether Nox2 to IL-1R1 during endocytosis following IL-1β stimulation (14, 15). Our data demonstrating that Rac1 is constitutively present in lipid rafts suggest that IL-1R1, Nox2, and Rac1 may be closely associated on the plasma membrane. Given that the Rac1 GEF Vav1 is recruited to lipid rafts following stimulation only with IL-1β and that it is required for NFκB activation, we speculate that Vav1 recruitment may limit Rac1 activation (and hence also Nox2-dependent reactive oxygen species production) to newly forming redoxosomes.
In conclusion, the redox pathway that activates NFκB following IL-1β stimulation is dependent on caveolin-1-dependent lipid raft-mediated endocytosis. Both Nox2 and IL-1R1 constitutively localize to lipid rafts and enter the redoxosome from the plasma membrane, whereas Nox2 activation within endosomal lipid rafts likely depends on the Rac1 GEF Vav1. Together, these findings provide new insights into the spatial and temporal regulation of IL-1β signaling and activation of NFκB through redoxosomes.
Acknowledgment
We gratefully acknowledge Dr. Christine Blaumueller for editorial assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK067928 and R01 DK051315 from NIDDK and Grant F30 ES015950 from NIEHS. This work was also supported by the University of Iowa Gene Transfer Vector Core, which is funded through the Center for Gene Therapy (National Institutes of Health Grant P30 DK54759), and by the Roy J. Carver Chair in Molecular Medicine.
- IL-1β
- interleukin-1β
- IL-1R1
- IL-1β receptor 1
- EEA1
- early endosomal antigen 1
- GEF
- guanine exchange factor
- GFP
- green fluorescent protein
- wtVav1
- wild-type Vav1
- dnVav1
- dominant-negative Vav1
- PBS
- phosphate-buffered saline
- MES
- 2-(N-morpholino)ethanesulfonic acid
- CTX
- cholera toxin B subunit.
REFERENCES
- 1.Alcamí A., Smith G. L. (1992) Cell 71, 153–167 [DOI] [PubMed] [Google Scholar]
- 2.Borish L., Mascali J. J., Dishuck J., Beam W. R., Martin R. J., Rosenwasser L. J. (1992) J. Immunol. 149, 3078–3082 [PubMed] [Google Scholar]
- 3.Chen X. L., Xia Z. F., Wei D., Ben D. F., Wang Y. J. (2005) Zhonghua Wai Ke Za Zhi 43, 185–188 [PubMed] [Google Scholar]
- 4.Fan H., Williams D. L., Breuel K. F., Zingarelli B., Teti G., Tempel G. E., Halushka P. V., Cook J. A. (2006) Front. Biosci. 11, 2264–2274 [DOI] [PubMed] [Google Scholar]
- 5.Lin H. W., Basu A., Druckman C., Cicchese M., Krady J. K., Levison S. W. (2006) J. Neuroinflamm. 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Bernuth H., Puel A., Ku C. L., Yang K., Bustamante J., Chang H. H., Picard C., Casanova J. L. (2005) Clin. Infect. Dis. 41, Suppl. 7, S436–S439 [DOI] [PubMed] [Google Scholar]
- 7.Wilson C. A., Jacobs C., Baker P., Baskin D. G., Dower S., Lernmark A., Toivola B., Vertrees S., Wilson D. (1990) J. Immunol. 144, 3784–3788 [PubMed] [Google Scholar]
- 8.Yamada J., Dana M. R., Sotozono C., Kinoshita S. (2003) Exp. Eye Res. 76, 161–167 [DOI] [PubMed] [Google Scholar]
- 9.Kabe Y., Ando K., Hirao S., Yoshida M., Handa H. (2005) Antioxid. Redox Signal. 7, 395–403 [DOI] [PubMed] [Google Scholar]
- 10.Arnalich F., Garcia-Palomero E., López J., Jiménez M., Madero R., Renart J., Vázquez J. J., Montiel C. (2000) Infect. Immun. 68, 1942–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verstrepen L., Bekaert T., Chau T. L., Tavernier J., Chariot A., Beyaert R. (2008) Cell. Mol. Life Sci. 65, 2964–2978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunne A., O'Neill L. A. (2003) Sci. STKE 2003, re3. [DOI] [PubMed] [Google Scholar]
- 13.Martin M. U., Wesche H. (2002) Biochim. Biophys. Acta 1592, 265–280 [DOI] [PubMed] [Google Scholar]
- 14.Li Q., Harraz M. M., Zhou W., Zhang L. N., Ding W., Zhang Y., Eggleston T., Yeaman C., Banfi B., Engelhardt J. F. (2006) Mol. Cell. Biol. 26, 140–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oakley F. D., Abbott D., Li Q., Engelhardt J. (2009) Antioxid. Redox Signal., 11, 1313–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller F. J., Jr., Filali M., Huss G. J., Stanic B., Chamseddine A., Barna T. J., Lamb F. S. (2007) Circ. Res. 101, 663–671 [DOI] [PubMed] [Google Scholar]
- 17.Mumbengegwi D. R., Li Q., Li C., Bear C. E., Engelhardt J. F. (2008) Mol. Cell. Biol. 28, 3700–3712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lassègue B., Sorescu D., Szöcs K., Yin Q., Akers M., Zhang Y., Grant S. L., Lambeth J. D., Griendling K. K. (2001) Circ. Res. 88, 888–894 [DOI] [PubMed] [Google Scholar]
- 19.Kreuzer J., Viedt C., Brandes R. P., Seeger F., Rosenkranz A. S., Sauer H., Babich A., Nürnberg B., Kather H., Krieger-Brauer H. I. (2003) FASEB J. 17, 38–40 [DOI] [PubMed] [Google Scholar]
- 20.Li Q., Spencer N. Y., Oakley F. D., Buettner G. R., Engelhardt J. (2009) Antioxid. Redox Signal., 11, 1249–1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legler D. F., Micheau O., Doucey M. A., Tschopp J., Bron C. (2003) Immunity 18, 655–664 [DOI] [PubMed] [Google Scholar]
- 22.Vilhardt F., van Deurs B. (2004) EMBO J. 23, 739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shao D., Segal A. W., Dekker L. V. (2003) FEBS Lett. 550, 101–106 [DOI] [PubMed] [Google Scholar]
- 24.Pike L. J. (2004) Biochem. J. 378, 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Q., Sanlioglu S., Li S., Ritchie T., Oberley L., Engelhardt J. F. (2001) Antioxid. Redox Signal. 3, 415–432 [DOI] [PubMed] [Google Scholar]
- 26.Burns K., Clatworthy J., Martin L., Martinon F., Plumpton C., Maschera B., Lewis A., Ray K., Tschopp J., Volpe F. (2000) Nat. Cell Biol. 2, 346–351 [DOI] [PubMed] [Google Scholar]
- 27.Charvet C., Auberger P., Tartare-Deckert S., Bernard A., Deckert M. (2002) J. Biol. Chem. 277, 15376–15384 [DOI] [PubMed] [Google Scholar]
- 28.Han C. H., Nisimoto Y., Lee S. H., Kim E. T., Lambeth J. D. (2001) J. Biochem. 129, 513–520 [DOI] [PubMed] [Google Scholar]
- 29.Han C. H., Lee M. H. (2000) J. Vet. Sci. 1, 19–26 [PubMed] [Google Scholar]
- 30.Choudhury A., Marks D. L., Proctor K. M., Gould G. W., Pagano R. E. (2006) Nat. Cell Biol. 8, 317–328 [DOI] [PubMed] [Google Scholar]
- 31.Teixeira A., Chaverot N., Schröder C., Strosberg A. D., Couraud P. O., Cazaubon S. (1999) J. Neurochem. 72, 120–128 [DOI] [PubMed] [Google Scholar]
- 32.Leclerc P. C., Auger-Messier M., Lanctot P. M., Escher E., Leduc R., Guillemette G. (2002) Endocrinology 143, 4702–4710 [DOI] [PubMed] [Google Scholar]
- 33.Song K. S., Li S., Okamoto T., Quilliam L. A., Sargiacomo M., Lisanti M. P. (1996) J. Biol. Chem. 271, 9690–9697 [DOI] [PubMed] [Google Scholar]
- 34.Smart E. J., Anderson R. G. (2002) Methods Enzymol. 353, 131–139 [DOI] [PubMed] [Google Scholar]
- 35.Carter G. C., Bernstone L., Sangani D., Bee J. W., Harder T., James W. (2009) Virology 386, 192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ros-Baro A., Lopez-Iglesias C., Peiro S., Bellido D., Palacin M., Zorzano A., Camps M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12050–12055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doherty G. J., McMahon H. T. (2009) Annu. Rev. Biochem. 78, 857–902 [DOI] [PubMed] [Google Scholar]
- 38.Puri C., Tosoni D., Comai R., Rabellino A., Segat D., Caneva F., Luzzi P., Di Fiore P. P., Tacchetti C. (2005) Mol. Biol. Cell 16, 2704–2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Harder T., Scheiffele P., Verkade P., Simons K. (1998) J. Cell Biol. 141, 929–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakahira K., Kim H. P., Geng X. H., Nakao A., Wang X., Murase N., Drain P. F., Wang X., Sasidhar M., Nabel E. G., Takahashi T., Lukacs N. W., Ryter S. W., Morita K., Choi A. M. (2006) J. Exp. Med. 203, 2377–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lambeth J. D. (2004) Nat. Rev. Immunol. 4, 181–189 [DOI] [PubMed] [Google Scholar]
- 42.Roepstorff K., Rasmussen I., Sawada M., Cudre-Maroux C., Salmon P., Bokoch G., van Deurs B., Vilhardt F. (2008) J. Biol. Chem. 283, 7983–7993 [DOI] [PubMed] [Google Scholar]
- 43.Welch H. C., Coadwell W. J., Ellson C. D., Ferguson G. J., Andrews S. R., Erdjument-Bromage H., Tempst P., Hawkins P. T., Stephens L. R. (2002) Cell 108, 809–821 [DOI] [PubMed] [Google Scholar]
- 44.Park H. S., Lee S. H., Park D., Lee J. S., Ryu S. H., Lee W. J., Rhee S. G., Bae Y. S. (2004) Mol. Cell. Biol. 24, 4384–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zuo L., Ushio-Fukai M., Ikeda S., Hilenski L., Patrushev N., Alexander R. W. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 1824–1830 [DOI] [PubMed] [Google Scholar]
- 46.Mizrahi A., Molshanski-Mor S., Weinbaum C., Zheng Y., Hirshberg M., Pick E. (2005) J. Biol. Chem. 280, 3802–3811 [DOI] [PubMed] [Google Scholar]
- 47.Smith R. M., Harada S., Smith J. A., Zhang S., Jarett L. (1998) Cell. Signal. 10, 355–362 [DOI] [PubMed] [Google Scholar]
- 48.Villalba M., Bi K., Rodriguez F., Tanaka Y., Schoenberger S., Altman A. (2001) J. Cell Biol. 155, 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hebeis B., Vigorito E., Kovesdi D., Turner M. (2005) Blood 106, 635–640 [DOI] [PubMed] [Google Scholar]
- 50.Miletic A. V., Graham D. B., Montgrain V., Fujikawa K., Kloeppel T., Brim K., Weaver B., Schreiber R., Xavier R., Swat W. (2007) Blood 109, 3360–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brissoni B., Agostini L., Kropf M., Martinon F., Swoboda V., Lippens S., Everett H., Aebi N., Janssens S., Meylan E., Felberbaum-Corti M., Hirling H., Gruenberg J., Tschopp J., Burns K. (2006) Curr. Biol. 16, 2265–2270 [DOI] [PubMed] [Google Scholar]
- 52.Sverdlov M., Shajahan A. N., Minshall R. D. (2007) J. Cell. Mol. Med. 11, 1239–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang A. Y., Yi F., Zhang G., Gulbins E., Li P. L. (2006) Hypertension 47, 74–80 [DOI] [PubMed] [Google Scholar]