Abstract
Saccharomyces cerevisiae Rad52 protein promotes homologous recombination by nucleating the Rad51 recombinase onto replication protein A-coated single-stranded DNA strands and also by directly annealing such strands. We show that the purified rad52-R70A mutant protein, with a compromised amino-terminal DNA binding domain, is capable of Rad51 delivery to DNA but is deficient in DNA annealing. Results from chromatin immunoprecipitation experiments find that rad52-R70A associates with DNA double-strand breaks and promotes recruitment of Rad51 as efficiently as wild-type Rad52. Analysis of gene conversion intermediates reveals that rad52-R70A cells can mediate DNA strand invasion but are unable to complete the recombination event. These results provide evidence that DNA binding by the evolutionarily conserved amino terminus of Rad52 is needed for the capture of the second DNA end during homologous recombination.
INTRODUCTION
Homologous recombination (HR)2 represents an error-free pathway for the elimination of DNA breaks and DNA interstrand cross-links from chromosomes, and it also provides a means for the restart of stalled or damaged DNA replication forks. Moreover, HR plays a crucial role in the proper segregation of homologous chromosomes in meiosis I (for review, see Refs. 1 and 2). Accordingly, a deficiency in HR or its deregulation leads to meiotic aneuploidy, genomic instability, and the tumor phenotype (for review, see Refs. 1–3).
HR is often initiated via the formation of DNA double-strand breaks (DSBs). After DSB formation, the ends of the break are nucleolytically processed to yield single-stranded DNA (ssDNA) tails (for review, see Ref. 4). The ubiquitous single-strand DNA-binding protein RPA first decorates the ssDNA. RPA is subsequently replaced by the Rad51 recombinase to form a right-handed helical polymer, referred to as the presynaptic filament. This nucleoprotein complex conducts a search for a homologous DNA sequence and invades the latter to form a nascent DNA joint called the displacement loop, or D-loop (for review, see Ref. 5). After invasion, the 3′ end of the invading strand primes DNA synthesis, which serves to replace the genetic information lost during the end-processing reaction and to expand the D-loop.
The D-loop intermediate can be resolved by two mechanistically distinct pathways. In the synthesis-dependent single-strand annealing pathway, the invading DNA strand with its newly synthesized DNA extension is displaced from the information donor and subsequently annealed to the ssDNA tail that adjoins the second end of the break. The synthesis-dependent single-strand annealing mode of HR does not have the potential of generating crossover recombinants. In the DNA double-strand break repair pathway, the D-loop is maintained and recruits the second ssDNA tail by annealing, to assemble a double Holliday junction. The resolution of the double Holliday junction by a resolvase then yields a mixture of crossover and noncrossover recombinants (1).
The Rad52 protein of Saccharomyces cerevisiae is needed for two distinct stages of the HR process. As a recombination mediator, Rad52 helps nucleate Rad51 onto RPA-coated ssDNA to seed the assembly of the presynaptic filament (5). Later in the HR process Rad52 promotes the capture of the second DNA strand via its DNA annealing activity to help complete the synthesis-dependent single-strand annealing or double-strand break repair reaction (2). Biochemical studies have revealed two distinct DNA binding domains in ScRad52, located within the amino and carboxyl termini of the protein (6, 7). Rad52 orthologs from different species exhibit the highest degree of relatedness in their amino-terminal region (8–11). Because deletion of the carboxyl-terminal DNA binding domain has no perceptible effect on the generation of single-end-invasion intermediates in meiotic recombination (12), one can infer that it is dispensable for second DNA end capture.
In this study we have employed the rad52-R70A mutant, impaired in the amino-terminal DNA binding activity, to address the role of this activity in the known HR functions of S. cerevisiae Rad52. We provide biochemical evidence that this mutation does not affect Rad52 recombination mediator attribute but specifically impairs its ability to carry out the annealing of RPA-coated complementary DNA strands. Accordingly, rad52-R70A cells can initiate but fail to complete the mating type switching reaction that is triggered by an HO endonuclease-made DSB. These results, thus, identify the critical role fulfilled by the evolutionarily conserved amino-terminal DNA binding domain of Rad52 in HR.
EXPERIMENTAL PROCEDURES
Construction of Plasmids
Full-length His6-RAD52 harboring codons 34–504 of RAD52 was ligated into the NcoI site of the pET11d vector. His6-rad52–327Δ encompassing codons 34–327 of RAD52 was ligated into the BamHI and EcoRI sites of the pRSETc vector. The various mutants (i.e. R70A, Y96A) were constructed using the QuikChange XL site-directed mutagenesis kit (Stratagene). Rad59 was cloned into pMal vector, making use of the EcoRI and PstI sites, thus fusing the Rad59 protein to the maltose-binding protein (MBP).
Purification of His6-tagged Rad52, rad52–327Δ, and Mutant Variants
Plasmids encoding RAD52, rad52–327Δ, and the R70A and Y96A mutant variants were used to transform the Escherichia coli Rosetta strain, and protein expression was induced by 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 4 h. Full-length His6-tagged Rad52 and the R70A and Y96A mutants were purified to near homogeneity as described previously (13). For the purification of rad52–327Δ and the R70A and Y96A mutant derivatives, clarified lysate prepared from 20 g of cell paste (14) was mixed with 2.5 ml of nickel-nitrilotriacetic acid-agarose (Ni-NTA)(Qiagen) for 3 h at 4 °C. The beads were washed with 20 ml of buffer K (20 mm K2HPO4, pH 7.4, 1 mm 2-mercaptoethanol, 0.01% Igepal, and 10% glycerol) containing 1 m KCl and 10 mm imidazole followed by 20 ml of buffer K containing 300 mm KCl and 10 mm imidazole. The bound His6-rad52–327Δ or mutant was eluted with 5 ml of 200 mm imidazole in buffer K containing 300 mm KCl. The elution fraction was diluted with 3 volumes of buffer K and applied onto a Source S column (0.5 ml), which was developed with a 10-ml gradient from 100 to 600 mm KCl in buffer K. The fractions containing His6-rad52–327Δ or mutant were combined (1.5 ml total; 220 mm KCl) and applied onto a 0.5-ml macrohydroxyapatite (Bio-Rad) column that was eluted with a 10-ml gradient from 0 to 240 mm KH2PO4 in buffer K. The macrohydroxyapatite fractions (2.5 ml total; 120 mm KH2PO4) that contained the peak of rad52–327Δ or mutant were combined, dialyzed against buffer K containing 500 mm KCl for 2 h, concentrated to 4 mg/ml in an Amicon Ultra-4 concentrator (Millipore), and stored in small portions at −80 °C. The overall yield of highly purified rad52–327Δ or mutant was ∼0.5 mg.
Purification of MBP-Rad59
The plasmid that encodes MBP-Rad59 was used to transform the E. coli Rosetta strain, and protein expression was induced by 0.4 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 37 °C. For protein purification, clarified extract made from 20 g of cell paste in 80 ml of cell breakage buffer (50 mm Tris-HCl, pH 7.5, 10% sucrose, 10 mm EDTA, 300 mm KCl, and 1 mm 2-mercaptoethanol) containing protease inhibitors (phenylmethylsulfonyl fluoride and benzamidine hydrochloride at 1 mm and aprotinin, chymostatin, leupeptin, and pepstatin A at 5 μg/ml each) was mixed with 2 ml of amylose beads (New England Biolabs) for 2 h at 4 °C. The beads were washed with 40 ml of buffer K containing 500 mm KCl, and the bound MBP-Rad59 was eluted with 3 ml of 10 mm maltose in buffer K containing 300 mm KCl. The elution fractions containing the MBP-tagged protein were combined, concentrated to 1 ml in an Amicon Ultra-4 concentrator, and then fractionated in a 35-ml Sephacryl S300 column in K buffer containing 300 mm KCl, collecting 0.7-ml fractions at 0.1 ml/min. The fractions (2.8 ml total) that contained the peak of MBP-Rad59 were concentrated to 15 mg/ml and stored in small portions at −80 °C. The overall yield of highly purified MBP-Rad59 was 2.7 mg.
Purification of Other Proteins
Rad51 and RPA were expressed and purified as described previously (14).
DNA Substrates
The ϕX174 viral (+) strand was purchased from New England Biolabs, and the replicative form (about 90% supercoiled form and 10% nicked circular form) was from Invitrogen. Oligonucleotide 1 used in the ssDNA annealing and DNA binding experiments has the sequence 5′-AAATGAACATAAAGTAAATAAGTATAAGGATAATACAAAATAAGTAAATGAATAAACATAGAAAATAAAGTAAAGGATATAAA-3′. Oligonucleotide 2 is the exact complement of Oligonucleotide 1. These oligonucleotides were purified and labeled with [γ-32P]ATP and T4 polynucleotide kinase (15) for use in DNA binding and single-strand annealing experiments. The 90-mer and 22-mer oligonucleotides used in the ssDNA binding assays were composed of poly(dT) species (Integrated DNA Technologies).
Affinity Pulldown Assays
In the pulldown experiment in Fig. 3, purified His-tagged Rad52 (4 μm) was incubated with purified MBP-tagged Rad59 (4 μm) in 30 μl of buffer K with 150 mm KCl at 4 °C for 30 min before being mixed with 12.5 μl of amylose beads (New England Biolabs) for 30 min to capture MBP-Rad59 and any associated Rad52 or mutant. The beads were then washed twice with 200 μl of the same buffer, and the bound proteins were eluted with 30 μl of 3% SDS. The supernatant containing unbound proteins, the KCl wash, and the SDS eluates harboring the captured proteins, 7 μl each, were analyzed by 10% SDS-PAGE with Coomassie Blue staining. In the pulldown experiment in Fig. 5A, purified His-tagged Rad52 (4 μm) was incubated with purified Rad51 (4 μm) and as before mixed with 7.5 μl of Ni-NTA-agarose beads (Qiagen) for 30 min. The beads were then washed twice with 200 μl of the same buffer, and the bound proteins were eluted with 30 μl of 3% SDS. The various fractions, 7 μl each, were analyzed as above.
FIGURE 3.
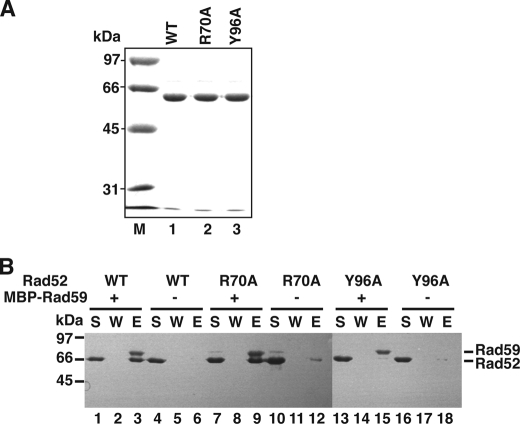
Interaction of mutant rad52 proteins with Rad59. A, full-length Rad52, rad52-R70A, and rad52-Y96A proteins, 2 μg each, were analyzed by SDS-PAGE with Coomassie Blue staining. B, in this affinity pulldown assay, purified full-length Rad52 (wild-type (WT)), rad52-R70A, or rad52-Y96A were incubated with MBP-tagged Rad59 before being mixed with amylose beads to capture MBP-Rad59 and any associated Rad52. The various supernatants (S) containing unbound proteins, the wash (W), and the SDS eluates (E) harboring the captured proteins were analyzed by SDS-PAGE with Coomassie Blue staining. We note that the faint Rad59 band in lane 10 of B likely stemmed from a slight spillover of the sample in lane 9 while loading the gel.
FIGURE 5.
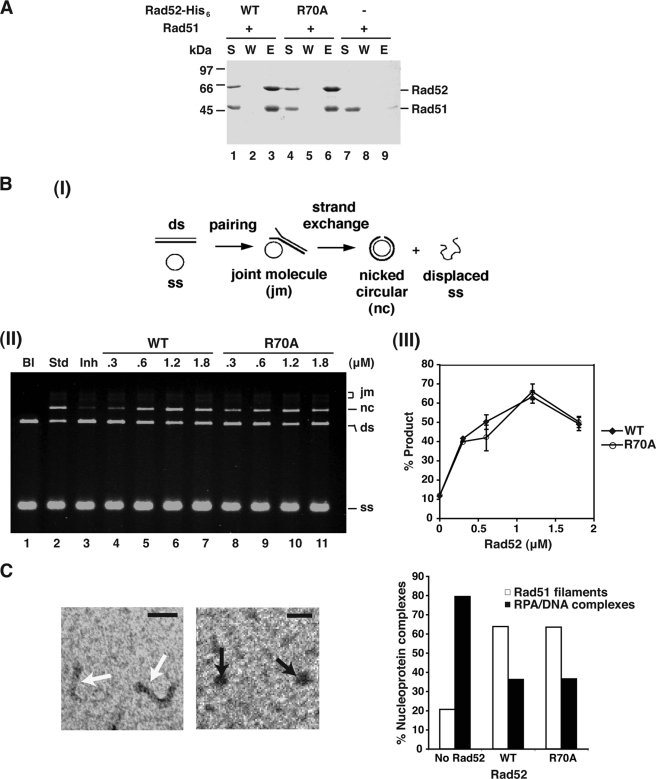
rad52-R70A interacts with Rad51 and possesses recombination mediator activity. A, purified full-length His6-tagged Rad52 (WT) or rad52-R70A were incubated with Rad51 before Ni-NTA beads were added to capture protein complexes. The beads were washed with buffer and treated with SDS to elute bound Rad52 or rad52-R70A and associated Rad51. The supernatant containing unbound Rad52 or rad52-R70A (S), the wash (W), and the SDS eluate (E) were analyzed by 10% SDS-PAGE followed by staining with Coomassie Blue. B, schematic of the DNA strand exchange assay is shown in panel I. The standard reaction (Std; lane 2) involved preincubating the ssDNA with Rad51 to allow for presynaptic filament assembly before RPA addition. Co-incubating the ssDNA with Rad51 and RPA resulted in severe inhibition of the DNA strand exchange reaction (Inh; lane 3). The inclusion of the indicated amounts of Rad52 (lanes 4–7) or rad52-R70A (lanes 8–11) during the incubation of ssDNA with Rad51 and RPA led to DNA strand exchange restoration. The plot of products sum of nc and jm) is included. C, in the electron microscopy analysis, the proportion of Rad51 presynaptic filaments (white arrows; left panel) versus RPA-ssDNA nucleoprotein complexes (black arrows; middle panel) among a population of 400 randomly picked nucleoprotein complexes was determined without or with Rad52 or rad52-R70A (right panel). The scale bar represents 50 nm.
DNA Binding Assay
Varying amounts of the indicated rad52–327Δ or Rad52 species were incubated with 32P-labeled Oligonucleotide 1 (1.36 μm nucleotides) or dsDNA (1.36 μm base pairs, obtained by hybridizing Oligonucleotide 1 to Oligonucleotide 2) or a specified length of poly(dT) oligonucleotide (1.36 μm nucleotides) at 37 °C in 10 μl of buffer D (40 mm Tris-HCl, pH 7.8, 50 mm KCl, 1 mm dithiothreitol, and 100 μg/ml bovine serum albumin) for 10 min. After the addition of gel loading buffer (50% glycerol, 20 mm Tris-HCl, pH 7.4, 0.5 mm EDTA, 0.05% orange G), the reaction mixtures were resolved in 12% native polyacrylamide gels in TAE buffer (40 mm Tris-HCl, pH 7.4, 0.5 mm EDTA) at 4 °C. The gels were dried, and the DNA species were quantified using Quantity One software in the Personal Molecular Imager FX (Bio-Rad). To show that the DNA substrate remained intact, we treated a reaction mixture containing the highest concentration of the rad52–327Δ or Rad52 species with 0.5% SDS and 0.5 mg/ml proteinase K at 37 °C for 5 min before the analysis.
Single-stranded DNA Annealing Assays
Oligonucleotide 1 (3.6 μm nucleotides) and radiolabeled Oligonucleotide 2 (3.6 μm nucleotides) were incubated in separate tubes at 37 °C for 2 min in the absence or presence of RPA (0.6 μm) in 24 μl of buffer D. Rad52 or rad52-R70A (1.2 μm) was added in 2 μl to the tube containing Oligonucleotide 1 and then mixed with Oligonucleotide 2. The completed reactions (50 μl) were incubated at 25 °C, and at the indicated times 9 μl of the annealing reactions was removed and treated with 0.5% SDS, 0.5 mg/ml proteinase K, and an excess of unlabeled Oligonucleotide 2 (20 μm) at 25 °C for 5 min in a total volume of 15 μl. The various samples (6 μl) were resolved in 12% native polyacrylamide gels run in TAE buffer. DNA annealing was quantified as the portion of the 32P-labeled Oligonucleotide 2 that had been converted into the double-stranded form.
DNA Strand Exchange Reaction
In the standard reaction, Rad51 (10 μm) was incubated with ssDNA (30 μm nucleotides) in 10 μl of buffer R (35 mm potassium-MOPS, pH 7.2, 1 mm dithiothreitol, 50 mm KCl, 2.5 mm ATP, and 3 mm MgCl2) for 5 min at 37 °C. After the addition of RPA (1.2 μm) in 0.5 μl, the reaction mixtures were incubated at 37 °C for another 5 min before the incorporation of linear double-stranded DNA (30 μm nucleotides) in 1 μl followed by 1 μl of 50 mm spermidine hydrochloride. After 120 min of incubation at 37 °C, the reaction was terminated by adding an equal volume of 1% SDS containing 1 mg/ml proteinase K followed by a 10-min incubation at 37 °C. The deproteinized samples (12 μl) were run on 0.9% agarose gels in TAE buffer, stained with ethidium bromide for 30 min, and then destained for at least 4 h in a large volume of water. Images were recorded and analyzed in a Bio-Rad Quantity One software system. To examine recombination mediator activity, reaction mixtures containing Rad51 and RPA with or without the indicated amounts of Rad52 or rad52-R70A were incubated on ice for 10 min before ϕX ssDNA was added. After a 10-min incubation at 37 °C, linear double-stranded DNA and spermidine were incorporated. The reaction mixtures were incubated and analyzed as above.
Electron Microscopy
Electron microscopy was conducted as described (16). Briefly, reaction mixtures containing a 150-mer oligonucleotide (7.2 μm nucleotides), Rad51 (2.4 μm), and RPA (280 nm) in 12 μl of buffer (35 mm potassium-MOPS, pH 7.2, 2.5 mm ATP, 3 mm MgCl2, and 50 mm KCl) were incubated in the absence or presence of Rad52 or rad52-R70A (280 nm) for 10 min at 37 °C. After a 10-fold dilution with buffer, a 3-μl aliquot of the reaction was applied to a 400-mesh grid coated with carbon film that had been glow-discharged in air. The grid was stained with 2% uranyl acetate for 30 s and rinsed with water. The samples were examined with a Tecnai 12 Biotwin electron microscope (FEI) equipped with a tungsten filament at 100 keV. Digital images were captured with a Morada (Olympus Soft Imaging Solutions) charge-coupled device camera at a nominal magnification of 87,000–135,000×.
Chromatin Immunoprecipitation
JKM179 strain (ho hml::ADE1 MATα hmr::ADE1 ade1–110 leu2,3-112 lys5 trp1::hisG ura3–52 ade3::GAL10::HO) (17, 18) harboring a RAD52 deletion was transformed with the centromeric vector pRS416, pRS416 containing the wild-type RAD52 gene (a kind gift from Lorraine Symington), or pRS416 containing rad52-R70A. All these yeast strains harbor the HO endonuclease gene under the control of the GAL10 promoter, which permits the galactose induction of HO expression. Chromatin immunoprecipitation (ChIP) was carried out as described (19). Briefly, cells were grown in yeast extract peptone supplemented with 3% glycerol for 6 h until the mid-log phase, and 2% galactose was added to induce HO expression. At the designated times the cells were harvested from 45 ml of the cultures, cross-linked with 1% formaldehyde for 20 min, and then quenched with 125 mm glycine for 5 min. After lysing the cells with glass beads and sonication to shear the chromatin, the cell lysate was incubated with anti-Rad51 antibodies (our laboratory stock) or anti-Rad52 antibodies (our laboratory stock) and Dynabeads protein G (Invitrogen). After a series of washes, the immunoprecipitates were incubated at 65 °C to reverse the protein-DNA cross-link. Radioactive semiquantitative PCR was performed to amplify the DNA. The primers used for amplifying PHO5 were 5′-AAATTAGCACGTTTTCGCA-3′ and 5′-GGTAATCTCGAATTTGCTTGCTC-3′, and those for MAT Z were 5′-TCCCCATCGTCTTGCTCT-3′ and 5′-GCATGGGCAGTTTACCTTTAC-3′ (17, 20). The PCR products were resolved on 10% native polyacrylamide gels and quantified by phosphorimaging (Bio-Rad) using the Quantity One software (Bio-Rad).
Molecular Analysis of Mating Type Switching
Yeast media containing twice the amount of leucine were prepared as described previously (21). SC−Ura−lactate medium was prepared as described by Sugawara and Haber (19). Galactose (20%) containing less than 0.01% glucose was prepared at room temperature and filter-sterilized. The extraction buffer comprised 2% SDS, 50 mm EDTA, and 100 mm Tris, pH 7.5. The strains used, W1588-4C (MATa ade2-1 can1–100 his3–11,15 leu2-3,112 trp1 ura3-1 RAD5 RAD52) (22), J788 (MATa ade2-1 can1–100 his3–11,15 leu2–3,112 trp1 ura3-1 RAD5 rad52-R70A) (23), and UM270–14C (MATa ade2-1 can1–100 his3–11,15 leu2-3,112 trp1 ura3-1 RAD5 rad52::HIS5) (this study), are derivatives of W303 (24) except that they are RAD5. Plasmid pJH132, kindly provided by James Haber, is a CEN4 ARS1-based vector containing a GAL10::HO fusion and a URA3 marker for selection.
Detection of mating type-switching intermediates and products by PCR was essentially performed as described in Sugawara and Haber (19). Strains were transformed with pJH132 and grown in liquid SC−Ura−lactate for 2 days at 30 °C with vigorous aeration to a final cell density of 3 × 106 to 1 × 107 cells/ml. To induce expression of the HO, galactose was added as a 20% (w/v) solution to a final concentration of 2%. After induction, HO expression was maintained for the remaining time of the experiment. At the indicated time points, samples of 50 ml were collected, and media were removed by centrifugation for 5 min at 4 °C. The cells were resuspended in 400 μl of extraction buffer, frozen in liquid nitrogen, and stored at −80 °C. Cells were thawed on ice, and genomic DNA was extracted as described in Sugawara and Haber (19). PCR analysis of individual samples was performed as follows. The activity of the HO endonuclease was assessed by using the primer set (pB and pD), which amplifies a fragment that spans the HO-cut site at the MAT locus; the strand invasion intermediate and the gene conversion product were detected by using the primer sets (pA and pB) and (pC and pD), respectively (19). A final primer set, LeuBst_fwd1 (CGGAACCGGCTTTTCATATAGA) and LeuEcoRI_R (TTTAACTGCATCTTCAATGGCCT), was used to amplify an 1187-bp section of leu2–3,112, to use as an internal standard to allow comparison between samples obtained at different time points and from different strains. PCR quantification was performed three times for each sample.
RESULTS
The rad52-R70A and Y96A Mutations Affect DNA Binding
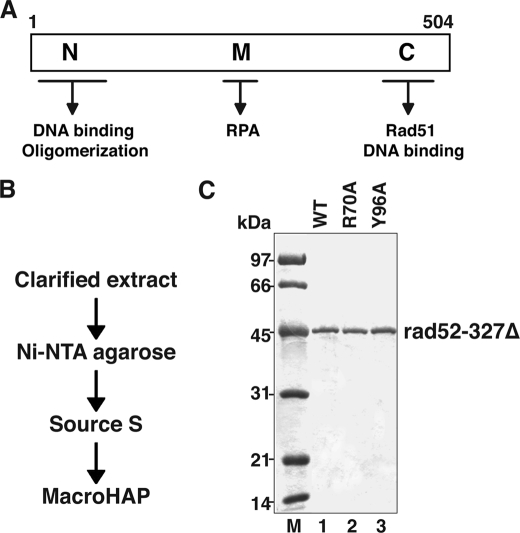
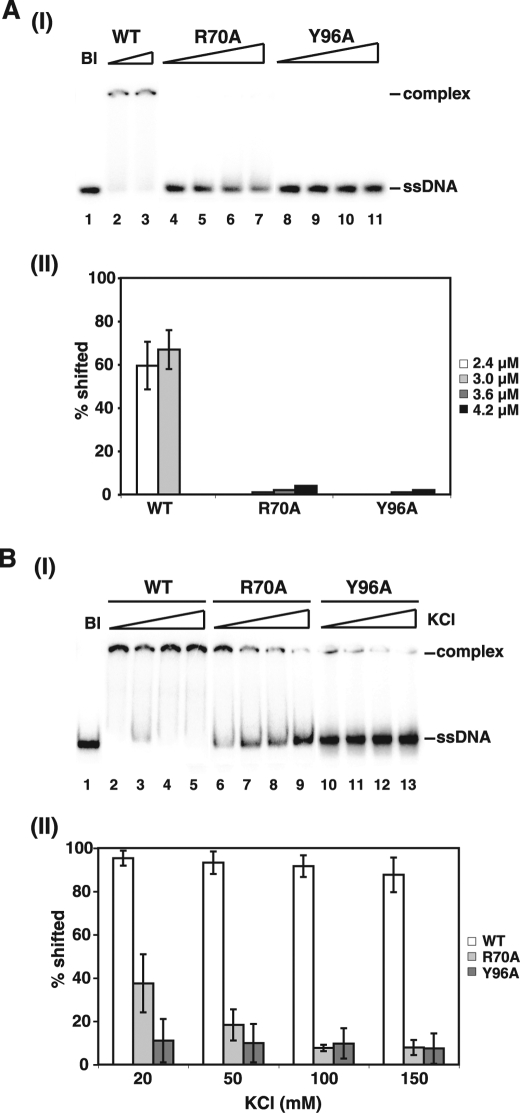
To assess the role of the amino-terminal DNA binding domain in the Rad52 functional attributes, we focused on two mutants, R70A and Y96A, which were generated in a previous study (25) and predicted to be affected for DNA binding based on a biochemical study done with variants of human Rad52 mutated for the equivalent residues (26). Because of the fact that ScRad52 possesses an additional DNA binding domain in its carboxyl terminus (7) (Fig. 1A), we first introduced the aforementioned mutations into a truncated form of ScRad52-rad52–327Δ that lacks the carboxyl-terminal DNA binding region. To facilitate protein purification, a His6 affinity tag was engineered into the amino terminus of rad52–327Δ and the corresponding R70A and Y96A mutant variants. All three proteins were then purified to near homogeneity (Fig. 1, B and C). The ability of the Rad52 species to bind ssDNA was tested in a gel-shift assay employing a radiolabeled 22-mer oligo(dT) as the DNA substrate and a low concentration (20 mm) of KCl in the incubation buffer. Using this assay, we found that neither the rad52–327Δ,R70A nor the rad52–327Δ,Y96A mutant protein had appreciable DNA binding activity (Fig. 2A). Interestingly, significant binding of a 90-mer oligo(dT) substrate by rad52–327Δ,R70A occurred at 20 mm KCl, but unlike rad52–327Δ, this DNA binding activity of the mutant protein is highly sensitive to the level of KCl in the reaction buffer (Fig. 2B). Little binding of this longer DNA substrate was seen with the Y96A mutant protein (Fig. 2B). Thus, the R70A mutation confers a salt-sensitive DNA binding deficiency, whereas the Y96A impairs the DNA binding attribute of the amino-terminal domain of Rad52.
FIGURE 1.
Purification of rad52–327Δ and mutants. A, functional domains in the S. cerevisiae Rad52 protein are shown. B, shown is the purification scheme for amino-terminal His6-tagged rad52–327Δ or variants that harbor the R70A or Y96A mutation. C, purified rad52–327Δ and the R70A and Y96A mutants, 1 μg each, were analyzed by 10% SDS-PAGE and Coomassie Blue staining.
FIGURE 2.
DNA binding ability of rad52–327Δ and mutant variants assessed with oligonucleotide substrates. DNA binding by His6-tagged rad52–327Δ or variants that harbor the R70A or Y96A mutation was evaluated by mobility shift of either radiolabeled 22-mer oligo(dT) (A) or 90-mer oligo(dT) (B). The series of experiments in A were done at 20 mm KCl, whereas those in B were done with different KCl concentrations (20, 50, 100, and 150 mm). A no-protein blank (Bl) was included. The protein concentrations were 2.4, 3.0, 3.6, and 4.2 μm in A and 1 μm in B. The results were plotted in panel II of A and B. We note that the slight smearing of the ssDNA band in lanes 4–7 in A probably stemmed from an unstable rad52-R70A-ssDNA nucleoprotein complex that had dissociated rapidly.
The rad52-Y96A but Not R70A Mutation Affects Complex Formation with Rad59
The amino-terminal region of Rad52 is oligomeric (27) and known to interact with the HR protein Rad59 (28). By gel filtration analysis alongside purified rad52–327Δ, we have verified that both rad52–327Δ,R70A and rad52–327Δ,Y96A retain the native oligomeric structure (data not shown). To further understand the role of the amino-terminal DNA binding domain in Rad52-dependent functions, we introduced the R70A or Y96A point mutation into full-length amino-terminal His6-tagged Rad52 and purified it alongside the wild-type protein to near homogeneity (Fig. 3A). Making use of the MBP tag on the Rad59 protein in an affinity pulldown assay, we have found that, although the rad52-R70A mutant associates with Rad59 just as avidly as the wild-type counterpart, the rad52-Y96A variant is impaired in this interaction (Fig. 3B). For this reason further experimentation was conducted with the R70A mutant solely.
The rad52-R70A Mutation Has No Effect on Recombination Mediator Activity
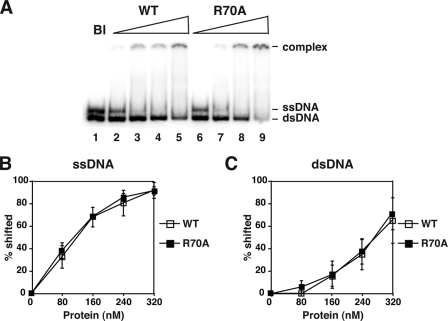
Interestingly, even though the rad52–327Δ,R70A mutant protein exhibits a salt-sensitive DNA binding deficiency (Fig. 2B), the full-length rad52-R70A mutant protein possesses an affinity for both ssDNA and dsDNA comparable with that of wild-type Rad52, even at 150 mm KCl (Fig. 4, B and C). Like the wild-type protein, the rad52-R70A mutant shows a strong preference for ssDNA over dsDNA (Fig. 4, B and C) (6, 7). Overall, the results provide evidence that the amino-terminal DNA binding deficiency engendered by the R70A mutation is effectively masked by the carboxyl-terminal DNA binding activity.
FIGURE 4.
DNA binding by Rad52 and the rad52-R70A mutant. A, the mixture of radiolabeled 80-mer dsDNA and 80-mer ssDNA substrates was incubated with increasing concentrations (80, 160, 240, 320 nm) of full-length Rad52 or rad52-R70A protein and analyzed. A no-protein blank (Bl) was included. B and C, graphical representation of the results.
Because the Rad51 interaction domain lies within the carboxyl-terminal region of Rad52 (29–31), we expected the rad52-R70A mutant protein to retain the ability to interact with Rad51. In congruence with this, results from an affinity pulldown experiment exploiting the His6 tags on the two Rad52 protein species demonstrate that rad52-R70A is just as adept as the wild-type protein in Rad51 interaction (Fig. 5A). We next used a well established DNA strand exchange system that involves plasmid DNA substrates (Fig. 5B, panel I) to ask whether the R70A mutation has a negative impact on the recombination mediator attribute of Rad52. In this system, when Rad51 is preincubated with ssDNA to assemble the presynaptic filament before RPA and the homologous linear duplex are added, strand invasion products (DNA joint molecules and nicked circular duplex) are readily formed (Fig. 5B, panel II). However, if Rad51 and RPA are co-incubated with ssDNA followed by the addition of duplex DNA, a much lower level of products is formed because of the competition of RPA for ssDNA binding. The latter reaction protocol allows for testing of the Rad52 recombination mediator activity. As expected, Rad52 restored product formation to the uninhibited level (Fig. 5B, panels II and III). Interestingly, the results showed that the rad52-R70A mutant is just as capable as the wild-type counterpart in restoring DNA strand exchange efficiency (Fig. 5B, panels II and III).
We also used electron microscopy to further characterize the recombination mediator activity of rad52-R70A. Herein, the ability of Rad52 or rad52-R70A to promote the assembly of Rad51 presynaptic filaments on a 150-mer oligonucleotide was compared and quantified by electron microscopy. Specifically, we randomly picked 400 nucleoprotein complexes from each of the reactions that contained Rad51, RPA, and DNA and without or with either Rad52 or rad52-R70A and then determined the relative distributions of Rad51 presynaptic filaments and RPA-ssDNA complexes within these populations. The results from our analyses provided clear validation for the premise that rad52-R70A possesses the wild-type level of recombination mediator activity (Fig. 5C).
rad52-R70A Is Proficient in the DSB Recruitment of Rad51 in Vivo
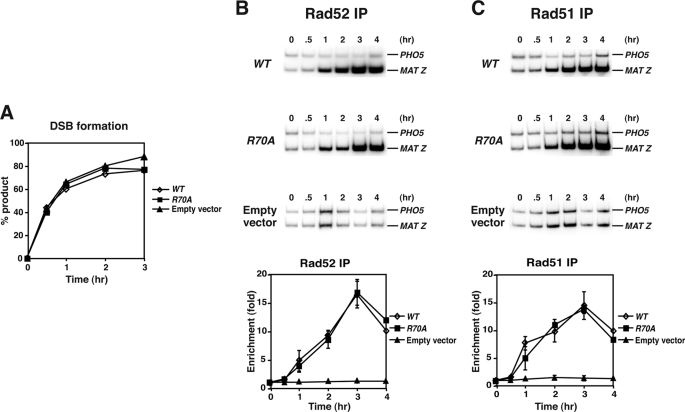
To obtain in vivo evidence for the proficiency of rad52-R70A as recombination mediator, we employed ChIP to examine the recruitment of Rad51 protein to the HO endonuclease-induced DSB at the MAT locus on chromosome 3 in rad52Δ yeast cells transformed with a centromeric RAD52 or rad52-R70A plasmid or the empty vector. The parent yeast strain used in the ChIP experiments has the HO gene under the control of the galactose-inducible GAL promoter, and it cannot repair the HO-induced break by HR because of the lack of a homologous donor sequence (17). The DSB recruitment of HR proteins was quantified by radioactive PCR amplification of the MAT Z sequence and the internal control PHO5 after immunoprecipitation of cross-linked nucleoprotein complexes with the appropriate affinity-purified antibodies (7, 20).
As expected, there was no difference in the kinetics of HO break induction in all the plasmid-bearing yeast strains (Fig. 6A). The ChIP results showed that the rad52-R70A mutant protein is recruited just as rapidly and to the same extent as Rad52 to the HO-induced break (Fig. 6B), and importantly, the rad52-R70A mutant strain is just as adept as the wild type in recruiting Rad51 to the DSB (Fig. 6C). Thus, the ChIP analysis provides good complementary evidence that the amino-terminal DNA binding domain of Rad52 is not critical for its recombination mediator function.
FIGURE 6.
DSB recruitment of Rad51 occurs normally in rad52-R70A cells. A, shown are the kinetics of DSB induction. Donorless rad52Δ cells harboring GAL10-HO and also the empty pRS416 vector or pRS416 plasmid expressing wild-type RAD52 (WT) or rad52-R70A were grown in the presence of galactose to induce HO expression and a DSB at MAT. The induction of the HO break was quantified by PCR. B and C, recruitment of Rad52 or rad52-R70A (B) and Rad51 (C) to the HO-induced break in rad52Δ cells harboring the pRS416 vector or pRS416 expressing RAD52 or rad52-R70A is shown. The MAT-Z target sequence and control PHO5 sequence were amplified by radioactive PCR, and the results were quantified and graphed.
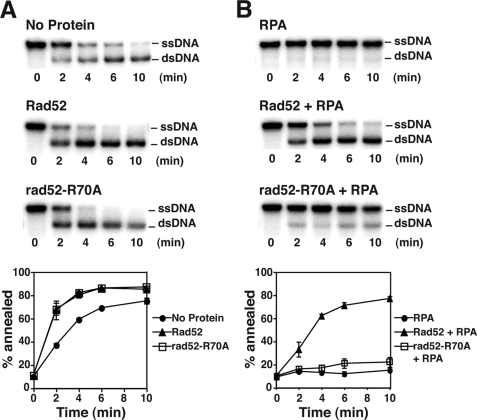
Annealing of RPA-coated DNA Strands Is Impaired by the R70A Mutation
A previous study by Lettier et al. (23) has provided evidence for a deficiency of rad52-R70A cells in performing the deletion type of single-strand annealing reaction between direct repeats. Being guided by this genetic study, we carried out single-strand DNA annealing reactions with Rad52 or rad52-R70A using complementary DNA oligonucleotide substrates that were either naked or precoated with RPA. The results showed that although rad52-R70A anneals naked DNA strands just as efficiently as wild-type Rad52 (Fig. 7A), it is notably deficient in the annealing of RPA-coated DNA strands (Fig. 7B). Thus, within the context of the full-length protein, compromising the Rad52 amino-terminal DNA binding activity adversely affects the ability of Rad52 to anneal RPA-coated ssDNA strands.
FIGURE 7.
rad52-R70A is defective in the annealing of RPA-coated ssDNA strands. A, the ability of either Rad52 or rad52-R70A to anneal a radiolabeled 83-mer oligonucleotide to its complement was tested. B, the annealing of RPA-coated DNA strands was tested. The background rate of DNA annealing was also determined in both cases. The results were quantified and graphed.
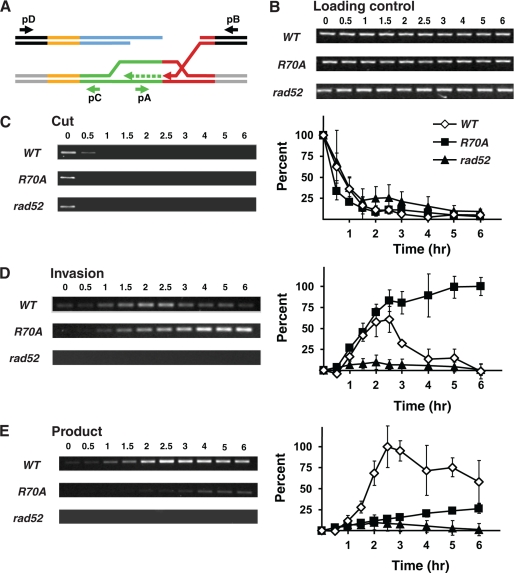
Completion of Gene Conversion Is Impaired in rad52-R70A Cells
The biochemical analyses described above provided evidence for an involvement of the Rad52 amino-terminal DNA binding activity in the annealing of RPA-coated DNA strands. HO-induced mating type switching is impaired in rad52-R70A cells compared with wild-type cells (23). Accordingly, we employed the mating type switching reaction as a model to further characterize rad52-R70A strains so as to pinpoint the specific defect during the gene conversion process engendered by this rad52 mutation. Specifically, we employed PCR-based analyses to monitor the progression of events after the formation of a DSB at the mating type locus, MAT, focusing on the kinetics of DNA strand invasion and of product formation during the switching of mating type by gene conversion of MAT using the information donors at HML and HMR (19, 32). This analysis reveals that the kinetics of formation of the PCR product detecting the strand invasion step is similar in the rad52-R70A and wild-type strain (Fig. 8D). In contrast, product formation in the rad52-R70A mutant is clearly compromised, and the timing for completion of the residual gene conversion is delayed as compared with the wild-type control (Fig. 8E). Because the HO endonuclease was present during the entire analysis after its initial induction, the strand invasion product in the wild-type strain decreases after 2.5 h, reflecting that the first round of mating type switching is completed, and a new one is initiated as the HO endonuclease cuts the switched MAT locus again. Importantly, in rad52-R70A cells, the strand invasion product accumulates and does not turn over as the mutant strain only inefficiently completes mating type switching.
FIGURE 8.
The final step of gene conversion is compromised in rad52-R70A cells. A, shown is a diagram showing strand invasion during mating type switching from MATa to MATα. Strands in black and gray are specific to MAT and HMLα loci, respectively. Red and orange strands indicate homologous MAT and HMLα strands (Z and WX, respectively). Blue strands indicate MATa specific sequences (Ya), and green strands represent HMLα specific sequences (Yα) interchanged during switching. Primers pB and pD bind sequences specific to the MAT locus and primers pA and pD bind sequences specific to HMLα (and MATα); see Refs. 19 and 32 for more detailed figures of primer positions. Genomic DNA samples from WT, rad52-R70A, and rad52Δ strains were collected before and after induction of the HO endonuclease at the indicated time points. B, DNA samples were PCR-analyzed for the presence of a leu2 fragment serving as internal loading control. C, PCR using primers pB and pD amplifies a section of the MAT locus containing the HO cut site (left panel). A quantitative representation of the data is shown (right panel). Error bars are based on the results of three independent PCR analyses. For each strain the band intensity at the zero time point (before induction) was set to 100%. D, PCR detection of the strand invasion intermediate during mating type switching using primers pA and pD (left panel). A quantitative representation of the data is shown (right panel). All samples were normalized to the intensity of the PCR band, set to 100% obtained, by the rad52-R70A strain at the 6-h time point. Curve details are as described in C. E, PCR detection of the gene conversion product using primers pC and pD (left panel). A quantitative representation of data is shown (right panel). All samples were normalized to the intensity of the PCR band, set to 100%, obtained by the wild-type strain at the 2.5-h time point.
Together, the above results provide further corroborative biological evidence that the amino-terminal DNA binding defect in Rad52 does not affect the initial stages of the HR reaction but, rather, impairs completion of the reaction because of a defect in the ability of Rad52 to anneal RPA-coated ssDNA strands.
DISCUSSION
In the Rad51-dependent recombination reaction, Rad52 plays an important role at two distinct stages of the process. Its early role at the presynaptic stage is to promote the assembly of the Rad51 presynaptic filament by mediating removal of RPA from the ssDNA tails that results from the resection of the ends of a DSB. Thus, Rad52 facilitates Rad51-catalyzed DNA strand invasion to form the D-loop intermediate in this manner. This recombination mediator or presynaptic role of Rad52 has been well characterized and studied. Fewer mechanistic details are available regarding the postsynaptic role of Rad52, wherein it has been postulated to mediate the annealing of the displaced DNA strand to the other end of the DSB (33, 34). Moreover, Rad52 is also involved in the single-strand annealing pathway of HR that is independent of Rad51.
Our findings that the R70A mutation impairs the amino-terminal DNA binding activity of Rad52 and its ability to anneal RPA-coated ssDNA strands have allowed us to address the role of the conserved DNA annealing activity in HR. In concordance with this observation is the fact that rad52-R70A cells are deficient in the single-strand annealing reaction (22). Previously, cytological analysis has determined that the rad52-R70A mutant protein associates with DSBs normally and can efficiently promote the DSB recruitment of Rad51 as well (23). Importantly, through a variety of in vitro studies using purified proteins and also chromatin immunoprecipitation, we have furnished evidence that the recombination mediator function of Rad52 is intact in the rad52-R70A mutant, suggesting that the DNA annealing activity present in the Rad52 amino terminus is required at a later step in DSB repair. In support of this view, in vivo studies monitoring a DSB-induced, site-specific gene conversion event at MAT by PCR analyses have established that rad52-R70A cells are just as capable as wild-type cells in catalyzing DNA strand invasion but that the completion of the HR event is greatly reduced in the mutant. Knowing that the rad52-R70A mutant protein is impaired for its ability to anneal RPA-coated single DNA strands, we attribute the gene conversion defect seen in our in vivo studies to an inability of mutant cells to anneal the DNA strand liberated from the D-loop structure with the ssDNA tail associated with the second DNA end.
Interestingly, the rad52-R70A mutant was originally identified as a mutant that exhibits sensitivity to γ-irradiation without affecting spontaneous interchromosomal heteroallelic HR (25). Accordingly, our results suggest that spontaneous HR depends on DNA strand invasion but that the DNA strand annealing activity of Rad52 is actually dispensable in this regard.
In summary, the studies reported herein have provided insights into the biochemical and biological roles of the amino-terminal DNA binding domain of the S. cerevisiae Rad52 protein in homologous recombination. Our results should be useful toward delineating the role of the highly conserved amino-terminal domain of Rad52 orthologs in recombination processes in other eukaryotes.
Acknowledgment
We thank Martin Engelhard Kornholt for excellent technical help.
This work was supported, in whole or in part, by National Institutes of Health Grants PO1CA92584, ES07061, ES10252, and GM50237. This work was also supported by the Danish Research Council for Technology and Production Sciences (to U. M.) and the Technical University of Denmark (to S. C. L. H.).
- HR
- homologous recombination
- DSB
- double-strand break
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA
- RPA
- replication protein A
- Sc
- S. cerevisiae
- MBP
- maltose-binding protein
- MOPS
- 3-(N-morpholino) propanesulfonic acid
- Ni-NTA
- nickel-nitrilotriacetic acid
- ChIP
- chromatin immunoprecipitation
- D-loop
- displacement loop
- WT
- wild type.
REFERENCES
- 1.Symington L. S. (2002) Microbiol. Mol. Biol. Rev. 66, 630–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sung P., Klein H. (2006) Nat. Rev. Mol. Cell Biol. 7, 739–750 [DOI] [PubMed] [Google Scholar]
- 3.Pierce A. J., Stark J. M., Araujo F. D., Moynahan M. E., Berwick M., Jasin M. (2001) Trends Cell Biol. 11, S52–S59 [DOI] [PubMed] [Google Scholar]
- 4.Raynard S., Niu H., Sung P. (2008) Genes Dev. 22, 2903–2907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.San Filippo J., Sung P., Klein H. (2008) Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 6.Mortensen U. H., Bendixen C., Sunjevaric I., Rothstein R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 10729–10734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seong C., Sehorn M. G., Plate I., Shi I., Song B., Chi P., Mortensen U., Sung P., Krejci L. (2008) J. Biol. Chem. 283, 12166–12174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muris D. F., Bezzubova O., Buerstedde J. M., Vreeken K., Balajee A. S., Osgood C. J., Troelstra C., Hoeijmakers J. H., Ostermann K., Schmidt H. (1994) Mutat. Res. 315, 295–305 [DOI] [PubMed] [Google Scholar]
- 9.Bezzubova O. Y., Schmidt H., Ostermann K., Heyer W. D., Buerstedde J. M. (1993) Nucleic Acids Res. 21, 5945–5949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostermann K., Lorentz A., Schmidt H. (1993) Nucleic Acids Res. 21, 5940–5944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bendixen C., Sunjevaric I., Bauchwitz R., Rothstein R. (1994) Genomics 23, 300–303 [DOI] [PubMed] [Google Scholar]
- 12.Lao J. P., Oh S. D., Shinohara M., Shinohara A., Hunter N. (2008) Mol. Cell 29, 517–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song B., Sung P. (2000) J. Biol. Chem. 275, 15895–15904 [DOI] [PubMed] [Google Scholar]
- 14.Van Komen S., Macris M., Sehorn M. G., Sung P. (2006) Methods Enzymol. 408, 445–463 [DOI] [PubMed] [Google Scholar]
- 15.Trujillo K. M., Sung P. (2001) J. Biol. Chem. 276, 35458–35464 [DOI] [PubMed] [Google Scholar]
- 16.Seong C., Colavito S., Kwon Y., Sung P., Krejci L. (2009) J. Biol. Chem. 284, 24363–24371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugawara N., Wang X., Haber J. E. (2003) Mol. Cell 12, 209–219 [DOI] [PubMed] [Google Scholar]
- 18.Moore J. K., Haber J. E. (1996) Mol. Cell. Biol. 16, 2164–2173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugawara N., Haber J. E. (2006) Methods Enzymol. 408, 416–429 [DOI] [PubMed] [Google Scholar]
- 20.Wolner B., van Komen S., Sung P., Peterson C. L. (2003) Mol. Cell 12, 221–232 [DOI] [PubMed] [Google Scholar]
- 21.Sherman F. (1991) Getting Started with Yeast, pp. 1–21, Academic Press, San Diego, CA [Google Scholar]
- 22.Zhao X., Muller E. G., Rothstein R. (1998) Mol. Cell 2, 329–340 [DOI] [PubMed] [Google Scholar]
- 23.Lettier G., Feng Q., de Mayolo A. A., Erdeniz N., Reid R. J., Lisby M., Mortensen U. H., Rothstein R. (2006) PLoS Genet. 2, e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas B. J., Rothstein R. (1989) Cell 56, 619–630 [DOI] [PubMed] [Google Scholar]
- 25.Mortensen U. H., Erdeniz N., Feng Q., Rothstein R. (2002) Genetics 161, 549–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kagawa W., Kurumizaka H., Ishitani R., Fukai S., Nureki O., Shibata T., Yokoyama S. (2002) Mol. Cell 10, 359–371 [DOI] [PubMed] [Google Scholar]
- 27.Shinohara A., Shinohara M., Ohta T., Matsuda S., Ogawa T. (1998) Genes Cells 3, 145–156 [DOI] [PubMed] [Google Scholar]
- 28.Cortés-Ledesma F., Malagón F., Aguilera A. (2004) Genetics 168, 553–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krejci L., Song B., Bussen W., Rothstein R., Mortensen U. H., Sung P. (2002) J. Biol. Chem. 277, 40132–40141 [DOI] [PubMed] [Google Scholar]
- 30.Milne G. T., Weaver D. T. (1993) Genes Dev. 7, 1755–1765 [DOI] [PubMed] [Google Scholar]
- 31.Ogawa T., Shinohara A., Ikeya T. (1995) Adv. Biophys. 31, 93–100 [DOI] [PubMed] [Google Scholar]
- 32.White C. I., Haber J. E. (1990) EMBO J. 9, 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugiyama T., Kantake N., Wu Y., Kowalczykowski S. C. (2006) EMBO J. 25, 5539–5548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nimonkar A. V., Sica R. A., Kowalczykowski S. C. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3077–3082 [DOI] [PMC free article] [PubMed] [Google Scholar]