Abstract
Oxidative stress can damage the active site cysteine of the antioxidant enzyme peroxiredoxin (Prx) to the sulfinic acid form, Prx-SO2−. This modification leads to inactivation. Sulfiredoxin (Srx) utilizes a unique ATP-Mg2+-dependent mechanism to repair the Prx molecule. Using selective protein engineering that involves disulfide bond formation and site-directed mutagenesis, a mimic of the enzyme·substrate complex has been trapped. Here, we present the 2.1 Å crystal structure of human Srx in complex with PrxI, ATP, and Mg2+. The Cys52 sulfinic acid moiety was substituted by mutating this residue to Asp, leading to a replacement of the sulfur atom with a carbon atom. Because the Srx reaction cannot occur, the structural changes in the Prx active site that lead to the attack on ATP may be visualized. The local unfolding of the helix containing C52D resulted in the packing of Phe50 in PrxI within a hydrophobic pocket of Srx. Importantly, this structural rearrangement positioned one of the oxygen atoms of Asp52 within 4.3 Å of the γ-phosphate of ATP bound to Srx. These observations support a mechanism where phosphorylation of Prx-SO2− is the first chemical step.
INTRODUCTION
Reactive oxygen species, such as hydrogen peroxide (H2O2) and peroxynitrite, have been recognized as compounds that cause significant damage to cells by acting on DNA, RNA, lipids, and proteins when present at elevated levels. Reactive oxygen species readily modify reactive protein sulfhydryls to higher oxidation states (1). Peroxiredoxins (Prxs),2 the primary enzyme family involved in breaking down low levels of peroxides, utilize a conserved, “peroxidatic” Cys residue, Cys-SPH, as the primary site of oxidation by peroxides (2–4). The resulting Cys sulfenic acid intermediate (Cys-SPOH) promotes the formation of a disulfide bond. For the 2-Cys subgroup of Prxs (PrxI–IV in humans), the “resolving” Cys residue, Cys-SRH, from the C terminus of the adjacent monomer of the homodimer participates in the disulfide bond, Prx-SP–SR-Prx. During oxidative stress, however, the peroxide levels can overwhelm the Prx system and hyperoxidize the Cys-SPOH intermediate to a Cys sulfinic acid, Cys-SPO2−. In this context, the 2-Cys Prxs are unique in that they can become inactivated by the peroxide substrate and subsequently are repaired by an enzyme known as sulfiredoxin (Srx) (5–9). This reversible peroxide-sensitive switch has been shown to regulate the activation of transcription factors in Schizosaccharomyces pombe (10, 11). Moreover, formation of Prx-SO2− results in cell cycle arrest that resumes after the reversal of the hyperoxidized state (12).
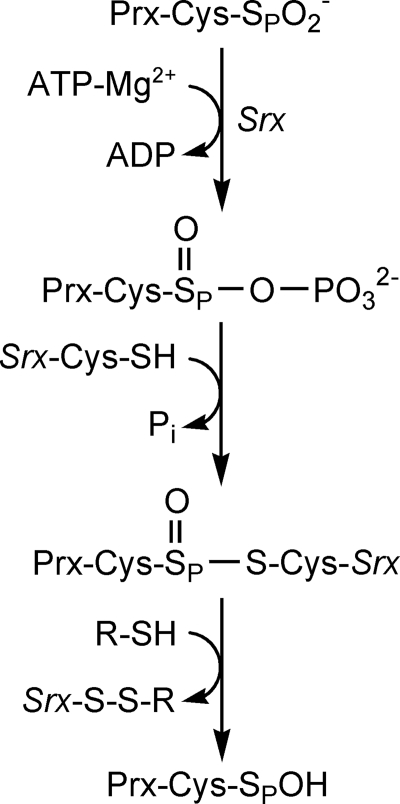
Srx catalyzes the reduction of Prx-SO2− utilizing ATP, Mg2+, an active site Cys in Srx, and a thiol reductant such as glutathione (GSH) or thioredoxin (Trx) (8, 13, 14). Biochemical and structural analyses support a mechanism (Fig. 1) where the ATP molecule, bound tightly to the nucleotide binding motif of Srx, is brought in close proximity to the sulfinic acid moiety of Prx (15, 16). The direct attack of the Cys-sulfinic acid on the γ- phosphate of ATP generates a sulfinic phosphoryl ester intermediate, which is followed by the thiol attack (Cys99 in human Srx) to form a Srx-Prx thiosulfinate species (17, 18). The thiosulfinate is further reduced by GSH or thioredoxin to regenerate the active forms of Prx and Srx.
FIGURE 1.
Mechanism of reduction of Prx-SO2− by Srx. Attack of the γ-phosphate of ATP by Prx-SO2− results in the formation of a sulfinic acid phosphoryl ester intermediate. The attack of Cys99 in Srx generates a Srx-Prx thiosulfinate intermediate that can be resolved by GSH or Trx to generate active Prx-SOH.
Major structural rearrangements within the Prx molecule are required for the reductive chemistry facilitated by Srx to occur (16, 19). These changes include: (i) the displacement of the conserved YF motif occluding the Prx active site from ATP-bound Srx; (ii) the unfolding of the Prx active site helix to place a conserved Phe residue within hydrophobic pocket of Srx; and (iii) the unfolding of the C terminus of Prx onto the noncatalytic, backside surface of Srx. The structural changes that must occur within the Prx active site to foster an attack on ATP molecule and the direct observation of the phosphorylated intermediate, however, remain elusive.
In this report, we describe the 2.1 Å resolution crystal structure of a quaternary complex between Prx, Srx, ATP and Mg2+. A novel protein engineering approach was used to enhance the interactions between Srx and Prx and to generate a stable mimic of the enzyme·substrate complex. The structure provides insight into the conformational changes within the Prx-SO2− molecule that enable it to be recognized as a substrate by Srx, resulting in its reduction. The crystal structure supports the proposal that Prx-SO2− repair proceeds directly through a sulfinic acid phosphoryl ester intermediate, formed by the direct transfer of the γ-phosphate from ATP to Prx-SO2−.
EXPERIMENTAL PROCEDURES
Protein Preparation and Engineering
Mutant forms of hSrx (C99A and N43C) and hPrxI (C52D, C71S, C83E, A86E, C173S, and K185C) were generated using the QuikChange site-directed mutagenesis kit from Stratagene. A truncated version of the human Srx mutant, residues 32–137, was expressed from a pET19 (Novagen) vector derivative containing a PreScission protease (GE Healthcare) cleavage site between Srx and the N-terminal His tag. The proteins were purified using nickel affinity and size-exclusion chromatography after the removal of the His tag (16, 20). The PrxI mutant was expressed from a pET17b (Novagen) vector in C41(DE3) Escherichia coli with an added, noncleavable His6 C-terminal tag, and purified using nickel affinity in the presence of 5 mm β-mercaptoethanol. The protein was subsequently treated with 5 mm EDTA and 2 mm dithiothreitol to disrupt mixed disulfides before size-exclusion chromatography (Superdex 200) was used to further purify the protein and to remove the reducing agents. The fractions corresponding to the dimeric form of PrxI were pooled, and 1 mm Ellman's reagent, 5,5′-dithiobis(2-nitrobenzoic acid), was added. Excess 5,5′-dithiobis(2-nitrobenzoic acid) and generated TNB2+ were removed from the TNB-linked PrxI using the size exclusion column. Srx was titrated into the PrxI solution until no further release of TNB2+ was observed at 412 nm (1.5-fold excess Srx over PrxI). The resulting disulfide-bonded Srx·PrxI complex, i.e. Cys43 of Srx linked to Cys185 of PrxI, was passed over the size-exclusion column to remove excess Srx and TNB2+. The complex was concentrated, aliquoted, flash frozen with liquid nitrogen, and stored at −80 °C.
Crystallization
The engineered Srx·Prx apocomplex was crystallized by the vapor diffusion method. Equal volumes of protein (20.6 mg ml−1 in 20 mm HEPES, pH 7.5, 100 mm NaCl) and well solutions (7–9.5% PEG 6000, 100 mm HEPES, pH 7.6) were mixed and incubated at 20 °C for a week as sitting drops. The ligands, 40 mm ATP and 8 mm Mg2+ in 12% PEG 6000 and 200 mm HEPES pH 7.6, were added slowly to the crystal drop over the course of several h and incubated overnight. A cryoprotectant consisting of the ATP soak solution with 25% ethylene glycol was added to the crystal drop before cryocooling the crystal in nitrogen gas at 100 K.
Data Collection and Structure Determination
The data set was collected on an in-house Rigaku/MSC MicroMax-007 generator with a Saturn-92 CCD detector. Diffraction intensities were integrated using d*Trek (Rigaku/MSC, Woodlands, TX) and scaled to 2.1 Å resolution. Cross-validation was performed with 4.9% of the reflections that were set aside. The space group of the crystal was P212121 with unit cell dimensions a = 57.3, b = 92.4, and c = 131.9 Å. Initial phases for the complex were obtained by the molecular replacement program PHASER (27.4–2.5 Å data used) using the apo Srx structure (PDB code 1XW3) and the conserved structural regions of the human PrxII (residues 3–45 and 70–168 of PDB code 1QQ2) as search models (19–21). Only one unique solution, i.e. not related to others by a 2-fold rotation axis, was found (rotation and translation Z-scores ranging from 6–25 and 26–43, respectively; overall LLG, 5036; Rwork, 36.2% following rigid body refinement). The asymmetric unit contained two monomers of Srx and one dimeric Prx. The molecular replacement solution was refined with CNS using alternating cycles of simulated annealing and positional and isotropic B-factor refinement (22). Model building was performed with COOT (23). Water molecules were identified with a |Fo − Fc| map contoured at 3σ and added with COOT. The final cycles of refinement were performed with REFMAC5 (24). All reflection data were used during refinement. The structure was validated using the MOLPROBITY server, which reported 99.8% of the residues present in the Ramachandran favored regions and 0.2% in the allowable regions (25). The data collection and refinement statistics for the structure are summarized in Table 1. All structural figures were generated with PyMOL (DeLano Scientific).
TABLE 1.
Crystallographic data and refinement statistics
| ATP·Mg2+·Srx·PrxI-CO2− | |
|---|---|
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 57.3, 92.4, 131.9 |
| α, β, γ (°) | 90, 90, 90 |
| Wavelength (Å) | 1.54 |
| Resolution (Å)a | 30.0-2.1 (2.18-2.10) |
| Rmerge (%) | 8.2 (26.5) |
| I/σI | 13.2 (5.3) |
| Completeness (%) | 99.9 (99.5) |
| Redundancy | 13.7 (10.3) |
| Wilson B (Å2) | 26.9 |
| Refinement | |
| Resolution (Å) | 27.4-2.1 |
| Rwork/Rfree (%) | 22.4/27.3 |
| No. of reflections used (work/free) | 39,439/2090 |
| No. of atoms | |
| Protein | 4450 |
| Ligands | 66 |
| Water | 323 |
| R.m.s.b deviations | |
| Bond lengths (Å) | 0.014 |
| Bond angles (°) | 1.47 |
| Average B-factor (Å2) | |
| Protein | 23.9 |
| Ligands | 24.7 |
| Solvent | 25.4 |
| Ramachandran analysis | |
| Favored regions (%) | 99.8 |
| Allowed regions (%) | 0.2 |
a Numbers in parentheses are for the highest resolution shell.
b R.m.s., root mean square.
RESULTS
Engineering and Crystallization of the Srx·Prx Complex with ATP and Mg2+
In our previous crystallographic study, we imitated the thiosulfinate intermediate between Srx and Prx through the introduction of a disulfide bond between Cys99 of Srx and Cys52-SPH of human PrxI (16). The presence of this engineered disulfide bond would, however, preclude the binding of the ATP molecule and prevent any potential conformational changes within the active site of both proteins. Therefore, a different approach would be required to evaluate the structural changes and geometric relationships underlying the repair of Prx-SO2−; in particular, the attack of the γ-phosphate of ATP.
Initial attempts to crystallize hyperoxidized PrxI-SO2− with Srx in the presence or absence of ATP and Mg2+ were unsuccessful. We rationalized that Srx within the complex was most likely still able to hydrolyze ATP over the lengthy time period to grow crystals, and the forces holding together the catalytic complex were insufficient to facilitate crystal packing. To address these issues, we substituted the sulfinic acid moiety of the Prx molecule with the carboxyl analogue (Prx-CO2−) by mutating Cys52 to Asp and inactivated Srx by mutating the active site Cys99 to Ala (8, 15). Moreover, the interaction between Srx and Prx was stabilized by introducing an engineered disulfide bond between two residues previously identified to be in proximity on the backside interface far from the active site; Asn43 of Srx and Lys185 of PrxI (16). It is important to note that the latter approach requires that all other Cys residues with the Srx and Prx molecules are mutated. In an effort to increase diffraction quality, the dimeric form of PrxI was generated by introducing a single charged residue, C83E, at the dimer-dimer interface. Although this approach worked previously (16), the resulting crystals diffracted to 3.5–4.0 Å resolution with cell parameters consistent with a decameric form of PrxI. In hindsight, these unexpected observations are consistent with the known stabilization of the decameric form when the Prx active site is partially unfolded (3).
Therefore, to further destabilize the decameric form, we introduced a second mutation, A86E, adjacent to C83E at the dimer-dimer interface. This variant of the Srx·Prx complex readily formed well diffracting crystals amenable to soaking with excess ATP and Mg2+. The dimeric structure of the quaternary ATP·Mg2+·Srx·PrxI-CO2− complex was solved by molecular replacement using the wild-type Srx and PrxI molecules as the search models with the appropriate modifications and refined to 2.1 Å resolution (Table 1), as described in the “Experimental Procedures.” The overall folds of PrxI and Srx molecules are in agreement with previously reported structural data with root mean square deviations of the common α-carbon atoms of 0.7 Å and 0.9 Å, respectively.
Overall Structure and Consequences of the Engineered Disulfide Bond
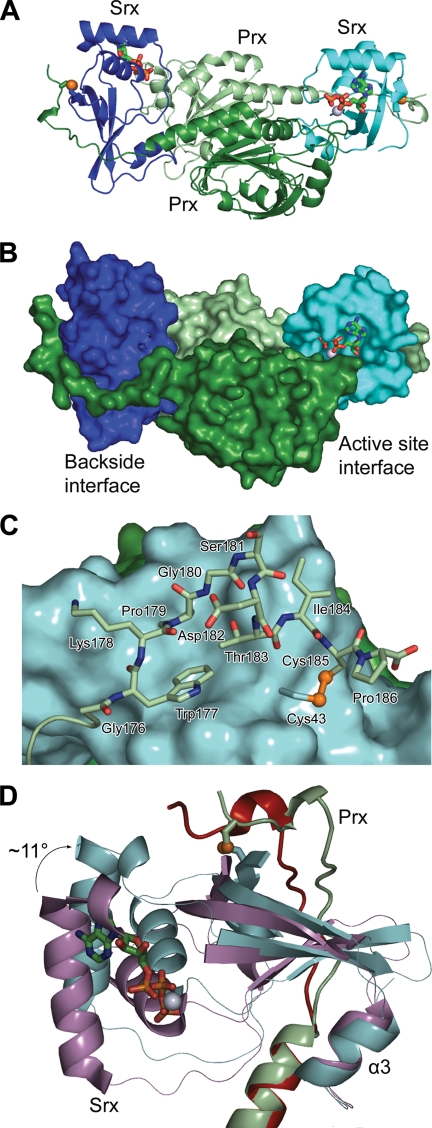
The 2-fold symmetric Srx·PrxI complex was obtained with each Srx molecule (e.g. chain X) sandwiched between the active site surface of one PrxI molecule (e.g. chain A, ∼550 Å2 buried) and the C-terminal residues (172–186; residues 187–199 were not visible in the electron density) extending from the α5-helix of the adjacent PrxI molecule (e.g. chain B, ∼920 Å2) (Fig. 2, A and B). The engineered disulfide bond is positioned on the backside interface of both Srx molecules, between Cys185 of PrxI and Cys43 of Srx, as designed (Fig. 2C). Importantly, this disulfide bond did not interfere with the previously observed packing of the C terminus of PrxI onto Srx (16). This interaction is facilitated by the packing of the conserved Trp177 and Pro179 of PrxI into the predominantly hydrophobic pocket of Srx composed of residues Val44, Pro45, Val48, His82, Phe93, and Leu117.
FIGURE 2.
The Srx·PrxI enzyme·substrate complex containing the backside, engineered disulfide bond. A, overall structure of the ATP·Mg2+·Srx·Prx-CO2− complex. A cartoon representation is shown with monomers of the PrxI dimer in dark green (molecule A) and light green (molecule B), and the two monomers of Srx in cyan (molecule X) and blue (molecule Y). One ATP molecule and one Mg2+ ion (green carbon atoms and gray sphere, respectively) are bound in each active site. The engineered disulfide bond is highlighted with the sulfur atoms as yellow spheres. B, surface representation of the Srx·PrxI complex, illustrating active site and backside interfaces. In this view the ATP molecule and Mg2+ ion can still be seen. C, backside interaction of Srx (cyan surface) with the predominantly conserved C-terminal residues of PrxI (light green stick). The engineered disulfide bond between Cys185 in PrxI and Cys43 in Srx is shown. D, rotation of Srx relative to the Prx molecule in the presence of ATP-Mg2+. The Prx dimer of the ATP·Mg2+·Srx·Prx-CO2− complex was superimposed onto the Prx dimer within the original, active site-based Prx-S–S-Srx disulfide-bonded complex (PDB code 2RII) (16). The Srx and Prx molecules of the new complex are colored as above, and the Srx and Prx molecules of the former complex are colored violet and red, respectively.
In contrast to the conservation of the backside Srx-Prx interface and the Prx dimer interactions, the relationship of each Srx active site relative to its Prx substrate molecule changed significantly (Fig. 2D). Whereas the C-terminal, α3-helix of Srx rests against the PrxI dimer interface in both structures, the active site of Srx within the quaternary complex has moved away from the Prx molecule resulting in an ∼11° tilt of the Srx molecule. We attribute this movement of Srx to increased breathing of the complex; a direct result of moving the stabilizing intermolecular disulfide bond from the active site to the backside of Srx. As described below, the PrxI active site has also rearranged to enable the Asp-based mimic of the sulfinic acid moiety to approach the ATP molecule.
Srx-Prx Active Site Interactions
Unambiguous electron density was observed around the ATP and Mg2+ molecules bound to Srx and the active site motif of PrxI (Fig. 3A). The binding mode of ATP is similar to that observed in our recently solved Srx·ATP·Mg2+ crystal structure (15). In short, the adenosine ring of ATP hydrogen bonds with Ser64 and Thr68 located on the α1-helix of Srx. The phosphate binding pocket of Srx, consisting of His100 and Arg101 within the conserved motif P96(G/S)GCHR101, provides hydrogen-bonding interactions with the β- and γ-phosphate groups, respectively. Lys61 from α1 complements these interactions by hydrogen bonding with the α-phosphate group. The Mg2+ ion is coordinated by the three phosphate groups of ATP and three water molecules in an octahedral fashion. The average Mg2+-oxygen distance in the coordination sphere is 2.1 Å. The C99A mutation in Srx is located behind and below the γ-phosphate of ATP.
FIGURE 3.
The Srx-Prx interface in complex with ATP and Mg2+. A, stereoview of a simulated-annealing, omit |Fo − Fc| electron density map (gray) contoured at 3σ in the active site of Srx (cyan) and the substrate Prx (dark green). The ATP molecule and Mg2+ ion are colored as described previously; water molecules are shown as red spheres. B, hydrogen-bonding interactions (yellow dashed lines) between the ATP molecule, Mg2+ ion, Srx, and Prx.
Although the binding mode of ATP changed little, the Prx active site motif (D47FTFVCPTEI56), previously present as a helix in the hyperoxidized state, unfolded locally to enable Phe50 to pack within a hydrophobic Srx pocket constituted by Leu52, Leu82, Phe96, Val118, Val127, and Tyr128. This rearrangement allowed the substrate mimic Asp52 to approach the ATP molecule and to interact with a water molecule of the Mg2+ ion hydration sphere. This interaction, and the observation that one oxygen atom of the Prx·Asp·CO2− moiety is located 4.3 Å from γ-phosphate of ATP (Fig. 3B), suggests that the carboxyl oxygen atom is slightly out of register to perform a direct in-line attack. This is not surprising as the planar carboxyl group cannot fully represent the electronic and geometric properties of the nonplanar sulfinic acid moiety. Nonetheless, this structure of the engineered substrate mimic reasonably approximates how the sulfinic acid group of Prx-SO2− could approach the ATP, resulting in its phosphorylation.
Another interesting observation from the new structure is that the conserved GGLG motif of Prx is within hydrogen bonding distance of the ATP molecule, although with suboptimal bonding angles. The backbone carbonyl groups of Lys92 and Lys93 interact with two water molecules of the Mg2+ ion hydration sphere. At this time, it is unclear whether or not these interactions play any role in catalysis.
DISCUSSION
This study describes a crystallographic approach to stabilize the interaction of two proteins by the introduction of an engineered intermolecular disulfide bond. Several studies have shown that disulfide bonds can be engineered to facilitate crystal growth for protein·protein and protein·DNA complexes, to enhance protein stability, and to trap catalytically relevant complexes within the DsbA-DsbB-DsbC system and other disulfide oxidoreductases (26–34). In the thioredoxin reductase system from E. coli, for example, an intermolecular disulfide was generated between Cys135 of thioredoxin reductase and Cys32 of Trx, stabilizing this important transient intermediate of the enzyme mechanism (29). We used a similar approach to solve the original Srx·Prx complex by mimicking the thiosulfinate intermediate with a disulfide bond (16). The presence of this engineered disulfide bond at the interface between the two active sites, however, would prevent the binding of ATP-Mg2+. As such, the current study selectively introduced a stabilizing disulfide bond between the proteins far from the active site on the backside interface. This allowed for the trapping of ATP-Mg2+ and additional structural changes within the Prx active site. We propose that this approach may be generally useful for stabilizing protein·protein complexes and trapping reaction intermediates where the structure of at least one component has been determined previously.
Another key element of the approach used to generate the Srx·Prx substrate complex with its ATP and Mg2+ cofactors was to substitute the Prx sulfinic acid moiety with an Asp residue, i.e. replace the sulfur atom with an oxygen atom. We have shown previously that this mutant in human PrxII, C51D, is readily phosphorylated by Srx, an observation reminiscent to the phosphorylation of Glu in the glutamine synthase reaction (15, 35).
The ATP·Mg2+·Srx·Prx-CO2− crystal structure reveals the geometric relationships underlying the first step of the unique Srx repair mechanism. Two alternatives for the first step have been proposed; either direct transfer of the γ-phosphate of ATP to the Prx molecule or transfer of the phosphate group to Prx with Srx acting as a phosphorylated intermediary (8, 13). One oxygen atom of Prx-Asp-CO2− is located in close proximity (Fig. 3) to the γ-phosphate of ATP, suggesting that the sulfinic acid moiety of the wild-type enzyme would be oriented favorably for an in-line attack. Importantly, the C99A mutation also supports that Cys99 is not positioned correctly for the alternative phosphotransferase role. Although the sulfinic phosphoryl ester intermediate has yet to be isolated, the quaternary ATP·Mg2+·Srx·Prx-CO2− crystal structure in combination with previous mechanistic studies supports that repair proceeds directly through this species (15, 17, 18).
Crystallographic and biochemical studies of the Srx-Prx interaction have provided us with the following understanding of the Srx repair mechanism. The inherent decreased mobility of the Prx C-terminal tail, occluding the Prx active site, is slowly displaced by Srx (Fig. 4A). The active site interfaces alone are insufficient for prolonged interactions and require the packing of the C-terminal tail of Prx onto the backside surface of Srx for stabilization (15, 20). As a result, Srx provides a hydrophobic surface for the binding of Phe50 of PrxI. This promotes the unfolding of the Prx active site helix and reorients Prx-Cys-SO2− to approach the phosphate of ATP for in-line attack (Fig. 4B). The active site Cys99 in Srx is now in position to attack the sulfinic phosphoryl ester intermediate to generate a thiosulfinate intermediate with Prx (Fig. 4C) (17, 18). This complex is further reduced by GSH or Trx to release enzymatically active Prx-SOH and Srx.
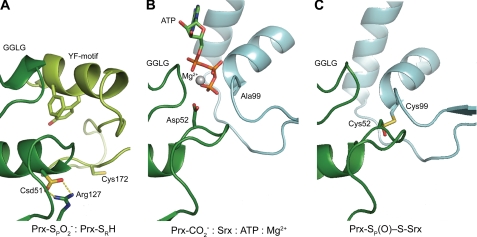
FIGURE 4.
Structural intermediates of the Srx reaction. A, human PrxII in the hyperoxidized, fully folded state, the substrate of Srx (PDB code 1QMV) (19). The sulfinic acid moiety of the Prx molecule, Csd51 (Cys sulfinic acid; dark green) is buried in the active site, salt-bridged to Arg127 and covered by the YF motif from the adjacent monomer in the dimer (light green). The residue numbering scheme is one residue shorter for PrxII. B, Asp52 mimic of PrxI-SO2− approaching the γ-phosphate of ATP in the first step of the reaction presented herein. C, model of the Srx-Prx thiosulfinate intermediate. The thiosulfinate was modeled on the disulfide-bonded Srx-Prx structure (PDB code 2RII) (16, 18).
The unfolding of the Prx active site helix observed within this new Srx·Prx complex structure may also provide some insight into the conformational changes involved in the catalytic conversion of H2O2 to water by 2-Cys Prxs. The 18 structural studies of 2-Cys Prxs to date have revealed two stable conformations (36). All states that do not contain an active site disulfide have Cys52 or its equivalent within a “fully folded” helix (3, 4). The ATP·Mg2+·Srx·Prx-CO2− complex represents the first high resolution structure of a nondisulfide-bonded 2-Cys Prx with Cys52 in the “locally unfolded” state. Although the locally unfolded state in Prx is normally promoted by the covalent disulfide bond with the resolving Cys residue, the unfolding of the helix in the Srx·Prx complex is triggered by the binding of Phe50 of Prx into a hydrophobic surface pocket of Srx. Additional work is still needed to determine whether or not a similar unfolding of the Prx active site is a prerequisite for peroxidase activity.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM072866 (to W. T. L.). This work was also supported by American Heart Association Postdoctoral Fellowship 0725399U (to T. J. J.).
The atomic coordinates and structure factors (code 3HY2) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Prx
- peroxiredoxin
- Cys-SPH
- peroxidatic Cys residue
- Cys-SRH
- resolving Cys residue
- Cys-SPOH
- Cys sulfenic acid
- Cys-SPO2−
- Cys sulfinic acid
- GSH
- glutathione
- Srx
- sulfiredoxin
- Trx
- thioredoxin
- PDB
- Protein Data Bank.
REFERENCES
- 1.Giles G. I., Jacob C. (2002) Biol. Chem. 383, 375–388 [DOI] [PubMed] [Google Scholar]
- 2.Hofmann B., Hecht H. J., Flohé L. (2002) Biol. Chem. 383, 347–364 [DOI] [PubMed] [Google Scholar]
- 3.Wood Z. A., Schröder E., Robin, Harris J., Poole L. B. (2003) Trends Biochem. Sci. 28, 32–40 [DOI] [PubMed] [Google Scholar]
- 4.Wood Z. A., Poole L. B., Karplus P. A. (2003) Science 300, 650–653 [DOI] [PubMed] [Google Scholar]
- 5.Woo H. A., Chae H. Z., Hwang S. C., Yang K. S., Kang S. W., Kim K., Rhee S. G. (2003) Science 300, 653–656 [DOI] [PubMed] [Google Scholar]
- 6.Woo H. A., Jeong W., Chang T. S., Park K. J., Park S. J., Yang J. S., Rhee S. G. (2005) J. Biol. Chem. 280, 3125–3128 [DOI] [PubMed] [Google Scholar]
- 7.Chevallet M., Wagner E., Luche S., van Dorsselaer A., Leize-Wagner E., Rabilloud T. (2003) J. Biol. Chem. 278, 37146–37153 [DOI] [PubMed] [Google Scholar]
- 8.Biteau B., Labarre J., Toledano M. B. (2003) Nature 425, 980–984 [DOI] [PubMed] [Google Scholar]
- 9.Jönsson T. J., Lowther W. T. (2007) Subcell Biochem. 44, 115–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vivancos A. P., Castillo E. A., Biteau B., Nicot C., Ayté J., Toledano M. B., Hidalgo E. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 8875–8880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bozonet S. M., Findlay V. J., Day A. M., Cameron J., Veal E. A., Morgan B. A. (2005) J. Biol. Chem. 280, 23319–23327 [DOI] [PubMed] [Google Scholar]
- 12.Phalen T. J., Weirather K., Deming P. B., Anathy V., Howe A. K., van der Vliet A., Jönsson T. J., Poole L. B., Heintz N. H. (2006) J. Cell Biol. 175, 779–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeong W., Park S. J., Chang T. S., Lee D. Y., Rhee S. G. (2006) J. Biol. Chem. 281, 14400–14407 [DOI] [PubMed] [Google Scholar]
- 14.Chang T. S., Jeong W., Woo H. A., Lee S. M., Park S., Rhee S. G. (2004) J. Biol. Chem. 279, 50994–51001 [DOI] [PubMed] [Google Scholar]
- 15.Jönsson T. J., Murray M. S., Johnson L. C., Lowther W. T. (2008) J. Biol. Chem. 283, 23846–23851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jönsson T. J., Johnson L. C., Lowther W. T. (2008) Nature 451, 98–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roussel X., Béchade G., Kriznik A., Van Dorsselaer A., Sanglier-Cianferani S., Branlant G., Rahuel-Clermont S. (2008) J. Biol. Chem. 283, 22371–22382 [DOI] [PubMed] [Google Scholar]
- 18.Jönsson T. J., Tsang A. W., Lowther W. T., Furdui C. M. (2008) J. Biol. Chem. 283, 22890–22894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schröder E., Littlechild J. A., Lebedev A. A., Errington N., Vagin A. A., Isupov M. N. (2000) Structure 8, 605–615 [DOI] [PubMed] [Google Scholar]
- 20.Jönsson T. J., Murray M. S., Johnson L. C., Poole L. B., Lowther W. T. (2005) Biochemistry 44, 8634–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCoy A. J. (2007) Acta Crystallogr D. Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr D. Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 23.Emsley P., Cowtan K. (2004) Acta Crystallogr D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 25.Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) Nucleic Acids Res. 32, W615–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stirnimann C. U., Rozhkova A., Grauschopf U., Grütter M. G., Glockshuber R., Capitani G. (2005) Structure 13, 985–993 [DOI] [PubMed] [Google Scholar]
- 27.Rozhkova A., Stirnimann C. U., Frei P., Grauschopf U., Brunisholz R., Grütter M. G., Capitani G., Glockshuber R. (2004) EMBO J. 23, 1709–1719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsumura M., Matthews B. W. (1991) Methods Enzymol. 202, 336–356 [DOI] [PubMed] [Google Scholar]
- 29.Lennon B. W., Williams C. H., Jr., Ludwig M. L. (2000) Science 289, 1190–1194 [DOI] [PubMed] [Google Scholar]
- 30.Inaba K., Murakami S., Suzuki M., Nakagawa A., Yamashita E., Okada K., Ito K. (2006) Cell 127, 789–801 [DOI] [PubMed] [Google Scholar]
- 31.Haebel P. W., Goldstone D., Katzen F., Beckwith J., Metcalf P. (2002) EMBO J. 21, 4774–4784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banatao D. R., Cascio D., Crowley C. S., Fleissner M. R., Tienson H. L., Yeates T. O. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 16230–16235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verdine G. L., Norman D. P. (2003) Ann. Rev. Biochem. 72, 337–366 [DOI] [PubMed] [Google Scholar]
- 34.Corn J. E., Berger J. M. (2007) Structure 15, 773–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Purich D. L. (2002) Methods Enzymol. 354, 1–27 [DOI] [PubMed] [Google Scholar]
- 36.Karplus P. A., Hall A. (2007) Subcell Biochem. 44, 41–60 [DOI] [PubMed] [Google Scholar]