Abstract
Benzo[a]pyrene-7,8-diol-9,10-epoxide (B[a]PDE), the major metabolite of B[a]P, has been well recognized as one ubiquitous carcinogen, but the molecular mechanism involved in its carcinogenic effect remains obscure. In the present study, we found that bronchial epithelial cells (Beas-2B) and hepatocytes treated with B[a]PDE presented a significant increase of cyclin D1 expression. Moreover, Akt, p70s6k, and MAPKs including JNK, Erks, and p38 were notably activated in B[a]PDE-treated Beas-2B cells, whereas NF-κB, NFAT, and Egr-1 were not. Our results demonstrated that JNK and Erks were required in B[a]PDE-induced cyclin D1 expression because the inhibition of JNK or Erks by a selective chemical inhibitor or dominant negative mutant robustly impaired the cyclin D1 induction by B[a]PDE. Furthermore, we found that overexpression of the dominant negative mutant of p85 (regulatory subunit of phosphatidylinositol 3-kinase) or Akt dramatically suppressed B[a]PDE-induced JNK and Erk activation as well as cyclin D1 expression, suggesting that cyclin D1 induction by B[a]PDE is via the phosphatidylinositol 3-kinase/Akt/MAPK-dependent pathway. In addition, we clarified that p70s6k is also involved in B[a]PDE-induced cyclin D1 expression because rampamycin pretreatment dramatically reduced cyclin D1 induction by B[a]PDE. More importantly, we demonstrated that up-regulated cyclin D1 by B[a]PDE plays a critical role in oncogenic transformation and tumorigenesis of Beas-2B cells. These results not only broaden our knowledge of the molecular mechanism of B[a]PDE carcinogenicity but also lead to the further study of chemoprevention of B[a]PDE-associated human cancers.
INTRODUCTION
Polycyclic aromatic hydrocarbons are ubiquitous environmental chemical carcinogens existing in cigarette smoke, charred foods, and exhaust gas from incomplete combustion of fossil fuels (1, 2). Benzo[a]pyrene (B[a]P),3 as the first identified carcinogenic component of polycyclic aromatic hydrocarbons, is the most extensively studied carcinogen in cigarette smoke and has been regarded as a critical mediator of lung cancer for a long time (3, 4). B[a]P is metabolized by cytochrome P450 enzyme to B[a]PDE, which is highly cytotoxic, mutagenic, and carcinogenic (5, 6). Carcinogenesis is a multistage process that consists of initiation, promotion, and progression (7). Initiation is a rapid and irreversible course, whereas promotion is a long term process requiring chronic exposure to a certain compound with tumor promotion activity (8). The carcinogenicity of B[a]PDE has been reported to be associated with its covalent binding to some critical targets in DNA and the subsequent DNA damage, which lead to the activation of oncogenes or inactivation of tumor suppressor genes (9). For example, B[a]PDE-DNA hydrophobic adduct found in lung tissue from smokers was verified to be able to induce p53 mutation, which is well accepted to mediate cancer initiation during tumor development (10). It has been also demonstrated that topical application of a low dose of B[a]PDE alone for a long period is able to cause cancer, implicating that B[a]PDE also possesses tumor promotion activity (11). In addition to its tumor initiation activity via DNA adduction and consequent gene mutation, the tumor promotion activity of B[a]PDE is also considered to be rather important for its carcinogenicity, but the underlying molecular mechanism is far from being fully understood (11).
The precise regulation of cell cycle transition is critical for maintaining cellular homeostasis, whereas abnormal expression of regulatory factors, which drives uncontrolled cell cycle progression, is involved in tumor promotion (12, 13). Previously, we found that B[a]PDE exposure could markedly induce cyclin D1 transcription in mouse epidermal Cl 41 cells (14). Cyclin D1, which complexes with and activates cyclin-dependent kinases, is an essential regulatory molecule in G1/S phase transition of cells exposed to various growth factors and stresses including chemical carcinogen. Overexpression of cyclin D1 has been observed in a number of human cancers including lung cancer, hepatocarcinoma, and bladder cancer, etc. Cyclin D1 knockdown was reported to inhibit tumorigenesis of human cancer cells in nude mice (15). Accumulating evidence has demonstrated that the carcinogenic effect of cyclin D1 is much more sophisticated than simply accelerating the cell cycle progression (16). As known to all, lung is the primary target of B[a]PDE from cigarette smoke. Therefore, it is important to study whether B[a]PDE is able to up-regulate cyclin D1 expression in pulmonary cells, and if so, what would be the molecular mechanism and the tumorigenic effect.
MATERIALS AND METHODS
Reagents and Cell Lines
Human bronchial epithelial Beas-2B cells, human hepatocyte L02 cells and their stable transfectants were cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS) at 37 °C in a humidified atmosphere with 5% CO2. B[a]PDE was purchased from Eagle-Picher Industries, Inc., Chemsyn Science Laboratories (Lenexa, KS). Antibodies against phospho-Akt (Thr308), phospho-Akt (Ser473), Akt, phospho-p70s6k (Thr389), phospho-p70s6k (Thr421/Ser424), p70s6k, cyclin A, cyclin D1, and cyclin E were purchased from Cell Signaling Technology (Beverly, MA). Egr-1, p38, JNK, and Erk antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Wortmannin, a PI-3K inhibitor, and rapamycin, an mTOR/p70s6k pathway inhibitor, were purchased from Calbiochem (La Jolla, CA). Specific inhibitor of MEK/Erks (PD98059), p38 (SB202190), and JNKs (SP600125) were obtained from Calbiochem-Merck (Darmstadt, Germany).
Plasmids
The cyclin D1 promoter-driven luciferase reporter (cyclin D1-luc) construct was obtained by inserting a 1.23-kb EcoRI-PvuII fragment of the cyclin D1 gene promoter from −1095 to +135 relative to the translation initiation site, into the pA3LUC vector as described previously (17). The NFAT-luciferase reporter plasmid and NF-κB-luciferase reporter plasmid were constructed as described previously (18). Plasmids expressing dominant negative mutants of Akt, Erk, JNK, and PI-3K (DN-Akt, DN-Erks, DN-JNK, and Δp85) were described in our previous studies (19–21). The specific small interference RNA (siRNA) targeting human cyclin D1 was inserted into pSuppressor vector as described previously (22).
Stable Transfection
Beas-2B cells were transfected with hygromycin B-resistant plasmid together with cyclin D1-luciferase reporter plasmid, or plasmid expressing Δp85, DN-Akt, DN-JNK, DN-Erk, or siCyclin D1, according to the manual of Lipofectamine 2000 reagent. 400 μg/ml hygromycin was utilized for the selection of stable transfectants. The stable transfectants: Beas-2B cyclin D1-luc mass1 cells, L02 cyclin D1-luc mass1 cells, Beas-2B DN-Akt mass1 cells, Beas-2B DN-JNK mass1 cells, Beas-2B DN-Erk mass1 cells, Beas-2B Δp85 mass1 cells, Beas-2B cyclin D1 siRNA mass1 cells, and their control cells were cultured in hygromycin-free 10% FBS DMEM for at least 2 days before each experiment.
Gene Reporter Assays
8 × 103 viable cells suspended in 100 μl of 10% FBS DMEM were seeded into each well of 96-well plates. After the cell density reached 80–90% confluence, the cells were exposed to B[a]PDE for various time periods and then lysed with 50 μl of lysis buffer. The luciferase activity was measured using the Promega Luciferase assay reagent with a luminometer (BioTek SynergyTM 2 multi-mode microplate reader). The results were expressed as cyclin D1 luciferase activity relative to medium control (relative cyclin D1 induction). Student's t test was used to determine the significance, and the differences were considered significant at p < 0.05.
RT-PCR
The cells were cultured in a 6-well plate until they reached 85–90% confluence. The cell culture medium was replaced with 0.1% FBS DMEM and cultured for 24 h. The cells were then exposed to various concentrations of B[a]PDE for 12 h. The cells were washed once with ice-cold phosphate-buffered saline, and whole RNA was extracted with TRIzol® reagent following the manufacturer's instructions (Invitrogen). The cDNA was synthesized from 1 μg of RNA by using a first strand synthesis system for RT-PCR (Invitrogen). The PCR was performed by using 2 μl of synthesized cDNA and the specific human cyclin D1 primers (sense, 5′-agctcctgtgctgcgaagtggaaac-3′; antisense, 5′-agtgttcaatgaaatcgtgcggggt-3′) or the β-actin primers (sense, 5′-gcgagaagatgacccagatcat-3′; antisense, 5′-gctcaggaggagcaatgatctt-3′). The relative mRNA level of cyclin D1 was normalized to the internal reference β-actin that was co-amplified in the same reaction for each sample.
Western Blot Assays for Cell Culture
2 × 105 Beas-2B cells or its stable transfectants were cultured in each well of 6-well plates till 70–80% confluence. The culture medium was replaced with 0.1% FBS DMEM. After being cultured for 24 h, the cells were exposed to B[a]PDE for various time periods. The cells were then washed once with ice-cold phosphate-buffered saline and extracted with SDS sample buffer. The extracts of cells were separated on polyacrylamide-SDS gels, transferred to nitrocellulose membrane, and probed with rabbit or mouse-specific antibodies. The protein band, specifically bound to the primary antibody, was detected using an IRDye 800CW-conjugated secondary antibody and LI-COR imaging systems (LI-COR Biosciences, Lincoln, NE).
Cell Cycle Assays
Beas-2B siCyclin D1 mass1 cells and the control cells were seeded in 6-well plate until they reached 85–90% confluence. After serum starvation for 24 h, the cells were exposed to 0.5 μm B[a]PDE for 24 h, then harvested, and fixed in 70% ice-cold ethanol overnight. The fixed cells was then centrifuged (1500 rpm, 3 min), suspended in suspension buffer (100 mm sodium citrate and 0.1% Triton X-100), and incubated for 15 min at room temperature. The cells were incubated with RNase A (10 mg/ml) (Sigma) for 10 min at room temperature, and DNA was stained with propidium iodide (50 mg/ml) for at least 1 h at 4 °C. The DNA content was determined by flow cytometry (Beckman Coulter, San Diego, CA) and EXPO 32 software.
Colony Formation Assay
Beas-2B stable transfectants were cultured in each well of six-well plates to 30–40% confluence with normal culture medium. The cells were treated with 0.5 μm B[a]PDE for 1 day and then recovered in fresh medium for 2 days. After repeated treatment with B[a]PDE for 12 weeks, the cells (1 × 103) were plated in 10% FBS DMEM on 96-well plates precoated with 100 μl of Matrigel (Becton Dickinson) and cultured for 2 weeks. The colony formation was assessed by counting the colony number under microscope, and representative views were photographed.
Tumorigenicity Assays
Six-week-old male nude mice were injected subcutaneously with 4 × 106 of Beas-2B cells with or without B[a]PDE long term treatment, respectively. Tumor growth was monitored by caliper measurement once or twice a week for at least 6 weeks. The tumor volume (cm3) was calculated using the following formula: 0.5 × (L × W2), where L is tumor length, and W is tumor width. Six weeks after the tumor cells inoculation, the mice were sacrificed by CO2 asphyxiation, and the xenograft tumors were excised for immunohistochemistry. These procedures were approved by the Animal Care and Use Committee (Shanghai, China).
Immunohistochemistry
The tumors were excised, fixed in formalin, embedded in paraffin, and cut into 5-mm-thick slices for staining. The sections were subjected to incubation with anti-cyclin D1 antibody or anti- proliferating cell nuclear antigen antibody (Cell Signaling Technology, Inc.) at 4 °C overnight and then with horseradish peroxidase-conjugated secondary antibody at 37 °C for 30 min. The sections were finally incubated with diaminobenzidine and counterstained with hematoxylin for detection. The slides were viewed by a Nikon Eclipse E600 microscope (Nikon Corporation, Tokyo, Japan), and representative views were photographed.
Statistical Analysis
The significance of the difference between the treated and untreated groups was determined using the Student's t test, and the statistical significance was set at p < 0.01 or p < 0.05.
RESULTS
B[a]PDE Enhances Cyclin D1 Expression in Bronchial Epithelial Cells and Hepatocytes
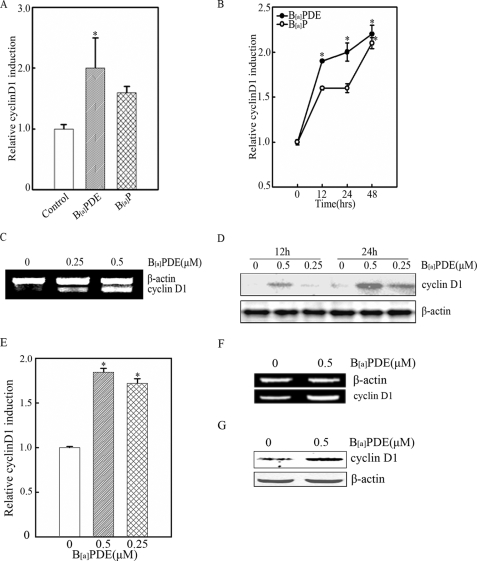
Epidemiological studies have shown that occupational and nonoccupational exposures to B[a]P are associated with an increased risk of lung cancer (3, 4). Previous studies have documented that cyclin D1 is constitutively overexpressed in human lung cancer (16). It is therefore interesting to determine whether cyclin D1 is inducible in human bronchial epithelial cells exposed to B[a]P. To address this question, Beas-2B cells were exposed to B[a]P and its major metabolite B[a]PDE. As shown in Fig. 1 (A and B), treatment of Beas-2B cells with B[a]PDE or B[a]P led to a marked increase of cyclin D1 transcription in gene reporter assay, and the induction of cyclin D1 by B[a]PDE was more obvious and rapid than that by B[a]P. It is well accepted that B[a]P exerts its carcinogenicity mainly through its intermediate metabolite B[a]PDE. Hence, our following studies were focused on B[a]PDE. RT-PCR (Fig. 1C) and Western blot assay (Fig. 1D) were also performed, and the results were consistent with the data from gene reporter assay. Considering liver is the primary target organ for ingested B[a]P, we also examined the cyclin D1 expression in hepatocytes exposed to B[a]PDE, and the results verified that B[a]PDE was able to up-regulate cyclin D1 expression in L02 hepatocytes (Fig. 1, E–G).
FIGURE 1.
B[a]PDE-induced cyclin D1 expression in bronchial epithelial cells and hepatocytes. A, Beas-2B cyclin D1-luc cells were treated with 1 μm B[a]PDE or B[a]P for 12 h and subjected to luciferase assay. B, Beas-2B cyclin D1-luc cells were exposed to 0.5 μm B[a]PDE or B[a]P for indicated time periods and subjected to luciferase assay. C and D, Beas-2B cells were exposed to B[a]PDE as indicated for 12 h and subjected to RT-PCR or Western blot assay. E, L02 cyclin D1-luc mass1 cells were exposed to 0.5 μm B[a]PDE for 24 h followed by luciferase assay. F and G, the cells were exposed to 0.5 μm B[a]PDE dosages and subjected to RT-PCR and Western blot assay.
Akt, p70s6k, and MAPKs Are Activated by B[a]PDE, Whereas NFAT, NF-κB, and Egr-1 Are Not
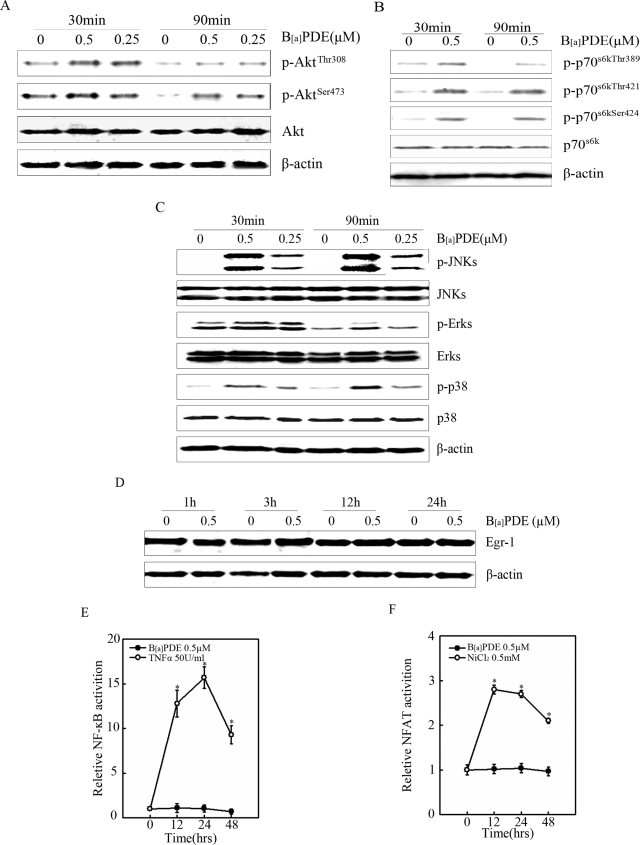
Numerous signaling cascades including PI-3K/Akt, MAPKs, IKK/NF-κB, p70s6k, and Egr-1 have been documented to participate in the regulation of cyclin D1 expression in different cell types (23, 24). Our previous results also revealed that in mouse epidermal Cl41 cells, B[a]PDE treatment resulted in marked activation of NFAT, which was responsible for B[a]PDE-induced cyclin D1 transcription in Cl41 cells. In present study, we found that phosphorylation of Akt, p70s6k, and MAPKs (Fig. 2, A–C) were significant enhanced in Beas-2B cells upon B[a]PDE exposure, whereas no obvious activation of Egr-1, NF-κB, or NFAT was observed (Fig. 2, D–F). These data demonstrated that Akt, p70s6k, and MAPKs might be implicated in B[a]PDE-induced cyclin D1 expression.
FIGURE 2.
Akt, p70s6k, and MAPKs were activated by B[a]PDE. A–D, Beas-2B cells were treated with various dosages of B[a]PDE for the indicated time periods and subjected to Western blot assay. E, Beas-2B NF-κB-luc cells were treated with B[a]PDE for indicated time periods and subjected to luciferase assay. Tumor necrosis factor α was used as a positive control. F, Beas-2B NFAT-luc cells were treated with B[a]PDE for indicated time periods and subjected to luciferase assay. Nickel chloride was used as a positive control.
JNK and Erks Are Required in B[a]PDE-induced Cyclin D1 Expression, but p38 Is Not
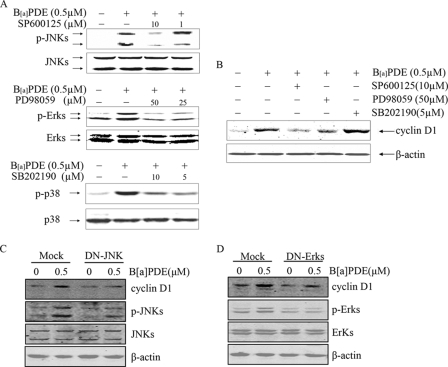
It has been well demonstrated that MAPK family, consisting of JNK, Erks, and p38, modulates the activation of signaling cascades by phosphorylating various transcription factors, which in turn regulate the target gene expression (25, 26). To clarify the role of JNK, Erks, and p38 in B[a]PDE-induced cyclin D1 expression in Beas-2B cells, specific inhibitors of these MAPK family members were used in the present study (Fig. 3A). As shown in Fig. 3B, pretreatment of the cells with SP600125 or PD98059 markedly inhibited cyclin D1 induction by B[a]PDE, but SB202190 pretreatment did not show any inhibitory effect. These results demonstrated that JNK and Erks were required in B[a]PDE-induced cyclin D1 expression, whereas p38 was not. To further confirm this finding, Beas-2B cells ectopically expressing DN-JNK or DN-Erks were treated with B[a]PDE. As shown in Fig. 3 (C and D), the impairment of JNK or Erk activation by dominant negative mutant dramatically attenuated cyclin D1 induction by B[a]PDE.
FIGURE 3.
JNK and Erks were required in B[a]PDE-induced cyclin D1 expression, but p38 was not. A, Beas-2B cells with or without chemical inhibitor pretreatment were exposed to B[a]PDE for 30 min and then subjected to Western blot analysis. B, Beas-2B cells pretreated with chemical inhibitors or not were exposed to B[a]PDE for 24 h and then subjected to Western blot analysis. C and D, Beas-2B DN-JNK mass1 cells and Beas-2B DN-Erk mass1 cells were exposed to 0.5 μm of B[a]PDE for 24 h and then subjected to Western blot analysis.
PI-3K/Akt/p70s6k-dependent Pathway Is Required for Cyclin D1 Induction by B[a]PDE
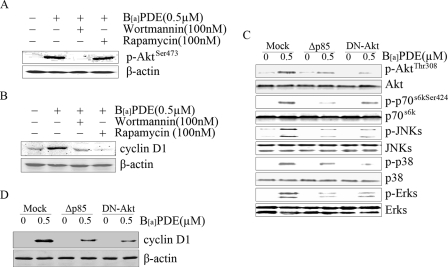
To explore whether Akt and p70s6k was involved in B[a]PDE-induced cyclin D1 expression, wortmannin, a specific inhibitor of PI-3K (which is a upstream mediator of Akt), and rampamycin, a specific inhibitor of the mTOR/p70s6k pathway, were utilized in this study. As shown in Fig. 4 (A and B), wortmannin pretreatment not only blocked Akt phosphorylation but also impaired cyclin D1 induction in Beas-2B cells upon B[a]PDE exposure, indicating that the PI-3K/Akt cascade is involved in B[a]PDE-induced cyclin D1 expression. Akt was reported to act as an upstream mediator of MAPKs in Cl41 cells exposed to B[a]PDE (27). We therefore investigated the correlation between the PI-3K/Akt cascade and MAPKs in B[a]PDE-induced cyclin D1 expression. Overexpression of Δp85 or DN-Akt not only inhibited B[a]PDE-elicited JNK and Erks activation but also impaired B[a]PDE-induced cyclin D1 expression (Fig. 4, C and D), which suggests that cyclin D1 induction by B[a]PDE is via the PI-3K/Akt/JNK- and Erk-dependent pathway. Moreover, the blockage of p70s6k phosphorylation resulted from Δp85 or DN-Akt overexpression, together with the impairment of cyclin D1 induction caused by rampamycin pretreatment, implicated that p70s6k is a downstream mediator of the PI-3K/Akt cascade in B[a]PDE response and participates in cyclin D1 induction as well.
FIGURE 4.
PI-3K/Akt/p70s6k signaling cascade was involved in B[a]PDE-induced cyclin D1 expression. Beas-2B cells with or without the pretreatment of chemical inhibitors were exposed to B[a]PDE for 30 min (A) or 24 h (B) and then subjected to Western blot analysis. Beas-2B Δp85 mass1 cells or Beas-2B DN-Akt mass1 cells were exposed to B[a]PDE for 30 min (C) or 24 h (D) and then subjected to Western blot assay.
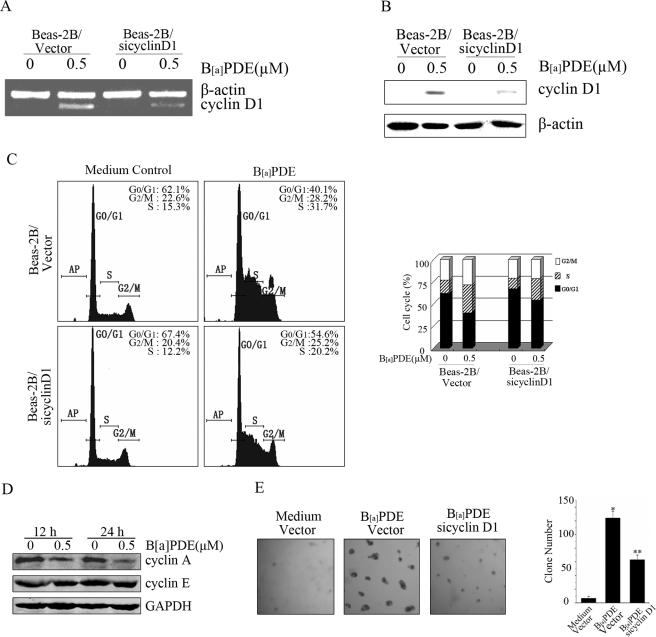
Increased Cyclin D1 by B[a]PDE Promotes Malignant Transformation of Beas-2B Cells
To further investigate the oncogenic effect of up-regulated cyclin D1 by B[a]PDE in pulmonary cells, we established cyclin D1 siRNA stable-transfected Beas-2B cells. As shown in Fig. 5 (A and B), cyclin D1 siRNA was able to specifically inhibit B[a]PDE-induced cyclin D1 expression. Fluorescence-activated cell sorter assay showed that B[a]PDE exposure significantly facilitated G1/S cell cycle transition. More importantly, B[a]PDE-triggered increase of S phase cells was dramatically reduced in cyclin D1 siRNA stable-transfected Beas-2B cells, which further confirmed the role of cyclin D1 in B[a]PDE-induced cell cycle progression (Fig. 5C). It has been accepted that induction of cyclin A or cyclin E may account for the enhanced G1/S cell cycle transition as well. But our results showed that expression of cyclin A was decreased, and cyclin E expression was not changed in B[a]PDE-treated Beas-2B cells, which excluded the role of cyclin A or cyclin E in B[a]PDE-triggered G1/S cell cycle transition (Fig. 5D). To test the potential role of cyclin D1 in B[a]PDE-induced cell transformation, colony formation assay was performed in the current study. We found that long term repeated exposure of Beas-2B cells to B[a]PDE was able to induce notable anchorage-independent cell growth, which could be significantly reduced by cyclin D1 knockdown using its specific siRNA (Fig. 5E). These results strongly indicate that induction of cyclin D1 is at least partially responsible for B[a]PDE-induced cell transformation.
FIGURE 5.
Increased cyclin D1 by B[a]PDE promotes oncogenic transformation. A and B, Beas-2B-Vc mass1 cells and Beas-2B siCyclin D1 mass1 cells were exposed to 0.5 μm B[a]PDE for 12 h and then subjected to RT-PCR assay or Western blot assay. C, Beas-2B-Vc mass1 cells and Beas-2B siCyclin D1 mass1 cells were starved for 24 h and then exposed to 0.5 μm of B[a]PDE for 12 h followed by flow cytometry assay. D, Beas-2B cells were treated with 0.5 μm B[a]PDE for the indicated time periods and subjected to Western blot assay. E, Beas-2B-Vc mass1 cells and Beas-2B siCyclin D1 mass1 cells were repeatedly treated with 0.5 μm of B[a]PDE for 12 weeks and then subjected to colony formation assay as described under “Materials and Methods.”
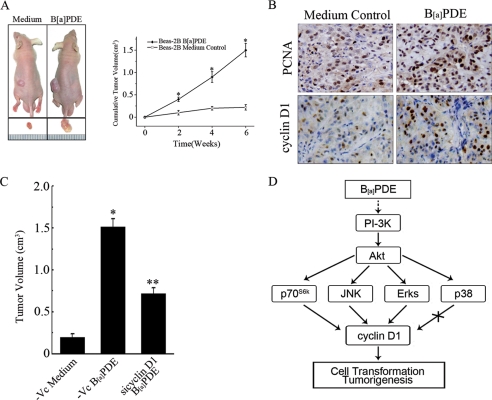
B[a]PDE-induced Cyclin D1 Up-regulation Facilitates Tumorigenesis of Beas-2B Cells
It is well accepted that the capacity of anchorage-independent growth of transformed cells in soft agar reflects their tumorigenicity in vivo. To evaluate the tumorigenicity of Beas-2B cells that have undergone long term B[a]PDE exposure, the nude mice were subcutaneously injected with B[a]PDE-treated Beas-2B cells or Beas-2B control cells, respectively. The results showed that long term repeated exposure of B[a]PDE robustly increased the tumorigenicity of Beas-2B cells (Fig. 6A). Immunohistochemistry revealed that the expression of proliferating cell nuclear antigen and cyclin D1 in xenograft tumor of B[a]PDE-treated cells was significantly higher than that in control tumor (Fig. 6B). More importantly, the size of xenograft tumors of cyclin D1 siRNA stable-transfected cells was significantly reduced compared with those of control cells (Fig. 6C), which further confirmed that cyclin D1 is involved in B[a]PDE-induced tumorigenesis. Taken together, these data demonstrated that cyclin D1 induction by B[a]PDE, via the PI-3K/Akt/MAPK and p70s6k-dependent pathway, is able to promote cell transformation and tumorigenesis (Fig. 6D).
FIGURE 6.
B[a]PDE-induced cyclin D1 up-regulation facilitates tumorigenesis. A, Beas-2B cells with or without B[a]PDE long term treatment were subcutaneously inoculated into the nude mice. A representative picture was taken in the fourth week post inoculation. The tumor dimensions were measured as described under “Materials and Methods.” B, xenografted tumors were excised and subjected to immunohistochemical staining. C, Beas-2B-Vc mass1 cells and Beas-2B siCyclin D1 mass1 cells repeatedly treated with B[a]PDE were injected subcutaneously into each spot of nude mice separately. The tumor volume was calculated as described above. D, illusion of the signaling pathway involved in B[a]PDE-induced cyclin D1 expression.
DISCUSSION
It has been well accepted that B[a]PDE exposure is closely associated with an increased risk of human lung cancer (2, 5, 9). However, the molecular mechanism of the B[a]PDE-induced carcinogenesis is far from being fully understood. In the current study, we found that B[a]PDE is able to induce cyclin D1 expression in human bronchial epithelial cells and hepatocytes. We also demonstrated that JNK, Erk, and p70s6k are involved in cyclin D1 induction by B[a]PDE in Beas-2B cells. Further study elucidated that PI-3K/Akt is the upstream mediator of both MAPK and p70s6k in B[a]PDE response and is required in B[a]PDE-induced cyclin D1 up-regulation. More importantly, we demonstrate here that the increased cyclin D1 by B[a]PDE facilitates cell transformation and tumorigenesis of bronchial epithelial cells, which may be at least partially responsible for B[a]PDE carcinogenicity in human lung cancer.
Cyclin D1 is an important regulator of cell cycle progression (G1 to S phase) and acts as a sensor of cell cycle machinery in response to growth factor signaling or extracellular stress (28) but its functions are more sophisticated than simple acceleration of cell cycle transition. Actually, cyclin D1 overexpression has been linked to the promotion and progression of cancer. Roy and Thompson (29) emphasized the importance of cyclin D1 not only in cell growth regulation (normal and abnormal) but also in mammary gland development and carcinogenesis (15). Cyclin D1 has been reported to distinguish benign and premalignant breast lesions from any form of human breast carcinoma (30). Gautschi et al. (16) concluded in their review that cyclin D1 overexpression is not a consequence but a pivotal element in the process of malignant transformation of pulmonary cells. Moreover, cyclin D1 is implicated in tumor vascularization by mediating vascular endothelial growth factor-stimulated growth of vascular endothelial cells (31). Cyclin D1 accumulation is regulated at multiple levels including transcription, post-translational activation, and intracellular localization throughout the cell cycle (32). Our previous study indicated that cyclin D1 transcription can be up-regulated by B[a]PDE or B[a]PDE plus ionizing radiation through NFAT3-involved pathway in mouse epidermal Cl 41 cells (14). In the current study, B[a]PDE-induced cyclin D1 expression was first verified at both RNA and protein levels in human bronchial epithelial Beas-2B cells.
In general, JNK and p38, which are usually activated by various extracellular stimuli such as oxidative stress or UV irradiation, play critical roles in the mediation of apoptosis (33), whereas Erk is required in cell growth and division (34). However, the functions of these MAPK members may be diverse depending on the cell types and stimuli. Previous studies have documented that MAPKs play a pivotal role in the regulation of cyclin D1 expression. Kashima et al. reported that activation of the MAPK pathway followed by up-regulation of cyclin D1 and cyclin E was involved in estrogen-induced overgrowth of endometrial carcinoma cells (35). Rac/Erk-dependent cyclin D1 induction has been clarified to correlate with heregulin-promoted cell proliferation of breast cancer (36). The MEK/Erk pathway was reported to up-regulate cyclin D1 transcription, as well as to facilitate the assembly of cyclin D1 with cyclin-dependent kinase 4 in murine fibroblast (37). p38 activation was also reported to promote heregulin-induced cell cycle progression and cyclin D1 expression in breast cancer cells (38). Moreover, MAPK was recognized as an essential mediator of gene expression in cells upon B[a]PDE exposure. Our previous study has verified that the MAPK/activator protein-1-dependent pathway is responsible for COX-2 induction by B[a]PDE in mouse epidermal Cl 41 cells (39). In the current study, we showed that JNK, Erk, and p38 were all activated by B[a]PDE in Beas-2B cells, but only Erk and JNK were involved in the B[a]PDE-induced cyclin D1 expression.
Besides the MAPK pathway, the PI-3K/Akt cascade also exhibits distinct activation in response to various extracellular stimuli (40). Aberrant activation of Akt is frequently observed in many types of human cancers (41). Constitutive activation of PI-3K/Akt signaling has been regarded as an indispensable event during cancer development, although the detailed mechanism has not been fully characterized (42). Accumulating evidence has indicated that activation of both MAPK and PI-3K/Akt pathways are usually necessary, yet insufficient alone, to promote cell survival, cell growth, and carcinogenesis (38). It was reported that inhibition of PI-3K in normal T-cells attenuated the activation of Erk signaling pathway (43). A previous study by Li et al. (27) also showed that Akt may act as upstream regulator of JNKs in Cl 41 cells upon B[a]PDE exposure. A recent study demonstrated that cyclin D1 is a downstream target of the PI-3K/Akt cascade and participates in the cell transformation caused by arsenite in human keratinocytes (44). In present study, we demonstrated that B[a]PDE-induced cyclin D1 expression is via the PI-3K/Akt/MAPK- and p70s6k-dependent pathway.
p70s6k is one of the important downstream protein kinases of the PI-3K/Akt pathway. Previous studies showed the activation of p70s6k is critical for the oncogenic properties of the PI-3K/Akt pathway (45). Tumors and tumor-derived cell lines in which the PI-3K pathway is aberrantly activated usually exhibit an increased sensitivity to rapamycin or rapamycin analogues (46–48). Other studies have also suggested the biological significance of the PI-3K/Akt/p70s6k pathway in thyroid tumor growth (49), NO-induced cyclin D1 expression (50), and the proliferation of multiple myeloma cells (51). Currently, our results demonstrated that p70s6k was involved in B[a]PDE-induced cyclin D1 expression in human pulmonary cells.
There is a growing amount of evidence indicating the essential role of cyclin D1 in the oncogenic transformation of cells (52). Ouyang et al. (44) reported that cyclin D1 induction by arsenite, via the PI-3K/Akt-dependent pathway, is responsible for arsenite-induced malignant transformation of human keratinocyte. Our current work elucidated that cyclin D1 is a downstream target of the PI-3K/Akt/MAPK cascade and participates in the cell transformation caused by B[a]PDE in human bronchial epithelial cells. Although the precise mechanism of cyclin D1-mediated cell transformation is still elusive, it is accepted that chronic checkpoint activation triggered by the nuclear cyclin D1-cyclin-dependent kinase complex might provide the cells with not only a growth advantage but also the tendency of genomic instability (52).
In summary, our study demonstrates that the PI-3K/Akt pathway, as well as downstream MAPK and p70s6k, is involved in B[a]PDE-induced cyclin D1 expression, and the increased cyclin D1 is required for the transformation and tumorigenesis of human bronchial epithelial cells exposed to B[a]PDE. These results not only broaden our understanding on the molecular mechanism of B[a]PDE carcinogenicity but also lead to the development of chemoprevention and therapeutic strategies for B[a]PDE-associated human cancers.
This work was supported in part by National Natural Science Foundation of China Grants 30530790, 30921006, and 2008ZX10002; National High Tech Research and Development Program Grant 2006AA02A412; and Shanghai Key Basic Science Foundation Grants 07DJ14006 and 06DJ14009.
- B[a]P
- benzo[a]pyrene
- B[a]PDE
- (±)-anti-benzo[a]pyrene-7,8-diol-9,10-epoxide
- MAPK
- mitogen-activated protein kinase
- Erk
- extracellular signal-regulated kinase
- JNK
- c-Jun N-terminal kinase
- NFAT
- nuclear factor of activated T-cells
- NF-κB
- nuclear factor-κB
- PI-3K
- phosphatidylinositol 3-kinase
- siRNA
- small interfering RNA
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- MEK
- MAPK/Erk kinase
- DN
- dominant negative
- RT
- reverse transcription.
REFERENCES
- 1.Baird W. M., Hooven L. A., Mahadevan B. (2005) Environ. Mol. Mutagen. 45, 106–114 [DOI] [PubMed] [Google Scholar]
- 2.Boström C. E., Gerde P., Hanberg A., Jernström B., Johansson C., Kyrklund T., Rannug A., Törnqvist M., Victorin K., Westerholm R. (2002) Environ. Health Perspect. 110, (Suppl. 3) 451–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexandrov K., Cascorbi I., Rojas M., Bouvier G., Kriek E., Bartsch H. (2002) Carcinogenesis 23, 1969–1977 [DOI] [PubMed] [Google Scholar]
- 4.Wynder E. L., Graham E. A. (1950) J. Am. Med. Assoc. 143, 329–336 [DOI] [PubMed] [Google Scholar]
- 5.Phillips D. H. (1983) Nature 303, 468–472 [DOI] [PubMed] [Google Scholar]
- 6.Smith L. E., Denissenko M. F., Bennett W. P., Li H., Amin S., Tang M., Pfeifer G. P. (2000) J. Natl. Cancer Inst. 92, 803–811 [DOI] [PubMed] [Google Scholar]
- 7.Klaunig J. E., Kamendulis L. M. (2004) Annu. Rev. Pharmacol. Toxicol. 44, 239–267 [DOI] [PubMed] [Google Scholar]
- 8.Huang C., Ma W. Y., Ryan C. A., Dong Z. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 11957–11962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krawczak M., Cooper D. N. (1998) Mutagenesis 13, 319–320 [DOI] [PubMed] [Google Scholar]
- 10.Hollstein M., Sidransky D., Vogelstein B., Harris C. C. (1991) Science 253, 49–53 [DOI] [PubMed] [Google Scholar]
- 11.Albert R. E., Miller M. L., Cody T., Andringa A., Shukla R., Baxter C. S. (1991) Carcinogenesis 12, 1273–1280 [DOI] [PubMed] [Google Scholar]
- 12.Massagué J. (2004) Nature 432, 298–306 [DOI] [PubMed] [Google Scholar]
- 13.Kastan M. B., Bartek J. (2004) Nature 432, 316–323 [DOI] [PubMed] [Google Scholar]
- 14.Ding J., Zhang R., Li J., Xue C., Huang C. (2006) Mol. Cell Biochem. 287, 117–125 [DOI] [PubMed] [Google Scholar]
- 15.Arber N., Doki Y., Han E. K., Sgambato A., Zhou P., Kim N. H., Delohery T., Klein M. G., Holt P. R., Weinstein I. B. (1997) Cancer Res. 57, 1569–1574 [PubMed] [Google Scholar]
- 16.Gautschi O., Ratschiller D., Gugger M., Betticher D. C., Heighway J. (2007) Lung Cancer 55, 1–14 [DOI] [PubMed] [Google Scholar]
- 17.Yan Y. X., Nakagawa H., Lee M. H., Rustgi A. K. (1997) J. Biol. Chem. 272, 33181–33190 [DOI] [PubMed] [Google Scholar]
- 18.Ding J., Li J., Xue C., Wu K., Ouyang W., Zhang D., Yan Y., Huang C. (2006) J. Biol. Chem. 281, 24405–24413 [DOI] [PubMed] [Google Scholar]
- 19.Huang C., Ma W. Y., Dong Z. (1996) Mol. Cell Biol. 16, 6427–6435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Watts R. G., Huang C., Young M. R., Li J. J., Dong Z., Pennie W. D., Colburn N. H. (1998) Oncogene 17, 3493–3498 [DOI] [PubMed] [Google Scholar]
- 21.Huang C., Li J., Ma W. Y., Dong Z. (1999) J. Biol. Chem. 274, 29672–29676 [DOI] [PubMed] [Google Scholar]
- 22.Ouyang W., Ma Q., Li J., Zhang D., Liu Z. G., Rustgi A. K., Huang C. (2005) Cancer Res. 65, 9287–9293 [DOI] [PubMed] [Google Scholar]
- 23.Huang C., Li J., Ding M., Wang L., Shi X., Castranova V., Vallyathan V., Ju G., Costa M. (2001) Mol. Cell Biochem. 222, 29–34 [PubMed] [Google Scholar]
- 24.Zhang X., Jin B., Huang C. (2007) Curr. Cancer Drug Targets 7, 305–316 [DOI] [PubMed] [Google Scholar]
- 25.Hagemann C., Blank J. L. (2001) Cell Signal. 13, 863–875 [DOI] [PubMed] [Google Scholar]
- 26.Karin M., Hunter T. (1995) Curr. Biol. 5, 747–757 [DOI] [PubMed] [Google Scholar]
- 27.Li J., Tang M. S., Liu B., Shi X., Huang C. (2004) Oncogene 23, 3932–3944 [DOI] [PubMed] [Google Scholar]
- 28.Winston J. T., Pledger W. J. (1993) Mol. Biol. Cell 4, 1133–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roy P. G., Thompson A. M. (2006) Breast 15, 718–727 [DOI] [PubMed] [Google Scholar]
- 30.Weinstat-Saslow D., Merino M. J., Manrow R. E., Lawrence J. A., Bluth R. F., Wittenbel K. D., Simpson J. F., Page D. L., Steeg P. S. (1995) Nat. Med. 1, 1257–1260 [DOI] [PubMed] [Google Scholar]
- 31.Yasui M., Yamamoto H., Ngan C. Y., Damdinsuren B., Sugita Y., Fukunaga H., Gu J., Maeda M., Takemasa I., Ikeda M., Fujio Y., Sekimoto M., Matsuura N., Weinstein I. B., Monden M. (2006) Clin. Cancer Res. 12, 4720–4729 [DOI] [PubMed] [Google Scholar]
- 32.Gladden A. B., Diehl J. A. (2005) J. Cell Biochem. 96, 906–913 [DOI] [PubMed] [Google Scholar]
- 33.Boldt S., Weidle U. H., Kolch W. (2002) Carcinogenesis 23, 1831–1838 [DOI] [PubMed] [Google Scholar]
- 34.Johnson G. L., Lapadat R. (2002) Science 298, 1911–1912 [DOI] [PubMed] [Google Scholar]
- 35.Kashima H., Shiozawa T., Miyamoto T., Suzuki A., Uchikawa J., Kurai M., Konishi I. (2008) Endocr. Relat. Cancer 16, 113–122 [DOI] [PubMed] [Google Scholar]
- 36.Yang C., Klein E. A., Assoian R. K., Kazanietz M. G. (2008) Biochem. J. 410, 167–175 [DOI] [PubMed] [Google Scholar]
- 37.Cheng M., Sexl V., Sherr C. J., Roussel M. F. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 1091–1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neve R. M., Holbro T., Hynes N. E. (2002) Oncogene 21, 4567–4576 [DOI] [PubMed] [Google Scholar]
- 39.Ouyang W., Ma Q., Li J., Zhang D., Ding J., Huang Y., Xing M. M., Huang C. (2007) Mol. Carcinog. 46, 32–41 [DOI] [PubMed] [Google Scholar]
- 40.Cheng G. Z., Park S., Shu S., He L., Kong W., Zhang W., Yuan Z., Wang L. H., Cheng J. Q. (2008) Curr. Cancer Drug Targets 8, 2–6 [PubMed] [Google Scholar]
- 41.Tokunaga E., Oki E., Egashira A., Sadanaga N., Morita M., Kakeji Y., Maehara Y. (2008) Curr. Cancer Drug Targets 8, 27–36 [DOI] [PubMed] [Google Scholar]
- 42.Samuels Y., Ericson K. (2006) Curr. Opin. Oncol. 18, 77–82 [DOI] [PubMed] [Google Scholar]
- 43.Eder A. M., Dominguez L., Franke T. F., Ashwell J. D. (1998) J. Biol. Chem. 273, 28025–28031 [DOI] [PubMed] [Google Scholar]
- 44.Ouyang W., Luo W., Zhang D., Jian J., Ma Q., Li J., Shi X., Chen J., Gao J., Huang C. (2008) Environ. Health Perspect. 116, 1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ouyang W., Li J., Ma Q., Huang C. (2006) Carcinogenesis 27, 864–873 [DOI] [PubMed] [Google Scholar]
- 46.Manning B. D., Cantley L. C. (2003) Biochem. Soc. Trans. 31, 573–578 [DOI] [PubMed] [Google Scholar]
- 47.Noh W. C., Mondesire W. H., Peng J., Jian W., Zhang H., Dong J., Mills G. B., Hung M. C., Meric-Bernstam F. (2004) Clin. Cancer Res. 10, 1013–1023 [DOI] [PubMed] [Google Scholar]
- 48.Baldo P., Cecco S., Giacomin E., Lazzarini R., Ros B., Marastoni S. (2008) Curr. Cancer Drug Targets 8, 647–665 [DOI] [PubMed] [Google Scholar]
- 49.Furuya F., Hanover J. A., Cheng S. Y. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1780–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pervin S., Singh R., Hernandez E., Wu G., Chaudhuri G. (2007) Cancer Res. 67, 289–299 [DOI] [PubMed] [Google Scholar]
- 51.Pene F., Claessens Y. E., Muller O., Viguié F., Mayeux P., Dreyfus F., Lacombe C., Bouscary D. (2002) Oncogene 21, 6587–6597 [DOI] [PubMed] [Google Scholar]
- 52.Pontano L. L., Diehl J. A. (2008) Cell Div. 3, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]