Abstract
CUL4A and CUL4B, which are derived from the same ancestor, CUL4, encode scaffold proteins that organize cullin-RING ubiquitin ligase (E3) complexes. Recent genetic studies have shown that germ line mutation in CUL4B can cause mental retardation, short stature, and other abnormalities in humans. CUL4A was observed to be overexpressed in breast and hepatocellular cancers, although no germ line mutation in human CUL4A has been reported. Although CUL4A has been known to be involved in a number of cellular processes, including DNA repair and cell cycle regulation, little is known about whether CUL4B has similar functions. In this report, we tested the functional importance of CUL4B in cell proliferation and characterized the nuclear localization signal (NLS) that is essential for its function. We found that RNA interference silencing of CUL4B led to an inhibition of cell proliferation and a prolonged S phase, due to the overaccumulation of cyclin E, a substrate targeted by CUL4B for ubiquitination. We showed that, unlike CUL4A and other cullins that carry their NLS in their C termini, NLS in CUL4B is located in its N terminus, between amino acid 37 and 40, KKRK. This NLS could bind to importin α1, α3, and α5. NLS-deleted CUL4B was distributed in cytoplasm and failed to promote cell proliferation. Therefore, the nuclear localization of CUL4B mediated by NLS is critical for its normal function in cell proliferation.
INTRODUCTION
Cullins function as a “scaffold” in cullin-RING-based E3 ubiquitin ligases (CRLs).3 CRLs constitute a major subclass of RING finger E3s that regulate diverse cellular processes, including cell cycle progression, transcription, signal transduction, and development (1, 2). Cullins are evolutionarily conserved from yeast to mammals. Although sequence homology spans the entire protein, the C terminus, characterized by the ∼200-amino acid cullin homology domain, is most conserved. Humans encode seven cullin members, CUL1, CUL2, CUL3, CUL4A, CUL4B, CUL5, and CUL7 (2). CUL4A and CUL4B are derived from one ancestor, CUL4, which exists in Schizosaccharomyces pombe (Pcu4), Xenopus laevis, Caenorhabditis elegans, Drosophila melanogaster, and Arabidopsis thaliana but is absent in Saccharomyces cerevisiae. CUL4A and CUL4B also exist in other higher organisms, including zebrafish and the mouse (3). The protein sequences between human CUL4A and CUL4B are 83% identical, with CUL4B having a unique N terminus of 149 amino acids (supplemental Fig. S1). CUL4A CRL complexes were shown to contain Rbx1 and the adaptor protein DDB1. DDB1 interacts with WD-40 repeat motif-containing proteins that determine the substrate specificity of the CUL4A ubiquitin ligase complex (3–7).
Loss of Cul4 in Drosophila cells leads to G1 arrest that is associated with an increase in the cyclin-dependent kinases (CDK) inhibitor Dacapo (8). Recent genetic studies have shown that mutations in CUL4B gene can cause an X-linked mental retardation syndrome (9, 10). However, no germ line mutation in human CUL4A has been reported, although overexpression of CUL4A was observed in breast and hepatocellular cancers (11–13).
Cul4b knock-out mice have not been reported. Cul4a knock-out mice were shown to have great variations in phenotypes by different groups (14, 15), from embryonic lethality associated with homozygous deletion of exon 1 (14) to lack of remarkable abnormalities in mutants with homozygous deletion of exons 17–19 (15). Deletion of exon 1 in Cul4a was recently shown to disrupt the function of Pcid2 on the complementary strand that is essential for development (15). However, it should be noted that homozygous deletion of exons 4–8 in Cul4a was also shown to cause severe proliferation defects in embryonic fibroblasts and hepatocytes and was associated with an increase in genome instability (16). Therefore, the function of CUL4A in development remains to be studied further.
Although many proteins, including CDT1, p21, p27, p53, c-Jun, HOXA9, H3, and CHK1, have been reported to be the substrates of CUL4A-DDB1 ubiquitin ligases (17, 18), much less is known about the targets of CUL4B. Recent studies showed that CUL4B can accumulate in the nucleus (19). Moreover, CUL4B can mediate the nuclear transportation of DDB1 independent of DDB2 (19). However, a functional nuclear localization signal (NLS) in CUL4B has not been identified. In this study, by tagging CUL4B with enhanced green fluorescent protein (EGFP) in a series of constructs containing deletions and point mutations, we identified a functional NLS and characterized its interaction with nuclear import receptor proteins. We demonstrated that lack of NLS rendered CUL4B nonfunctional in the proteolysis of cyclin E, which, when overaccumulated, prolongs S phase progression.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfections
HeLa, HEK293, C6, C2C12, U2OS, PC3, CRL1730, U87, U251, and MCF7 cell lines were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum plus penicillin and streptomycin, and PC12 cells were cultured as described previously (20). All cells were cultured at 37 °C in a humidified atmosphere of 5% CO2.
For cell synchronization at G0 phase, cells were starved in Dulbecco's modified Eagle's medium with 0.2% fetal bovine serum for 48 h. After starvation, cells were stimulated to initiate a new cell cycle in fresh medium containing 10% fetal bovine serum for 20 h and then were used for a BrdUrd incorporation assay and fluorescence-activated cell sorting analysis. For synchronization of the cell cycle at the G1/S boundary, a double-thymidine block was performed. Briefly, cells were treated with 2 mm thymidine for 18 h, followed by a 10-h release in fresh medium without thymidine and successive retreatment with the drug for 16 h. Transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Plasmids
pcDNA3.1/Myc-His A-CUL4B was made by subcloning a PCR-amplified CUL4B fragment using HeLa cDNA as template in frame into the pcDNA3.1/Myc-His A vector (Invitrogen) between the BamHI and XhoI sites. pECFP-C1-CUL4B was constructed by ligating the KpnI and ApaI fragment from pcDNA3.1/Myc-His A-CUL4B in frame to the pEGFP-C1 vector (Clontech).
pEGFP-C1-CUL4B 1–36, 1–40, 1–50, and 1–115 were generated by ligating PCR-amplified fragments from pEGFP-C1-CUL4B into XhoI and BamHI sites of pEGFP-C1 vector, respectively. To construct pEGFP-C1-CUL4B 37–895, 41–895, and 150–895, different PCR fragments were subcloned into the pEGFP-C1-CUL4B vector in which the N-terminal fragment of CUL4B was removed by digestion with KpnI and ScaI. pEGFP-C1-CUL4B 1–729 was generated by ligating the PCR fragment into the pEGFP-C1-CUL4B in which the C-terminal fragment of CUL4B was removed by digestion with HhaI and ApaI. Point mutations and internal deletion mutants within the full-length protein were generated by site-directed mutagenesis using overlap extension PCR (21). Products of the extension PCR were subcloned into the pEGFP-C1-CUL4B vector in which the N-terminal fragment of CUL4B was removed by digestion with KpnI and ScaI.
The RNAi-resistant expression vectors pEGFP-C1-CUL4B(R), pEGFP-C1-CUL4B 150–895(R), and pEGFP-C1-CUL4BΔ37–40(R) used in rescue experiments were constructed by introducing three silent point mutations to the region of the CUL4B that is targeted by the RNAi. Site-directed mutagenesis was generated by overlap extension PCR described above. The products of the extension PCR were subcloned into the pEGFP-C1-CUL4B, pEGFP-C1-CUL4B 150–895, and pEGFP-C1-CUL4BΔ37–40 vectors in which the C-terminal fragment of CUL4B was removed by digestion with HhaI and ApaI. All primer sequences are listed in supplemental Table S1.
The plasmids containing importins α1, α3, α5, and β were kindly provided by Dr Anne Carine Østvold (22). Importins α1, α3, and β were cloned into pGEX-2T (Amersham Biosciences) vector, and importin α5 was cloned into pGEX5-X-3 (Amersham Biosciences).
Immunofluorescence
Cells were seeded onto coverslips in the wells of 24-well plates. After washing with phosphate-buffered saline (PBS), cells were fixed with 4% paraformaldehyde for 20 min, washed twice with PBS, and permeabilized with 0.2% Triton X-100 for 20 min and then blocked with 5% goat serum in PBS for 1 h at room temperature. Cells were then incubated overnight at 4 °C with the primary antibodies at the appropriate dilution. The antibodies used were mouse monoclonal anti-Myc (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and mouse polyclonal anti-CUL4B (Abcam). Cells were then washed with PBS and incubated for 60 min at 37 °C with rhodamine-conjugated secondary antibody (Jingmei). After washing with PBS three times, cells were further stained with 4,6-diamidino-2-phenylindole for 10 min. The fluorescence signal was examined with a DP71 fluorescence microscope (Olympus). Cells transfected with the EGFP fusion plasmids were observed directly with an Axiovert 200 inverted fluorescence microscope (Zeiss).
Micronucleus Assay
Micronulei were scored as previously reported (23). Briefly, cells were seeded onto coverslips in the wells of 6-well plates and grown for 48 h. At least 1000 cells/sample were scored for analysis. All experiments were repeated three times.
Whole, Cytoplasmic, and Nuclear Protein Extraction
Total cellular extracts were obtained using cell lysis buffer for Western and IP (Beyotime) according to the manufacturer's instructions. Cytoplasmic and nuclear extracts were prepared using a nuclear and cytoplasmic protein extraction kit (Beyotime) following the manufacturer's instructions. Briefly, cells were washed with ice-cold PBS and then lysed using cell lysis buffer containing 10 mm Hepes, pH 7.9, 10 mm KCl, 0.1 mm EDTA, 1 mm dithiothreitol, 0.4% IGEPAL, 1 mm phenylmethanesulfonyl fluoride and incubated on ice for 20 min. After centrifugation, supernatants (corresponding to cytoplasmic extracts) were collected. The nuclear pellets were further washed in ice-cold cell lysis buffer and resuspended in nuclear extraction buffer (0.4 m NaCl, 20 mm Hepes, pH 7.9, 1 mm EDTA, 1 mm dithiothreitol, 1 mm phenylmethanesulfonyl fluoride). After vigorously shaking for 30 min at 4 °C, the nuclear extract was collected by centrifuged. Whole cell lysates and cytoplasmic and nuclear extracts were quantified using a BCA protein assay kit (Beyotime).
Western Blotting
Western blotting was performed as described previously (20). In brief, equal amounts (30–60 μg) of extracts were subjected to 12% SDS-polyacrylamide gel for electrophoresis, transferred to a polyvinylidene difluoride membrane (Amersham Biosciences), followed by incubation with specific primary antibodies at the appropriate dilution overnight at 4 °C. The primary antibodies used include mouse anti-α-tubulin (Santa Cruz Biotechnology, Inc.), mouse anti-p53 (Santa Cruz Biotechnology, Inc.), mouse anti-p21 (Santa Cruz Biotechnology, Inc.), mouse anti-β-actin (Sigma), mouse anti-GST (Sigma), rabbit anti-GFP (Sigma), rabbit anti-H2A (Santa Cruz Biotechnology, Inc.), rabbit anti-p27 Kip1 (Abcam), rabbit anti-cyclin D1 (Abcam), rabbit anti-cyclin E (Abcam), rabbit anti-CHK1 (Abcam), rabbit anti-CDK2 (Abcam), rabbit anti-CDT1 (Santa Cruz Biotechnology, Inc.), mouse anti-CDC6 (Santa Cruz Biotechnology, Inc.), rabbit anti-E2F1 (Cell Signaling Technology), rabbit anti-CDC25C (Santa Cruz Biotechnology, Inc.), and rabbit anti-WEE1 (Santa Cruz Biotechnology, Inc.). Polyclonal anti-CUL4B antibodies were generated by immunizing rabbits with a synthetic peptide corresponding to residues 16–44 of human CUL4B (AAQEVRSATDGNTSTTPPTSAKKRKLNSS). After they were washed with TBST, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Chemiluminescence detection was performed by the ECL PLUS kit (Amersham Biosciences).
Reverse Transcription-PCR and Real-time PCR Assay
Total RNA from cultured cells was isolated using TRIzol reagent (Invitrogen) and treated with RQ1 RNase-free DNase (Promega) to eliminate genomic DNA contamination. Total RNA was transcribed to generate cDNA using the avian myoblastosis virus reverse transcriptase (Promega). Real-time quantitative PCR was performed using the TaqManTM 7500 instrument (PE Applied Biosystems). Human glyceraldehyde-3-phosphate dehydrogenase was used as endogenous control. The levels of CUL4A and CUL4B mRNA were measured by a TaqMan probe-based assay using TaqMan Universal PCR Master Mix (Applied Biosystems). The primers and TaqMan probes were designed using the Primer Express program version 2.0 (Applied Biosystems). The mRNA levels of cyclin E was measured by a SYBR Green I assay using SYBR Green Universal PCR Master Mix (Applied Biosystems). The primers used for cyclin E were from the primer bank of Harvard Medical School (available on the World Wide Web). All primer sequences are listed in supplemental Table S2. For the SYBR Green I assay, melting curve analysis was carried out after amplification to confirm amplification of specific transcripts. Each reaction was run in triplicate and in parallel. All cDNA was measured in triplicate. The relative amounts of transcripts were calculated by the relative quantification (ΔΔCt) study method by using 7500 System SDS Software version 1.2 (Applied Biosystems).
GST Pull-down Experiments
Plasmids expressing GST or GST fusion proteins were transformed into Escherichia coli BL21. The bacteria were grown for 14 h at 37 °C followed by the addition of 0.7 mm isopropyl-1-thio-β-d-galactopyranoside for 6 h at 30 °C. The bacteria were harvested and lysed by sonication. Then GST and GST fusion proteins were incubated with glutathione-Sepharose 4B (Amersham Biosciences) for 4 h at 4 °C followed by extensive washing with PBS. Afterward, GST and GST fusion proteins immobilized on glutathione-Sepharose 4B beads were incubated with transfected or wild-type HeLa cells lysates (3 mg) in cell lysis buffer overnight at 4 °C with gentle agitation. The beads were collected by centrifugation and washed six times with cell lysis buffer. After removing the supernatant in the final wash, the samples were resuspended in 2× SDS loading buffer and boiled for 5 min, and the proteins retained on the beads were analyzed by Western blotting.
RNA Interference
For knockdown of CUL4B expression in cells, we used the BLOCK-iTTM Pol II miR RNAi expression vector kit (Invitrogen) according to the manufacturer's instructions. Single-stranded DNA oligomers were designed using Invitrogen RNAi Designer (available on the World Wide Web), annealed, and cloned into the pcDNA6.2-GW/EmGFP-miR vector, which contains a GFP expression cassette. The oligomer sequences are as follows: 5′-TGCTGAATATTTCCCGGAACATTCTGGTTTTGGCCACTGACTGACCAGAATGTCGGGAAATATT-3′ and 5′-CCTGAATATTTCCCGACATTCTGGTCAGTCAGTGGCCAAAACCAGAATGTTCCGGGAAATATTC-3′. The resulting construct was transformed into E. coli TOP10, and the correct expression vector was verified by DNA sequencing. The control vector pcDNA 6.2-GW/EmGFP-miR-neg encodes an mRNA not to target any known vertebrate gene.
For stable knockdown, cells were incubated in the selective medium containing 10 μg/ml blasticidin (Invitrogen) 24 h after transfection. Three weeks after selection, cells resistant to blasticidin were isolated, expanded, and tested by real-time PCR and Western blotting analysis.
For cyclin E knockdown, small interfering RNA (siRNA) duplexes were used. The cyclin E and negative control siRNA duplexes were purchased from GenePharma, and the oligonucleotide sequences were as follows: cyclin E, 5′-CCAUGGAAUUAAUGAUUAUdTdT-3′; negative control, 5′-UUCUCCGAACGUGUCACGUdTdT-3′.
Cell Growth and Apoptosis Assays
Effect of CUL4B silencing on HeLa cell proliferation was measured by MTT assay (24). Briefly, cells were seeded into each well of the 96-well culture plates, and 20 μl of MTT reagent (5 mg/ml) was added at the 24, 48, 72, 96, and 120 h time points, respectively. The plate was incubated at 37 °C in 5% CO2. 4 h later, the medium was replaced with 100 μl of DMSO, and the plate was vortexed for 30 min at room temperature. The absorbance of each well was measured at 490 nm. Each experiment was repeated three times and carried out in a set of 10 wells.
Apoptotic cells were detected in situ by a TUNEL assay using an in situ cell death detection kit (TMR red) according to the manufacturer's instructions (Roche Applied Science), as previously described (23). At least 600 cells from five different fields were examined in each experiment, and the cell death was expressed as a percentage of TUNEL-positive cells. All experiments were repeated three times.
BrdUrd Incorporation and Flow Cytometry Assay
Cells grown on coverslips were labeled with 50 μm BrdUrd (Sigma) in normal medium for 45 min before being harvested. Afterward, cells were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, denatured with 2 m HCl, and neutralized with 0.1 m sodium borate (pH 8.5). After blocking with 5% goat serum, cells were incubated with anti-BrdUrd monoclonal antibody (Sigma) followed by incubation with rhodamine-conjugated secondary antibody. Cells were counterstained with 4,6-diamidino-2-phenylindole. At least 500 cells from five different microscopic views were examined for each sample, and the BrdUrd incorporation is presented as the percentage of positive nuclei relative to total cell number. All experiments were repeated three times.
The cell cycle distribution was analyzed by flow cytometry as previously described (23). Briefly, cells were trypsinized, collected, and fixed in 70% ice-cold ethanol at −20 °C for at least 14 h. Fixed cells were then washed with PBS and stained with propidium iodide (5 μg/ml) in PBS containing 0.2% Tween 20 and 2.5 μg/ml RNase for 30 min at 37 °C. The cell cycle distribution was analyzed by measuring DNA content using a FACSCalibur flow cytometer (BD Biosciences).
Statistical Analysis
The statistical significance of the differences in data from the two groups was assessed by a two-tailed unpaired t test using SPSS13.0 for Windows. Results were considered statistically significant where p was <0.05.
RESULTS
Both Tagged and Endogenous CUL4B Accumulate in the Nuclei
In order to examine the subcellular localization of CUL4B protein, the EGFP-CUL4B expression construct was generated by fusing the entire open reading frame of human CUL4B gene to the C-terminal of EGFP. Western blotting analysis with anti-GFP antibody indicated that the EGFP-CUL4B fusion protein was properly expressed in the transiently transfected HeLa cells (Fig. 1A). Consistent with the previous report, wild type CUL4B tagged with EGFP was detected exclusively in the nucleus (Fig. 1B, left), whereas EGFP alone was diffusely distributed in both cytoplasm and nucleus (Fig. 1B, right). Similar results were obtained using other cell lines, including PC12, HEK293, MCF7, C6, PC3, U2OS, and C2C12 cells (Fig. 1C). To exclude the position effect of GFP fusion on the nuclear localization of the CUL4B fusion protein, we also examined the subcellular localization of CUL4B-Myc-His fusion protein in which the tagged protein was at the C-terminal end of CUL4B in HeLa, HEK293, and U2OS cell lines by using anti-Myc antibody. No staining was detected when the cells were transfected with pcDNA3.1A or when secondary antibody was used alone, demonstrating the specificity of the immunofluorescence staining of Myc fusion protein. Like EGFP-CUL4B, CUL4B-Myc-His fusion protein predominantly distributed in nucleus in different cell lines examined (Fig. 1D).
FIGURE 1.
Subcellular distribution of CUL4B. A, detection of the protein expression of the full-length EGFP-CUL4B and EGFP empty constructs. HeLa cells were transiently transfected with each construct and allowed to recover for 48 h. The protein expression was confirmed by Western blotting employing anti-GFP antibody. B, localization of the EGFP-CUL4B and EGFP in HeLa cells. HeLa cells were transiently transfected with respective constructs and were grown on coverslips for 48 h after transfection before they were fixed and counterstained with 4,6-diamidino-2-phenylindole (DAPI) for nuclei (blue). C, localization of the EGFP-CUL4B and EGFP in different cell lines. D, distribution of CUL4B-Myc-His fusion protein in HeLa, U2OS, and HEK293 cells. HeLa, U2OS, and HEK293 cells were transfected with CUL4B-Myc-His construct and immunostained with mouse monoclonal anti-Myc antibody, followed by detection with rhodamine-conjugated anti-mouse IgG secondary antibody. E, distribution of endogenous CUL4B in different cell lines. Cells grown on coverslips were fixed and immunostained with polyclonal anti-CUL4B antibody, followed by detection with rhodamine-conjugated anti-rabbit IgG secondary antibody. F, subcellular localization of endogenous CUL4B examined by cell fractionation and Western blotting. HeLa and HEK293 cells were separated into nuclear (N) and cytoplasmic (C) fractions. Equal amounts of proteins (60 μg) were loaded onto SDS-PAGE (12%) gels, and the protein expression was analyzed with anti-CUL4B antibody, anti-H2A antibody, and anti-α-tubulin antibody, respectively.
To confirm that the subcellular localization of the CUL4B fusion protein was not due to the overexpression of heterogeneous protein, we next examined the localization of endogenous CUL4B with anti-CUL4B antibody in different cell lines, including HEK293, CRL1730, HeLa, MCF7, U2OS, U87, and U251 cells. As shown in Fig. 1E, endogenous CUL4B protein was detected mainly in the nucleus. However, different from the distribution of EGFP-CUL4B fusion protein, weak cytoplasmic staining was also observed for the endogenous CUL4B. To further confirm the subcellular localization of endogenous CUL4B, Western blotting analyses were performed using subcellular fractions prepared from HeLa and HEK293 cells. In agreement with the immunofluorescence results, CUL4B was predominantly in the nuclear fraction (Fig. 1F). Probing with antibodies against H2A and α-tubulin demonstrated the purity of the cytosolic and nuclear fractions. Taken together, these results indicate that both tagged and endogenous CUL4B are mainly distributed in the nuclei.
The N Terminus, but Not the C-terminal Region, Is Required for CUL4B Nuclear Localization
The C-terminal regions of cullin proteins contain the ROC1 binding domain and NEDD8 conjugation and modification sites and are highly conserved among cullin family members. It was previously shown that the C-terminal region in human CUL1, CUL2, and CUL5 as well as that in mouse CUL4A were required for the nuclear localization of the cullins (25). Therefore, we first determined whether the C terminus of human CUL4B was required for its nuclear localization. To this end, a C-terminal deletion construct pEGFP-C1-CUL4B 1–729 was generated and transiently expressed in HeLa cells. As shown in Fig. 2, deletion of 166 amino acids (the sequence 730–895) in C terminus did not impair the nuclear accumulation of fused EGFP-CUL4B, indicating that the C-terminal region of CUL4B does not contain the signal for its nuclear transportation.
FIGURE 2.
The N terminus of CUL4B is sufficient to mediate nuclear localization of EGFP-CUL4B. N-terminal and C-terminal deletion mutants derived from the pEGFP-C1-CUL4B are depicted schematically. The different constructs were introduced into HeLa cells, and the subcellular distribution of fusion proteins was visualized under a fluorescence microscope.
Structurally, CUL4B distinguishes itself from the other members of the cullin family in its possession of a unique N terminus containing 149 amino acid residues. Therefore, the unique N terminus of CUL4B could be responsible for its nuclear localization. To test this notion, an N-terminal deletion construct (pEGFP-C1-CUL4B 150–895) lacking the N-terminal amino acids 1–149 was generated and transfected into HeLa cells. As shown in Fig. 2, CUL4B lacking the N-terminal 149 amino acids failed to accumulate in nucleus, suggesting that the unique N terminus of CUL4B contains a functional NLS. To further determine the contribution of the N terminus in nuclear transport, we fused a fragment of 115 amino acids in the N terminus of CUL4B to the C terminus of EGFP and examined the subcellular distribution of this fusion protein in HeLa cells. As shown in Fig. 2, the N-terminal fragment (pEGFP-C1-CUL4B 1–115) of CUL4B was capable of directing EGFP to the nucleus. Similar results were also obtained with HEK293 cells (data not shown). These results suggest that the N terminus of CUL4B is necessary and sufficient for nucleus targeting. Taken together, we concluded that the CUL4B protein is distinctly regulated in its nuclear importation and that its unique N terminus contains the functional NLS.
Identification of a Functional NLS of CUL4B
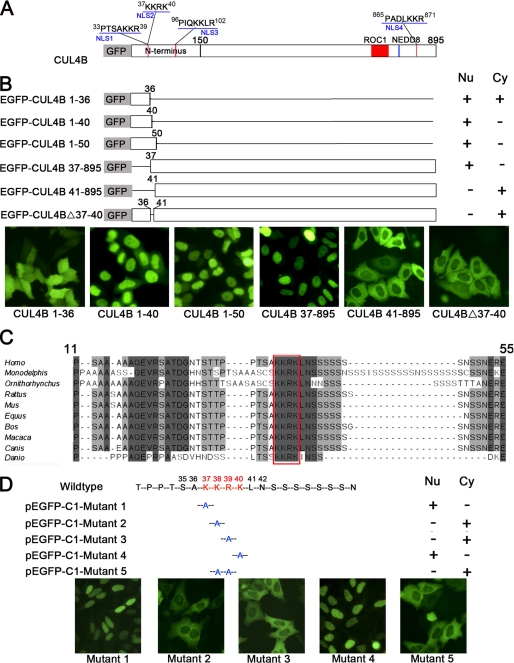
We used a web-based computer program, PSORT II (available from the PSORT web site), to search for the functional NLS in CUL4B. There were four putative NLS regions containing multiple positively charged amino acid residues located at amino acids 33–39, 37–40, 96–102, and 865–871, designated as NLS1, NLS2, NLS3, and NLS4, respectively (Fig. 3A). Because the amino acids 865–871 lie outside the region mapped by deletion experiments, we focused on the three NLSs in the N terminus. A series of C-terminal truncation constructs in which different fragments of CUL4B were fused to the C terminus of EGFP were generated and transiently expressed in HeLa cells (Fig. 3B). Western blotting analysis of cell extracts prepared from transiently transfected HeLa cells using an anti-GFP antibody indicated that EGFP-CUL4B mutant proteins were properly expressed and were stable (data not shown).
FIGURE 3.
Mapping of the functional NLS on human CUL4B by deletion mutagenesis and site-directed mutagenesis. A, schematic representation of CUL4B and its four putative NLSs (red) predicted by the computer program PSORTII. B, schematic diagram of EGFP-CUL4B fusion proteins and their respective subcellular localization. Six deletion constructs were generated and transiently transfected into HeLa cells. 48 h after transfection, the subcellular localizations of EGFP-tagged proteins were visualized under a fluorescence microscope. C, the NLS from residue 37 to 40 is highly conserved. A sequence alignment of the human CUL4B with orthologous proteins in nine species was carried out using the ClustalW alignment program. Missing amino acids are denoted by hyphens. Identical and similar residues are highlighted in black and gray, respectively. The NLS of CUL4B is boxed, and residue numbers corresponding to the human sequence are also shown. D, determination of the essential amino acids of NLS by mutational analysis. HeLa cells were transiently transfected with each of five mutant constructs derived from pEGFP-C1-CUL4B by site-directed mutagenesis and examined by fluorescence microscopy.
As shown in Fig. 3B, the construct containing the first 50 amino acids of N terminus was predominantly localized in the nucleus (Fig. 3B, pEGFP-C1-CUL4B 1–50), thus excluding NLS3 as a functional NLS. This result indicated that the region regulating nuclear localization of CUL4B resides in the first 50 amino acids, where NLS1 and NLS2 were located.
NLS1 and NLS2 overlap by three amino acids (KKR), suggesting that there could only be one functional NLS. In addition, alignment of this region with orthologous proteins from other species showed that 37KKRK40 is highly conserved among all species examined (Fig. 3C) and corresponds to the classic consensus sequence K(K/R)X(K/R) of a monopartite NLS, whereas 33PTSAKKR39 is less conserved and does not correspond to the classic NLS. This result suggested that 37KKRK40 probably acts as a functional NLS. As expected, the functionality of 33PTSAKKR39 as an NLS was excluded by localization data obtained from EGFP-CUL4B fusion protein, which, although devoid of the first 36 amino acids, retained nuclear localization (Fig. 3B, pEGFP-C1-CUL4B 37–895). EGFP-CUL4B fusion proteins lacking either the first 40 amino acids or only residues 37–40, on the other hand, were exclusively localized in the cytoplasm (Fig. 3B, pEGFP-C1-CUL4B 41–895 and pEGFP-C1-CUL4BΔ37–40). Furthermore, the construct containing 37KKRK40 was capable of translocating EGFP to nucleus (Fig. 3B, pEGFP-C1-CUL4B 1–40), whereas the construct lacking this region was not (Fig. 3B, pEGFP-C1-CUL4B 1–36).
In order to determine the contribution of specific amino acids within this region, site-directed mutagenesis was performed to generate five mutant constructs. As shown in Fig. 3D, substitutions K37A and K40A did not impair the nuclear localization of EGFP-CUL4B. However, substitution K38A or R39A alone disrupted nuclear localization. As expected, the double mutant K38A/R39A showed the same distribution pattern as the K38A and R39A single mutants. Thus, Lys38 and Arg39 are most crucial in mediating nuclear localization of CUL4B.
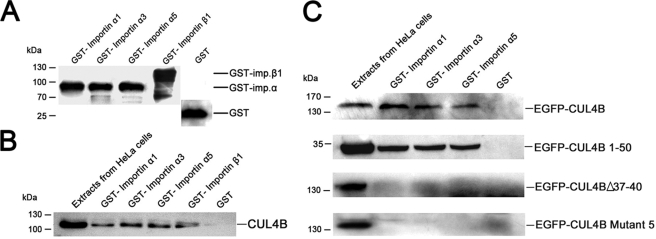
The CUL4B Protein Interacts Directly with Importin α Proteins
In eukaryotic cells, proteins containing a classical NLS are recognized and imported by a heterodimeric receptor consisting of importin α/β. Importin α proteins have an NLS-binding site and function as adapter molecules to mediate the interaction between the cargo and importin βs (26–28). In humans, there are at least six isoforms of importin αs, and these can be divided into three main subfamilies: importin α1-like subfamily, importin α3-like subfamily, and importin α5 subfamily (29). The classical monopartite NLS in CUL4B closely resembles that of the SV40 large T antigen (126PKKKRRV132), which is known to bind to all importin isoforms (30, 31). To further test the notion that nuclear transport of CUL4B protein is NLS-mediated, we performed an in vitro GST pull-down assay to test the ability of CUL4B to interact with importin α and β proteins. Three importin αs (importin α1, α3, and α5), representing each of the subfamilies of importin αs, and importin β1 were expressed in fusion with GST in bacteria (Fig. 4A). The GST protein alone was utilized as a negative control. As shown in Fig. 4B, endogenous CUL4B was able to interact with all tested importin α proteins and with importin β alone. The binding of CUL4B to three importin αs was also confirmed using extracts from HeLa cells stably transfected with pEGFP-C1-CUL4B (Fig. 4C). To investigate whether the binding is NLS-dependent, the beads were incubated with extracts from HeLa cells transiently transfected with pEGFP-C1-CUL4B 1–50, pEGFP-C1-CUL4BΔ37–40, and pEGFP-C1-CUL4B mutant 5, respectively, and the bound proteins were detected with anti-GFP antibodies. As shown in Fig. 4C, EGFP-CUL4B 1–50 fusion protein was recognized by all importin αs. EGFP-CUL4BΔ37–40 and EGFP-CUL4B mutant 5, on the other hand, did not bind these importin αs. Thus, the nuclear transport of CUL4B is mediated by importin α via its binding to 37KKRK40.
FIGURE 4.
Binding of CUL4B to different GST-importin αs in vitro. A, expression analysis of different GST-importin fusion proteins expressed in E. coli. 5% of the input amount of GST and GST fusion proteins used in the GST pull-down assay was subjected to SDS-PAGE and then detected with monoclonal anti-GST antibody. B and C, CUL4B interacts with GST-importin fusion proteins in a GST pull-down assay. GST and GST fusion proteins expressed in E. coli were immobilized on glutathione-Sepharose beads. After extensive washing, the washed beads were incubated with equal amounts of protein extracts from untransfected HeLa cells (B) and extracts from HeLa cells transfected with pEGFP-C1-CUL4B, pEGFP-C1-CUL4B 1–50, pEGFP-CUL4BΔ37–40, and pEGFP-CUL4B mutant 5, respectively (C). The retained proteins were analyzed by Western blotting with anti-CUL4B (B) or anti-GFP (C) antibodies.
Silencing of CUL4B Results in Cell Proliferation Defects
Previous studies have shown that CUL4 is involved in the regulation of key cell cycle regulators, such as p53, p21, p27, cyclin E, and CHK1 (8, 17, 32–37). In addition, CUL4A is reported to be overexpressed in breast cancer and hepatocellular carcinomas (11–13). These results prompted us to examine whether CUL4B inhibition similarly affects cell proliferation. We employed CUL4B-specific RNAi to stably knock down endogenous CUL4B expression in HeLa cells and determined changes in cell proliferation. After three weeks of selection with blasticidin, colonies stably expressing RNAi 2055, designated as miCUL4B, or control oligonucleotides, designated as miNeg, were selected, and the CUL4B expression in these colonies was determined by real-time PCR and Western blotting. CUL4B was markedly decreased at mRNA and protein levels in miCUL4B, but not in miNeg, when compared with wild type HeLa cells (Fig. 5, A and B). Because of the high degree of homology between CUL4A and CUL4B, the mRNA of CUL4A was also determined by real-time PCR analysis. As shown in Fig. 5A, there was no difference in the expression of CUL4A between miCUL4B and miNeg HeLa cells, confirming the specificity of RNAi against CUL4B.
FIGURE 5.
CUL4B down-regulation inhibits cell proliferation. A and B, validation of the effect and specificity of RNAi against CUL4B in stably transfected cells. A, RNA was extracted from wild-type HeLa cells and each of the stably transfected cell lines. The expression of CUL4A and CUL4B mRNA was quantified by real-time PCR. The assay was performed in triplicate, and relative means ± S.E. are shown. B, equal amounts (60 μg) of protein lysates were subjected to SDS-PAGE (12%) and then detected using the antibodies against CUL4B and β-actin, respectively. C, RNAi of CUL4B inhibits cell proliferation. Equal numbers of the indicated cells were plated on 96-well plates, and cell proliferation was determined by MTT assay at different time points. The data in each time point are the mean results ± S.D. of the averaged values from 10 replicates. These experiments were performed in triplicate. *, p < 0.001 versus miNeg. D, effects of CUL4B RNAi on DNA synthesis. The indicated cells were synchronized by serum starvation and then stimulated to progress through the cell cycle by the addition of fresh medium containing 10% fetal bovine serum. After 20 h of treatment, before being fixed, cells were labeled with 50 μm BrdUrd for 45 min. Shown were the percentages of BrdUrd-labeled cells. Results represent means ± S.D. *, p < 0.01 versus miNeg. E, effect of CUL4B RNAi on cell cycle distribution of HeLa cells. Cells were synchronized and restimulated to enter the cell cycle as described above. Cell cycle distribution was determined by flow cytometric analysis of DNA content. The x and y axes show DNA content and cell number, respectively. The results shown are representative of three independent experiments with similar results. Bars, means ± S.D. *, p < 0.05 versus miNeg. F, effects of CUL4B RNAi on S phase progression. The indicated cells were synchronized by a double thymidine block and then released to progress through the cell cycle by replating in fresh drug-free medium. For each time point, the cells were labeled with BrdUrd for 45 min. The percentage of BrdUrd-labeled cells is shown.
RNAi of CUL4B led to a significantly decreased growth rate of HeLa cells, with an about 70% decrease at the 120 h time point, as measured by an MTT assay (Fig. 5C). The TUNEL assay showed no difference in apoptosis between miCUL4B and miNeg cells (data not shown), suggesting that the inhibition of cell proliferation may be mediated by cell cycle arrest. Fluorescence-activated cell sorting analysis of cell cycle distribution and a BrdUrd incorporation assay were employed to evaluate the cell cycle progression in miCUL4B cells. Consistent with the results obtained with the MTT assay, miCUL4B cells showed a significant decrease in the percentage of BrdUrd-positive cells compared with the control group (Fig. 5D). Interestingly, although BrdUrd-positive cells decreased, the percentage of cells in S phase increased about 10% in miCUL4B cells (Fig. 5E).
We then proceeded to perform a detailed analysis of the effects of CUL4B silencing on S phase progression. Cells were synchronized by a double-thymidine block and then released to progress into S phase, and at different time points the rate of BrdUrd incorporation was measured. As shown in Fig. 5F, the highest rate of BrdUrd incorporation in miCUL4B cells was 71.7%, compared with 83.2% in the control cells. Moreover, the rate of BrdUrd incorporation peaked at around 7.5 h, which was 2.5 h later than that of the miNeg cells, suggesting that the progression through the S phase was prolonged in the CUL4B RNAi cells. Taken together, these results indicate that constitutive knockdown of CUL4B inhibits the proliferation of HeLa cells by prolonging S phase progression.
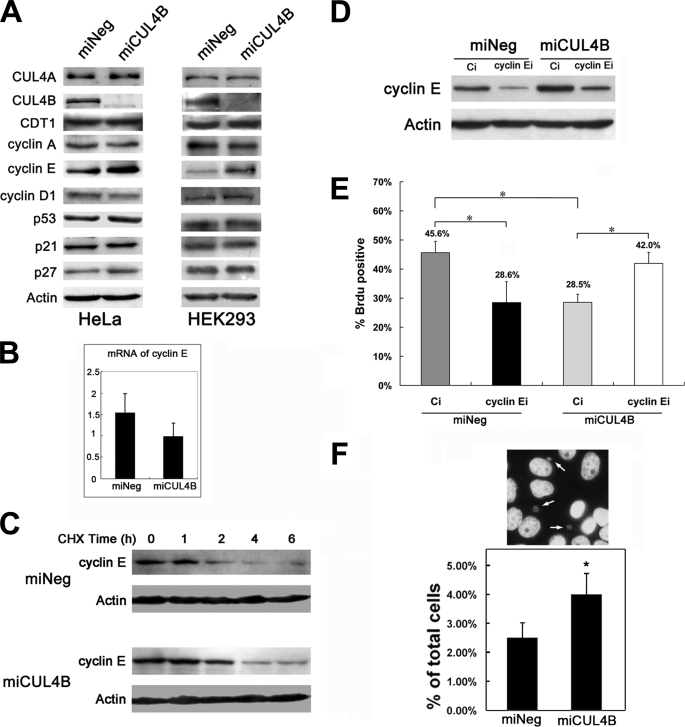
Reduced CUL4B Expression Causes an Accumulation of Cyclin E
It has been known that mammalian cell cycle progression is tightly controlled by CDKs, CDK inhibitors, and tumor suppressors. To understand the molecular basis for CUL4B depletion-induced S phase prolongation in HeLa cells, we examined the expression levels of several cell cycle regulators by Western blotting analysis. Down-regulation of CUL4B resulted in an accumulation of cyclin E as compared with control cells (Fig. 6A). Similar results were observed with HEK293 cells (Fig. 6A).
FIGURE 6.
CUL4B knockdown results in accumulation of cyclin E. A, protein levels of representative CUL4 targets, including cyclin E. CUL4B knockdown causes an elevation in cyclin E. Cells were harvested, and equal amounts (60 μg) of total cell lysates of the indicated asynchronous cells were subjected to SDS-PAGE (12%) and detected with the indicated antibodies. B, in parallel, the mRNA levels of cyclin E in each group of asynchronous HeLa cells were measured by real-time PCR. C, increased half-life of cyclin E in CUL4B RNAi cells. Cells were treated with cycloheximide (50 μg/ml) and harvested at the indicated time points. Equal amounts (60 μg) of total cell lysates of the indicated cells were analyzed by Western blotting assays. D, RNAi of cyclin E reduced the overaccumulation of cyclin E caused by the RNAi of CUL4B. The indicated cells were transfected with cyclin E siRNA (cyclin Ei) or control siRNA (Ci). 48 h after transfection, cyclin E protein expression was analyzed by Western blotting. E, alleviation of the cell proliferation defects in CUL4B and cyclin E co-silenced cells. The indicated cells were transfected with cyclin E siRNA or control siRNA. 24 h after transfection, cells were synchronized at the G1/S boundary by a double-thymidine block and then were released into the cell cycle. 1.5 h later, cells were analyzed by a BrdUrd incorporation assay. Results represent means ± S.D. from three independent experiments. *, p < 0.001. F, RNAi of CUL4B increases micronuclei formation in HeLa cells. The results are shown as the percentage of cells showing micronuclei formation. Bars, means ± S.D. *, p < 0.01 versus miNeg.
We further performed real-time PCR to determine whether the enhanced cyclin E accumulation was due to increased cyclin E transcription. There was no corresponding increase of cyclin E mRNA in miCUL4B-transfected HeLa cells (Fig. 6B), suggesting that the miCUL4B cells were impaired in cyclin E degradation. To test this hypothesis, we measured the half-life of cyclin E proteins. Cells treated with cycloheximide, which blocks new protein synthesis, were harvested for analysis of the cyclin E levels by Western blotting at various time points. We observed that silencing of CUL4B resulted in a significant increase in the half-life of cyclin E compared with that of control cells (Fig. 6C). Taken together, these results indicate that CUL4B probably participates in the proteosomal degradation of cyclin E.
The fact that inhibition of cell proliferation caused by CUL4B RNAi was associated with cyclin E accumulation suggests that CUL4B might regulate cell proliferation through a mechanism involving modulation of cyclin E levels. To investigate this, we reduced the expression of cyclin E using siRNA and measured the rate of BrdUrd incorporation. As shown in Fig. 6D, the cyclin E protein levels were significantly reduced in cells transfected with small interfering RNA against cyclin E compared with cells transfected with control siRNA, to a level close to that of miNeg cells. Correspondingly, although RNAi of cyclin E in miNeg cells caused BrdUrd-positive cells to drop from 45.6 to 28.6%, consistent with a previous report that RNAi of cyclin E inhibited DNA synthesis (38), cyclin E silencing increased BrdUrd-positive cells from 28.5 to 42.0% in miCUL4B cells (Fig. 6E). Thus, the overaccumulation of cyclin E directly contributed to the reduction of proliferation of CUL4B RNAi cells.
Although recent studies have shown that CUL4A regulates the ubiquitination and degradation of p21, p53, p27, CHK1, and CDT1 (8, 17, 32–37, 39, 40), the expression levels of these proteins remained unchanged after CUL4B knockdown (Fig. 6A and supplemental Fig. S2), suggesting that CUL4A and CUL4B may regulate cell proliferation via distinct mechanisms. It is also possible that residual activity of CUL4B in CUL4B RNAi cells is sufficient for the ubiquitination of other substrates.
A defect in the degradation of cyclin E was reported to induce genetic instability (41). To test whether the accumulation of cyclin E in response to the reduction in CUL4B could have a similar consequence, we examined the formation of micronuclei in miCUL4B and miNeg cells. The percentage of micronuclei-containing cells was moderately increased in miCUL4B cells as compared with control cells (Fig. 6F), suggesting that lack of CUL4B may lead to increased genomic instability.
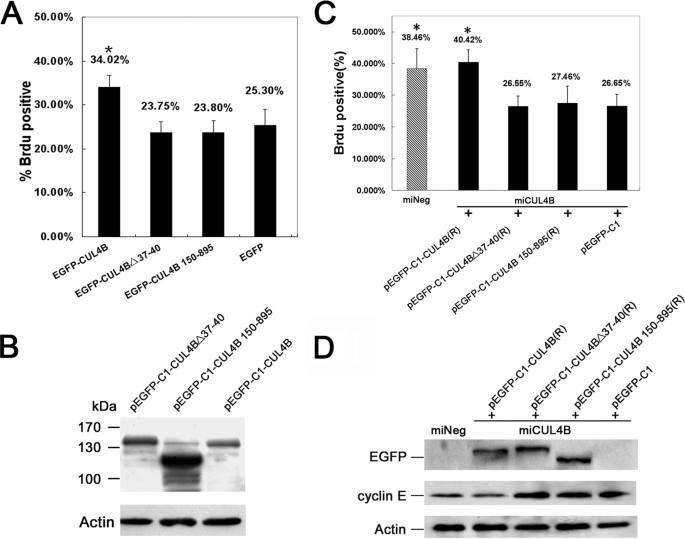
The Nuclear Localization of CUL4B Is Required for Its Regulation of Cell Proliferation
We further determined whether nuclear localization of CUL4B was required for its functions in regulation of cell proliferation. Four constructs (pEGFP-C1-CUL4B, pEGFP-C1-CUL4B 150–895, pEGFP-C1-CUL4BΔ37-40, and pEGFP-C1 empty vector) were each transiently transfected into HeLa cells, and the effect on cell growth was analyzed by a BrdUrd incorporation assay. As shown in Fig. 7A, overexpression of full-length CUL4B significantly increased the number of BrdUrd-positive cells compared with cells transfected with pEGFP-C1, consistent with the finding that down-regulation of CUL4B inhibits cell proliferation (Fig. 5D). In contrast, constructs in which the NLS or N-terminal region had been deleted did not have the growth-promoting effect. These differential effects of pEGFP-C1-CUL4B, pEGFP-C1-C1-CUL4B 150–895, and pEGFP-C1-Del 37–40 on cell proliferation were not due to differential expression of the three proteins in the transfectants (Fig. 7B). These results indicate that the nuclear localization of CUL4B was required for its regulation of cell proliferation.
FIGURE 7.
Nuclear localization of CUL4B was required for cell proliferation. A, full-length CUL4B promotes cell proliferation. HeLa cells were transfected with the indicated constructs. At 48 h post transfection, cells were fixed and analyzed by BrdUrd incorporation assay. Results represent means ± S.D. from three independent experiments. *, p < 0.01 versus wild-type HeLa cells transfected with pEGFP. B, expression of CUL4B in transfected cells. Equal amounts (60 μg) of total cell lysates of HeLa cells transiently transfected with pEGFP-C1-CUL4B, pEGFP-C1-CUL4BΔ37–40, and pEGFP-C1-CUL4B 150–895 were subjected to SDS-PAGE (12%) and detected with anti-GFP antibody. C, Introduction of exogenous full-length CUL4B rescues the proliferation inhibition caused by silencing of the endogenous CUL4B. CUL4B-down-regulated HeLa cells were transfected with the indicated plasmids. At 48 h post-transfection, cells were fixed and analyzed by a BrdUrd incorporation assay. Results represent means ± S.D. from three independent experiments. *, p < 0.01 versus CUL4B-down-regulated HeLa cells transfected with pEGFP-C1. D, introduction of exogenous full-length CUL4B reduces the accumulation of cyclin E caused by knockdown of the endogenous CUL4B. Equal amounts (60 μg) of total cell lysates of miCUL4B HeLa cells transiently transfected with pEGFP-C1-CUL4B, pEGFP-C1-CUL4BΔ37–40, pEGFP-C1-CUL4B 150–895, pEGFP-C1, and miNeg HeLa cells were subjected to SDS-PAGE (12%) and detected with anti-GFP or anti-cyclin E antibody.
The above conclusion was substantiated by rescue experiments. When RNAi-resistant expression vectors pEGFP-C1-CUL4B(R), pEGFP-C1-CUL4B 150–895(R), pEGFP-C1-CUL4BΔ37–40(R), and pEGFP-C1 (empty vector) were transiently transfected into miCUL4B HeLa cells, respectively, only pEGFP-C1-CUL4B was capable of restoring the normal level of cyclin E and the normal rate of DNA synthesis that were impaired in CUL4B-depleted HeLa cells (Fig. 7, C and D).
DISCUSSION
In this study, we determined the subcellular localization of human CUL4B protein. We found that both tagged and endogenous CUL4B accumulate in the nuclei. We demonstrated that, unlike CUL4A and other cullins in which the NLS is located in the C terminus, the NLS of CUL4B is located in the N terminus. We also provided evidence showing that CUL4B could bind to importin α1, α3, and α5 via its NLS, and its nuclear localization is required for cell proliferation. When CUL4B was down-regulated or when its NLS was absent, cyclin E became overaccumulated and inhibited proliferation by causing a prolongation in S phase.
Like CUL4B, CUL4A is also required for cellular proliferation. However, although previous reports showed that CUL4A targets p53, p21, p27, CHK1 and CDT1 (8, 17, 32–37, 39, 40), knockdown of CUL4B had no effect on the levels of those proteins (Fig. 6A and supplemental Fig. S2). Consistent with a previous report (8), the accumulation of cyclin E is greatly enhanced when CUL4B is down-regulated, indicating the involvement of CUL4B in the degradation of cyclin E. Cyclin E is thought to act as a positive regulator of cell cycle progression by promoting S phase entry and DNA replication via the induction of S phase-specific genes (42). Cyclin E is tightly regulated by ubiquitin-mediated proteolysis system, and its overexpression could increase cell proliferation through accelerated S phase entry (43). However, several studies also showed that elevated cyclin E levels may have no effect or even opposite effects on cell proliferation. Although the depletion of hCDC4 resulted in an increase in cyclin E, the proliferation of hCDC4−/− cells was indistinguishable from that of parental cells (41). Accumulation of cyclin E was found to reduce or stop cell divisions in Drosophila (44). Constitutive overexpression of cyclin E led to a decrease in cell proliferation by inducing S phase arrest and a reduced rate of DNA synthesis (45). In the present study, we showed that the increased accumulation of cyclin E is responsible for the S phase prolongation when CUL4B is down-regulated. Together with other reports, we can conclude that CUL4B plays a critical role in maintaining cyclin E at a level that favors normal cell cycle progression.
Nuclear proteins, including transcription factors, are synthesized in the cytoplasm and need to be transported into the nucleus to fulfill their function (46). Nuclear transport usually requires the presence of an NLS in the cargo and the nuclear transport receptors (karyopherins), which recognize and bind to NLS (47). Generally, NLSs are composed of short stretches of highly basic amino acids, either as single stretches of amino acids (monopartite) or in the form of two short stretches separated by a spacer region (biopartite). CUL4B possesses four potential basic NLSs, but only one of them functions in mediating the nuclear localization of CUL4B. Importantly, this functional NLS, 37KKRK40, is highly conserved and corresponds to the classic consensus sequence K(K/R)X(K/R) of a monopartite NLS.
By participating in the ubiquitination of key cell cycle regulators, such as Cdt1, p53, p27, and Chk1, CUL4A was believed to play a critical role in regulating cell cycle progression (17, 35, 40, 48). It may promote genome integrity by preventing DNA synthesis in presence of DNA damage via the degradation of Cdt1 (49). On the other hand, CUL4A probably also contributes to the resumption of DNA synthesis and cell cycle progression by facilitating ubiquitin-mediated degradation of CHK1 (17). Interestingly, mice deficient in CUL4A were protected from UV-induced skin carcinogenesis because DNA repair capacity was augmented (15). However, another recent report showed that cells lacking CUL4A exhibited abnormalities in the formation of centrosomes and spindles and had increased formation of micronuclei (16). Such cells were even more sensitive to UV irradiation. Thus, the precise role of CUL4A in the maintenance of genomic stability remains to be clarified. It will be of interest to test whether and how the S phase prolongation caused by RNAi of CUL4B is associated with changes in DNA damage response.
In summary, CUL4B is primarily a nuclear protein, and, unlike other cullins, its nuclear localization is determined by a functional NLS, 37KKRK40, in its N terminus. This NLS is required for CUL4B to participate in the degradation of cyclin E and in the regulation of cell cycle progression.
Supplementary Material
Acknowledgment
We thank Dr. Anne Carine Østvold (Center for Cellular Stress Responses, University of Oslo, Norway) for the kind gift of importin α1, α3, α5, and β vectors.
This work was supported by National Science Foundation Research Grant 30830065, National Basic Research Program of China grant 2007CB512001, and National High-tech Research and Development Program of China Grant 2006AA02A406.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1 and S2.
- CRL
- cullin-RING-based E3 ubiquitin ligase
- CDK
- cyclin-dependent kinase
- NLS
- nuclear localization signal
- EGFP
- enhanced green fluorescent protein
- BrdUrd
- bromodeoxyuridine
- RNAi
- RNA interference
- PBS
- phosphate-buffered saline
- GST
- glutathione S-transferase
- siRNA
- small interfering RNA
- MTT
- 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide
- TUNEL
- terminal dUTP nick end labeling.
REFERENCES
- 1.Petroski M. D., Deshaies R. J. (2005) Nat. Rev. Mol. Cell. Biol. 6, 9–20 [DOI] [PubMed] [Google Scholar]
- 2.Bosu D. R., Kipreos E. T. (2008) Cell Division 3, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Higa L. A., Zhang H. (2007) Cell Division 2, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angers S., Li T., Yi X., MacCoss M. J., Moon R. T., Zheng N. (2006) Nature 443, 590–593 [DOI] [PubMed] [Google Scholar]
- 5.Higa L. A., Wu M., Ye T., Kobayashi R., Sun H., Zhang H. (2006) Nat. Cell Biol. 8, 1277–1283 [DOI] [PubMed] [Google Scholar]
- 6.He Y. J., McCall C. M., Hu J., Zeng Y., Xiong Y. (2006) Genes Dev. 20, 2949–2954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin J., Arias E. E., Chen J., Harper J. W., Walter J. C. (2006) Mol. Cell 23, 709–721 [DOI] [PubMed] [Google Scholar]
- 8.Higa L. A., Yang X., Zheng J., Banks D., Wu M., Ghosh P., Sun H., Zhang H. (2006) Cell Cycle 5, 71–77 [DOI] [PubMed] [Google Scholar]
- 9.Tarpey P. S., Raymond F. L., O'Meara S., Edkins S., Teague J., Butler A., Dicks E., Stevens C., Tofts C., Avis T., Barthorpe S., Buck G., Cole J., Gray K., Halliday K., Harrison R., Hills K., Jenkinson A., Jones D., Menzies A., Mironenko T., Perry J., Raine K., Richardson D., Shepherd R., Small A., Varian J., West S., Widaa S., Mallya U., Moon J., Luo Y., Holder S., Smithson S. F., Hurst J. A., Clayton-Smith J., Kerr B., Boyle J., Shaw M., Vandeleur L., Rodriguez J., Slaugh R., Easton D. F., Wooster R., Bobrow M., Srivastava A. K., Stevenson R. E., Schwartz C. E., Turner G., Gecz J., Futreal P. A., Stratton M. R., Partington M. (2007) Am. J. Hum. Genet. 80, 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zou Y., Liu Q., Chen B., Zhang X., Guo C., Zhou H., Li J., Gao G., Guo Y., Yan C., Wei J., Shao C., Gong Y. (2007) Am. J. Hum. Genet. 80, 561–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L. C., Manjeshwar S., Lu Y., Moore D., Ljung B. M., Kuo W. L., Dairkee S. H., Wernick M., Collins C., Smith H. S. (1998) Cancer Res. 58, 3677–3683 [PubMed] [Google Scholar]
- 12.Schindl M., Gnant M., Schoppmann S. F., Horvat R., Birner P. (2007) Anticancer Res. 27, 949–952 [PubMed] [Google Scholar]
- 13.Yasui K., Arii S., Zhao C., Imoto I., Ueda M., Nagai H., Emi M., Inazawa J. (2002) Hepatology 35, 1476–1484 [DOI] [PubMed] [Google Scholar]
- 14.Li B., Ruiz J. C., Chun K. T. (2002) Mol. Cell. Biol. 22, 4997–5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu L., Lee S., Zhang J., Peters S. B., Hannah J., Zhang Y., Yin Y., Koff A., Ma L., Zhou P. (2009) Mol. Cell 34, 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kopanja D., Stoyanova T., Okur M. N., Huang E., Bagchi S., Raychaudhuri P. (2009) Oncogene 28, 2456–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leung-Pineda V., Huh J., Piwnica-Worms H. (2009) Cancer Res. 69, 2630–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Waning D. L., Li B., Jia N., Naaldijk Y., Goebel W. S., HogenEsch H., Chun K. T. (2008) Blood 112, 320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerrero-Santoro J., Kapetanaki M. G., Hsieh C. L., Gorbachinsky I., Levine A. S., Rapić-Otrin V. (2008) Cancer Res. 68, 5014–5022 [DOI] [PubMed] [Google Scholar]
- 20.Xia Y., Jiang B., Zou Y., Gao G., Shang L., Chen B., Liu Q., Gong Y. (2008) Biochem. Biophys. Res. Commun. 368, 438–444 [DOI] [PubMed] [Google Scholar]
- 21.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 22.Grundt K., Haga I. V., Huitfeldt H. S., Ostvold A. C. (2007) Biochim. Biophys. Acta 1773, 1398–1406 [DOI] [PubMed] [Google Scholar]
- 23.Liu Z., Liu Q., Xu B., Wu J., Guo C., Zhu F., Yang Q., Gao G., Gong Y., Shao C. (2009) Mutat. Res. 662, 75–83 [DOI] [PubMed] [Google Scholar]
- 24.Mosmann T. (1983) J. Immunol. Methods 65, 55–63 [DOI] [PubMed] [Google Scholar]
- 25.Furukawa M., Zhang Y., McCarville J., Ohta T., Xiong Y. (2000) Mol. Cell. Biol. 20, 8185–8197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goldfarb D. S., Corbett A. H., Mason D. A., Harreman M. T., Adam S. A. (2004) Trends Cell Biol. 14, 505–514 [DOI] [PubMed] [Google Scholar]
- 27.Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. (2007) J. Biol. Chem. 282, 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chook Y. M., Blobel G. (2001) Curr. Opin. Struct. Biol. 11, 703–715 [DOI] [PubMed] [Google Scholar]
- 29.Lischka P., Sorg G., Kann M., Winkler M., Stamminger T. (2003) J. Virol. 77, 3734–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fagerlund R., Kinnunen L., Köhler M., Julkunen I., Melén K. (2005) J. Biol. Chem. 280, 15942–15951 [DOI] [PubMed] [Google Scholar]
- 31.Yoneda Y. (2000) Genes Cells 5, 777–787 [DOI] [PubMed] [Google Scholar]
- 32.Banks D., Wu M., Higa L. A., Gavrilova N., Quan J., Ye T., Kobayashi R., Sun H., Zhang H. (2006) Cell Cycle 5, 1719–1729 [DOI] [PubMed] [Google Scholar]
- 33.Kim Y., Starostina N. G., Kipreos E. T. (2008) Genes Dev. 22, 2507–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishitani H., Shiomi Y., Iida H., Michishita M., Takami T., Tsurimoto T. (2008) J. Biol. Chem. 283, 29045–29052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miranda-Carboni G. A., Krum S. A., Yee K., Nava M., Deng Q. E., Pervin S., Collado-Hidalgo A., Galic Z., Zack J. A., Nakayama K., Nakayama K. I., Lane T. F. (2008) Genes Dev. 22, 3121–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abbas T., Sivaprasad U., Terai K., Amador V., Pagano M., Dutta A. (2008) Genes Dev. 22, 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li B., Jia N., Kapur R., Chun K. T. (2006) Blood 107, 4291–4299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li K., Lin S. Y., Brunicardi F. C., Seu P. (2003) Cancer Res. 63, 3593–3597 [PubMed] [Google Scholar]
- 39.Hu J., McCall C. M., Ohta T., Xiong Y. (2004) Nat. Cell Biol. 6, 1003–1009 [DOI] [PubMed] [Google Scholar]
- 40.Senga T., Sivaprasad U., Zhu W., Park J. H., Arias E. E., Walter J. C., Dutta A. (2006) J. Biol. Chem. 281, 6246–6252 [DOI] [PubMed] [Google Scholar]
- 41.Rajagopalan H., Jallepalli P. V., Rago C., Velculescu V. E., Kinzler K. W., Vogelstein B., Lengauer C. (2004) Nature 428, 77–81 [DOI] [PubMed] [Google Scholar]
- 42.Möröy T., Geisen C. (2004) Int. J. Biochem. Cell Biol. 36, 1424–1439 [DOI] [PubMed] [Google Scholar]
- 43.Simone C., Resta N., Bagella L., Giordano A., Guanti G. (2002) Mol. Pathol. 55, 200–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Doronkin S., Djagaeva I., Beckendorf S. K. (2003) Dev. Cell 4, 699–710 [DOI] [PubMed] [Google Scholar]
- 45.Spruck C. H., Won K. A., Reed S. I. (1999) Nature 401, 297–300 [DOI] [PubMed] [Google Scholar]
- 46.Cyert M. S. (2001) J. Biol. Chem. 276, 20805–20808 [DOI] [PubMed] [Google Scholar]
- 47.Suntharalingam M., Wente S. R. (2003) Dev. Cell 4, 775–789 [DOI] [PubMed] [Google Scholar]
- 48.Nag A., Bagchi S., Raychaudhuri P. (2004) Cancer Res. 64, 8152–8155 [DOI] [PubMed] [Google Scholar]
- 49.Higa L. A., Mihaylov I. S., Banks D. P., Zheng J., Zhang H. (2003) Nat. Cell Biol. 5, 1008–1015 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.