Abstract
Autosomal dominant mutations in the SFTPC gene are associated with idiopathic pulmonary fibrosis, a progressive lethal interstitial lung disease. Mutations that cause misfolding of the encoded proprotein surfactant protein C (SP-C) trigger endoplasmic reticulum (ER)-associated degradation, a pathway that segregates terminally misfolded substrate for retrotranslocation to the cytosol and degradation by proteasome. Microarray screens for genes involved in SP-C ER-associated degradation identified MKS3/TMEM67, a locus previously linked to the ciliopathy Meckel-Gruber syndrome. In this study, MKS3 was identified as a membrane glycoprotein predominantly localized to the ER. Expression of MKS3 was up-regulated by genetic or pharmacological inducers of ER stress. The ER lumenal domain of MKS3 interacted with a complex that included mutant SP-C and associated chaperones, whereas the region predicted to encode the transmembrane domains of MKS3 interacted with cytosolic p97. Deletion of the transmembrane and cytosolic domains abrogated interaction of MKS3 with p97 and resulted in accumulation of mutant SP-C proprotein; knockdown of MKS3 also inhibited degradation of mutant SP-C. These results support a model in which MKS3 links the ER lumenal quality control machinery with the cytosolic degradation apparatus.
INTRODUCTION
Newly translated secretory and membrane proteins achieve their native conformations following translocation into the endoplasmic reticulum (ER)2 and subsequent interaction with molecular chaperones and folding enzymes. Mutations that prevent folding to a stable conformer increase protein load, leading to activation of the unfolded protein response. The unfolded protein response is an integrated response to ER stress that 1) decreases translation, thereby slowing entry of new substrate into the ER; 2) enhances ER folding capacity by increasing transcription of ER chaperones; and 3) promotes degradation of folding-incompetent proteins by transcriptional up-regulation of genes involved in ER-associated degradation (ERAD) (1–3). The ERAD pathway segregates terminally misfolded proteins from folding-competent proteins for retrotranslocation to the cytosol and degradation by proteasome (4, 5). Failure to clear misfolded proteins leads to formation of cytotoxic protein aggregates and/or activation of apoptotic pathways.
Surfactant protein C (SP-C) is an integral membrane protein expressed by alveolar type II epithelial cells in the lung. Mutations in the gene encoding SP-C (SFTPC) are associated with familial and sporadic forms of interstitial lung disease that are frequently fatal (6, 7). Mutant SP-C is rapidly cleared by ERAD; however, when the ERAD pathway is saturated or proteasome is inhibited, the mutant protein forms cytotoxic aggregates (8–11). Overexpression of mutant SP-C in type II epithelial cells of transgenic mice results in cell death, lung dysmorphogenesis, and perinatal lethality, confirming the importance of rapid elimination of the misfolded protein (12).
To identify candidate genes involved in ERAD of mutant SP-C, HEK293 cells were transiently transfected with plasmids encoding wild-type or mutant SP-C and analyzed by microarray (13). Among the 50 most highly up-regulated genes in cells expressing mutant SP-C was MGC26979/TMEM67, subsequently identified as MKS3. Genotype-phenotype analyses indicated that recessive mutations at the MKS3 locus were associated with Meckel-Gruber syndrome, a lethal disease characterized by polycystic kidneys, malformations of the liver and central nervous system, and polydactyly (14, 15); however, the function of MKS3 and its role in pathogenesis are not known. The link between mutant SP-C and elevated expression of MKS3 led us to test the hypothesis that MKS3 is a component of the ERAD machinery.
EXPERIMENTAL PROCEDURES
Reagents
The following antibodies were used in this study: mouse anti-FLAG and rabbit anti-GRP78/BiP antibodies (Sigma); rabbit anti-hemagglutinin (HA) antibody (Santa Cruz Biotechnology); rabbit anti-p97/valosin-containing protein (VCP) and rabbit anti-glyceraldehyde-3-phosphate dehydrogenase antibodies (Bethyl Laboratories); rabbit anti-BiP antibody (Cell Signaling Technology); mouse anti-KDEL antibody (Stressgen); rabbit anti-calreticulin antibody (Affinity Bioreagents); mouse anti-calnexin antibody (Abcam); sheep anti-α1-antitrypsin antibody (MP Biomedicals); rabbit anti-pro-SP-C and mouse anti-actin antibodies (Seven Hills Bioreagents); and Alexa 488-labeled goat anti-rabbit IgG, Alexa 555-labeled goat anti-guinea pig IgG, and wheat germ agglutinin (WGA)-conjugated Oregon Green 488 (Molecular Probes). Anti-MKS3 antibody was generated by injecting guinea pigs with recombinant peptide (see “cDNA Constructs” and supplemental Fig. S1). The (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) (MTS) reduction assay and Dual-Luciferase reporter assay kits were purchased from Promega. MG-132 was purchased from EMD Biosciences, dithiobis(succinimidyl propionate) (DSP) from Pierce, tunicamycin and thapsigargin from Sigma, and endoglycosidase H from New England Biolabs.
cDNA Constructs
Human MKS3 constructs (full-length wild-type MKS3, MKS3-(1–501), MKS3-(1–744), and MKS3-(776–924)) were cloned from HEK293 cell cDNA by PCR using primers based on the human cDNA sequence (NM_153704.3); site-directed mutagenesis of wild-type (WT) MKS3 was used to generate MKS3(P394L) and MKS3(Q376P). MKS3-(776–924) was cloned into pET-28 (Novagen) and expressed in bacteria, and recombinant protein was purified by nickel-nitrilotriacetic acid chromatography as described by the manufacturer (Qiagen) for generation of antibody. All other MKS3 constructs were cloned into p3×FLAG-CMV-14 (Sigma) for expression in HEK293 cells or murine embryonic fibroblasts (MEFs). Spliced XBP-1 was generated by PCR as described by Iwakoshi et al. (16) using tunicamycin-treated isolated mouse alveolar type II epithelial cells (17) and cloned into pcDNA3.1 (Invitrogen). Dominant-negative XBP-1 was generated from the spliced XBP-1 construct as described by Lee et al. (18). The MKS3 promoter-luciferase construct was generated by cloning the 5′-flanking region of the MKS3 gene (−97 to −1064 bp) into pGL3 (Promega). Generation of WT SP-C, mutant SP-C constructs (SP-CΔexon4 and SP-C(L188Q)), and HA-tagged ERdj4 and ERdj5 constructs have been described previously (9, 13). The α1-antitrypsin and PiZ variant (cloned into pcDNA3.1) were kindly provided by Dr. Rick Sifers (Baylor College of Medicine).
Cell Culture and Expression of Constructs
HEK293 cells stably expressing WT SP-C, SP-CΔexon4, or SP-C(L188Q) were generated and cultured as described previously (9, 13). Constructs were transiently expressed in HEK293 cells following transfection with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Where indicated, cells were treated with 5 μm MG-132 for the last 4 h of culture and/or with 1 mm DSP for the final 30 min of culture as described previously (9, 12, 13). Pulse-chase experiments were performed 24 h after transfection of constructs as described previously (13). To induce ER stress, tunicamycin or thapsigargin was added to the medium to a final concentration of 2 μg/ml or 300 nm, respectively, for the indicated time periods. Targeting and non-targeting MKS3 small interfering RNA (siRNA) pools were transfected at a final concentration of 75 nm as directed by the manufacturer (Dharmacon).
MEFs were generated from day 13 embryos resulting from heterozygous crosses of wpk+/− rats (the generous gift of Dr. Vincent Gattone, Indiana University School of Medicine). MEFs were isolated and cultured as described by Guo and Zheng (19) and transfected using Lipofectamine LTX (Invitrogen). All experiments involving the use of animals were approved by the Institutional Animal Care and Use Committee of the Cincinnati Children's Research Foundation.
Analyses of Cell Lysates
HEK293 cells and MEFs were analyzed 24, 36, or 48 h after transfection. Immunoprecipitation of 35S-labeled cells and endoglycosidase H treatment of captured proteins were performed as described previously (21). We have also previously described conditions for co-immunoprecipitation, Western blotting, luciferase assays, and real-time PCR (9, 13). Analyses of MKS3 membrane association by detergent solubilization and proteinase K digestion was performed using the procedure described by Schuberth and Buchberger (22).
Microscopy Analyses
Confocal, fluorescence, and phase microscopies were performed as described previously (9). Briefly, HEK293 cells grown on 40-mm square glass coverslips were fixed with 4% paraformaldehyde in phosphate-buffered saline (pH 7.4) at room temperature for 10 min, followed by post-fixation with fresh fixative at 4 °C overnight. Localization of MKS3 and BiP was demonstrated by indirect immunofluorescence using guinea pig anti-MKS3 and rabbit anti-GRP78/BiP antibodies. To determine whether MKS3 localized to the plasma membrane and Golgi apparatus, fixed and permeabilized HEK293 cells were first incubated with guinea pig anti-MKS3 antibody and Alexa 555-labeled goat anti-guinea pig IgG, followed by staining with 5 μg/ml WGA-conjugated Oregon Green 488 in phosphate-buffered saline (23) at room temperature for 1 h. Stacks of immunofluorescence images were acquired by sequential acquisitions with a 0.15-μm step distance using a Nikon C1si confocal microscope.
For ultrastructural analyses, rat lungs, mouse kidneys, and HEK293 cells were cut into 1–2 mm3 blocks; fixed with 4% paraformaldehyde/lysine/sodium m-periodate, 0.1% glutaraldehyde, and 0.1% CaCl2 in 0.2 m HEPES (pH 7.2) at 4 °C overnight; and cryoprotected with buffered polyvinyl pyrrolidone/sucrose. Localization of MKS3 was visualized by cryo-immunogold labeling using 10 nm of protein A-gold as described previously (24). Compartment-specific localization of MKS3 to rat alveolar type II epithelial cells (n = 16 cells, 3049 total gold counts), mouse ciliated epithelial cells of proximal convoluted tubules (n = 36, 3723 total gold counts), and HEK293 cells (n = 61 cells, 7227 total gold counts) was individually assessed by gold quantification using the relative labeling index described by Mayhew et al. (25). Compartments with a relative labeling index greater than unity and significantly different by χ2 analysis (degrees of freedom = 7, p ≤ 0.001) were considered to be specifically labeled with anti-MKS3 antibody.
RESULTS
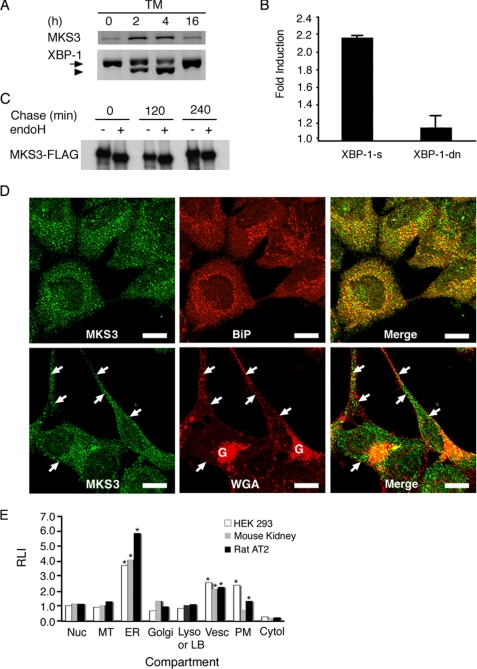
The results of previous microarray analyses indicated that expression of MKS3 is up-regulated by ∼5-fold in response to mutant SP-C (13). Quantitative PCR analyses confirmed that MKS3 mRNA was increased by >2-fold in cells expressing two different SP-C mutations, SP-CΔexon4 and SP-C(L188Q) (supplemental Fig. S1A). Treatment of untransfected HEK293 cells with tunicamycin (Fig. 1A) or thapsigargin (data not shown), pharmacological inducers of ER stress, significantly increased expression of MKS3 mRNA. Identification of two putative unfolded protein response element consensus sites in the 5′-flanking sequence (−232 to −252 bp and −605 to −625 bp) suggested that the MKS3 gene might be a target of XBP-1, a key transcriptional regulator of multiple genes involved in the ER stress response. Cotransfection of spliced XBP-1 enhanced expression of an MKS3 promoter-luciferase construct in HEK293 cells (Fig. 1B), whereas cotransfection of a dominant-negative XBP-1 construct had little effect on reporter expression. Taken together, these data are consistent with the hypothesis that expression of MKS3 is increased in response to ER stress.
FIGURE 1.
MKS3 is an ER stress response gene. A, HEK293 cells were treated with tunicamycin (TM) for the indicated number of hours. Reverse transcription-PCR was performed for MKS3 and XBP-1. The arrow indicates unspliced XBP-1, and the arrowhead indicates spliced (active) XBP-1. B, HEK293 cells were transiently transfected with an MKS3 promoter (−97 to −1064)-luciferase construct, pRL-TK vector encoding Renilla luciferase, and plasmid encoding spliced (XBP-1-s) or dominant-negative (XBP-1-dn) forms of XBP-1. Firefly luciferase activity was normalized to Renilla luciferase activity. Results are expressed as -fold induction of luciferase activity relative to vector-transfected cells and represent the average of three independent experiments. C, HEK293 cells were transfected with MKS3-FLAG. 24 h later, cells were labeled with [35S]Met/Cys for 30 min, after which the labeling medium was replaced with chase medium. Cell lysates were immunoprecipitated with anti-FLAG antibody, treated with endoglycosidase H (endoH), and analyzed by SDS-PAGE/autoradiography. D, endogenous MKS3 in HEK293 cells was detected by confocal microscopy using antibody directed against recombinant human MKS3 (see supplemental Fig. S1D). Anti-BiP antibody was used as a marker for ER (upper panels), whereas WGA was used to stain the plasma membrane and Golgi apparatus (lower panels). The arrows indicate colocalization of MKS3 and WGA at the plasma membrane. Scale bars = 10 μm. E, HEK293 cells, rat lung, and mouse kidney were prepared for cryoultramicrotomy and immunogold labeling with anti-MKS3 antibody/protein A-gold. Subcellular localization of MKS3 was assessed by quantitation of gold particles and is expressed as the relative labeling index (RLI). Cytol, cytosol; LB, lamellar body; Lyso, lysosome; MT, mitochondria; Nuc, nucleus; PM, plasma membrane; vesc, vesicle. The asterisks identify subcellular compartments with significant accumulation of MKS3 as defined under “Experimental Procedures.”
To determine whether MKS3 was localized to the ER, pulse-chase studies were performed in transiently transfected HEK293 cells. MKS3-FLAG remained endoglycosidase H-sensitive throughout the chase period (Fig. 1C), consistent with prolonged ER retention. Extraction of MKS3-FLAG from cell lysates required Triton X-100 (supplemental Fig. S1B), suggesting membrane association, as predicted previously (14, 26). Treatment of isolated microsomes with proteinase K digested the FLAG tag in the absence of detergent (supplemental Fig. S1C). These data indicate that MKS3 is an integral membrane glycoprotein with a C-terminal cytosolic tail. Polyclonal antibodies generated against recombinant protein corresponding to a segment of the predicted cytosolic domain (residues 776–924) of human MKS3 detected a single protein (∼120 kDa) in HEK293 cell lysates (supplemental Fig. S1D). Confocal microscopy demonstrated extensive colocalization of endogenous MKS3 and BiP (Fig. 1D, upper panels) and calreticulin (data not shown) in HEK293 cells; a small amount of MKS3 colocalized with WGA at the plasma membrane (Fig. 1D, lower panels). Immunogold labeling confirmed that endogenous MKS3 predominantly localized to the ER in HEK293 cells, mouse kidney, and rat alveolar type II epithelial cells (Fig. 1E); MKS3 was also detected on the plasma membrane, as reported previously (26), and in peripheral vesicles (Fig. 1, D and E, and supplemental Fig. S1E). Collectively, these data indicate that a significant proportion of endogenous MKS3 is localized to the ER.
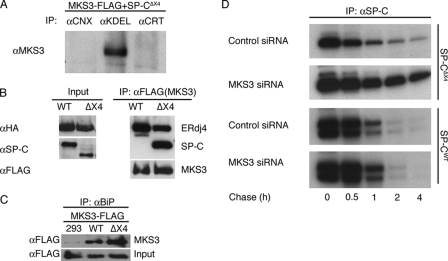
The results of preliminary experiments in HEK293 cells transiently transfected with MKS3-FLAG demonstrated that MKS3 did not co-precipitate with the ER chaperones calnexin and calreticulin (Fig. 2A); however, MKS3 did co-precipitate with an unidentified “KDEL”-containing protein(s) (Fig. 2A). Coupled with the observation that MKS3 expression was significantly increased in the presence of mutant SP-C, these results suggest that MKS3 may associate with specific ER resident proteins involved in quality control of misfolded proteins. To test this hypothesis, HEK293 cells were cotransfected with MKS3-FLAG and WT SP-C or SP-CΔexon4, followed by immunoprecipitation for MKS3 and Western blotting to identify SP-C. SP-CΔexon4 is rapidly degraded via a proteasome-dependent pathway in HEK293 cells, whereas WT SP-C is exported from the ER to lysosomes via the Golgi (27). MKS3 co-precipitated with mutant SP-C (SP-CΔexon4) but not with WT SP-C (Fig. 2B and supplemental Fig. S2). MKS3 also bound cotransfected ER chaperones involved in SP-C ERAD (13), including ERdj4 (Fig. 2B) and ERdj5 (supplemental Fig. S2); the reverse co-precipitation experiments confirmed the specificity of the interaction (data not shown). Importantly, immunoprecipitation of endogenous BiP, a KDEL-containing ER resident chaperone, captured MKS3, and interaction of BiP and MKS3 was enhanced in the presence of mutant SP-C (Fig. 2C). These results suggest that MKS3 is a component of an ER chaperone complex involved in detection and/or disposal of terminally misfolded SP-C. This hypothesis predicts that knockdown of MKS3 would lead to accumulation of mutant SP-C proprotein in the ER. Even though complete knockdown of MKS3 protein was not achieved within the 48-h treatment window (supplemental Fig. S3), siRNA treatment significantly slowed degradation of SP-CΔexon4 proprotein (Fig. 2D). MKS3 siRNA did not affect loss of WT SP-C proprotein, which is exported from the ER to the lysosome (27). Overall, these results indicate that MKS3 selectively associates with mutant SP-C and that this event is linked to turnover of the misfolded substrate.
FIGURE 2.
MKS3 interacts with mutant SP-C and ER chaperones. A, HEK293 cells were cotransfected with plasmids encoding SP-CΔexon4 (SP-CΔX4) and MKS3-FLAG. 48 h later, cells were treated with DSP to cross-link interacting proteins. Cell lysates were immunoprecipitated (IP) with antibodies directed against calnexin (CNX), KDEL, or calreticulin (CRT) and analyzed by SDS-PAGE, followed by Western blotting with antibody directed against MKS3. B, HEK293 cells stably expressing WT SP-C or SP-CΔexon4 (ΔX4) were cotransfected with plasmids encoding ERdj4-HA or MKS3-FLAG. 24 h later, cells were treated with MG-132 to prevent proteasome-mediated degradation of SP-CΔexon4 and with DSP to cross-link interacting proteins. Cell lysates were immunoprecipitated with anti-FLAG antibody and analyzed by SDS-PAGE/Western blotting with anti-SP-C, anti-HA, or anti-FLAG antibodies. C, untransfected HEK293 cells or HEK293 cells stably expressing WT SP-C or SP-CΔexon4 were transfected with MKS3-FLAG. 24 h later, cells were treated with MG-132 and DSP as described above, and lysates were immunoprecipitated with anti-BiP antibody, followed by SDS-PAGE/Western blotting with anti-FLAG antibody. D, HEK293 cells stably expressing WT SP-C or SP-CΔexon4 were treated with MKS3 siRNA or control siRNA (see supplemental Fig. S3) for 48 h. Cells were labeled with [35S]Met/Cys for 30 min, after which the labeling medium was replaced with chase medium. Lysates were immunoprecipitated with anti-SP-C antibody and analyzed by SDS-PAGE/autoradiography.
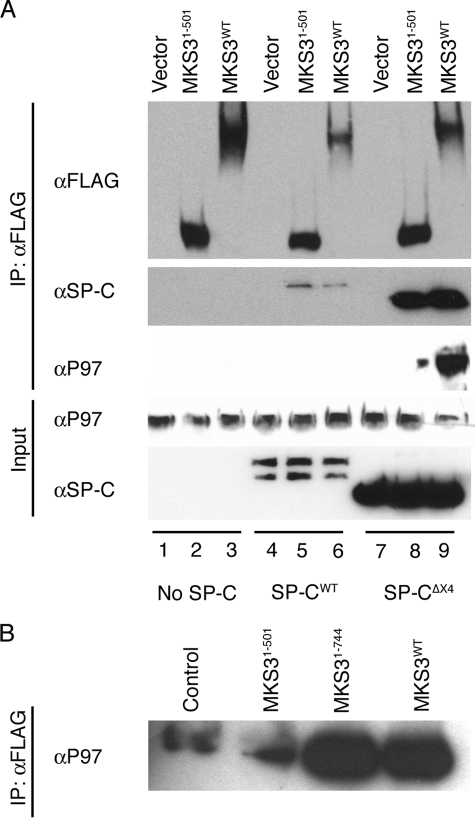
To further investigate the potential role of MKS3 in degradation of mutant SP-C, a truncated form of MKS3 encompassing the predicted lumenal domain of the protein (MKS3-(1–501)) was expressed in HEK293 cells. MKS3-(1–501) colocalized with calreticulin (supplemental Fig. S4A) and co-precipitated with mutant SP-C, ERdj4, and ERdj5 but did not interact with WT SP-C (supplemental Fig. S4B), similar to full-length MKS3. Thus, the soluble lumenal domain of MKS3 is sufficient for association with the ER chaperone complex bound to mutant SP-C. To determine whether truncation of MKS3 altered association of the lumenal complex with the cytosolic degradation machinery, SP-C was transiently expressed in HEK293 cells with MKS3-(1–501) or WT MKS3. Pulldown experiments with full-length MKS3 identified a complex that included mutant SP-C and cytosolic p97 (Fig. 3A, lane 9). Interaction of MKS3 with endogenous p97 was abrogated in the absence of SP-CΔexon4 expression. Pulldown experiments with MKS3-(1–501) detected mutant SP-C, as expected, but associated very weakly with p97 (Fig. 3A, lane 8). An MKS3 construct that included the predicted transmembrane domains but not the cytosolic domain (MKS3-(1–744)) also bound p97, implicating the region encompassing the transmembrane domains as essential for MKS3/p97 interaction (Fig. 3B). Thus, MKS3 appears to link the quality control complex in the ER lumen with the degradation machinery in the cytosol.
FIGURE 3.
MKS3 interacts with cytosolic p97. A, HEK293 cells stably expressing WT SP-C or SP-CΔexon4 (SP-CΔX4) were transiently transfected with full-length MKS3 or a truncated construct encoding the predicted lumenal domain of MKS3 (MKS3-(1–501)) (see supplemental Fig. S4). 24 h after transfection, cells were treated with MG-132 and DSP, and cell lysates were immunoprecipitated (IP) with anti-FLAG antibody. MKS3 complexes were analyzed by SDS-PAGE/Western blotting with anti-FLAG, anti-SP-C, and anti-p97 antibodies. B, the experiment in A was repeated with an additional construct (MKS3-(1–744)) that included the region encoding the transmembrane domains but not the putative cytosolic domain. Control refers to untransfected HEK293 cell lysate Western-blotted with anti-p97 antibody.
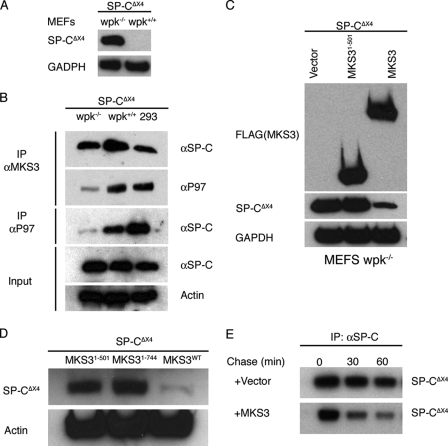
Autosomal recessive mutations in the MKS3 gene are associated with multi-organ disease in humans and the wpk rat. To determine whether ERAD was altered in the presence of mutant MKS3, two disease-associated mutant MKS3 proteins (Q376P and P394L, associated with disease in humans and wpk rats, respectively) were expressed in HEK293 cells. The turnover of mutant MKS3 in transfected HEK293 cells was very similar to that of the WT protein (supplemental Fig. S5A); in contrast, the stability of MKS3-(1–501) was increased. Similar to MKS3-(1–501), both Q376P and P394L co-precipitated with SP-CΔexon4, ERdj4, and ERdj5 (data not shown). We next considered the possibility that the relatively high concentration of endogenous WT MKS3 in HEK293 cells (supplemental Fig. S5B) modified the turnover and/or activity of mutant MKS3. MEFs were prepared from embryonic wpk−/− rats and WT littermates harvested at embryonic day 13. Western blot (supplemental Fig. S5B) and immunofluorescence (supplemental Fig. S5C) analyses demonstrated that MKS3(P394L) protein was greatly reduced in wpk−/− MEFs, consistent with destabilization. Subsequently, MEFs were transiently transfected with SP-CΔexon4 or WT SP-C. Mutant SP-C accumulated in wpk−/− but not wpk+/+ MEFs (Fig. 4A). Immunoprecipitation of endogenous MKS3 in wpk−/− cells pulled down SP-CΔexon4 but very little p97 (Fig. 4B). Consistent with this result, immunoprecipitation of endogenous p97 recovered very little SP-CΔexon4 in wpk−/− cells compared with WT MEFs and HEK293 cells (Fig. 4B). Cotransfection of WT MKS3 protein reduced accumulation (Fig. 4, C and D) and accelerated degradation (Fig. 4E) of mutant SP-C. Importantly, neither MKS3-(1–501) (Fig. 4C) nor MKS3-(1–744) (Fig. 4D) reduced accumulation of SP-CΔexon4, indicating that the cytosolic domain of MKS3 is required for restoration of degradation. Collectively, these results indicate that mutant MKS3(P394L) can associate with mutant SP-C in the ER lumen but that association with the cytosolic degradation machinery is dramatically reduced, leading to accumulation of misfolded substrate in the ER lumen.
FIGURE 4.
ERAD is decreased in MEFs expressing mutant MKS3. A, MEFs were isolated from day 13 wpk+/+ and wpk−/− embryos and transiently transfected with SP-CΔexon4 (SP-CΔX4). 36 h later, cells were analyzed by SDS-PAGE/Western blotting with anti-SP-C antibody. B, MEFs and HEK293 cells were transiently transfected with SP-CΔexon4. wpk+/+ MEFs and HEK293 cells were treated with MG-132 to promote accumulation of mutant SP-C; all cells were treated with DSP to cross-link interacting proteins. Cell lysates were immunoprecipitated (IP) for endogenous MKS3 or p97 and analyzed by SDS-PAGE/Western blotting with anti-SP-C and anti-p97 antibodies. C, wpk−/− MEFs were transiently cotransfected with SP-CΔexon4 and vector, MKS3-(1–501), or full-length MKS3. 48 h later, cell lysates were analyzed by SDS-PAGE/Western blotting with anti-FLAG, anti-SP-C, and anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibodies. D, the experiment in C was repeated with MKS3-(1–744). E, wpk−/− MEFs were transiently cotransfected with SP-CΔexon4 and vector or full-length MKS3. 24 h later, cells were labeled with [35S]Met/Cys for 30 min, after which the labeling medium was replaced with chase medium. Cell lysates were immunoprecipitated with anti-SP-C antibody and analyzed by SDS-PAGE/autoradiography.
To determine whether MKS3 is required for degradation of a different ERAD substrate, wpk−/− MEFs were transfected with α1-antitrypsin (AAT) or a disease-associated mutant form of AAT (PiZ). Mutant (52 kDa) but not WT (55 kDa) AAT accumulated in MEFs (supplemental Fig. S6A). AAT and PiZ were also cotransfected into HEK293 cells with MKS3-FLAG. MKS3 co-precipitated with PiZ but not WT AAT (supplemental Fig. S6B). Thus, MKS3 promotes ERAD of at least two different mutant proteins.
DISCUSSION
Meckel-Gruber syndrome is categorized as a ciliopathy, and consistent with a cilium-related function, the MKS3 protein was shown previously to be localized to the plasma membrane and cilia of epithelial cells (26). Based on topological similarity to Frizzled receptors, it was suggested that MKS3 may function in non-canonical Wnt signaling (14, 26). The results of this study demonstrate that a significant pool of MKS3 protein is also localized to the ER, where it may play a role in ERAD. Both genetic and pharmacological inducers of ER stress resulted in up-regulation of MKS3 expression in human embryonic kidney cells. Stress-induced expression of MKS3 may be mediated in part by XBP-1, a transcriptional regulator of multiple genes involved in ERAD. Stable interaction of MKS3 with an ER lumenal complex composed of chaperones and misfolded substrate, as well as with p97, a cytosolic ATPase involved in extraction of misfolded protein from the ER membrane for degradation by proteasome, provides additional evidence linking MKS3 to ERAD. Consistent with this conclusion, knockdown of MKS3 resulted in accumulation of misfolded protein. Taken together, these data suggest that MKS3 is a previously unrecognized component of the ERAD pathway.
This study focused primarily on the role of MKS3 in proteasome-mediated turnover of mutant SP-C. SP-C is a non-glycosylated protein with a single transmembrane domain and a relatively short cytosolic domain. Two disease-associated mutations, SP-CΔexon4 and SP-C(L188Q), have been extensively studied (6, 28–30). Both mutant proteins are ERAD substrates (9–12) that form lumenal complexes with BiP, ERdj4, and ERdj5 (13) and, as shown in this study, interact with MKS3. Unlike SP-C, AAT is a soluble secreted glycoprotein; however, similar to mutant SP-C, mutant AAT associated with MKS3 and accumulated when the MKS3/p97 interaction was abrogated. Deletion analyses of MKS3 indicated that the lumenal domain was sufficient for interaction with the SP-C·chaperone complex in the ER. The region encompassing the transmembrane domains facilitated interaction of MKS3 with cytosolic p97; however, this interaction was not sufficient to restore degradation of mutant SP-C in wpk−/− MEFs. Only WT MKS3 restored SP-C ERAD in wpk−/− MEFs, consistent with an important function for the cytosolic domain of MKS3. These results support a model in which MKS3 links the ER lumenal quality control machinery with the cytosolic degradation complex. However, the precise function of MKS3 and its relationship to the retrotranslocon remain unclear.
The results of this study raise the question of whether the ERAD function of MKS3 is related to its role in ciliogenesis. Although direct evidence supporting this hypothesis is lacking, a recent study of polycystin-2 (PC2) suggests a possible link (31). Like MKS3, PC2 localizes to both the primary cilial membrane and the ER. Mutations in PC2 are associated with autosomal dominant polycystic kidney disease (32), a ciliopathy with phenotypic similarities to Meckel-Gruber syndrome. WT PC2 is degraded by ERAD, and turnover is accelerated in response to ER stress (31). Interestingly, expression of PC2 is elevated in Taz−/− mice in association with polycystic kidney disease and emphysema (33). TAZ (transcriptional co-activator with PDZ-binding motif) is a component of an E3 ligase complex involved in ubiquitination and subsequent degradation of PC2. Thus, both overexpression and underexpression of PC2 are associated with polycystic kidney disease, and ERAD may play an important role in ciliogenesis by limiting expression of PC2. Likewise, MKS3 may promote turnover of WT and mutant ciliogenic proteins via ERAD; this function is not specific for ciliogenic proteins because neither SP-C nor AAT is involved in ciliogenesis. Alternatively, it is possible that the ERAD function of MKS3 is unrelated to ciliogenesis.
Mutations in MKS3 are associated with a range of ciliopathies, including Joubert syndrome (34), COACH syndrome (35), and Bardet-Biedl syndrome (36), in addition to Meckel-Gruber syndrome (14, 15). There are very little data regarding the effect of mutations on protein stability/function or the function of the WT protein in ciliogenesis. Dawe et al. (26) reported that MKS3 was undetectable in the kidney tissue of a patient homozygous for the Q376P mutation; however, when transfected into RCC4 or HEK293 cells, the mutant and WT proteins were expressed at similar levels. We confirmed this finding in HEK293 cells and also showed that the P394L mutation, associated with polycystic kidney disease in wpk rats (14), was similarly stable when expressed in HEK293 cells. However, in MEFs isolated from wpk−/− rats, expression of MKS3(P394L) was greatly reduced compared with MEFs isolated from WT littermates. These results suggest that the primary effect of the P394L mutation is accelerated turnover of the mutant protein, which, in turn, leads to loss of function. Consistent with this hypothesis, complete loss of MKS3 in bpck/bpck mutant mice is associated with polycystic kidney disease, similar to that in wpk rats (37, 38). Importantly, the effect of the mutation on protein stability is masked in the presence of WT MKS3.
MKS3 is constitutively expressed in a wide range of cells and is up-regulated in response to ER stress. The lumenal domain of MKS3 interacts with an ER quality control complex, and the region encompassing the transmembrane domains interacts with cytosolic p97, providing an essential link between the two compartments involved in ERAD.
Supplementary Material
Acknowledgments
We gratefully acknowledge Ann Maher for secretarial assistance and Angela Keiser for generating the anti-MKS3 antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants P01-HL61646 and R01-HL086492 from NHLBI.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and additional references.
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- SP-C
- surfactant protein C
- HA
- hemagglutinin
- WGA
- wheat germ agglutinin
- DSP
- dithiobis(succinimidyl propionate)
- WT
- wild-type
- MEFs
- murine embryonic fibroblasts
- siRNA
- small interfering RNA
- AAT
- α1-antitrypsin
- PC2
- polycystin-2.
REFERENCES
- 1.Yoshida H. (2007) FEBS J. 274, 630–658 [DOI] [PubMed] [Google Scholar]
- 2.Ron D., Walter P. (2007) Nat. Rev. Mol. Cell Biol. 8, 519–529 [DOI] [PubMed] [Google Scholar]
- 3.Schröder M., Kaufman R. J. (2005) Annu. Rev. Biochem. 74, 739–789 [DOI] [PubMed] [Google Scholar]
- 4.Nakatsukasa K., Brodsky J. L. (2008) Traffic 9, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nogee L. M., Dunbar A. E., 3rd, Wert S. E., Askin F., Hamvas A., Whitsett J. A. (2001) N. Engl. J. Med. 344, 573–579 [DOI] [PubMed] [Google Scholar]
- 7.Nogee L. M. (2004) Annu. Rev. Physiol. 66, 601–623 [DOI] [PubMed] [Google Scholar]
- 8.Wang W. J., Mulugeta S., Russo S. J., Beers M. F. (2003) J. Cell Sci. 116, 683–692 [DOI] [PubMed] [Google Scholar]
- 9.Bridges J. P., Xu Y., Na C. L., Wong H. R., Weaver T. E. (2006) J. Cell Biol. 172, 395–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mulugeta S., Nguyen V., Russo S. J., Muniswamy M., Beers M. F. (2005) Am. J. Respir. Cell Mol. Biol. 32, 521–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulugeta S., Maguire J. A., Newitt J. L., Russo S. J., Kotorashvili A., Beers M. F. (2007) Am. J. Physiol. Lung Cell. Mol. Physiol. 293, L720–L729 [DOI] [PubMed] [Google Scholar]
- 12.Bridges J. P., Wert S. E., Nogee L. M., Weaver T. E. (2003) J. Biol. Chem. 278, 52739–52746 [DOI] [PubMed] [Google Scholar]
- 13.Dong M., Bridges J. P., Apsley K., Xu Y., Weaver T. E. (2008) Mol. Biol. Cell 19, 2620–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith U. M., Consugar M., Tee L. J., McKee B. M., Maina E. N., Whelan S., Morgan N. V., Goranson E., Gissen P., Lilliquist S., Aligianis I. A., Ward C. J., Pasha S., Punyashthiti R., Malik Sharif S., Batman P. A., Bennett C. P., Woods C. G., McKeown C., Bucourt M., Miller C. A., Cox P., Algazali L., Trembath R. C., Torres V. E., Attie-Bitach T., Kelly D. A., Maher E. R., Gattone V. H., 2nd, Harris P. C., Johnson C. A. (2006) Nat. Genet. 38, 191–196 [DOI] [PubMed] [Google Scholar]
- 15.Khaddour R., Smith U., Baala L., Martinovic J., Clavering D., Shaffiq R., Ozilou C., Cullinane A., Kyttala M., Shalev S., Audollent S., d'Humieres C., Kadhom N., Esculpavit C., Viot G., Boone C., Oien C., Encha-Razavi F., Batman P. A., Bennett C. P., Woods C. G., Roume J., Lyonnet S., Genin E., Le Merrer M., Munnich A., Gubler M. C., Cox P., Macdonald F., Vekemans M., Johnson C. A., Attie-Bitach T. (2007) Hum. Mutat. 28, 523–524 [DOI] [PubMed] [Google Scholar]
- 16.Iwakoshi N. N., Lee A. H., Vallabhajosyula P., Otipoby K. L., Rajewsky K., Glimcher L. H. (2003) Nat. Immunol. 4, 321–329 [DOI] [PubMed] [Google Scholar]
- 17.Rice W. R., Conkright J. J., Na C. L., Ikegami M., Shannon J. M., Weaver T. E. (2002) Am. J. Physiol. Lung Cell. Mol. Physiol. 283, L256–L264 [DOI] [PubMed] [Google Scholar]
- 18.Lee A. H., Iwakoshi N. N., Glimcher L. H. (2003) Mol. Cell. Biol. 23, 7448–7459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo F., Zheng Y. (2004) Oncogene 23, 5577–5585 [DOI] [PubMed] [Google Scholar]
- 20.Deleted in proof
- 21.Lin S., Phillips K. S., Wilder M. R., Weaver T. E. (1996) Biochim. Biophys. Acta 1312, 177–185 [DOI] [PubMed] [Google Scholar]
- 22.Schuberth C., Buchberger A. (2005) Nat. Cell Biol. 7, 999–1006 [DOI] [PubMed] [Google Scholar]
- 23.Tartakoff A. M., Vassalli P. (1983) J. Cell Biol. 97, 1243–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ueno T., Linder S., Na C. L., Rice W. R., Johansson J., Weaver T. E. (2004) J. Biol. Chem. 279, 16178–16184 [DOI] [PubMed] [Google Scholar]
- 25.Mayhew T. M., Mühlfeld C., Vanhecke D., Ochs M. (2009) Ann. Anat. 191, 153–170 [DOI] [PubMed] [Google Scholar]
- 26.Dawe H. R., Smith U. M., Cullinane A. R., Gerrelli D., Cox P., Badano J. L., Blair-Reid S., Sriram N., Katsanis N., Attie-Bitach T., Afford S. C., Copp A. J., Kelly D. A., Gull K., Johnson C. A. (2007) Hum. Mol. Genet. 16, 173–186 [DOI] [PubMed] [Google Scholar]
- 27.Conkright J. J., Apsley K. S., Martin E. P., Ridsdale R., Rice W. R., Na C. L., Yang B., Weaver T. E. (2009) Am. J. Respir. Cell Mol. Biol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas A. Q., Lane K., Phillips J., 3rd, Prince M., Markin C., Speer M., Schwartz D. A., Gaddipati R., Marney A., Johnson J., Roberts R., Haines J., Stahlman M., Loyd J. E. (2002) Am. J. Respir. Crit. Care Med. 165, 1322–1328 [DOI] [PubMed] [Google Scholar]
- 29.Soraisham A. S., Tierney A. J., Amin H. J. (2006) J. Perinatol. 26, 67–70 [DOI] [PubMed] [Google Scholar]
- 30.Chibbar R., Shih F., Baga M., Torlakovic E., Ramlall K., Skomro R., Cockcroft D. W., Lemire E. G. (2004) Mod. Pathol. 17, 973–980 [DOI] [PubMed] [Google Scholar]
- 31.Liang G., Li Q., Tang Y., Kokame K., Kikuchi T., Wu G., Chen X. Z. (2008) Hum. Mol. Genet. 17, 1109–1119 [DOI] [PubMed] [Google Scholar]
- 32.Torres V. E., Harris P. C. (2006) Nat. Clin. Pract. Nephrol. 2, 40–55 [DOI] [PubMed] [Google Scholar]
- 33.Tian Y., Kolb R., Hong J. H., Carroll J., Li D., You J., Bronson R., Yaffe M. B., Zhou J., Benjamin T. (2007) Mol. Cell. Biol. 27, 6383–6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baala L., Romano S., Khaddour R., Saunier S., Smith U. M., Audollent S., Ozilou C., Faivre L., Laurent N., Foliguet B., Munnich A., Lyonnet S., Salomon R., Encha-Razavi F., Gubler M. C., Boddaert N., de Lonlay P., Johnson C. A., Vekemans M., Antignac C., Attie-Bitach T. (2007) Am. J. Hum. Genet. 80, 186–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brancati F., Iannicelli M., Travaglini L., Mazzotta A., Bertini E., Boltshauser E., D'Arrigo S., Emma F., Fazzi E., Gallizzi R., Gentile M., Loncarevic D., Mejaski-Bosnjak V., Pantaleoni C., Rigoli L., Salpietro C. D., Signorini S., Stringini G. R., Verloes A., Zabloka D., Dallapiccola B., Gleeson J. G., Valente E. M. (2009) Hum. Mutat. 30, E432–E442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leitch C. C., Zaghloul N. A., Davis E. E., Stoetzel C., Diaz-Font A., Rix S., Alfadhel M., Lewis R. A., Eyaid W., Banin E., Dollfus H., Beales P. L., Badano J. L., Katsanis N. (2008) Nat. Genet. 40, 443–448 [DOI] [PubMed] [Google Scholar]
- 37.Cook S. A., Collin G. B., Bronson R. T., Naggert J. K., Liu D. P., Akeson E. C., Davisson M. T. (2009) J. Am. Soc. Nephrol. 20, 753–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gattone V. H., 2nd, Tourkow B. A., Trambaugh C. M., Yu A. C., Whelan S., Phillips C. L., Harris P. C., Peterson R. G. (2004) Anat. Rec. A Discov. Mol. Cell Evol. Biol. 277, 384–395 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.