Abstract
The Rev protein is a key regulator of human immunodeficiency virus type 1 (HIV-1) gene expression. Rev is primarily known as an adaptor protein for nuclear export of HIV RNAs. However, Rev also contributes to numerous other processes by less well known mechanisms. Understanding the functional nature of Rev requires extensive knowledge of its cellular interaction partners. Here we demonstrate that Rev interacts with members of a large family of multifunctional host cell factors called hnRNPs. Rev employs amino acids 9–14 for specific binding to the heterogeneous nuclear ribonucleoproteins (hnRNP) A1, Q, K, R, and U. In addition, Rev interacts with hnRNP E1 and E2 by a different mechanism. The set of hnRNPs recognized by the N terminus of Rev feature RGG boxes. Exemplary testing of hnRNP A1 revealed a critical role of arginine residues within the RGG box for interaction with Rev. Finally, we demonstrate that expression levels of hnRNP A1, Q, K, R, and U influence HIV-1 production by persistently infected astrocytes, linking these hnRNPs to HIV replication. The novel interaction of HIV-1 Rev with functionally diverse hnRNPs lends further support to the idea that Rev is a multifunctional protein and may be involved in coupling HIV replication to diverse cellular processes and promoting virus-host cell interactions.
INTRODUCTION
During human immunodeficiency virus (HIV)3 replication, the transcripts that encode viral structural proteins and the viral RNA genome contain introns and would normally be eliminated by the host cell. The production of these RNAs and their utilization is achieved by strictly controlled alternative splicing mechanisms and the regulatory activities of the HIV trans-activator protein Rev (reviewed in Refs. 1–3).
Rev is an RNA-binding protein that specifically binds a recognition element (RRE) within intron-containing HIV RNAs. One of the best studied functions of Rev is the recruitment of cellular factors that mediate nuclear export of Rev-bound RNAs (2, 3). Rev has also been shown to influence splicing (4), stability (3, 5, 6), and translation (7–9) of the viral RNAs as well as their packaging (10, 11). These regulatory activities of the Rev protein render it a key player in the HIV replication cycle. Detailed and sophisticated studies of the Rev protein identified at least three functional domains in Rev (2): (i) an arginine rich-motif that functions both as nuclear localization signal and RNA-binding sequence (AA 35–50), (ii) a bipartite multimerization domain (AA 12–29 and AA 52–60), and (iii) a nuclear export signal (AA 75–83). Host factors shown to bind to these functional domains include B23 and Importin β for the nuclear localization signal and CRM1/Exportin-1 and eIF-5A for the nuclear export signal (2, 12). In addition, Rev has been shown to interact with several RNA helicases (13–15).
However, the overall number of cellular interactor proteins identified for Rev is still surprisingly small, compared with, for example, Tat (16). The many activities of Rev (17) and evidence for host-cell regulation of Rev activities (18, 19) also suggest that the current knowledge of interactions of Rev with host-cell factors is still incomplete. This is further supported by the fact that no cellular interaction partners have been assigned to several regions of Rev that are known to be significant for its activity (20, 21). One of these unexplored regions is the N-terminal end of Rev.
In this study we demonstrate for the first time interaction of Rev with a large group of multifunctional proteins called hnRNPs. We show that the N terminus of Rev contains a specific region for recognition of a subgroup of hnRNPs, thus describing a novel function for this region of Rev. We also present evidence linking HIV production in persistently infected cells with expression levels of hnRNP A1, Q, R, K, and U, respectively. In silico evaluation of the functional context of Rev-interacting hnRNPs by a systems-oriented approach suggests that these hnRNPs may link Rev to a larger spectrum of biological processes than previously anticipated.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
Eukaryotic Expression Plasmids
The plasmid pC-hnRNP A1-CYN was generated by replacing the srev sequence in pC-sRev-CFP-YFP-N (22) with a cDNA sequence encoding hnRNP A1 (sequences amplified by reverse transcription-PCR from U138MG cells), using the SacII and NheI restriction sites. The construct pC-CFP-YFP-N (22) was used for control experiments (expression of CYN).
Prokaryotic Expression Plasmids
The vector system pASK-IBA3plus (IBA GmbH, Göttingen, Germany) was used for production of bacterial recombinant proteins. This vector is inducible with anhydrotetracycline. BsaI restriction sites were added to the 5′- and 3′-ends of the srevgfp, Δ2–8srevgfp, Δ2–14srevgfp, gfp, hnRNP A1, and hnRNP A1 RGG mutant sequences by PCR, using pCsRevsg143 (18), pFRED143 (24), or pC-hnRNP A1-CFP-YFP-N as templates. The srev sequence encodes for two additional amino acids (AS) after the starting methionine due to introduction of an NheI restriction site. PCR products were cloned in-frame with the StrepTagII sequence into pASK-IBA3plus resulting in the construction of the plasmids pASK-IBA3plus-sRevGFP-StrepTagII, pASK-IBA3plus-Δ2–8RevGFP-StrepTagII, pASK-IBA3plus-Δ2–14RevGFP-StrepTagII, pASK-IBA3plus-GFP-StrepTagII, pASK-IBA3plus-hnRNP A1-StrepTagII, and pASK-IBA3plus-hnRNP A1 mut-StrepTagII. All PCR amplification products were verified by sequence analysis (Sequiserve, Vaterstetten, Germany) before further use.
Peptides
10 mg of synthetic peptide (Fig. 1A) consisting of AA 1–14 of Rev (MAGRSGDSDEELIR) and the StrepTagII sequence (WSHPQFEK) was purchased from Metabion (Martinsried, Germany) at a purity of 80% (Mr 2576).
FIGURE 1.
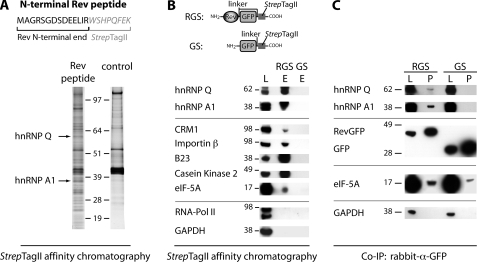
Interaction of Rev with hnRNPs A1 and Q. hnRNP A1 and Q were identified as novel interactors of Rev by affinity chromatography of astrocytic cell lysates (U138MG) with immobilized Rev baits (A and B) and by co-immunoprecipitation (IP) assays (C). A, SDS-PAGE analysis of proteins captured from cell lysates with a bait peptide containing Rev AA 1–14 fused to the StrepTagII sequence. hnRNP Q and A1 were identified by MALDI-TOF mass spectrometry. B, Western blot analysis of proteins in cell lysates (L) and elution fraction (E) of affinity chromatography assays, using either Rev-containing bait proteins (RevGFP-StrepTagII = RGS) or bait proteins lacking the Rev sequence (GFP-StrepTagII = GS). Specific Rev interaction was demonstrated for hnRNP Q and A1 and for 5 known Rev-interactors (CRM1, Importin β, B23, Casein kinase 2, and eIF-5A). RNA-polymerase II and GAPDH showed no interaction with Rev. C, Western blot analysis of proteins in lysates and immunoprecipitates (P) from human cells stably expressing either the RGS or GS bait protein. Immunoprecipitations were performed with antibodies against GFP, and proteins were detected with specific antibodies. eIF-5A served as a control for a known Rev interactor and GAPDH as a control for a non-interacting protein.
Cell Lines, Cell Culture, and Plasmid Transfections
85HG66 is an astrocytoma cell line (25). U138MG is an astrocytoma cell line obtained from American Type Culture Collection (ATCC HTB-14). TH4-7-5 cells are chronically HIV-1-infected 85HG66 cells (25). Stable RevGFP-StrepTagII and GFP-StrepTagII expressing U138 cell lines were generated after transfection of the cells with 500 ng of the respective plasmid and subsequent selection using G418 (Invitrogen). GFP-positive cells were enriched by fluorescence-activated cell sorting. All cells were cultured under standard conditions in Dulbecco's modified Eagle's medium containing 2 mm Glutamax (Invitrogen), 10% fetal calf serum (Biochrom AG, Berlin, Germany), 100 mg/ml of penicillin, and 100 units/ml of streptomycin. Cell transfections were performed with the FuGeneHD reagent following the manufacturer's instructions (Roche Diagnostics).
Recombinant Protein Expression in Escherichia coli
Recombinant proteins were produced in E. coli strain BL21codonplus RP (Stratagene). T7 polymerase expression was induced by 1 mm isopropyl 1-thio-β-d-galactopyranoside at an A550 value of 0.6 at 34 °C. Expression of the protein of interest was induced by addition of 200 ng/ml of anhydrotetracycline at an A550 value of 0.8 followed by incubation at 34 °C for 3 h. Cultures were harvested by centrifugation at 3000 × g and resuspended in lysis buffer (100 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, 0.5 mg/ml of lysozyme and protease inhibitors) at 4 °C. After 30 min at 37 °C, the culture suspension was sonicated on ice. A 20-min centrifugation at 3,000 × g (4 °C) and 30 min at 20,000 × g (4 °C) led to clarification of the lysate from cellular debris. Finally, the cell lysate was filtered (pore size of 5 μm).
StrepTagII Affinity Chromatography
StrepTagII Affinity Chromatography with a Peptide as Bait
Peptide was bound on a StrepTactin-Sepharose column (IBA GmbH) and excess peptide was washed away, using buffers and protocols provided by the manufacturer. A control column lacking the peptide was treated in the same manner. All following steps were performed at 4 °C. 1 × 108 U138 astrocytoma cells were lysed in 15 ml of freshly prepared lysis buffer (50 mm Tris, pH 7.5, 150 mm NaCl, 2 mm EDTA, 0.1% Triton X-100, 5% glycerol, 2 mm dithiothreitol, protease inhibitor Mini complete (Roche Diagnostics)). Half of the lysate was allowed to run over the column with bound peptide and the other half over the control column. After washing the columns with buffer W, peptides and bound proteins were eluted using buffer E (contains the desthiobiotin competitor), and concentrated using an Amicon Ultra-15 spin column (Millipore, Schwalbach, Germany) with an exclusion size of 3 kDa. This step removed the synthetic peptide but retained most of the protein in the concentrate. The concentrated elution fraction was then applied on pre-cast polyacrylamide gels (Invitrogen). Gels were colloidal stained and bands were excised for mass spectrometry that appeared only in the lane of the peptide column but not in that of the control column.
StrepTagII Affinity Chromatography with Recombinant Proteins as Bait
Bacterial lysates containing RevGFP-StrepTagII or GFP-StrepTagII were applied on the StrepTactin-Sepharose column (IBA GmbH), and bacterial proteins were washed away using washing buffer I (100 mm Tris-HCl, pH 8.0, 500 mm NaCl, 1 mm EDTA). Eukaryotic cell lysates (1 × 108 cells per affinity chromatography, lysed in 25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Triton X-100 supplemented with protease inhibitors and homogenized using a 0.55-μm syringe) or bacterial lysates containing recombinant proteins were added to the column. Unbound proteins were removed using washing buffer II (100 mm Tris-HCl, pH 8.0, 250 mm NaCl, 1 mm EDTA). Bait-associated proteins were eluted with a high salt containing elution buffer (100 mm Tris-HCl, pH 8.0, 1.5 m NaCl, 1 mm EDTA), leaving the StrepTagII-coupled “bait” proteins on the column. Elution fractions were concentrated using Amicon Ultra-15 spin columns (Millipore) with an exclusion size of 3 kDa and analyzed by Western blot.
Immunoprecipitation
For immunoprecipitation analysis, 1 × 107 cells were lysed using 2 ml of phosphobuffer (TBS, pH 7.4, 1% Triton X-100, 2 mm EDTA, 10 mm EGTA, 10 mm Na2P2O7, protease inhibitors) and homogenized using a 0.55-μm syringe. The supernatant of the centrifuged lysate was pre-cleared by incubating with 60 μl of immobilized Protein G (Perbio Science) for 1 h at 4 °C under rotary agitation. The bead pellet was discarded and the supernatant incubated with 8 μg of antibody for 1 h at 4 °C under continuous rotation. 60 μl of immobilized Protein G was added to the sample and incubated overnight at 4 °C under rotary agitation. Next the beads (Protein G-antibody-antigen) were washed five times with phosphate buffer without protease inhibitors. Finally, sample buffer was added to the beads, samples were boiled at 95 °C for 5 min and analyzed by Western blot.
SDS-PAGE, Western Blot Analysis, and Colloidal Staining
SDS-PAGE was carried out using 4–12% precast polyacrylamide gels and the MES buffer system of Invitrogen. Colloidal staining of polyacrylamide gels was carried out with the colloidal staining kit of Invitrogen. For Western blot analyses, the following primary antibodies were used: mouse:α-B23 (1:500, Abcam, Cambridge, UK), rabbit:α-casein kinase 2 (1:2500, AbD Serotech, Düsseldorf, Germany), mouse:α-CRM1 (1:1000, BD Biosciences, Heidelberg, Germany), mouse:α-eIF-5A (1:10000, BD Biosciences), mouse:α-GAPDH (1:10000, Chemicon/Millipore, Schwalbach, Germany), rat:α-GFP (1:50, Helmholtz Zentrum München), monoclonal mouse:α-hnRNP A1 (1:500, Sigma, Deisenhofen, Germany), polyclonal rabbit:α-hnRNP A1 (1:500, Acris Antibodies, Herford, Germany), mouse:α-hnRNP E1 (1:2000, Acris Antibodies), mouse:α-hnRNP E2 (1:300, Acris Antibodies), rabbit:α-hnRNP K (1:2000, Novus Biologicals, Littleton, CO), mouse:α-hnRNP Q (1:500, Sigma), rabbit:α-hnRNP R (1:500, Abcam), mouse:α-hnRNP U (1:2000, Abcam), rat:α-Importin β (1:1000, Biozol, Eching, Germany), rat:α-Rev 5C6 (1:50, Helmholtz Zentrum München), and mouse:α-RNA-Polymerase II 8WG16 (1:1000, Helmholtz Zentrum München). Secondary antibodies were: goat:α-mouse (1:10000, Dianova, Hamburg, Germany), goat:α-rabbit (1:10000, Dianova), and goat:α-rat (1:10000, Dianova). The nitrocellulose membrane was incubated for 2 h with the primary and for 1 h with the secondary antibody. Protein bands were detected by enhanced chemiluminescence (Perbio Science, Bonn, Germany).
Mass Spectrometry
Protein bands were stained with the colloidal Coomassie kit of Invitrogen and subsequently excised from the polyacrylamide gel. Trypsin digestion and peptide analysis by MALDI-TOF mass spectrometry were performed as described (26).
Analysis of the Influence of Various hnRNPs on HIV-1 Production by TH4-7-5 Cells
Cells were seeded at a density of 1 × 105 cells per well of a 12-well plate 1 day prior to transfection. To analyze the effect of overexpression of hnRNP A1, cells were transfected with 500 ng of plasmid-DNA/well. For knock-down of hnRNPs, cells were transfected with specific or nonsilencing siRNAs (Qiagen, Hilden, Germany) with RNAiFect transfection reagent (Qiagen) according to the manufacturer's protocol. 24 h after transfection, culture medium was renewed and cells were incubated for another 72 h. Cell culture supernatants (extracellular sample) were incubated with Triton X-100 at a final concentration of 0.5%. Cells were lysed in 5% Triton X-100 (intracellular sample), and lysates were diluted with phosphate-buffered saline to a final Triton X-100 concentration of 0.5%. Both samples were centrifuged for 5 min at 16,000 × g (room temperature), and cleared supernatants were subjected to the HIV-1-p24-Antigen-ELISA for quantification of p24 antigen using the manufacturer's protocol (Beckman Coulter, Krefeld, Germany).
Analysis of Cell Viability by the MTT Test
For detection of cell viability after siRNA treatment, cells were seeded into 96-well plates and transfected with the appropriate siRNA at day 1. After 24 h, culture medium was renewed and cells were incubated for a further 72 h. Next, cells were analyzed by the MTT test as previously described (27). In brief, cells were washed with phosphate-buffered saline, incubated with 0.5 mg/ml of MTT in serum-free medium for 4 h, and lysed with DMSO/SDS solution for absorbance measurement at 570 nm (reference wavelength 630 nm).
Determination of the Functional Context of hnRNPs by Statistical Analysis of GO Categories Annotated for Genes Co-cited with Individual hnRNPs
For each individual hnRNP gene, all co-cited genes were extracted from PubMed abstracts using BiblioSphere (Genomatix Software, Munich, version 7.20). Subsequently a ranking of GO terms from GO “biological process” associated with these co-cited genes were carried out, and results were ranked by z-scores. The top 10 GO categories were selected for further analysis.
We determined the non-redundant GO associations based on the gene lists associated with the GO categories as follows. We extracted the lists of all co-cited genes associated with the 10 top ranking GO categories. All lists were compared exhaustively for common gene IDs using GEMS Launcher's Compare lists function (Genomatix Software, version 4.8). If more than 50% of the gene IDs of the higher ranking list were also found in a lower ranking list, the GO categories were merged as follows: GO categories belonging to the same subtree were joined under the first common higher node unless this higher node was on the first or second level (immediately downstream of the general category biological process). In this case the original categories were retained.
If less than 50% gene IDs were in common, the GO categories were listed as independent. Finally, the relative percentage of the remaining GO categories was compiled to a pie chart using the Keynote program (Apple, Cupertino). Because GO categories equal or higher than level 2 of the main tree (biological process) were deleted from the process, the sum of all remaining categories is not necessarily 10 in all cases.
RESULTS
Rev Interacts with the hnRNPs Q and A1
To expand our knowledge of the Rev-interaction network, this study aimed to identify cellular factors that interact with the N-terminal region of Rev. Therefore we established an affinity chromatography assay, entailing the capture of Rev-interactor proteins from human cell lysates by Rev baits equipped with a StrepTagII sequence. Rev baits were attached to a StrepTactin-Sepharose matrix and exposed to lysates of human cells. Interactor proteins were eluted from the matrix as described under “Experimental Procedures,” separated by SDS-PAGE, and identified by mass spectrometry or immunoblotting.
Capture assays with a bait peptide containing amino acids 1–14 of Rev led to specific isolation of several proteins not detected in control assays performed without the Rev bait. MALDI-MS/MS analysis identified two of the specific interactor proteins as hnRNP Q and hnRNP A1, respectively (Fig. 1A). Because hnRNPs have been linked to HIV-1 gene expression and replication in the literature (28–30) we chose to investigate the Rev interacting capabilities of hnRNP A1 and Q in more detail.
To investigate whether these hnRNPs also interact with the complete Rev protein, we performed capture assays with a recombinant (bacterial) bait fusion protein containing full-length Rev, a GFP spacer, and the StrepTagII affinity tag (RevGFP-StrepTagII). The functionality of the recombinant Rev bait protein was tested beforehand in an in vitro nuclear export assay (described in Refs. 31 and 32) that demonstrated its ability to bind to RRE-RNA and to mediate the nuclear export of this RNA.4 Elution fractions of assays performed with the RevGFP-StrepTagII bait protein contained hnRNP Q and A1 (Fig. 1B), whereas these proteins were not detected in control assays with bait proteins lacking Rev sequences (i.e. GFP-StrepTagII). Elution fractions containing hnRNP Q and A1 also contained the known Rev interactors, CRM1, Importin β, B23, casein kinase 2, and eIF-5A (Fig. 1B). In contrast, GAPDH and RNA polymerase II were not detected in these fractions, although the latter proteins were clearly present in the input cell lysates (Fig. 1B). These data confirm the specific capture of both novel and known Rev interactors from human cell lysates by this assay. To verify interaction of Rev with hnRNP Q and A1 in the context of human cells, we performed co-immunoprecipitation assays with lysates of U138MG cells stably expressing either RevGFP-StrepTagII or GFP-StrepTagII. Western blot analyses showed the presence of hnRNP Q and A1 in precipitates containing the RevGFP-StrepTagII bait, but not in precipitates containing the GFP-StrepTagII control protein (Fig. 1C). Specific co-precipitation of hnRNP Q and A1 with RevGFP-StrepTagII was retained when cell lysates were treated with RNase prior to immunoprecipitation (see supplemental Fig. S1), indicating that RNA is not essential for interaction of Rev with these hnRNPs. The known Rev-interactor eIF-5A precipitated more efficiently with RevGFP-StrepTagII than with GFP-StrepTagII, whereas the negative control protein, GAPDH, was not detected in either of the precipitates (Fig. 1C).
The N-terminal Region of Rev Contains a Sequence (AA 9–14) for Interaction with Multiple hnRNPs
To further investigate the role of the N-terminal region of Rev for interaction with hnRNP A1 and Q, we performed capture assays with Rev bait proteins lacking 6 (Δ2–8RevGFP-StrepTagII) and 13 (Δ2–14RevGFP-StrepTagII) amino acids from the N-terminal region of the 117 AA Rev protein (Fig. 2). Both hnRNP Q and A1 interacted with the Rev bait protein lacking AA 2–8 in a similar manner as with the full-length Rev bait protein (Fig. 2). In contrast, the Rev bait protein lacking AA 2–14 showed strongly diminished interaction with these hnRNPs (Fig. 2). This indicates that the segment of Rev encompassing amino acids 9–14 of Rev is crucial for the interaction of Rev with hnRNP Q and A1. To investigate whether this region also mediates interaction of Rev with other hnRNPs, we examined the elution fractions of the different capture assays for the presence of several hnRNPs. These were selected on the basis of a previously reported connection with HIV infection (hnRNPs U, E1, and E2 (28, 29)) or similarities to hnRNP Q (hnRNP R (33)) and hnRNP E1/E2 (hnRNP K (34)). None of these or any other hnRNPs had previously been shown to interact with Rev.
FIGURE 2.
Identification of a short region in the N-terminal region of Rev that mediates specific interaction of Rev with multiple hnRNPs. Astrocytic (U138MG) lysates (L) were subjected to chromatography with three different Rev bait proteins (RGS) that contained either the full-length Rev sequence or lacked different segments of the N terminus of Rev (Δ2–8 and Δ2–14; sequences of the wild type Rev protein and the segments deleted in the mutants are indicated at the top). Parallel chromatography assays were performed with GFP-StrepTagII (GS), which lacks the entire Rev domain to control for interaction specificity. Western blot analysis of elution fractions (E) shows specific isolation of the hnRNPs A1, Q, R, U, and K by bait proteins containing full-length Rev and the Δ2–8 Rev truncation mutant, whereas isolation of these hnRNPs was dramatically reduced in assays with the Δ2–14 Rev bait. Elution fractions of assays with all three Rev bait proteins contained hnRNP E2 and hnRNP E1 (very low amounts), the known Rev interactor casein kinase 2, and the Rev bait proteins.
The results shown in Fig. 2 demonstrate that not only hnRNPs Q and A1 but also the hnRNPs R, U, and K bind to Rev and that the interaction strongly depends on Rev region AA 9–14. Furthermore, we observed that the interaction of Rev with hnRNP E2 was independent of Rev region AA 9–14 and that hnRNP E1, although it is closely related to hnRNP E2 (34) interacted with Rev to a much lesser extent than hnRNP E2. Together these results suggest that Rev uses different mechanisms to interact with different hnRNPs.
Arginine Residues within the RGG Box of hnRNP A1 Are Crucial for Binding to Rev
Interestingly, all five hnRNPs recognized by the Rev N-terminal region contained an RGG box (see supplemental Fig. S2), in contrast to hnRNP E1 and E2. One of the hallmarks of the RGG box is the presence of numerous arginine residues (35). We investigated the role of the arginine residues in the RGG box for interaction with Rev exemplarily for hnRNP A1. Therefore, we designed a mutant hnRNP A1 prey protein with 6 arginine to alanine exchanges within the RGG box (Fig. 3A). Capture assays confirmed the capability of a bacterial recombinant wild type hnRNP A1 prey protein to interact with the Rev bait protein (Fig. 3B). In contrast, the mutant hnRNP A1 protein failed to bind to the Rev protein (Fig. 3C). Western blot analysis demonstrated that the input lysates used in both assays contained similar levels of the wild type and mutant hnRNP A1 proteins, respectively (Fig. 3C). These results demonstrate that arginine residues within the RGG box of hnRNP A1 are crucial for interaction with Rev.
FIGURE 3.
Arginine residues in the RGG domain of hnRNP A1 are essential for Rev binding. A, the diagram shows the sequence of the RGG domain in wild type (wt) hnRNP A1 and the positions of the arginine to alanine exchanges in the hnRNP A1 RGG mutant (mut). B, lysates (L) of bacteria expressing wild type hnRNPA1 were subjected to affinity chromatography assays with the RevGFP-StrepTagII (RGS) bait protein or the GFP-StrepTagII (GS) control protein. Western blot analysis with antibodies against hnRNP A1 shows expression of hnRNP A1 in bacterial lysates (L) and specific capture of wild type hnRNP A1 by the Rev bait protein but not by the control protein. C, affinity chromatography assays with bacterial lysates expressing the wild type or the mutant hnRNP A1 protein. Western blot analysis with a polyclonal hnRNP A1 antibody confirms similar expression levels of the wild type and mutant hnRNP A1 proteins in bacterial lysates and demonstrates specific capture of the wild type but not the mutant hnRNP A1 protein. E, elution fractions.
Influence of Intracellular Expression Levels of hnRNP A1, Q, R, K, and U on HIV Production by Persistently Infected Cells
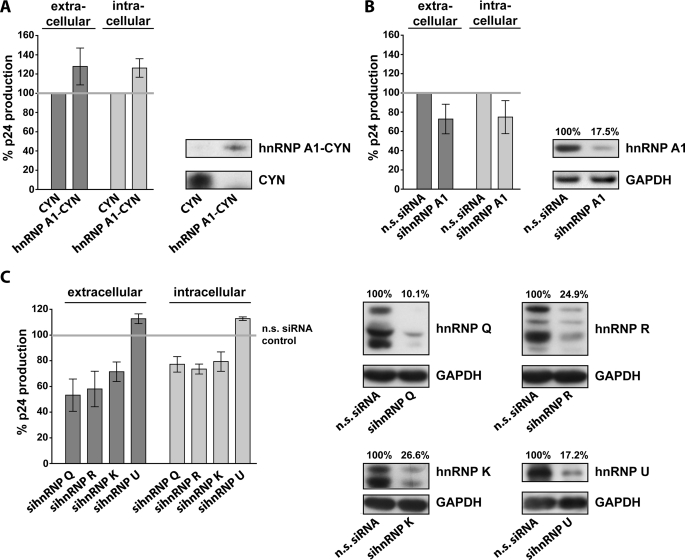
hnRNP A1 has been shown to influence HIV replication in acutely infected cells (30), suggesting a role of hnRNP A1 as regulator of HIV replication. To further pursue this hypothesis we investigated whether hnRNP A1 can influence HIV replication in persistently HIV-infected cells that represent potential virus reservoirs. As a model we used the persistently HIV-infected astrocytoma cell line TH4-7-5, a well established model for HIV reservoirs in the brain (19). To increase levels of hnRNP A1, cells were transiently transfected with plasmids (pC-hnRNP A1-CYN) that carry a gene for production of hnRNP A1 fused to a blue fluorescent tag. Cells were transfected in parallel with plasmids lacking hnRNP A1 sequences (pC-CYN) to control for unspecific effects. Transfection of TH4-7-5 cells with pC-hnRNP A1-CYN led to a moderate but distinct increase of Gag p24 production (Fig. 4A). In line with this observation, RNA interference experiments with siRNAs directed against hnRNP A1 showed that reduction of endogenous hnRNP A1 expression has an inverse effect on HIV gene expression, leading to a diminished p24 production compared with the non-silencing control (Fig. 4B). siRNA against hnRNP A1 had no effect on cell viability as measured by the MTT test (see supplemental Fig. S3).
FIGURE 4.
Influences of the expression levels of hnRNP A1, Q, R, K, and U on HIV production by persistently infected astrocytes. A, overexpression of the hnRNP A1 protein increased Gag p24 production by TH4-7-5 cells. TH4-7-5 cells were transfected with plasmids for expression of hnRNP A1 fused to a blue fluorescent tag (hnRNP A1-CYN) or with plasmids for expression of the unlinked blue fluorescent protein (CYN) as control. Gag production was monitored by measuring intra- and extracellular levels of Gag p24. Gag p24 levels are indicated relative to Gag levels in cells transfected with the CYN control plasmid. Expression of ectopic proteins was confirmed by Western blot analysis. B, siRNA-mediated knock-down of endogenous hnRNP A1 expression reduced Gag p24 produced by TH4-7-5 cells. Gag p24 levels are indicated relative to Gag levels in cells transfected with non-silencing (n.s.) siRNAs. Western blot analysis confirms knockdown of hnRNP A1. C, investigation of the effects of hnRNP Q, R, K, and U knock-down on HIV production by TH4-7-5 cells. Knock-down of hnRNP Q, R, and K reduced Gag p24 production by TH4-7-5 cells. Knock-down of hnRNP U slightly increased Gag production. Gag p24 levels are indicated relative to Gag levels in cells transfected with non-silencing siRNAs. Western blot analysis confirms knock-down of each hnRNP.
In addition, the effects of hnRNP Q, R, K, and U knockdown on HIV production were analyzed by RNA interference. Extra- and intracellular p24 productions were decreased by ≥20% compared with the non-silencing control using sihnRNP Q, R, and K (Fig. 4C). Incubation of the cells with sihnRNP U had no negative impact on HIV replication, but rather slightly increased p24 production (Fig. 4C). Again MTT tests with these siRNAs revealed no effects on cell viability (see supplemental Fig. S3).
DISCUSSION
This work presents for the first time an entirely novel property of the Rev protein, the interaction with cellular hnRNPs. We identify a short region at the N terminus of Rev for interaction of Rev with a group of hnRNPs that includes A1, Q, R, K, and U. In addition, we show robust interaction of Rev with at least one other hnRNP (E2) in a manner independent of the N-terminal hnRNP recognition region. This suggests that Rev can interact with different hnRNPs by different mechanisms. Four of these interactions were analyzed by co-immunoprecipitation (hnRNP A1, Q, R, and U), indicating that complex formations also occur inside the cell and are not due to unspecific in vitro protein-protein interactions. Specificity of Rev-hnRNP interactions in the affinity chromatography assays was confirmed by strongly diminished interaction of the hnRNPs with a bait protein lacking the Rev moiety. Importantly, elimination of AA 2–14 from Rev strongly reduced interaction of Rev with the hnRNPs A1, Q, R, U, and K, whereas a Rev bait protein lacking only AA 2–8 of Rev showed similarly efficient interaction with these hnRNPs as the full-length protein.
Large regions of the Rev protein are conserved among clades of the M-group (see supplemental Fig. S4). The AA 1–14 region itself contains a number of highly conserved amino acid positions. Two highly conserved positions located in the short hnRNP-recognition region (AA 9–14) are occupied by amino acids with negative charges. Together with a third, somewhat less well conserved acidic amino acid at position 11, these could form a negative “patch” within the hnRNP recognition region. It would be tempting to speculate that the interaction of Rev with the hnRNPs A1, Q, R, U, and K is mediated by electrostatic interactions between this negative patch in Rev and the hnRNPs, because the set of hnRNPs recognized by the N-terminal region of Rev all contain RGG boxes with numerous positively charged arginine residues (35). Furthermore, we demonstrate that arginine residues are essential for interaction of hnRNP A1 with Rev. However, it is unlikely that specific Rev-hnRNP binding is mediated solely by the negative patch in Rev, as it was shown in another study that hnRNP A1 does not interact with other proteins with up to 5 negative patches (i.e. hnRNP C1, E1, and K) (36)). Rather it appears that other amino acid residues in Rev may also play a role for the interactions of Rev with hnRNPs. The identification of these residues and their contributions to the biochemical and conformational determinants of Rev-hnRNP interactions is an interesting issue for future investigations.
In this study we also show that alterations of expression levels of hnRNP A1, Q, R, K, and U affect HIV production in stably infected astrocytes. This clearly demonstrates a link between these hnRNPs and HIV replication. Knock-down of hnRNP A1, Q, R, and K reduced HIV-1 production, suggesting that these hnRNPs can function as positive modulators of HIV-1 replication. In contrast knock-down of hnRNP U had a slight stimulatory effect, in agreement with the previously reported negative regulatory effects of an hnRNP U fragment (28). The overall relative moderate effects may be on the one hand a consequence of the multiple mechanisms for restriction of HIV replication in these cells (reviewed in Ref. 19). On the other hand, redundant influences of hnRNPs during HIV replication could reduce the effects. Moreover, hnRNPs are abundantly expressed in metazoa (37) and therefore difficult to knock-down completely. However, together with previous studies (29, 30, 38), these results lend further credence to the concept that hnRNPs can regulate HIV replication.
Previous studies demonstrate interaction of hnRNPs with HIV RNAs (reviewed in Refs. 1, 39, and 40). Our present work extends the potential HIV interaction partners of hnRNPs to the Rev protein and indicates that hnRNP-Rev interactions can occur in the absence of viral RNAs. The capacity of hnRNPs to interact with Rev may have manifold functional implications, because hnRNPs are involved in numerous cellular processes (reviewed in Refs. 34 and 41–43). To gain a better understanding of the functional context of each hnRNP, we generated a profile of the most significant biological processes associated with this hnRNP in the literature. This was done by identifying statistically overrepresented gene ontology categories in the set of genes co-cited with each hnRNP (for details see “Experimental Procedures”). The results of this in silico analysis are shown in Fig. 5A. Comparison of these profiles reveals common biological processes associated with different hnRNPs as well as specific biological processes associated with individual hnRNPs. hnRNPs A1, Q, R, and E2 all have the potential to link Rev to RNA processing/splicing or translation, providing further support for the reported involvement of Rev in these processes (see above). Interestingly, these profiles also include biological processes not associated with Rev so far (Fig. 5B). These include cytokine production (mainly hnRNP R), protein kinase cascades (hnRNP K), DNA repair/metabolism (hnRNP U), and cell cycle/apoptosis (hnRNP K). This suggests that interaction of Rev with hnRNPs may link Rev to novel biological processes, empowering Rev to influence HIV replication and pathogenesis by mechanisms that extend beyond its role in regulation of HIV expression. Thus this more systems-oriented view links Rev to a much larger spectrum of biological processes than previously anticipated and points to new directions for investigation of the roles of Rev for HIV replication and pathogenesis.
FIGURE 5.
In silico analysis of the functional context of Rev-interacting hnRNPs. A, the functional context of each hnRNP was determined by statistical evaluation of the GO annotations of co-cited genes (top 10 categories, for details see “Experimental Procedures”). The pie charts represent the major non-redundant biological processes associated with each hnRNP and the percentage of GO categories representing each biological process within this group. Short descriptions of the respective GO categories (biological process) are indicated with the color code in the center of the figure. Complete designations of the individual GO categories are provided in supplemental Table 1. B, major hypothetical links of the HIV-1 Rev protein to biological processes as inferred from hnRNP interactions. The hnRNPs associated with the respective biological processes as determined in A of the figure are shown superimposed on the links between the HIV-1 Rev protein and the biological processes. hnRNP E1 was excluded from the diagram because of its weak interaction with Rev (see Fig. 2).
Proteins of other viruses (e.g. herpes viruses and hepatitis C virus) have also been shown to interact with individual hnRNPs (23, 44) indicating that the interplay of hnRNPs with viral proteins is not limited to HIV Rev. This further underscores the significance of hnRNPs for virus-host interactions and advocates more detailed analysis of these interactions as potential targets for antiviral intervention.
We expect the manifold implications of our discovery of Rev-hnRNPs interactions to initiate numerous future studies. Further investigations of these interactions and potential interactions of Rev with other RNA-binding proteins (e.g. SR proteins) will promote the understanding of the integration of virus-host interactions with cellular processes and may provide novel insights for drug design and therapeutic intervention strategies.
Supplementary Material
Acknowledgments
We thank our colleagues from the Helmholtz Zentrum München, Elisabeth Kremmer for generating the monoclonal antibodies rat-anti-GFP and rat-anti-Rev 5C6, and Dirk Eick for providing the mouse anti-RNA polymerase II 8WG16 antibody. We thank Viola Baumgärtel, Center for Nanotechnology, for analysis of recombinant Rev proteins in the in vitro OSTR export assay.
This work was supported by Deutsche Forschungsgemeinschaft Grant 1710/1-3 (to R. B. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
V. Baumgärtel, unpublished data.
- HIV
- human immunodeficiency virus
- AA
- amino acid
- hnRNP
- heterogeneous nuclear ribonucleoprotein
- CFP
- cyan fluorescent protein
- YFP
- yellow fluorescent protein
- GFP
- green fluorescent protein
- MES
- 4-morpholineethanesulfonic acid
- MALDI-TOF
- matrix-assisted laser desorption ionization time-of-flight
- siRNA
- small interfering RNA
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- eIF
- eukaryotic initiation factor.
REFERENCES
- 1.Stoltzfus C. M., Madsen J. M. (2006) Curr. HIV Res. 4, 43–55 [DOI] [PubMed] [Google Scholar]
- 2.Kjems J., Askjaer P. (2000) Adv. Pharmacol. 48, 251–298 [DOI] [PubMed] [Google Scholar]
- 3.Felber B. K., Zolotukhin A. S., Pavlakis G. N. (2007) Adv. Pharmacol. 55, 161–197 [DOI] [PubMed] [Google Scholar]
- 4.Kammler S., Otte M., Hauber I., Kjems J., Hauber J., Schaal H. (2006) Retrovirology 3, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felber B. K., Hadzopoulou-Cladaras M., Cladaras C., Copeland T., Pavlakis G. N. (1989) Proc. Natl. Acad. Sci. U.S.A. 86, 1495–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolff H., Brack-Werner R., Neumann M., Werner T., Schneider R. (2003) Nucleic Acids Res. 31, 2839–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Groom H. C., Anderson E. C., Dangerfield J. A., Lever A. M. (2009) J. Gen. Virol. 90, 1141–1147 [DOI] [PubMed] [Google Scholar]
- 8.Arrigo S. J., Chen I. S. (1991) Genes Dev. 5, 808–819 [DOI] [PubMed] [Google Scholar]
- 9.D'Agostino D. M., Felber B. K., Harrison J. E., Pavlakis G. N. (1992) Mol. Cell. Biol. 12, 1375–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lever A. M. (2007) Adv. Pharmacol. 55, 1–32 [DOI] [PubMed] [Google Scholar]
- 11.Brandt S., Blissenbach M., Grewe B., Konietzny R., Grunwald T., Uberla K. (2007) PLoS Pathogens 3, e54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suhasini M., Reddy T. R. (2009) Curr. HIV Res. 7, 91–100 [DOI] [PubMed] [Google Scholar]
- 13.Nekhai S., Jeang K. T. (2006) Future Microbiol. 1, 417–426 [DOI] [PubMed] [Google Scholar]
- 14.Ma J., Rong L., Zhou Y., Roy B. B., Lu J., Abrahamyan L., Mouland A. J., Pan Q., Liang C. (2008) Virology 375, 253–264 [DOI] [PubMed] [Google Scholar]
- 15.Yedavalli V. S., Neuveut C., Chi Y. H., Kleiman L., Jeang K. T. (2004) Cell 119, 381–392 [DOI] [PubMed] [Google Scholar]
- 16.Ptak R. G., Fu W., Sanders-Beer B. E., Dickerson J. E., Pinney J. W., Robertson D. L., Rozanov M. N., Katz K. S., Maglott D. R., Pruitt K. D., Dieffenbach C. W. (2008) AIDS Res. Hum. Retroviruses 24, 1497–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Groom H. C., Anderson E. C., Lever A. M. (2009) J. Gen. Virol. 90, 1303–1318 [DOI] [PubMed] [Google Scholar]
- 18.Ludwig E., Silberstein F. C., van Empel J., Erfle V., Neumann M., Brack-Werner R. (1999) J. Virol. 73, 8279–8289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kramer-Hämmerle S., Rothenaigner I., Wolff H., Bell J. E., Brack-Werner R. (2005) Virus Res. 111, 194–213 [DOI] [PubMed] [Google Scholar]
- 20.Malim M. H., Böhnlein S., Hauber J., Cullen B. R. (1989) Cell 58, 205–214 [DOI] [PubMed] [Google Scholar]
- 21.Wolff H., Hadian K., Ziegler M., Weierich C., Kramer-Hammerle S., Kleinschmidt A., Erfle V., Brack-Werner R. (2006) Exp. Cell Res. 312, 443–456 [DOI] [PubMed] [Google Scholar]
- 22.Wolff H., Hartl A., Eilken H. M., Hadian K., Ziegler M., Brack-Werner R. (2006) BioTechniques 41, 688, 690,, 692 [DOI] [PubMed] [Google Scholar]
- 23.Wadd S., Bryant H., Filhol O., Scott J. E., Hsieh T. Y., Everett R. D., Clements J. B. (1999) J. Biol. Chem. 274, 28991–28998 [DOI] [PubMed] [Google Scholar]
- 24.Stauber R. H., Horie K., Carney P., Hudson E. A., Tarasova N. I., Gaitanaris G. A., Pavlakis G. N. (1998) BioTechniques 24, 462–466, 468–471 [DOI] [PubMed] [Google Scholar]
- 25.Brack-Werner R., Kleinschmidt A., Ludvigsen A., Mellert W., Neumann M., Herrmann R., Khim M. C., Burny A., Müller-Lantzsch N., Stavrou D., Erfle V. (1992) AIDS 6, 273–285 [PubMed] [Google Scholar]
- 26.Hauck S. M., Schoeffmann S., Deeg C. A., Gloeckner C. J., Swiatek-de Lange M., Ueffing M. (2005) Proteomics 5, 3623–3636 [DOI] [PubMed] [Google Scholar]
- 27.Mosmann T. (1983) J. Immunol. Methods 65, 55–63 [DOI] [PubMed] [Google Scholar]
- 28.Valente S. T., Goff S. P. (2006) Mol. Cell 23, 597–605 [DOI] [PubMed] [Google Scholar]
- 29.Woolaway K., Asai K., Emili A., Cochrane A. (2007) Retrovirology 4, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jablonski J. A., Caputi M. (2009) J. Virol. 83, 981–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tschödrich-Rotter M., Peters R. (1998) J. Microsc. 192, 114–125 [DOI] [PubMed] [Google Scholar]
- 32.Peters R. (2006) Methods Mol. Biol. 322, 259–272 [DOI] [PubMed] [Google Scholar]
- 33.Mourelatos Z., Abel L., Yong J., Kataoka N., Dreyfuss G. (2001) EMBO J. 20, 5443–5452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makeyev A. V., Liebhaber S. A. (2002) RNA 8, 265–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burd C. G., Dreyfuss G. (1994) Science 265, 615–621 [DOI] [PubMed] [Google Scholar]
- 36.Kim J. H., Hahm B., Kim Y. K., Choi M., Jang S. K. (2000) J. Mol. Biol. 298, 395–405 [DOI] [PubMed] [Google Scholar]
- 37.Reed R., Magni K. (2001) Nat. Cell Biol. 3, E201–204 [DOI] [PubMed] [Google Scholar]
- 38.Dowling D., Nasr-Esfahani S., Tan C. H., O'Brien K., Howard J. L., Jans D. A., Purcell D. F., Stoltzfus C. M., Sonza S. (2008) Retrovirology 5, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cochrane A. W., McNally M. T., Mouland A. J. (2006) Retrovirology 3, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bolinger C., Boris-Lawrie K. (2009) Retrovirology 6, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He Y., Rothnagel J. A., Epis M. R., Leedman P. J., Smith R. (2009) Mol. Carcinog. 48, 167–179 [DOI] [PubMed] [Google Scholar]
- 42.Carpenter B., MacKay C., Alnabulsi A., MacKay M., Telfer C., Melvin W. T., Murray G. I. (2006) Biochim. Biophys. Acta 1765, 85–100 [DOI] [PubMed] [Google Scholar]
- 43.Krecic A. M., Swanson M. S. (1999) Curr. Opin. Cell Biol. 11, 363–371 [DOI] [PubMed] [Google Scholar]
- 44.Kim C. S., Seol S. K., Song O. K., Park J. H., Jang S. K. (2007) J. Virol. 81, 3852–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.