Abstract
The formin protein formin-like 1 (FMNL1) is highly restrictedly expressed in hematopoietic lineage-derived cells and has been previously identified as a tumor-associated antigen. However, function and regulation of FMNL1 are not well defined. We have identified a novel splice variant (FMNL1γ) containing an intron retention at the C terminus affecting the diaphanous autoinhibitory domain (DAD). FMNL1γ is specifically located at the cell membrane and cortex in diverse cell lines. Similar localization of FMNL1 was observed for a mutant lacking the DAD domain (FMNL1ΔDAD), indicating that deregulation of autoinhibition is effective in FMNL1γ. Expression of both FMNL1γ and FMNL1ΔDAD induces polarized nonapoptotic blebbing that is dependent on N-terminal myristoylation of FMNL1 but independent of Src and ROCK activity. Thus, our results describe N-myristoylation as a regulative mechanism of FMNL1 responsible for membrane trafficking potentially involved in a diversity of polarized processes of hematopoietic lineage-derived cells.
INTRODUCTION
Formins represent a protein family indispensable for many fundamental actin-dependent processes, including migration, vesicle trafficking, morphogenesis, and cytokinesis (1). Because these polarized processes are also involved in inflammation, deregulated proliferation, and metastasis, formins have been suggested to represent attractive drug targets for inflammatory and malignant diseases. Formin-like 1 (FMNL1)3 is expressed restrictedly in hematopoietic lineage-derived cells and overexpressed in malignant cells of different origin. This restricted expression suggests FMNL1 to be an attractive target for novel immunotherapies in malignant and inflammatory diseases (2, 3). However, function and regulation of FMNL1 are less well characterized. Previous work has shown involvement of FMNL1 in the reorientation of the microtubule-organizing center toward the immunological synapse and cytotoxicity of T cells (4). The murine homolog FRL, which has 85% homology to the human counterpart, has been additionally shown to be involved in cell adhesion and motility of macrophages as well as Fcγ receptor-mediated phagocytosis (5, 6). To date, it is not clear how these different membrane-associated processes are regulated.
Formins are defined by a unique and highly conserved C-terminal formin homology (FH) 2 domain that mediates the effects on actin (7–11). The FH2 domain is proceeded by a proline-rich FH1 domain that binds with low micromolar affinity to profilin (12, 13). In a conserved subfamily of formins known as diaphanous-related formins (DRFs), the FH1 and FH2 domains are flanked by an array of regulatory domains at the N terminus and by a single C-terminal diaphanous autoregulatory domain (DAD) (14). The large N-terminal regulatory region includes a binding domain for small G proteins like Rho-GTPase followed by an adjacent diaphanous-inhibitory domain (DID) and a dimerization domain (13, 15–17). The DAD, which comprised only a small stretch of amino acid residues, binds to the DID. Interaction of DAD and DID is responsible for autoinhibition of DRFs. The mammalian diaphanous 1 (Dia1) as well as the macrophage-enriched murine formin-like protein 1 (FRL) are both regulated by autoinhibition in a DAD-dependent manner (5, 6, 18, 19).
In addition to the domain-specific functions, formin proteins seem to be intensively regulated by splicing. Splicing at the N terminus has been demonstrated to be involved in distinct protein regulation and function of Dia2 (20). The DAD domain is also a hotspot of splicing. Within this area, two splice variants have been characterized for FRL, although functional differences have not been observed (19). In contrast, abrogation of autoinhibition in mutants lacking the C terminus in Dia1 and FRL specifically induces peripheral and plasma membrane localization (5, 21). The exact mechanism of how DRFs locate to the plasma membrane is, however, currently unknown.
We identified a novel splice variant and constitutively active form of FMNL1 with distinct membrane localization. We demonstrate that this novel splice variant (FMNL1γ) directly mediates intensive blebbing that is independent of Src and ROCK activity. In contrast, FMNL1-mediated membrane trafficking and bleb formation are dependent on N-terminal myristoylation of FMNL1γ, potentially representing a general mechanism involved in diverse membrane-associated functions of FMNL.
EXPERIMENTAL PROCEDURES
Cells and Cell Lines
Peripheral blood mononuclear cells (PBMCs) from healthy donors as well as patients with chronic lymphocytic leukemia (CLL) were collected with donors' and patients' informed consent following the requirements of the local ethical board. Patients had diagnosis of CLL by morphology, flow cytometric analysis, and cytogenetics. PBMCs were obtained by density gradient centrifugation on Ficoll/Hypaque (Biochrom). PBMC subpopulations from healthy donors were isolated by negative or positive magnetic depletion (Invitrogen). A T cell clone (SK22) with specificity for the FMNL1-derived peptide PP2 was used to analyze polarized T cells (3). For polarization experiments, T2 cells were pulsed with the specific peptide followed by incubation with SK22 for 15 min before staining. Unspecific stimulation of PBMCs was induced by interleukin-2 (50 units/ml; Chiron Vaccines International) and OKT3 (30 ng/ml; ATCC) for 3 days. B cells and CLL cells were activated by soluble CD40-ligand (1 μg/ml; Tebu-bio). The following cell lines were used for expression and analysis of FMNL1 splice variants after adenoviral transfer: chronic myelogenous leukemia cell line K562 (ATCC CCL-243), breast carcinoma cell line MDA-MB 231 (CLS), the human fibrosarcoma cell line HT1080 (ATCC CCL-121), and HEK293T embryonal kidney cells (ATCC CRL-1573). In addition, 293A (Invitrogen) cells were used for production of adenoviral supernatant.
Detection of FMNL1 Splice Variants and Sequence Analysis
The DAD region of FMNL1 in CLL cells was analyzed using the following exon-overlapping primers: forward, GTGCTGCAGGAGCTAGACATG, and reverse, CCCTCTAGCCCCTCAGATCTG. Gel electrophoresis of PCR amplicons revealed PCR products of two different sizes, 1312 and 1489 (NCBI FJ534522), representing the C termini of FMNL1α and FMNL1γ, respectively. The PCR products were cloned into pCR 2.1 using the Topo cloning kit (Invitrogen) followed by transformation of Top10 Escherichia coli (Invitrogen). Growing clones were analyzed by sequencing (Sequiserve) revealing two different C-terminal sequences. In addition, the C terminus representing FMNL1β was derived from human lung carcinoma cDNA clone IMAGp958M162704Q2 (RZPD). The three splice variants are shown in Fig. 2B.
FIGURE 2.
Identification of a novel FMNL1 splice variant (FMNL1γ) containing an intron retention at the C terminus. A, schematic diagrams of FMNL1α, FMNL1β, FMNL1γ, and FMNL1ΔDAD constructs used in this study. B, amino acid sequences of the C termini of FMNL1α and β, corresponding to murine FRLα and β, as well as of the novel isoform FMNL1γ containing a C-terminal intron retention but sharing the final C-terminal amino acids with FMNL1α. The DAD sequence responsible for autoinhibition in the murine homolog is framed, and the identical C-terminal amino acids in FMNL1α and FMNL1γ are underlined. AA, amino acids. C, quantitative mRNA expression of overall FMNL1 (black bars) and the isoform FMNL1γ (gray bars) with specific exon-overlapping primers in wild type (WT) 293T cells and 293T cells transduced with the different FMNL1 isoforms. The relative quantitative expression compared with skeletal muscle was calculated using the Δ-Δ cycle threshold method. D–F, quantitative mRNA expression of general FMNL1 (black bars) and isoform-specific FMNL1γ (gray bars) in healthy tissue (D) and in diverse cell populations isolated from peripheral blood of healthy donors (E) and 20 patients with CLL (F). PBMC subpopulations were isolated by negative (CD4, CD8, CD19, and CD14) and positive (CD34) selection. Dendritic cells (DC) were generated by adherence and cytokine maturation (E).
Quantitative Reverse Transcription-PCR
To evaluate quantitative mRNA expression of FMNL1γ compared with overall FMNL1 expression, total RNA was extracted from normal PBMCs and malignant cells from patients with CLL and cell lines (22), and cDNA was synthesized by Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. In addition, cDNA derived from normal tissues pooled from different donors (Clontech) was used. Detection of splice variant-specific FMNL1 mRNA was conducted using the Light Cycler PCR Master Mix (Roche Applied Science). The following exon-overlapping primers were used for overall FMNL1 detection, resulting in a 299-bp amplicon: 5′-CAAGAACCCCAGAACCAAGGCTCTGG and 3′-CTGCAGGTCGTACGCCTCAATGGC. In addition, exon-overlapping FMNL1γ splice variant-specific primers were applied, resulting in a 254-bp amplicon: 5′-GCAGCAGAAGGAGCCACTCATTTATGAGAGC and 3′-GCACCGTCTTGATCACTGAGTGGGGGTGG. These primers resulted in high cycle threshold values (>26) in 293T cells overexpressing FMNL1α and β (see Fig. 2D). The amplicons were detected by fluorescence (530 nm) using a double-stranded DNA-binding dye, SYBR Green I with the Light Cycler instrument. 18 S rRNA was processed as a control for relative quantification. Relative quantitative expression was calculated using the Δ-Δ cycle threshold method (23). Skeletal muscle was used as reference tissue because it showed the highest cycle threshold for detection of FMNL1 and FMNL1γ (cycle threshold for overall FMNL1 expression in skeletal muscle, 25; for FMNL1γ expression in skeletal muscle, 30).
Cloning of FMNL1 Splice Variants and Mutants
DNA of different splice variants was cloned into pcDNA3.1 and a pEntr11/pACDC-derived entry clone containing the complete FMNL1β sequence using the single SbfI restriction site within FMNL1 to exchange the C termini with the sequences of FMNL1α and FMNL1γ. The pEntr11/pACDC-derived entry clone additionally contains the green fluorescent protein gene. This vector was used for recombination into pAD/PL-DEST adenoviral vector, digested with PacI, and transfected into a 293A adenoviral producer cell line according to the manufacturer's instructions (Invitrogen). Amplification of adenoviral stocks resulted in titers of 107–109 particles/ml. Virus supernatants were used to transiently infect different cell lines for transduction of FMNL1 splice variants. Transduction efficiency was analyzed by green fluorescent protein expression of infected cells and was 90–100% for HT1080 and MDA-MB 231 but only 20–40% for K562. In addition, pcDNA3.1 vectors containing different FMNL1 splice variants were transfected into 293T cells by calcium phosphate transfection as indicated, resulting in transfection rates of approximately 40–50%. Mutation of the hypothetical N-terminal myristoylation site was performed using the QuikChange mutagenesis kit (Stratagene) according to the manufacturer's instructions. The G2T/A4T mutations were performed at the N terminus (see Fig. 5A).
FIGURE 5.
FMNL1γ contains an N-terminal myristoylation site. A, schematic view of the N terminus of FMNL1. The predicted N-terminal myristoylation site was mutated on positions 2 and 4, glycine and alanine to threonine (gray), respectively. AA, amino acid. B and C, immunoprecipitation and metabolic labeling of FMNL1γ with [3H]myristic acid. 293T cells transfected with FMNL1γ or G2T/A4T were cultured for 16 h with [3H]myristic acid. The indicated proteins were immunoprecipitated with the FMNL1-specific monoclonal antibody 8A8 as well as the isotype control (iso), separated by SDS-PAGE followed by immunoblotting with anti-FMNL1 monoclonal antibody 6F2 (B) or fluorography (C). Molecular mass sizes are indicated in kDa.
Mutants lacking the DAD were generated for FMNL1 and the mutant G2TA4T by PCR and cloned in pcDNA3.1 using the following primers: FMNL1, 5′-ATTGCTAGCCACCATGGGCAACGCGGCCGGCAGCGCCGAGC; G2T/A4T, 5′-ATTGCTAGCCACCATGACCAACACGGCCGGCAGCGCCGAG; and FMNL1ΔDAD, 3′-ATTCTAGACTAGCTCTCATAAATGAGTGGCTCCTTCTGCTG. The FMNL1 lacking DAD was also cloned in pEntr11/pACDC-derived entry clone using the following primers: FMNL1, 5′-ATGGATCCCACCATGGGCAACGCGGCCGGCAGCGCCGAGCAGC; and FMNL1ΔDAD, 3′-AAGGATCCCTAGCTCTCATAAATGAGTGGCTCCTTCTGCTG.
Reagents and Antibodies
The previously described C-terminal FMNL1-specific rat anti-human antibody 6F2 recognizing the peptide KKEAAAQEAGADT was used to detect FMNL1 by immunofluorescence microscopy and Western blotting. Specificity for FMNL1 has been demonstrated previously by immunoprecipitation and mass spectrometry (3). We generated an additional N-terminal FMNL1-specific rat anti-human antibody (8A8) recognizing the peptide PAAPPPKQPAPPKQP which was used for immunoprecipitation of FMNL1. In addition, the following primary and secondary antibodies were used: rabbit anti-human CD3 polyclonal antibody (Dako), mouse anti-human α-tubulin monoclonal antibody (Santa Cruz Biotechnology), mouse anti-human γ-tubulin monoclonal antibody (Sigma), and rabbit anti-human myosin IIb antibody (Sigma). Appropriate secondary antibodies with minimal cross-reactivity and conjugation to Cy3, Cy5 (Jackson), and Alexa Fluor 488 (Invitrogen) were applied as indicated. Alexa Fluor 488-labeled phalloidin (Invitrogen) was used to stain actin. For immunoblotting, horseradish peroxidase-conjugated secondary antibodies to rat and mouse were used. For inhibition of different targets involved in blebbing the following reagents were used: blebbistatin (Calbiochem) at 1 μm, 50 μm and 100 μm; PP1 (Calbiochem) at 50 μm; latrunculin B (Calbiochem) at 1 μm, 10 μm, and 25 μm; Y-27632 (Sigma) at 90 μm; and nocodazole (Calbiochem) at 1 μm, 10 μm, 100 μm, and 200 μm. 2-Hydroxymyristic acid (Biozol) was used for inhibition of N-terminal myristoylation at a concentration of 1 mm.
Immunofluorescence
Nonadherent cells were dropped on poly-l-lysine-coated coverslips, and adherent cells were settled on sterilized coverslips overnight. Cells were fixed in 3% paraformaldehyde for 30 min. Cells were then washed (phosphate-buffered saline, 0.5% Nonidet P-40, 0.01% NaN3), blocked with 10% fetal calf serum for 20–30 min and stained with specific primary and secondary antibodies as indicated. DAPI (Molecular Probes) was used for nuclear staining. Glass coverslips were mounted on the cells in mounting medium (Molecular Probes) and investigated by Leica confocal microscopy.
Immunoprecipitation, Immunoblotting, and [3H]Myristic Acid Uptake Assay
293T cell were transfected with FMNL1γ or G2T/A4T and 24 h later biosynthetically labeled with [3H]myristic acid (0.2 mCi/ml) (PerkinElmer) followed by incubation for 16 h in Dulbecco's modified Eagle's medium supplemented with 5% heat-inactivated fetal bovine serum. Labeled cells were washed with phosphate-buffered saline and lysed in lysis buffer (20 mm Tris-HCl, 150 mm NaCl, pH 7.4, 150 mm Chaps, 1 mm phenylmethylsulfonyl fluoride, 1 mm Na3VO4, 1 mm NaF, 1× Complete). FMNL1γ and G2T/A4T proteins were immunoprecipitated with the FMNL1-specific antibody 8A8 and protein G-Sepharose 4 Fast Flow (Amersham Biosciences). Eluted proteins were split and forwarded for SDS-PAGE on two separated gels. One gel was used for immunoblotting to detect the presence of FMNL1 using the FMNL1-specific antibody 6F2. The other gel was treated with AmplifyTM fluorographic reagent (Amersham Biosciences) as indicated by the manufacturer and exposed for 12 days to Hyperfilm MP (Amersham Biosciences) at −80 °C using an intensifying screen.
RESULTS
Expression and Localization of Endogenous FMNL1 in Diverse Hematopoietic Lineage-derived Cells
We have previously demonstrated protein expression of endogenous human FMNL1 in different subtypes of PBMCs by Western blotting using the FMNL1-specific antibody 6F2 (3). Here, we investigated the localization of FMNL1 in these cells by confocal microscopy. FMNL1 showed a dot-like expression pattern in the cytoplasma of unstimulated and unspecifically stimulated T cells, B cells, and CLL cells (Fig. 1A). In T cells, FMNL1 has been reported to be involved in reorientation of the microtubule-organizing center toward the immunological synapse (4). We similarly observed polarization of FMNL1 toward the immunological synapse after targeting of FMNL1-PP2-specific T cells with peptide-pulsed T2 cells (Fig. 1B) (3). Function-associated localization of endogenous human FMNL1 was also observed at the phagocytic cup of monocytes during phagocytosis of cells and platelets (Fig. 1C) being in common with previous investigations of the function of murine FRL. Thus, our results confirm expression of human FMNL1 in different hematopoietic lineage-derived cells as well as involvement in diverse polarized and membrane-associated processes.
FIGURE 1.
FMNL1 is expressed in diverse hematopoietic lineage-derived cells and shows function-specific polarized localization. A, FMNL1 was visualized in unstimulated and stimulated PBMC subtypes as T cells, B cells, and malignant B cells from patients with CLL by immunofluorescence staining with the rat FMNL1-specific antibody 6F2 followed by Cy3-labeled goat anti-rat antibody. T cells were stimulated with interleukin-2 and OKT3, and B cells as well as CLL cells were activated using soluble CD40-ligand. Scale bars, 2 μm. B, to investigate FMNL1 expression in polarized T cells, the FMNL2-PP2-specific T cell clone SK22 was incubated for 15 min with T2 cells pulsed with the peptide FMNL1-PP2 at 10 μmol. Cells were then fixed and stained with DAPI for nuclear staining (blue), rabbit anti-human CD3 antibody followed by Cy5-labeled goat anti-rabbit antibody (yellow) for T cell identification, and rat FMNL1-specific antibody 6F2 followed by Cy3-labeled goat anti-rat antibody (red). The microtubule-organizing center was stained with mouse anti-human γ-tubulin antibody followed by Alexa Fluor 488-labeled goat anti-mouse antibody (green). Enrichment of FMNL1 and CD3 was observed around the microtubule-organizing center. Scale bar, 2 μm. C, FMNL1 is localized at the phagocytic cup (white arrows) of monocytes during phagocytosis of cells and platelets. FMNL1 (red) was stained as above, F-actin was stained by Alexa 488-conjugated phalloidin (green), and nuclei were stained with DAPI (blue). Scale bars, 2 μm. Fluorescent images were visualized at room temperature through a LEICA TCS SP2/405 confocal microscope using the HCX PL APO objective (63 × 1.4-0.6 OIL λBL) and the LEICA DC 300F camera. The LEICA confocal software was used for acquisition and analysis (A–C).
Identification and Cloning of Different FMNL1 Splice Variants
DRFs including FRL have been previously described to be regulated by autoinhibition, which is dependent on the C-terminal DAD (5). Using exon-specific primers for the C cDNA of CLL cells and human lung carcinoma, resulting in varying C-terminal amino acid sequences (Fig. 2A). Two splice variants are corresponding to the previously described murine splice variants, FRLα and FRLβ (6, 19). We additionally identified another C-terminal splice variant, FMNL1γ, containing an intron retention but sharing the final C terminus with FMNL1α (Fig. 2B). We selected splice variant-specific exon-overlapping primers (Fig. 2C) to quantify the mRNA expression of the novel splice variant FMNL1γ in different tissues and hematopoietic derived cells by quantitative reverse transcription-PCR. FMNL1γ shows a similar tissue-specific mRNA expression profile compared with the non-splice-specific expression of FMNL1 (Fig. 2, D and E), although some tissues such as bone marrow, fetal brain, and thymus show relatively higher expression of FMNL1γ compared with general FMNL1 (3). Of note, FMNL1γ is highly expressed in malignant cells of a subset of patients with CLL (Fig. 2F).
FMNL1γ and FMNL1 Lacking the C-terminal DAD Are Located at the Cell Membrane and Induce Membranous Bleb Formation
Because FMNL1γ differed in the DAD sequence representing an important region for FMNL1 regulation, we cloned all three different FMNL1 splice variants and investigated transfected 293T cells by confocal microscopy. 293T cells transfected with FMNL1α and FMNL1β showed mainly intracellular cytoplasmic distribution of FMNL1, whereas cells transfected with FMNL1γ showed a distinct membranous FMNL1 localization as well as extensive polarized membrane protrusions and blebs (Fig. 3A). Similar membrane protrusions were observed after adenoviral transduction of K562 cells with FMNL1γ but not with the other splice variants. However, in these cells the shape of the blebs was different, and a more prominent enrichment of FMNL1γ at the cell cortex has been observed (Fig. 3B). Similarly, 293T cells and K562 cells transfected or transduced with FMNL1 lacking the DAD domain (FMNL1ΔDAD, Fig. 2A) showed membranous and cortical localization of FMNL1 and extensive polarized membrane protrusions and blebs (Fig. 3, A and B), suggesting that deregulation of autoinhibition is responsible for membrane localization and blebbing observed in cells overexpressing the splice variant FMNL1γ. Increase of blebbing was less obvious in HT1080 and MDA-MB 231 cells after adenoviral transduction of FMNL1γ. However, these cell lines also demonstrated FMNL1γ localization at the cell membrane and especially enrichment of FMNL1γ at intracellular vesicles, whereas cells transduced with the other two splice variants had a more dispersed distribution within the cytoplasma (Fig. 3C). Again, similar localization was observed in HT1080 and MDA-MB 231 cells after adenoviral transduction of FMNL1ΔDAD (Fig. 3C). We additionally noticed colocalization of FMNL1γ with F-actin in blebs of FMNL1γ-transfected 293T cells as shown by phalloidin staining pointing to an induction of actin assembly activity by FMNLγ (Fig. 3D). Myosin IIb and α-tubulin were also polarized to the FMNL1γ-induced blebs (Fig. 3D). A significant increase of membrane blebbing in 293T and K562 cells after transduction with FMNL1γ and FMNL1ΔDAD compared with the other splice variants was quantified by independent counting experiments (Fig. 3, E and F).
FIGURE 3.
FMNL1γ and FMNL1ΔDAD show membranous and cortical localization and induce bleb formation. A, FMNL1γ and FMNL1ΔDAD induce extensive blebbing in 293T cells. 293T cells were transfected with the pcDNA3.1 vector containing three different isoforms of FMNL1 and analyzed by confocal microscopy after immunofluorescence staining with the rat FMNL1-specific antibody 6F2 followed by Cy3-labeled goat anti-rat antibody (red). Nuclei were stained with DAPI (blue). Scale bars, 2 μm. DIC, differential interference contrast. B, FMNL1γ and FMNL1ΔDAD induce blebbing in K562 cells. FMNL1 isoforms were adenovirally transduced into K562 cells and analyzed by confocal microscopy as described in A. Scale bars, 2 μm. C, FMNL1γ and FMNL1ΔDAD are enhanced in intracellular vesicles of HT1080 and MDA-MB 231 cells. FMNL1 isoforms were adenovirally transduced into HT1080 and MDA-MB 231 cells and analyzed by confocal microscopy as described above. Scale bars, 2 μm. D, F-actin, α-tubulin, and myosin IIb show polarized localization in FMNL1γ-induced blebs in 293T cells. 293T cells were transfected with the pcDNA3.1 vector containing FMNL1γ and stained with rat FMNL1-specific antibody 6F2 followed by Cy3-labeled goat anti-rat antibody (red). Myosin IIb was stained with a rabbit anti-myosin IIb antibody followed by Cy5-labeled goat anti-rabbit antibody (yellow). F-actin was stained with Alexa Fluor 488-conjugated phalloidin (green), and α-tubulin was stained with a mouse anti-α-tubulin antibody followed by Alexa Fluor 488-conjugated goat anti-mouse antibody (green). Nuclei were stained with DAPI (blue). Scale bars, 2 μm. E and F, cell blebbing was quantified by counting 100 FMNL1-transduced cells in 293T cells (E) and K562 cells (F) by independent counting experiments (n ≥ 3), indicating a significant increase of blebbing in cells expressing FMNL1γ and FMNL1ΔDAD (***, p < 0.001). Fluorescent images (A–D) were investigated as described in Fig. 1.
Blebbing Induced by FMNL1γ Is Independent of Src and ROCK but Depends on Actin, Myosin, and Tubulin Integrity
Plasma membrane blebbing has been previously shown to be induced by the formin FHOD1 when coexpressed with ROCK1 (24). Moreover, blebbing induced by FHOD1 requires Src activity (24). To investigate plasma membrane blebbing induced by FMNL1γ further, we transfected 293T cells with FMNL1γ and added different substances previously reported to inhibit membrane blebbing. Whereas the Src inhibitor PP1 and the ROCK inhibitor Y-27632 had only minimal effects on membrane blebbing, this was grossly reduced by addition of latrunculin B, blebbistatin, and nocodazole (Fig. 4, A and B and supplement Fig. S1), demonstrating that the mechanism of blebbing induced by FMNL1γ is not mediated by Src and ROCK but depends on actin, myosin, and tubulin integrity. Similar data were obtained by FMNL1ΔDAD-transfected 293T cells (data not shown).
FIGURE 4.
Blebbing induced by FMNL1γ is independent of ROCK and Src but depends on actin, myosin, and microtubules. A and B, 293T cells were transfected with the pcDNA3.1 vector containing FMNL1γ and treated with different bleb inhibitors and solvent controls. Cells were stained after 2 days with the rat FMNL1-specific antibody 6F2 followed by Cy3-labeled goat anti-rat antibody (red). Nuclei were stained with DAPI (blue). Scale bars, 2 μm. Blebbing cells were analyzed by confocal microscopy as described in A. Cell blebbing was quantified by counting 100 FMNL1γ-transduced cells in three independent experiments (B), classifying them as cells exhibiting extended blebs (white bars), limited blebs (black bars), or none (gray bars). DMSO, dimethyl sulfoxide. The S.D. of independent counting experiments is shown (n = 3; ***, p < 0.001).
FMNL1 Is Myristoylated at the N Terminus
Membrane localization of FMNL1γ was a predominant feature in 293T cells transfected with the splice variant FMNL1γ and FMNL1ΔDAD. The mechanism of how FMNL1 is located to the cell membrane remains elusive. Investigation of the FMNL1 structure by prediction analyses revealed a potential N-terminal myristoylation site (see the Eukaryotic Linear Motif (ELM) resource on the Internet). This N-terminal myristoylation motif is highly specific for the leukocyte-specific formins FMNL1, 2, and 3 in man and mouse but is not present in other formin proteins. To prove whether FMNL1γ is myristoylated at the N terminus, we mutated glycine at position 2 and alanine at position 4 to threonine within FMNL1γ (G2T/A4T) (Fig. 5A). 293T cells were transfected with FMNL1γ or the G2T/A4T mutant form and then metabolically labeled with [3H]myristic acid. Total protein of transfected 293T cells was immunoprecipitated by the FMNL1-specific antibody 8A8 detecting an N-terminal epitope not including the N-terminal myristoylation site. Immunoprecipitated probes were investigated by immunoblotting using the FMNL1-specific antibody 6F2 (Fig. 5B) as well as autoradiography to analyze incorporation of [3H]myristic acid (Fig. 5C). Both proteins, FMNL1γ and G2T/A4T, were sufficiently immunoprecipitated by the FMNL1-specific antibody 8A8 (Fig. 5B). However, only FMNL1γ, but not G2T/A4T, was myristoylated after transfection in 293T cells.
Membrane Localization and Blebbing of FMNL1 Are Mediated by N-terminal Myristoylation
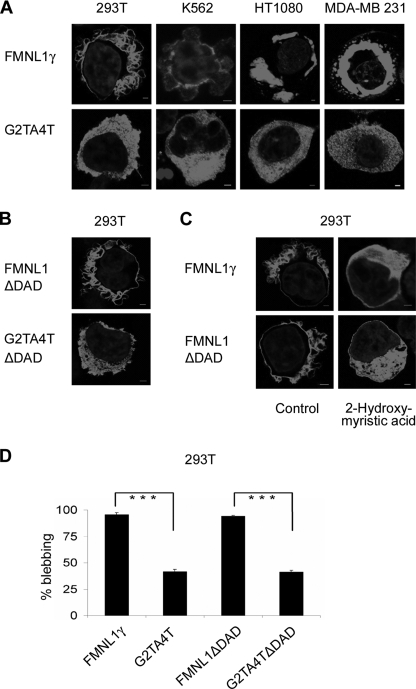
To investigate whether N-terminal myristoylation plays a role in FMNL1 function and membrane localization, cells were transfected with FMNL1γ or G2T/A4T. The mutant G2T/A4T, in fact, abolished membranous and cortical localization of FMNL1γ after transfection of the mutant splice variant in 293T cells as well as after genetic transfer of this mutant in other cell lines (Fig. 6A). Similarly, mutation of the N-myristoylation site in the FMNL1ΔDAD (G2T/A4TΔDAD) also abrogated membranous localization of FMNL1ΔDAD (Fig. 6B). Membrane localization of FMNL1γ and FMNL1ΔDAD could be also inhibited by adding 2-hydroxymyristate, a potent inhibitor of protein myristoylation (25), further confirming that plasma membrane localization of FMNL1 is dependent on N-terminal myristoylation (Fig. 6C). Blebbing was significantly reduced in 293T cells transfected with G2T/A4T or G2T/A4TΔDAD compared with FMNL1γ or FMNL1ΔDAD (Fig. 6D), indicating that membrane localization mediated by N-terminal myristoylation is important for bleb formation induced by FMNL1.
FIGURE 6.
Localization and function of FMNL1 are regulated by N-terminal myristoylation. A, localization of FMNLγ at the membrane or intracellular vesicle is abrogated by mutation of the N-terminal myristoylation motif. Different cell lines were transfected or adenovirally transduced with either FMNL1γ or the mutant G2T/A4T followed by staining with the rat FMNL1-specific antibody 6F2 and Cy3-labeled goat anti-rat antibody (red). Nuclei were stained with DAPI (blue). Scale bars, 2 μm. B, localization of FMNL1ΔDAD at the membrane is abrogated by mutation of the N-terminal myristoylation motif. 293T cell lines were transfected with either FMNL1ΔDAD or the mutant G2T/A4TΔDAD followed by staining with the rat FMNL1-specific antibody 6F2 and Cy3-labeled goat anti-rat antibody (red). Nuclei were stained with DAPI (blue). Scale bars, 2 μm. C, 293T cells transfected with the isoform FMNL1γ and FMNL1ΔDAD were treated with the inhibitor 2-hydroxymyristic acid and stained as described above. Scale bars, 2 μm. D, mutation of the N-terminal myristoylation motif reduced membrane blebbing induced by FMNL1γ or FMNL1ΔDAD in 293T cells. For investigation of cell blebbing, 100 transduced cells were analyzed. The bars show the S.D. of independent counting experiments (n = 3; ***, p < 0.001). Fluorescent images (A–C) were investigated as described previously.
DISCUSSION
Formin proteins have been described to promote the formation of actin networks and to be involved in regulation of essential cellular functions such as cell division, migration, adhesion, and intracellular trafficking. So far, little is known about the hematopoietic lineage-specific formin FMNL1. Here, we show expression and polarized localization of native human FMNL1 in different hematopoietic lineage-derived cells, confirming previous reports about different functional aspects of human FMNL1 and murine FRL (4–6).
A complex regulation network seems to be responsible for activation and different functions of formin proteins. A central regulative tool for DRFs is autoregulation, which can be released by interaction with active Rho-GTPases and potentially additional factors (1). Splicing has been also shown to result in differential functionality of forming proteins (20). We isolated a novel FMNL1 splice variant (FMNL1γ) from malignant cells of patients with CLL. This splice variant showed increased mRNA expression in bone marrow, thymus, fetal brain, and diverse hematopoietic lineage-derived cells. Moreover, we observed a high expression in malignant cells of a subset of CLL patients. Detailed analyses of systematically isolated and processed patient samples need to be performed to clarify whether overexpression of FMNL1γ in CLL samples is associated with an activated malignant cell status and/or patient prognosis and will be a focus of further studies. However, although a role of FMNL1γ in malignant transformation could not be ascertained in this study, this splice variant showed distinct functional properties similar to cells expressing a mutant of FMNL1 with deregulated autoinhibition caused by the lack of the autoinhibitory DAD sequence (FMNL1ΔDAD). FMNL1γ contains an intron retention of 58 amino acids at the C terminus but shares the last 30 amino acids with the previously described FMNL1α. This intron retention affects the DAD sequence and results in a positive charged polar amino acid lysine at position 1062 instead of the nonpolar amino acids leucine or isoleucine present in FMNL1α and FMNL1β. Because binding of the DAD peptide region to the DID armadillo repeat structure is mainly dependent on hydrophobic interactions as described for Dia1 (17, 26), this polar lysine may be responsible for release of autoinhibition of FMNL1γ, resulting in a constitutively activated form. In fact, the splice variant FMNL1γ was specifically located at the membrane and cortex and induced bleb formation. The shape and extent of blebs varied in different cell lines, indicating that cell type-specific interaction partners additionally play an important role in function and bleb shape. However, a similar effect on FMNL1 localization and function in diverse cell lines was observed in cells transfected with a FMNL1 mutant missing the DAD sequence, confirming that deregulation of autoinhibition is responsible for membrane localization and blebbing in FMNL1γ-transfected cells. FMNL1γ colocalized with F-actin in bleb protrusions, suggesting FMNL1γ induced actin assembly. Blebbing but not membrane localization of FMNL1γ was totally abrogated by latrunculin B, further confirming regulation of actin assembly and actin-mediated polarized bleb formation by FMNLγ. We additionally observed polarization of myosin IIb and α-tubulin to FMNL1γ-induced blebs in 293T cells. Moreover, bleb formation was significantly inhibited by blebbistatin and nocodazole, confirming a substantial role of myosin and microtubules in FMNL1γ-induced blebbing. These results are in common with previous reports demonstrating that bleb induction by the Leishmania parasite virulence factor HASPB depends on the integrity of F-actin and requires myosin II function as well as microtubule networks (27). Moreover, the formins FHOD1, mDia1, and mDia2 have been also previously associated with blebbing (24, 28, 29). However, in contrast to bleb formation induced by HASPB and FHOD1 (24, 27), blebbing induced by FMNL1γ is independent of Src and ROCK because the Src inhibitor PP1 and the ROCK inhibitor Y-27632 did not reduce blebbing in our experiments.
To evaluate the mechanism of membrane localization and bleb induction by FMNL1γ further, we analyzed the protein sequence motifs of FMNL1 and identified a myristoylation site at the N terminus of human FMNL1 and murine FRL with glycine at position 2 representing an essential requirement for myristoylation. In fact, N-terminal myristoylation of FMNL1γ but not the mutant G2T/A4T could be directly proved by [3H]myristic acid uptake. Moreover, myristoylation could be inhibited by 2-hydroxymyristate, a potent inhibitor of myristoylation (25), resulting in loss of membrane localization and confirming the significance of N-terminal myristoylation for the specific membranous localization of FMNL1. Loss of myristoylation by mutation of glycine at position 2 additionally significantly reduced blebbing induced by FMNL1γ and FMNL1ΔDAD, demonstrating that the induction of blebbing was dependent on N-terminal myristoylation of FMNL1. N-terminal myristoylation can facilitate anchoring of proteins to lipid membranes by hydrophobic interaction of myristate with the membrane bilayer. This motif is typically present in the SH4 domain of Src kinases, and myristoylation has been shown to be critical for malignant transformation of v-Src (30, 31). Moreover, it has been reported previously that expression of active SH4 domains is associated with cell blebbing (27). We therefore propose that activation of FMNL1 is regulated not only by binding of a Rho-GTPase but additionally by an increase of intracellular Ca2+ or a specific ligand binding. This may cause a myristoyl switch and therefore induces extrusion of the myristoyl group, enabling it to interact quickly and reversibly with lipid bilayer membranes as described previously for other myristoylated proteins (32, 33). Thus, by its N-terminal myristoylation site, FMNL1, in contrast to other formin proteins, may be able to perform expeditious cytoskeletal changes independent of Src and ROCK.
In conclusion, the newly identified splice variant FMNL1γ represents a constitutively active form of FMNL1 that localizes at the membrane and induces bleb formation. We identified N-terminal myristoylation as an important regulatory tool for FMNL1 necessary for bleb formation, enabling fast and reversible membrane localization compatible with diverse functions of hematopoietic lineage-derived cells. The identification of interaction partners of FMNL1 in diverse hematopoietic derived cells as well as the further characterization of splice variants will be highly interesting to identify key molecules regulating different FMNL1 functions, potentially revealing possibilities for specific therapeutic interaction in malignant and inflammatory diseases.
Supplementary Material
Acknowledgments
We thank Vigo Heissmeyer for vectors and technical support using the adenoviral system as well as fruitful discussions, Manuel Deutsch for technical support at the confocal microscope, and Rüdiger Laubender for statistical support.
This work was supported by Deutsche Forschungsgemeinschaft Grant KR2305/3-1, the Helmholtz Zentrum München, and the Life Science Stiftung.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- FMNL1
- formin-like 1
- Chaps
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid
- CLL
- chronic lymphocytic leukemia
- DAD
- diaphanous autoregulatory domain
- DID
- diaphanous inhibitory domain
- DRF
- diaphanous-related formin
- FH
- formin homology
- FRL
- murine formin-like protein 1
- PBMC
- peripheral blood mononuclear cell.
REFERENCES
- 1.Faix J., Grosse R. (2006) Dev. Cell 10, 693–706 [DOI] [PubMed] [Google Scholar]
- 2.Krackhardt A. M., Witzens M., Harig S., Hodi F. S., Zauls A. J., Chessia M., Barrett P., Gribben J. G. (2002) Blood 100, 2123–2131 [DOI] [PubMed] [Google Scholar]
- 3.Schuster I. G., Busch D. H., Eppinger E., Kremmer E., Milosevic S., Hennard C., Kuttler C., Ellwart J. W., Frankenberger B., Nössner E., Salat C., Bogner C., Borkhardt A., Kolb H. J., Krackhardt A. M. (2007) Blood 110, 2931–2939 [DOI] [PubMed] [Google Scholar]
- 4.Gomez T. S., Kumar K., Medeiros R. B., Shimizu Y., Leibson P. J., Billadeau D. D. (2007) Immunity 26, 177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seth A., Otomo C., Rosen M. K. (2006) J. Cell Biol. 174, 701–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yayoshi-Yamamoto S., Taniuchi I., Watanabe T. (2000) Mol. Cell Biol. 20, 6872–6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wallar B. J., Alberts A. S. (2003) Trends Cell Biol. 13, 435–446 [DOI] [PubMed] [Google Scholar]
- 8.Watanabe N., Higashida C. (2004) Exp. Cell Res. 301, 16–22 [DOI] [PubMed] [Google Scholar]
- 9.Zigmond S. H. (2004) Curr. Opin. Cell Biol. 16, 99–105 [DOI] [PubMed] [Google Scholar]
- 10.Higgs H. N., Peterson K. J. (2005) Mol. Biol. Cell 16, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Higgs H. N. (2005) Trends Biochem. Sci. 30, 342–353 [DOI] [PubMed] [Google Scholar]
- 12.Evangelista M., Blundell K., Longtine M. S., Chow C. J., Adames N., Pringle J. R., Peter M., Boone C. (1997) Science 276, 118–122 [DOI] [PubMed] [Google Scholar]
- 13.Watanabe N., Madaule P., Reid T., Ishizaki T., Watanabe G., Kakizuka A., Saito Y., Nakao K., Jockusch B. M., Narumiya S. (1997) EMBO J. 16, 3044–3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alberts A. S. (2001) J. Biol. Chem. 276, 2824–2830 [DOI] [PubMed] [Google Scholar]
- 15.Rose R., Weyand M., Lammers M., Ishizaki T., Ahmadian M. R., Wittinghofer A. (2005) Nature 435, 513–518 [DOI] [PubMed] [Google Scholar]
- 16.Otomo T., Otomo C., Tomchick D. R., Machius M., Rosen M. K. (2005) Mol. Cell 18, 273–281 [DOI] [PubMed] [Google Scholar]
- 17.Nezami A. G., Poy F., Eck M. J. (2006) Structure 14, 257–263 [DOI] [PubMed] [Google Scholar]
- 18.Li F., Higgs H. N. (2003) Curr. Biol. 13, 1335–1340 [DOI] [PubMed] [Google Scholar]
- 19.Harris E. S., Li F., Higgs H. N. (2004) J. Biol. Chem. 279, 20076–20087 [DOI] [PubMed] [Google Scholar]
- 20.Gasman S., Kalaidzidis Y., Zerial M. (2003) Nat. Cell Biol. 5, 195–204 [DOI] [PubMed] [Google Scholar]
- 21.Copeland S. J., Green B. J., Burchat S., Papalia G. A., Banner D., Copeland J. W. (2007) J. Biol. Chem. 282, 30120–30130 [DOI] [PubMed] [Google Scholar]
- 22.Chomczynski P., Sacchi N. (1987) Anal. Biochem. 162, 156–159 [DOI] [PubMed] [Google Scholar]
- 23.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 24.Hannemann S., Madrid R., Stastna J., Kitzing T., Gasteier J., Schönichen A., Bouchet J., Jimenez A., Geyer M., Grosse R., Benichou S., Fackler O. T. (2008) J. Biol. Chem. 283, 27891–27903 [DOI] [PubMed] [Google Scholar]
- 25.Paige L. A., Zheng G. Q., DeFrees S. A., Cassady J. M., Geahlen R. L. (1990) Biochemistry 29, 10566–10573 [DOI] [PubMed] [Google Scholar]
- 26.Lammers M., Meyer S., Kühlmann D., Wittinghofer A. (2008) J. Biol. Chem. 283, 35236–35246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tournaviti S., Hannemann S., Terjung S., Kitzing T. M., Stegmayer C., Ritzerfeld J., Walther P., Grosse R., Nickel W., Fackler O. T. (2007) J. Cell Sci. 120, 3820–3829 [DOI] [PubMed] [Google Scholar]
- 28.Kitzing T. M., Sahadevan A. S., Brandt D. T., Knieling H., Hannemann S., Fackler O. T., Grosshans J., Grosse R. (2007) Genes Dev. 21, 1478–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eisenmann K. M., Harris E. S., Kitchen S. M., Holman H. A., Higgs H. N., Alberts A. S. (2007) Curr. Biol. 17, 579–591 [DOI] [PubMed] [Google Scholar]
- 30.Cross F. R., Garber E. A., Pellman D., Hanafusa H. (1984) Mol. Cell. Biol. 4, 1834–1842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kamps M. P., Buss J. E., Sefton B. M. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 4625–4628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ames J. B., Ishima R., Tanaka T., Gordon J. I., Stryer L., Ikura M. (1997) Nature 389, 198–202 [DOI] [PubMed] [Google Scholar]
- 33.Seykora J. T., Myat M. M., Allen L. A., Ravetch J. V., Aderem A. (1996) J. Biol. Chem. 271, 18797–18802 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.