Abstract
It has long been predicted that the members of the hyaluronidase enzyme family have important non-enzymatic functions. However, their nature remains a mystery. The metabolism of hyaluronan (HA), their major enzymatic substrate, is also enigmatic. To examine the function of Hyal2, a glycosylphosphatidylinositol-anchored hyaluronidase with intrinsically weak enzymatic activity, we have compared stably transfected rat fibroblastic BB16 cell lines with various levels of expression of Hyal2. These cell lines continue to express exclusively the standard form (CD44s) of the main HA receptor, CD44. Hyal2, CD44, and one of its main intracellular partners, ezrin-radixin-moesin (ERM), were found to co-immunoprecipitate. Functionally, Hyal2 overexpression was linked to loss of the glycocalyx, the HA-rich pericellular coat. This effect could be mimicked by exposure of BB16 cells either to Streptomyces hyaluronidase, to HA synthesis inhibitors, or to HA oligosaccharides. This led to shedding of CD44, separation of CD44 from ERM, reduction in baseline level of ERM activation, and markedly decreased cell motility (50% reduction in a wound healing assay). The effects of Hyal2 on the pericellular coat and on CD44-ERM interactions were inhibited by treatment with the Na+/H+ exchanger-1 inhibitor ethyl-N-isopropylamiloride. We surmise that Hyal2, through direct interactions with CD44 and possibly some pericellular hyaluronidase activity requiring acidic foci, suppresses the formation or the stability of the glycocalyx, modulates ERM-related cytoskeletal interactions, and diminishes cell motility. These effects may be relevant to the purported in vivo tumor-suppressive activity of Hyal2.
INTRODUCTION
The turnover rate of hyaluronan (HA),2 the major unbranched glycosaminoglycan of the extracellular matrix, is surprisingly rapid: approximately one-third of total body HA is replaced daily (1). Although it is generally agreed that hyaluronidases play important roles in the degradation of HA, the specific functions of the six or seven members of the hyaluronidase family remain obscure (2, 3). Only three of the hyaluronidase genes are broadly expressed in somatic tissues: HYAL1, HYAL2, and HYAL3 (4), all of which are grouped on human chromosomal locus 3p21.3. Mice deficient in each of these genes have recently been described. However, Hyal3 knockout mice do not display a distinct phenotype (5), Hyal1 null animals develop a slowly progressive osteoarthritis without significant elevation of plasma or tissue levels of HA (6), and Hyal2−/− mice show skeletal and hematological anomalies as well as 10-fold increases in plasma HA (7). Some of these apparent anomalies are explained perhaps by the non-enzymatic functions of these proteins.
In vitro, the glycosylphosphatidylinositol-anchored Hyal2 has a much weaker hyaluronidase activity compared with Hyal1 or to the sperm hyaluronidase PH20 (8). Fragments of HA, ∼20 kDa in mass, may build up during Hyal2 functioning (8–10), which raises the possibility that Hyal2 participates in inflammatory events in which such HA fragments accumulate (11, 12). Intriguingly, sheep Hyal2 also functions as a cell-entry receptor for oncogenic ovine retroviruses (13), although this function is independent of any HA-degrading activity (8). Recently, expression of HYAL2 and HYAL1 genes has been shown to inhibit tumor growth in vivo without noticeable effect on growth in vitro, suggesting that these hyaluronidases control in some unknown manner tumor-host interactions (14). Hyal2 also anchors tumor growth factor-β1 to the cell surface and mediates some of its pro-apoptotic effects (15). Conversely, Hyal2 may facilitate cancer progression: Hyal2 overexpression in murine astrocytoma cells accelerates intracerebral, but not subcutaneous, tumor formation (16); the in vitro invasion capacity of several breast cancer cell lines correlates with Hyal2 expression (17, 18); and a cell surface proteomics study demonstrates that Hyal2 is enriched 13-fold in a highly invasive HT-1080 fibrosarcoma cell clone relative to a congenic variant with a low potential for intravasation and metastasis (19). A better understanding of all these divergent events necessitates more information regarding the action of Hyal2 at the cellular level.
We hypothesized that the main action of Hyal2 would be at the plasma membrane level where it interferes with pericellular HA or with CD44, the principal HA receptor. First, Hyal2 and CD44 have previously been shown to co-exist and interact within lipid rafts of the breast cancer cell line MDA-MB231 (17). Upon HA addition, the activity of the sodium-hydrogen exchanger NHE1 increases, the pH drops locally, and Hyal2 begins to degrade HA (17). Second, the membrane localization and enzymatic activity of Hyal2 have an absolute requirement for CD44 in transfected HEK293 cells. In that situation, the HA-degrading activity of Hyal2 is detected at pH 6.0–7.0 (10). Third, the pericellular HA-rich coat, or glycocalyx, that surrounds many types of cells disappears after exposure to either the HA-specific Streptomyces hyaluronidase or to HA oligosaccharides (20–22). This coat appears to be influenced by the level of Hyal2 expression.
To test this hypothesis in a non-cancerous cell line, given that our only effective antibodies were specific for rat Hyal2, we selected BB16 cells, which are v-src-transformed rat fibroblasts (23). These cells display a large pericellular glycocalyx coat and a high level of expression of CD44. In the current study, we have shown that high expression of Hyal2 is able to suppress anchoring of the pericellular coat of these cells almost entirely. This may occur through a destabilization of CD44, leading to a block in one of the main CD44 signaling systems, i.e. ezrin-radixin-moesin (ERM) activation. Loss of cell motility then ensues.
EXPERIMENTAL PROCEDURES
Cell Culture
Rat-1 cells transformed by the B77 subclone of Rous Sarcoma Virus (BB16) were kindly provided by Prof. Pierre Courtoy, Institute of Cellular Pathology, Brussels, Belgium (23). Cells were grown at 37 °C in Dulbecco's modified Eagle's medium (Cambrex) supplemented with 15 mm Hepes, 10 μg/ml streptomycin, 66 μg/ml penicillin, and 10% (v/v) fetal bovine serum (Cambrex) under 5% CO2.
Antibodies and Reagents
The rat Hyal2 cDNA (GenBankTM accession number AF034218, from 1997) was cloned in our laboratory, and polyclonal rabbit antibodies were generated. The P16 antibody used in the current study was raised against a 16-amino acid region in the body of rat Hyal2. The following commercial anti-CD44 antibodies were used: mouse monoclonal OX49 (BD Pharmingen) for immunoprecipitation and immunofluorescence, mouse monoclonal OX50 (BIOSOURCE) for CD44 blockade, and rabbit polyclonal HCAM (sc-7946, Santa Cruz Biotechnology, Santa Cruz, CA) for Western blotting. Goat polyclonal anti-lamin (sc-6214), mouse monoclonal anti-α-tubulin (sc-5286), and rabbit polyclonal anti-GFP (sc-8334) were from Santa Cruz Biotechnology. Rabbit polyclonal anti-ERM and anti-pERM (phosphorylated ERM) were from Cell Signaling Technology. Mouse monoclonal anti-β-actin, ethyl-N-isopropylamiloride (EIPA), wortmannin, LY294002, 4-methylumbelliferone (4-MU), Stains-All, Pronase, mitomycin C, and Streptomyces hyaluronidase were obtained from Sigma-Aldrich. Peroxidase-conjugated goat anti-rabbit and anti-mouse IgG and peroxidase-conjugated horse anti-goat IgG were purchased from Dako. Alexa Fluor 488-tagged goat anti-mouse and anti-rabbit IgG and Alexa Fluor 568-tagged goat anti-mouse and anti-rabbit antibodies were from Molecular Probes. 6- and 10-mer HA oligosaccharides were kindly donated by Seikagaku Corp. Three sources of high molecular mass HA were used: umbilical cord HA (∼2.5 × 106 Da, Sigma-Aldrich), Healon (∼1.6 × 106 Da, Amersham Biosciences), and preparations of smaller molecular mass HA (0.02–1.2 × 106 Da). The latter were a kind gift of Dr. Ove Wik, Amersham Biosciences.
Transfections
To obtain stable transfectants, BB16 cells were transfected with expression constructs containing cDNA inserts encoding rat Hyal2 or with the expression vector (pcDNA3.1+) alone, selected for Geneticin resistance, and isolated using cloning rings. Clones with a medium high expression level of Hyal2 (BB16Hy2+) were selected for most experiments of the present study. In some experiments, clones with intermediate levels of expression of Hyal2 were used. Clones transfected with the vector without Hyal2 insert (BB16mock) were used as controls. Transient transfections of BB16 cells with homemade EGFP-rat Hyal2 or commercial CD44 (RZPD, German Science Centre for Genome Research) were obtained using Lipofectamine 2000 (Invitrogen). For siRNA transfection, target sequences were selected and obtained from Eurogentec (Liege, Belgium). Three specific Hyal2 target sequences (Table 1) and one scrambled sequence (not shown) were used. Cells at 50–70% confluence in 6-well plates were transfected with 50 pmol of either the Hyal2 siRNA pool or the scrambled sequence, in 1.5 ml of complete medium, using Lipofectamine 2000. Cells were then incubated at 37 °C in 5% CO2 for 24 h, harvested, and processed for Western blotting, immunoprecipitation, and migration assays.
TABLE 1.
Sequences of the siRNA combination used in order to inhibit Hyal2 expression
| siRNA | Sequence |
|---|---|
| HYAL2 #1 sense | 5′-CUG-CUA-CAA-UCA-UGA-UUA-UdTdT-3′ |
| HYAL2 #1 antisense | 5′-AUA-AUC-AUG-AUU-GUA-GCA-GdTdT-3′ |
| HYAL2 #2 sense | 5′-CUC-CCA-GUC-UAC-GUC-UUC-AdTdT-3′ |
| HYAL2 #2 antisense | 5′-UGA-AGA-CGU-AGA-CUG-GGA-GdTdT-3′ |
| HYAL2 #3 sense | 5′-GAU-GUG-UAU-CGC-CGG-UUA-UdTdT-3′ |
| HYAL2 #3 antisense | 5′-AUA-ACC-GGC-GAU-ACA-CAU-CdTdT-3′ |
Western Blot Analysis
Cells were cultured until 80% confluence, washed 3× with phosphate-buffered saline (PBS), and solubilized in ice-cold RIPA buffer (50 mm Tris-HCl (pH 7.4), 120 mm NaCl, 1% Triton X-100, 0.1% SDS, 1% deoxycholate) with Complete Protease Inhibitor Mixture (Roche Applied Science), tyrosine phosphatase inhibitors (200 μm sodium molybdate, 500 μm sodium orthovanadate), and serine/threonine protein phosphatase inhibitors (1 μm microcystin-LR (Calbiochem) and 300 μm okadaic acid (Sigma)). In some cases, cell lysates were pretreated with endoglycosidase F (New England Biolabs). In other conditions, cells were pretreated with EIPA (20 μm) or with the HA-specific Streptomyces hyaluronidase (5 units/ml) for 16 h. Proteins (usually 5 μg) were analyzed using SDS-PAGE gel according to Laemmli (10% acrylamide/bisacrylamide) using a Mini Gel Protean set (Bio-Rad), followed by transfer to polyvinylidene difluoride membranes (Amersham Biosciences) with incubation for 1 or 2 h at room temperature with specific antibodies. Blots were visualized with horseradish peroxidase-conjugated anti-mouse or anti-rabbit IgG antibody (Dako), followed by an enhanced chemiluminescence Western blotting detection system (PerkinElmer Life Sciences). To detect CD44 shedding, cells were serum-starved for 16 h and medium was collected, centrifuged to remove unattached cells, filter sterilized, and then concentrated. 25-μl samples were analyzed on 10% SDS-PAGE.
Biotinylation
Cell surface biotinylation was performed as described (24) on cells seeded in 92-mm culture dishes using 2 mg/ml sulfo-NHS-SS-biotin and NeutrAvidinTM (Pierce).
Immunoprecipitations
To immunoprecipitate Hyal2, cells were labeled for 18 h with [35S]methionine, washed with ice-cold PBS and homogenized in the same buffer. The homogenate was centrifuged at low speed, and the pellet was resuspended in a solution containing 0.25 m sucrose, 3 mm imidazole (pH 7.4), 50 mm Tris, 150 mm NaCl, 0.1% Triton, 0.5% SDS, 0.5% deoxycholate, and Complete Protease Inhibitor Mixture. Suspensions were homogenized with a Dounce tissue (tight) grinder and protein content was measured by Bio-Rad Protein Assay. Cells lysates (500 μg of proteins) were preincubated with protein A-agarose (Roche Applied Science). Supernatants were incubated with P16 antibodies and then with protein A-agarose. Following incubation, immunoprecipitates were washed 5× with PBS containing 1% Triton, 0.1% SDS, and 0.5% deoxycholate and boiled with an equal volume of Laemmli sample buffer, followed by SDS-PAGE and Cyclone phosphorimager (Canberra Packard) analysis. For other immunoprecipitations, including CD44-Hyal2 co-immunoprecipitation, pellets were resuspended in RIPA buffer with protease inhibitors. For CD44-ERM and CD44-pERM co-immunoprecipitation, cells were lysed in a solution containing 10 mm Tris (pH 7.5), 2 mm EDTA, 1% Triton X-100, as well as protease and phosphatase inhibitors.
Immunofluorescence and Confocal Microscopy
Cells were fixed with 4% paraformaldehyde in PBS for 10 min. In some cases, they were permeabilized with 0.5% Triton X-100 in PBS for 5 min. After three washes in PBS, nonspecific binding sites were blocked by incubation for 30 min with 1% bovine serum albumin in PBS. Subsequently, cells were incubated with one or two primary antibodies for 2 h at room temperature in a wet chamber. Goat anti-rabbit or anti-mouse IgG labeled with Alexa Fluor 488 or Alexa Fluor 568 (Molecular Probes) was added for 1 h at room temperature in the dark. After each antibody incubation step cells were washed 5× with 1% bovine serum albumin in PBS and mounted in Mowiol. Series of optical sections were taken with a Zeiss LSM 510 confocal microscope and were projected to single images using LSM software.
Morphometric Analysis of the HA-rich Pericellular Matrix
The particle exclusion assay was described previously (25). In our experiments, 1.5 × 105 cells were plated in 35-mm tissue culture dishes and cultured in Dulbecco's modified Eagle's medium with 10% fetal bovine serum. After 24 h, 750 μl of a suspension of fixed and washed horse red blood cells (108 cells/ml) was added to the cells and allowed to settle for 15 min. The pericellular matrix was assessed from photographs by measuring the area delimited between the red blood cells and the cell membrane. A ratio of 1.0 indicates no matrix. To demonstrate the structural dependence of the pericellular matrix on HA and on CD44, cells were (a) treated with 5 units/ml HA-specific Streptomyces hyaluronidase for 1 h before adding OX50 antibodies and observing coat recovery; (b) incubated with 100 μm 4-MU, an inhibitor of HA synthesis, in serum-free medium for 16 h, or (c) treated with 6-mer or 10-mer HA oligosaccharides for 16 h. To assess the effect of phosphoinositide 3-kinase (PI3K) inhibition, cells were preincubated with either 50 nm wortmannin or 5 μm LY294002 for 6 h before coat estimation.
HA Assay
Cells were seeded in 92-well culture dishes. HA concentration was analyzed by a pseudo-RIA kit (Pharmacia HA Test) in the medium and in cell extracts following overnight digestion with 500 milliunits/ml Pronase at 55 °C in a 0.1 m Tris-HCl (pH 7.4) buffer containing 10 mm CaCl2. The homogenate was boiled for 10 min before protein (Bio-Rad kit) and HA assay.
Hyaluronidase Activity
Both cell extracts and media were tested for hyaluronidase activity at different pH levels using zymography, which was performed as recently published by our group (7). In addition, 25-μg protein samples from cell extracts were incubated in vitro with 15 μg of 2.5 × 106 Da HA for 16 h at 37 °C in a reaction buffer containing 100 mm formate at pH 3.7. The samples were mixed with 1/6 volume of loading buffer (7× TAE (40 mm Tris acetate, 1 mm EDTA, pH 8.0), 85% glycerol) and subjected to electrophoresis in 0.8% TAE-agarose gels at 50 V for 10 h. The size distribution of HA in samples was measured using a modification of the method described by Lee and Cowman (26). The gels were stained in 0.005% Stains-All in 50% ethanol overnight and photographed on a fluorescent light Transilluminator.
HA Binding Assay
Healon was iodinated with Na125I using IODO-BEADs (Pierce) according to the manufacturer's instructions, and HA-binding activity was determined using a modified version of a method previously described (27). The samples were dissolved in 200 μl of Hanks' balanced salt solution containing increasing concentrations of 125I-labeled HA (0 to 8 μg/250 μl) in the absence or presence of non-labeled HA (100 μg/250 μl). After shaking for at least 20 min, 250 μl of saturated (NH4)2SO4 was added, followed by 25 μl of nonfat milk. The samples were centrifuged at 9,000 × g for 5 min. The tubes were rinsed twice with 50% saturated (NH4)2SO4, and the pellets were dissolved in water and processed for scintillation counting. Specific binding was calculated by subtracting the background label measured in the presence of an excess of non-labeled HA.
Flow Cytometric Analysis
Purified HA (0.02–1.2 × 106 Da, Amersham Biosciences) was conjugated to fluorescein as described by de Belder and Wik (28). Cells were resuspended in 0.5 ml of PBS with 0.1% NaN3 and a 1:100 dilution of fluorescein-HA (final concentration, 50 μg/ml). The mixture was incubated for 1 h at 4 °C. Following washes with 0.1% NaN3 in PBS, cells were fixed with 2% paraformaldehyde in the same solution for 30 min. Samples were analyzed on a FACSCalibur (BD Biosciences).
Wound Healing Assay
Population-based movement of cells was measured by a wound healing assay as described previously (29). In this assay, cells are grown to confluency on plastic Petri dishes, then induced to re-populate a “wound” created by stripping a sharply defined channel using a sterile razor blade. To prevent growth during migration, cells were pre-treated with 4.5 μm mitomycin C for 2 h. These concentrations were optimized for the BB16 cell line as described (30). After band-stripping, cells were allowed to migrate into the wound for 6 h in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, supplemented or not with different hyaluronidases, 4-MU, or a PI3K inhibitor. Cells were then fixed with 3% formaldehyde in PBS for 20 min at 4 °C and stained with Crystal Violet. The number of cells that had colonized the wound margin was assessed in random microscopic fields (magnification, 320×; 15 fields per assay).
Statistical Analyses
Results are presented as means ± S.E. One- and two-way analyses of variance were used. When statistically significant differences were found (p < 0.05), individual comparisons were made using Dunn or Bonferroni tests with GraphPad Prism Version 5 software.
RESULTS
Development and Characterization of BB16 Clones Overexpressing Hyal2
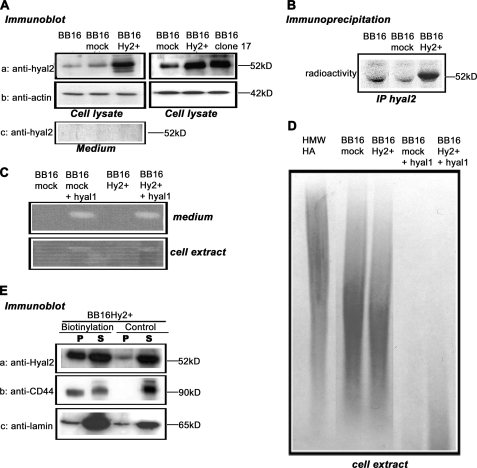
Experiments were performed on the rat fibroblastic v-src-transformed cell line BB16. Hyal2 protein was detected by Western blotting using a rabbit polyclonal antibody, P16, raised against a peptide sequence of rat Hyal2. Because Hyal2 is poorly expressed in BB16 cells (Fig. 1A), stable transfectants were established using either rat Hyal2 cDNA or an empty vector. For comparison purposes, a clone with high expression of Hyal2 (BB16Hy2+) and a mock transfected clone (BB16mock) were selected. Several other clones overexpressing Hyal2 were also obtained; the relative amounts of Hyal2 mRNA in these clones based on quantitative reverse transcription-PCR ranged from 1.3 to 8.0 (supplemental Fig. S1B). None of the clones secreted Hyal2 into the medium (shown in Fig. 1A for BB16Hy2+ cells). The specificity of the P16 antibody was shown by preincubation with the immunogenic peptide (data not shown). Hyal2 overexpression in BB16Hy2+ cells was confirmed by immunoprecipitation after metabolic labeling (Fig. 1B). BB16Hy2+ cells synthesized and secreted similar amounts of HA compared with BB16 cells (Table 2).
FIGURE 1.
Quantification of Hyal2 expression and its hyaluronidase activity. v-src-transformed rat fibroblasts (BB16) were stably transfected with either an empty plasmid (BB16mock clone) or a plasmid expressing Hyal2 (BB16Hy2+ clone and clone 17). A, Western blot analyses of cell lysates (5 μg of proteins) and concentrated media with anti-Hyal2 (P16) and anti-actin antibodies. B, immunoprecipitation of Hyal2 using P16 in cells labeled with [35S]methionine for 18 h. C, zymographic measurements of hyaluronidase activity at pH 3.7 in BB16mock and BB16Hy2+ cells, and in the same cells transiently transfected with rat Hyal1 cDNA (+Hyal1). D, measurements of HA-degrading activity in BB16mock and BB16Hy2+ cells, and in the same cells transiently transfected with rat Hyal1 cDNA (+Hyal1). Cell extracts were incubated with 2.5 × 106 Da HA for 16 h at pH 3.7, followed by agarose gel electrophoresis of the resulting incubate and detection of HA with Stains-All, allowing assessment of HA amount and size. The first lane is the high molecular mass HA incubated for 16 h at pH 3.7 with the buffer only. E, Western blots of BB16Hy2+ cells using anti-Hyal2 (P16) and anti-CD44 (OX49) antibodies following cell surface biotinylation and precipitation of the biotinylated proteins with streptavidin-Sepharose beads. The signal in the pellet (P) represents half the amount of Hyal2, CD44, or lamin present on the cell surface. The signal in the supernatant (S) represents 1/10th the amount of these proteins that was inaccessible to external biotin. A control experiment without biotinylation is shown. Anti-lamin antibodies were used to detect a typical nuclear protein.
TABLE 2.
Quantification of endogenous HA
HA was measured with a pseudo-RIA kit in cells without (BB16) or with (BB16Hy2+) Hyal2 overexpression. Means ± S.E. of six experiments in each group are shown.
| BB16 | BB16Hy2+ | p | |
|---|---|---|---|
| Cell-associated HA (ng/1000 cells) | 1.26 ± 0.19 | 1.66 ± 0.17 | NS |
| HA in culture medium (ng/1000 cells) | 3.68 ± 1.05 | 3.51 ± 0.88 | NS |
Hyal2 Has No Detectable Enzymatic Activity in Cell Extracts or in the Media
Neither BB16 nor BB16Hy2+ cells showed any hyaluronidase activity in zymography at pHs ranging from 3.7 (Fig. 1C) to 6.8 (data not shown), whether in the medium or in cell extracts. The partial HA-degrading activity detected in BB16 cell extracts at pH 3.7 did not significantly increase in any Hyal2 overexpression clone (Fig. 1D). Furthermore, this type of activity could not be detected at any significant level in cell membrane fractions at pH 4.0 or 6.5 (supplemental Fig. S2), contrary to what has been observed by Harada and Takahashi in CD44- and Hyal2-overexpressing HEK293 cells (10). This almost complete lack of hyaluronidase activity of Hyal2 should be contrasted with the effects of transient transfections of rat Hyal1 cDNA in the same cells, which led to strong HA-degrading activity (Fig. 1, C and D).
Hyal2 Is Partly Expressed at the Cell Surface
Prior studies have reported conflicting results regarding the subcellular localization of Hyal2, which has been assigned previously to lysosomes (9, 31), to the plasma membrane (13), and even to mitochondria (32) as well as to nuclei (15). Thus, the level of Hyal2 exposure on the outer cell surface of BB16 clones was evaluated by a cell surface biotinylation assay. A significant portion of Hyal2 was detected among biotin-labeled proteins in the streptavidin pellet of BB16Hy2+ cells, meaning Hyal2 was exposed at the outer plasma membrane during the process of biotinylation (Fig. 1E). However, for both Hyal2 and CD44, a well characterized HA receptor used as a control membrane protein, a good portion of the molecules remained inaccessible to external biotin (Fig. 1E).
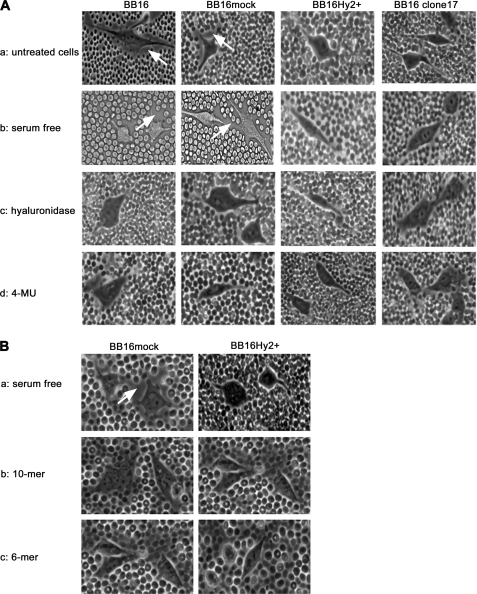
Hyal2 Interferes with the Pericellular HA-rich Matrix
A standard red blood cell exclusion assay indicated that BB16 and BB16mock cells were capable of producing and organizing a HA-rich pericellular matrix with or without exposure to 10% fetal bovine serum (Fig. 2 and Table 3). In contrast, BB16Hy2+ cells were almost devoid of such a pericellular coat. The ratio of combined cell and pericellular matrix areas relative to the cell area was ∼2.0 in cells expressing low levels of Hyal2 (BB16 and BB16mock), but fell to ∼1.2 in BB16Hy2+ cells. In these cell lines and in six other clones expressing intermediate levels of Hyal2, there was a strong correlation between individual Hyal2 mRNA levels and the reduction in coat size (supplemental Fig. S1D).
FIGURE 2.
Effect of Hyal2 overexpression and exposure to HA oligosaccharides on the pericellular coat. The latter was visualized by a particle exclusion assay in phase-contrast microscopy; representative images of cells without (BB16 and BB16mock) and with (BB16Hy2+ and clone17) Hyal2 overexpression are shown. Arrows highlight the external border of the coat in each cell type. A, effects of four different conditions: (a) usual culture medium containing 10% fetal bovine serum; (b) the same medium after 16-h serum deprivation; (c) following 1-h treatment with 5 units/ml Streptomyces hyaluronidase in serum-free medium; (d) following 16-h treatment with 100 μm 4-methylumbelliferone (4-MU) in serum-free medium. B, effects of: (a) control serum-free medium; and addition of (b) 10-mer or (c) 6-mer HA oligosaccharides at 100 μg/ml for 16 h in serum-free medium.
TABLE 3.
Effects of different treatments on pericellular HA coats
Coats were visualized using a particle exclusion assay in Hyal2 overexpressing (BB16Hy2+) cells and in control (BB16 and BB16mock) cells and expressed as the ratio of combined cell and pericellular matrix areas relative to the cell area. A value of 1.0 means no discernible coat. Results are presented as means ± S.E. of 17 to 81 cells in each group. p values in the last column result from comparisons of BB16mock and BB16Hy2+ cells in two-way analysis of variance with Bonferroni correction. Statistical analyses were performed in four separate settings: basal settings, coat inhibitors, potential Hyal2 activity inhibitors, and coat recovery after HA treatment. Statistical significance of the effect of treating either BB16mock or BB16Hy2+ cells with various agents was tested in one-way analysis of variance followed by Dunnett's test. The results are given in footnotes a–d. All other relevant comparisons in BB16mock and BB16Hy2+ cells were NS (not significant).
| BB16 | BB16mock | BB16Hy2+ | p | |
|---|---|---|---|---|
| Basal settings | ||||
| Untreated cells | 2.05 ± 0.05 | 2.08 ± 0.04 | 1.22 ± 0.07 | <0.001 |
| Scrambled RNA | 2.28 ± 0.11 | 1.09 ± 0.08 | <0.001 | |
| Hyal2 siRNA | 1.99 ± 0.07 | 1.71 ± 0.07a | NS | |
| Coat inhibitors | ||||
| Serum-free untreated cells | 2.01 ± 0.07 | 1.99 ± 0.05 | 1.18 ± 0.09 | <0.001 |
| HA10 oligomers, 100 μg/ml | 1.29 ± 0.02b | 1.11 ± 0.04 | NS | |
| HA6 oligomers, 100 μg/ml | 1.17 ± 0.09b | 1.07 ± 0.08 | NS | |
| Streptomyces HA (5 units/ml) | 1.08 ± 0.14 | 1.11 ± 0.05b | 1.14 ± 0.10 | NS |
| 4-MU, 100 μm | 1.21 ± 0.09 | 1.26 ± 0.06b | 1.19 ± 0.07 | NS |
| Potential Hyal2 activity inhibitors | ||||
| DMSO, 0.1% | 2.17 ± 0.07 | 1.08 ± 0.02 | <0.001 | |
| EIPA, 20 μm | 2.25 ± 0.08 | 1.86 ± 0.06c | NS | |
| CD44, transient overexpression | 2.14 ± 0.06 | 2.08 ± 0.05c | NS | |
| Coat recovery after HA treatment | ||||
| Coat recovery in 4 h | 2.08 ± 0.07 | 1.17 ± 0.04 | <0.001 | |
| Coat recovery with OX50 antibody | 1.37 ± 0.09d | 1.10 ± 0.03 | NS | |
| Coat recovery with OX50 and OX49 antibodies | 1.14 ± 0.08d | 1.06 ± 0.04 | NS | |
a p < 0.001 for Hyal2 siRNA versus scrambled siRNA in BB16Hy2+ cells.
b p < 0.001 for HA oligomers, HA, and 4-MU versus serum-free untreated BB16mock cells.
c p < 0.001 for CD44 transfection versus mock transfection and for EIPA versus DMSO vehicle in BB16Hy2+ cells.
d p < 0.001 for OX50 and OX50 plus OX49 versus controls in BB16mock cells previously treated with HA (coat recovery).
In an analysis for specificity, BB16Hy2+ cells were transiently transfected either with siRNA directed against Hyal2 or with a scrambled (control) small RNA: only in the former case did the pericellular coat reappear in a significant amount, reaching a value of ∼1.7 (Table 3). The effect of Hyal2 overexpression on the pericellular coat was compared with treatment of BB16mock cells with either a global HA synthase inhibitor (4-MU), Streptomyces hyaluronidase, or HA oligomers (competitive inhibitors of CD44-polymeric HA binding): all removed the coat to the same extent (Fig. 2 and Table 3). This suggests that HA synthesis and CD44 are both required for the continuous maintenance of a pericellular HA-rich matrix in BB16 cells. Considering the absence of any additional hyaluronidase activity in Hyal2 overexpression cells relative to control cells, the effect of Hyal2 on pericellular coat is thus likely to be predominantly a non-enzymatic phenomenon.
Hyal2 Reduces Specific HA Binding to the Cell Surface
Disruption of the pericellular coat may derive from defective HA binding to the cell surface. The binding of exogenous HA, either in isotopic (Table 4) or fluorescent (Table 5) form, was measured, therefore, and found to be reduced by ∼50% in BB16Hy2+ cells, without any significant change in the dissociation constant, the Kd (Table 4). Decreased HA binding in BB16Hy2+ cells was observed with HA of various sizes: not only 1.2 × 106 Da (Tables 4 and 5) but also 0.2 × 106 and 0.02 × 106 Da (data not shown). Thus, Hyal2 reduces the availability, but not the affinity, of HA binding sites, possibly through a direct interaction with HA receptors.
TABLE 4.
125I-Labeled HA binding
EDTA-isolated Hyal2 overexpressing (BB16Hy2+) cells and control BB16 cells were incubated with protease inhibitors and 125I-labeled HA at concentrations ranging from 0.1 to 8.0 μg/ml with either 100 μg/ml unlabeled HA or 1% PBS-bovine serum albumin for 20 min at room temperature. Radioactivity associated with the pellet was taken as a measure of bound 125I-labeled HA. Total and specific binding was calculated as well as the equilibrium dissociation constant (Kd) and maximum specific binding of labeled HA (Bmax). Means ± S.E. of six experiments in each group are shown.
| BB16 | BB16Hy2+ | p | |
|---|---|---|---|
| Kd (micrograms of HA/ml) | 3.55 ± 0.84 | 2.04 ± 0.43 | NS |
| Bmax (micrograms of HA/ml) | 0.21 ± 0.03 | 0.11 ± 0.01 | < 0.03 |
TABLE 5.
Fluorescein-HA binding
EDTA-isolated Hyal2 overexpressing (BB16Hy2+) cells and control (BB16mock) cells were incubated with 50 μg/ml fluorescein-HA for 1 h at 4 °C without or with 1000 μg/ml non-fluorescent HA, extensively washed, and analyzed using FACS. The threshold level of fluorescence of HA-positive cells was determined. The table shows the percentage of HA-positive cells as means ± S.E. of four experiments in each group.
| BB16mock | BB16Hy2+ | p | |
|---|---|---|---|
| % | % | ||
| No HA addition | 7.9 ± 1.0 | 7.1 ± 0.8 | NS |
| Fluorescein-HA | 95.4 ± 1.2 | 45.8 ± 1.4 | <0.001 |
| Fluorescein-HA plus excess of non-fluorescent HA | 23.6 ± 1.4 | 19.9 ± 1.1 | NS |
Effect of Hyal2 on Pericellular Coat Requires an Acidic Microenvironment
In the cancer cell experiments performed by Bourguignon et al. (17), Hyal2 was closely associated with both NHE1 and CD44, and Hyal2-mediated HA modification was shown to require an acidic microenvironment that could be suppressed by EIPA, a cell-impermeable NHE1 inhibitor. Therefore, BB16Hy2+ cells were incubated with EIPA. They were found to regain a thick coat (Table 3), suggesting that the action of Hyal2, or its interaction with other proteins, does indeed require acid foci at the pericellular surface.
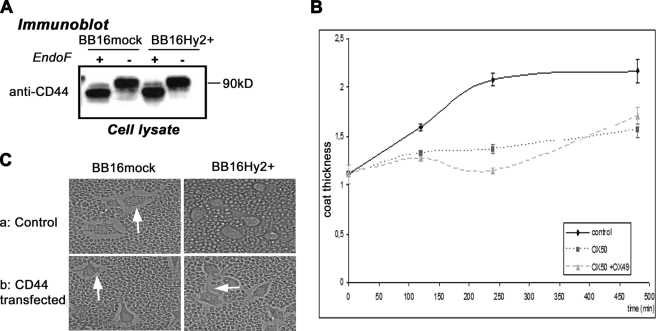
Effect of Hyal2 on Pericellular Coat Is CD44-dependent
CD44 is a usual prerequisite for pericellular coat assembly (21, 33). Therefore, we first examined CD44 expression levels and glycosylation patterns using Western blots with or without endoglycosidase F in BB16Hy2+ cells. No difference with BB16mock cells was found (Fig. 3A). It is known that the HA-binding capacity of CD44 differs among its many splice variants (34) or following changes in glycosylation (35, 36), or in sulfation (37, 38). Therefore, we also examined BB16Hy2+ cells for variant isoforms of CD44, using a sensitive reverse transcription-PCR technique. No variant was found (data not shown). Similarly, no sulfation of CD44 was revealed using [35S]sulfate (data not shown). We conclude that BB16 and BB16Hy2+ cells continue to express exclusively the standard form (CD44s) of CD44 without major post-translational variations.
FIGURE 3.
Implication of CD44 in coat recovery. A, Western blots of CD44 using HCAM antibody with and without endoglycosidase F treatment. B and C, coat thickness measured by the particle exclusion assay. In B, after treatment with 5 units/ml Streptomyces hyaluronidase for 1 h, BB16mock cells were left to recover without or with one (OX50) or two (OX50 + OX49) CD44 blocking antibodies. The inhibitory effects of one or two antibodies on coat formation were highly significant (two-way analysis of variance, p < 0.01, n = 5). In C, cells were transfected with plasmid only (control) or with rat CD44 cDNA and incubated for 24 h before phase contrast microscopy observation. Representative images are shown.
Then, using the CD44-blocking antibodies OX49 and OX50, we confirmed that CD44 is involved in the regeneration of a HA coat in BB16mock cells after exposure to hyaluronidase (Table 3 and Fig. 3B). In addition, a transient transfection with CD44 cDNA induced a complete reversal of the effect of Hyal2 overexpression on the pericellular coat of BB16Hy2+ cells (Table 3 and Fig. 3C).
In conclusion, although Hyal2 does not alter CD44 expression patterns, it can induce a decrease of CD44 binding capacity and a loss of pericellular coat that is rescued by overexpressing CD44. The next step was thus to determine if Hyal2 and CD44 interact directly.
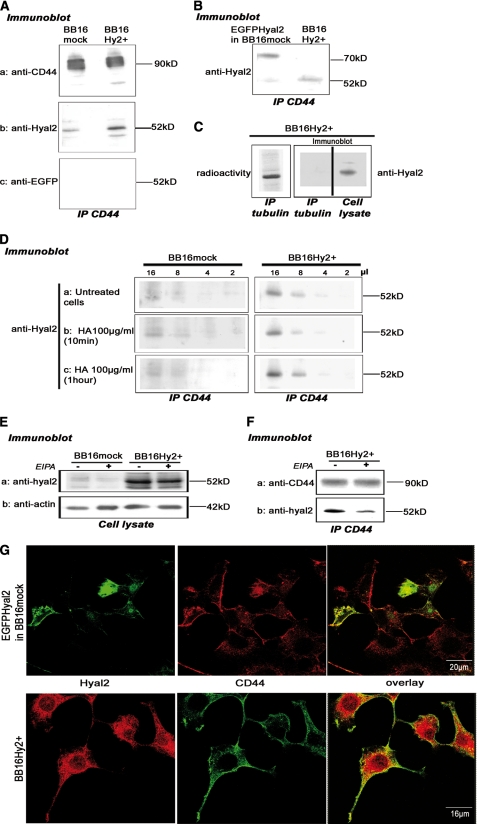
Hyal2 Is Closely Associated with CD44
This was revealed by co-immunoprecipitation of either Hyal2 or a Hyal2-EGFP construct with CD44 but not with, e.g. tubulin (Fig. 4, A–C). It was important to verify that Hyal2 does not accompany a non-relevant protein in the pellet, in view of the great hydrophobicity of Hyal2. On the other hand, Hyal2 immunoprecipitation using P16 requires such drastic conditions that co-immunoprecipitation of other proteins could not be established.3 HA addition at high concentration had no effect on the CD44-Hyal2 association (Fig. 4D). Conversely, EIPA reduced the amount of Hyal2 immunoprecipitating with CD44 (Fig. 4F) while leaving the total amount of Hyal2 unchanged (Fig. 4E). This suggests that an acidic pericellular microenvironment is needed to hold Hyal2 and CD44 in a strongly bound complex. The co-localization of Hyal2 and CD44 was also demonstrated using immunofluorescence and confocal microscopy (Fig. 4G).
FIGURE 4.
The Hyal2-CD44 interaction. A, detection of Hyal2 (with P16 antibody), CD44 (HCAM antibody, positive control), and EGFP (negative control) in CD44 (OX49 antibody) immunoprecipitates. B, Hyal2 was also detected using P16, at a higher molecular mass, in CD44 immunoprecipitates of BB16mock cells transiently transfected with a Hyal2-EGFP cDNA construct. C, as a control, tubulin was immunoprecipitated in BB16Hy2+ cells labeled with [35S]methionine; Hyal2 was not detected in the precipitate but only in the cell lysate. D, experiment performed as in A after incubation with 100 μg/ml high molecular mass HA for 10 or 60 min. Different volumes of CD44 immunoprecipitate (2–16 μl) were loaded in four consecutive lanes. E and F, where indicated, cells were treated for 16 h with 20 μm EIPA, an NHE1 inhibitor, and Hyal2 was then detected by Western blot in the cell lysate (E) and in CD44 immunoprecipitates (F). In the latter case, Western blot detection of CD44 in the precipitates was used as a positive control. G, confocal microscopy images of immunofluorescent detection of EGFP-Hyal2 (anti-GFP antibodies) transfected in BB16mock cells, Hyal2 (P16 antibodies) in BB16Hy2+ cells, and CD44 (OX49 antibodies) in both cell types. In the overlays, co-localization of CD44 with either Hyal2 or EGFP-Hyal2 is revealed by the yellow color.
Hyal2 and the Loss of Pericellular Coat Induce CD44 Shedding
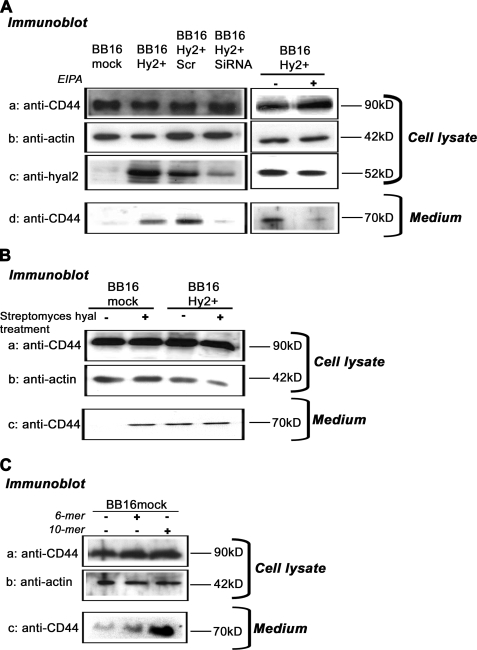
HA oligosaccharides are known to prevent polymeric HA binding to CD44 and to remove the pericellular coat (as shown in Table 3), like Hyal2. HA oligosaccharides also induce CD44 cleavage and shedding into the extracellular milieu (39). Thus, we examined CD44 cleavage in BB16Hy2+ cells. We observed a large amount of immunoreactive CD44 in the medium of BB16Hy2+ cells but not in BB16mock cells or Hyal2 siRNA-transfected cells (Fig. 5A). CD44 shedding in BB16Hy2+ cells was suppressed by treatment with EIPA (Fig. 5A) and was mimicked by coat disruption using Streptomyces hyaluronidase (Fig. 5B) or HA oligomers, in particular the 10-mer form (Fig. 5C).
FIGURE 5.
Shedding of CD44. A, CD44 was detected in Western blots (HCAM antibody) of cell lysates at a molecular mass of ∼90 kDa and in 10-fold concentrated medium at a molecular mass of ∼70 kDa. The presence of CD44 in the medium is a sign of shedding. Actin and Hyal2 detections were used as controls of loading and efficiency of siRNA transfection, respectively. In some experiments, BB16Hy2+ cells were transfected with a scrambled RNA or Hyal2 siRNA, and in other experiments they were treated with 20 μm ethylisopropylamiloride (EIPA) for 16 h. B and C, detection of CD44 shedding in cells treated with 5 units/ml Streptomyces hyaluronidase for 1 h (B), or with 100 μg/ml 6-mer or 10-mer HA oligosaccharides for 16 h (C).
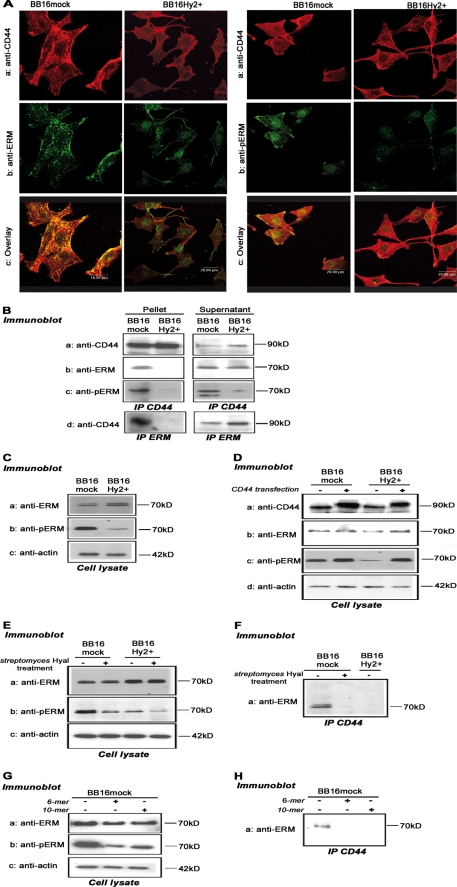
Hyal2 and the Loss of Pericellular Coat Sever the CD44-ERM Linkage and Reduce Levels of ERM
To determine if the effects of Hyal2 on CD44 (decreases in HA binding capacity, increases in shedding) were reflected in changes in intracellular CD44 associations, we examined ERM, one of the major CD44 partners, which links it to the actin cytoskeleton (40, 41). First, using confocal microscopy, ERM co-localized with CD44 in BB16mock but not in BB16Hy2+ cells (Fig. 6A). CD44 was detected both in the plasma membrane and in cytoplasmic vesicles; this pattern was unaffected by Hyal2 overexpression (Fig. 6A). The active form of ERM, i.e. phosphorylated ERM (pERM), also co-localized with CD44 in BB16mock cells. This overlap was difficult to discern in BB16Hy2+ cells, because the total amount of immunoreactive pERM was quite low (Fig. 6A).
FIGURE 6.
The Hyal2-CD44-ERM interaction. A, representative confocal microscopic images of immunofluorescent CD44 (red, OX49 antibody), ERM, or phosphorylated ERM (pERM, green), and their co-localization (yellow in the overlay images). B, co-immunoprecipitation of CD44 (OX49 antibody) and ERM with each other and with pERM. Supernatants of the antibody-antigen-beads complex are shown as controls. C, Western blots of ERM, pERM, and actin in cell lysates (5 μg of protein). D, the same blots, as well as CD44 detection, were repeated 24 h after transfection of a rat CD44 cDNA and compared with untransfected controls. E and F, Western blots of ERM, pERM, and actin in cell lysates, and detection of pERM in CD44 immunoprecipitates, following 1-h treatment with 5 units/ml Streptomyces hyaluronidase or vehicle. G and H, same detection steps following 16-h treatment with 100 μg/ml 6-mer and 10-mer HA oligosaccharides.
The most obvious conclusion from these observations is that Hyal2 disconnects ERM from CD44 and reduces the basal activation level of ERM, i.e. in the absence of added factors. This was confirmed by immunoprecipitation experiments. Almost no ERM or pERM could be detected in CD44 precipitates in BB16Hy2+ cells, contrary to an effective ERM-CD44 co-immunoprecipitation in BB16mock cells (Fig. 6B). Accordingly, CD44 was lost from ERM precipitates in BB16Hy2+ cells compared with mock cells (Fig. 6B). The greatly reduced pERM/ERM ratio in BB16Hy2+ cells suggested by immunofluorescence microscopy was confirmed in Western blots of cell lysates (Fig. 6C). Interestingly, this effect could be entirely rescued by transient transfection of CD44 in BB16Hy2+ cells (Fig. 6D), supporting the view that Hyal2 prevented ERM phosphorylation by acting on CD44. We invoked the possibility that this effect of Hyal2 was linked to disappearance of the pericellular coat. Therefore, BB16 cells were treated with Streptomyces hyaluronidase in order for them to shed their coat. Under those conditions, the cellular amount of immunoreactive pERM, but not ERM, decreased markedly (Fig. 6E), and ERM vanished from CD44 immunoprecipitates in BB16mock cells (Fig. 6F). The effect of exogenous hyaluronidase could be mimicked by HA oligomers (Fig. 6G). We concluded that an effective pericellular coat is needed for basal activation/phosphorylation of ERM through the intervention of CD44.
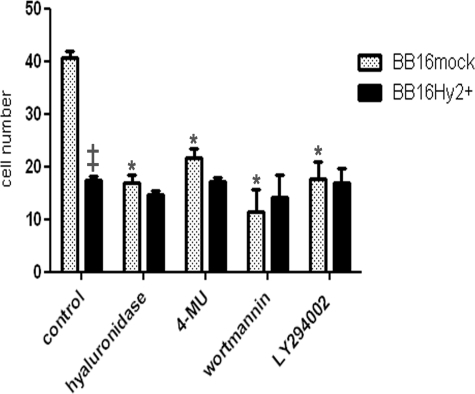
Hyal2 and the Loss of Pericellular Coat Reduce Cell Motility
One of the major consequences of a loss of pericellular coat is a reduction in cell motility, as shown previously in, e.g. vascular smooth muscle cells (22), proximal tubular epithelial cells (42), and prostate cancer cells (43). ERM phosphorylation has also been linked to increased movement, especially in T cells (44) and fibroblasts (45). Therefore, we measured cell motility in BB16mock and BB16Hy2+ cells: motility was reduced by >50% in the latter (Fig. 7). Treatment of BB16mock cells with hyaluronidase or 4-MU, which reduce the pericellular coat in the same proportion as does Hyal2 overexpression (see Table 3), had the same inhibitory effects on motility (Fig. 7). As expected, the PI3K inhibitors wortmannin and LY294002 also reduced BB16mock cell motility (Fig. 7) but without significant effect on pericellular coat (data not shown).
FIGURE 7.
Wound healing assays. Cells were grown to confluency, then induced to re-populate a wound created by a sterile razor blade. To prevent growth during migration, cells were pre-treated with 4.5 mm mitomycin C for 2 h. After band-stripping, they were allowed to migrate into the wound for 6 h in medium containing 10% fetal calf serum alone or supplemented with 5 units/ml Streptomyces hyaluronidase, a HA synthase inhibitor (4-MU at 100 μm), or PI3K inhibitors (50 nm wortmannin or 5 μm LY294002). Results are presented as means ± S.E. of the number of cells that had colonized the wound margin in random microscopic fields (*, p < 0.001 versus untreated BB16mock cells, n = 5 in each group; ‡, p < 0.001 versus BB16mock cells, n = 5).
DISCUSSION
This study explored the action of Hyal2 at the cellular level through a direct comparison of stable clones of the rat fibroblastic cell line BB16 with either low or high expression of Hyal2. Most observations in this report stem from BB16mock and BB16Hy2+ cells, which were transfected with an empty vector and a vector containing the full-length rat Hyal2 cDNA, respectively. Similar results were obtained using other clones overexpressing Hyal2 at various levels (Figs. 1 and 2 and supplemental Fig. S1). BB16mock cells did not differ in any way from the parent cell line. The behavior of BB16Hy2+ cells transiently transfected with Hyal2 siRNA reverted to that of the parent cell line or to BB16mock cells (Table 3, Fig. 6, and data not shown), bringing further support to the specificity and relevance of the observations presented in the current report. BB16 cells are well suited for a direct analysis of the action Hyal2: they have a weak basal hyaluronidase activity and display a large pericellular coat.
What is the cellular localization of Hyal2 in these cells? Although Hyal2 was accessible to biotin labeling at the cell surface (Fig. 1E), a large proportion of it was present within the cell, similar to CD44, suggesting rapid shuttling of Hyal2 and CD44 molecules between the plasma membrane and some as yet unidentified intracellular vesicles. A comparable pattern of expression of CD44 has been described in cells with a high level of HA uptake, such as alveolar macrophages (46).
The main result of the current study is the disappearance of the pericellular coat, or glycocalyx, in proportion to Hyal2 overexpression in BB16 cells. Endogenous Hyal2 was as effective as pharmacological interventions known to remove the pericellular coat, such as treatments with (a) HA-specific bacterial hyaluronidase, which cleaves HA molecules into small oligosaccharides, (b) 6-mer and 10-mer oligosaccharides, or (c) inhibitors of HA synthesis such as 4-MU (21, 22, 47). These treatments displace polymeric HA from its receptors. Hyal2, which can be up- or down-regulated in some inflammatory conditions or silenced in cancer cells, thus appears to constitute a key physiological control mechanism for the stability of the cell glycocalyx.
Hyal2 may prevent coat formation through different mechanisms. The two most obvious ones are enzymatic degradation of HA and interference with CD44-HA binding. Both were explored.
First, our study did not detect a well defined enzymatic activity for Hyal2. Previous studies have shown that a soluble form of recombinant Hyal2 has weak hyaluronidase activity at pH <4.0 (8) and that the enzymatic action of Hyal2 measured in cell or membrane preparations requires the presence of CD44 (10, 17). Although we did not measure any difference in the hyaluronidase activity of cell extracts, including cell membrane fractions, from BB16 versus Hyal2-overexpressing clones, we cannot exclude the possibility that Hyal2 has a highly localized HA-degrading activity, reducing the mean size of HA molecules that are attached specifically to the cell surface. This type of attachment requires HA synthases, HA receptors, aggregating proteoglycans, and covalent HA-protein linkages (42, 48, 49). Hyal2 itself may be part of this macromolecular complex. We were not able to measure a difference in size of HA molecules at the cell surface (data not shown) but were able to document that Hyal2 expression induced no change (no decrease) in global HA synthesis (Table 2) or in the mRNA levels of HA synthases measured using real-time reverse transcription-PCR (data not shown).
Second, we examined the Hyal2-CD44 connection, after confirmation that CD44 is a necessary component for coat formation in BB16 cells (Fig. 3 and Table 3). BB16 and BB16Hy2+ cells expressed standard CD44 (CD44s) but not alternatively spliced variant isoforms. Hyal2 was a strong partner of CD44: this was shown by co-immunoprecipitation, microscopic co-localization, and functional changes in CD44. Neither the total amount of CD44, nor the presence of variants, post-translational modification, or cellular localization was affected by Hyal2 overexpression. Nevertheless, CD44 lost half of its capacity to bind exogenous HA (Tables 4 and 5) and most of its intracellular connections with ERM (Fig. 6). The loss of HA binding, perhaps in association with direct effects of Hyal2 on the pericellular HA mesh, may explain the breakup of the pericellular coat. An important argument in this regard is the almost complete rescue of the glycocalyx in BB16Hy2+ cells following transient transfection of CD44 (Fig. 3C and Table 3). This would not be expected to occur if Hyal2 was to break up the coat purely through an enzymatic activity. Thus, we favor the hypothesis that the coat-disrupting effect of Hyal2 is mediated primarily through its interaction with CD44.
The loss of CD44-ERM connection in BB16Hy2+ cells was explored further. ERM binds to the cytoplasmic tail of CD44 (50) where it can be phosphorylated (51). This is followed by actin reorganization and increased cell motility and invasion (40). Perturbation of CD44-ERM interaction using actin or kinase inhibitors reduces HA binding in myeloid cells (41). We found that BB16 and BB16mock cells have a high proportion of pERM at baseline; this is apparently due to constitutive src activation and to a high level of expression of CD44. Hyal2 markedly reduced the amount of pERM (Fig. 6) and, accordingly, decreased BB16 cell motility by ∼50%, a level equivalent to that obtained with typical PI3K inhibitors like wortmannin and LY294002 (Fig. 7). Although maintenance of an effective CD44-ERM connection and persistence of high levels of pERM required the presence of a pericellular coat, the inhibitory effect of Hyal2 on ERM phosphorylation and on motility seemed to be transduced by its interaction with CD44, as shown by phenotypic rescue using CD44 overexpression (Fig. 6D).
Another effect of Hyal2 on CD44 was increased cleavage and shedding. This phenomenon has been described mostly in cancer cells, where it can be induced by HA oligosaccharides. It seems to play a critical role in tumor cell migration (39, 52). CD44 proteolytic cleavage can also occur in fibroblasts and monocytes following anti-CD44 addition (53), but its physiological role in inflammatory cells remains unclear (54). In the current study, CD44 shedding was induced by Hyal2 and by a loss of pericellular coat, and was linked to reduced cell motility. CD44 shedding could thus be considered as another sign of CD44 instability instead of a tumor promoting effect.
In the current study, the NHE1 inhibitor EIPA was able to protect the pericellular coat from Hyal2, to prevent CD44 shedding, and to decrease the level of Hyal2-CD44 co-immunoprecipitation. In previous studies, EIPA was shown to block the enzymatic action of Hyal2 at the cell surface (17). We surmise that acid foci, which may be located below the HA coat or in caveolae-like structures, may be required for Hyal2 to act or to interact with other transmembrane molecules such as CD44 (55).
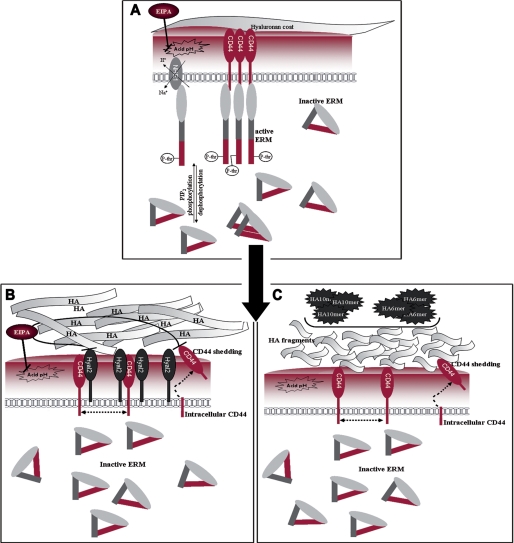
Overall, the mechanism underlying the effects of Hyal2 on CD44 remains speculative. One attractive possibility is a de-clustering of CD44 molecules following association with Hyal2. CD44 clustering or dimerization seems required for HA binding in some cell types, particularly in lymphocytes, as demonstrated by, e.g. the effect of multivalent but not monovalent CD44 antibodies (56, 57). CD44 clustering may also be required for ERM activation (41). Although we were not able to measure CD44 clusters in BB16 cells, the hypothesis that Hyal2 separates CD44 molecules at the cell surface may be validated in further studies. We speculate that this effect leads to loss of pericellular coat and lack of ERM activation. Our views are summarized in Fig. 8. At the end of the chain, the inhibitory effect of Hyal2 on cell motility may be relevant to its purported in vivo tumor-suppressive activity (14). Cancer cells deprived of their pericellular coat through hyaluronidase or HA oligosaccharide treatments or through inhibition of hyaluronan export lose much of their capacity to metastasize (58–61). Hyal2 may function similarly.
FIGURE 8.
Model of a novel interaction: CD44-Hyal2-ERM. A, basal situation in BB16 and BB16mock cells; EIPA = ethylisopropylamiloride, an inhibitor of the sodium-hydrogen exchanger-1 (NHE1). B, in the presence of high levels of Hyal2. C, in the presence of HA oligosaccharides. In B and C, HA fragments have been generated, CD44 molecules are de-clustered, and some of them are cleaved.
In summary, we suggest that Hyal2 is a critical modulator of the way cells interact with their microenvironment through their coat/glycocalyx and is involved in regulating their motility. Most probably this occurs through a direct interaction between Hyal2 and the main HA receptor, CD44, disturbing its intracellular link with ERM. Our results also support the growing speculation that Hyal2 as well as the other hyaluronidases have functions other than or in addition to their enzymatic activity. Additionally, it may be that Hyal3 (for which only one citation is available for any enzymatic activity (62)) and Hyal4 (for which no activity has ever been described) are hyaluronidase-like proteins for which non-enzymatic functions predominated in the course of the evolutionary development of this complex protein family.
Supplementary Material
Acknowledgments
We thank Prof. Pierre Courtoy (Institute of Cellular Pathology, Brussels, Belgium) for his kind gift of BB16 cells and advice, Dr. Ove Wik (Amersham Biosciences) for his gift of purified HA samples, and Seikagaku Corp. for their gift of HA oligosaccharides. We are very grateful to Prof. Robert Stern (University of California, San Francisco, CA) for his many advices and help with the manuscript.
This work was supported by The Academy of Finland, the Sigrid Juselius Foundation, and the EVO Funds of Kuopio University Hospital.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
C. Duterme, J. Mertens-Strijthagen, M. Tammi, and B. Flamion, unpublished data.
- HA
- hyaluronan
- ERM
- ezrin-radixin-moesin
- pERM
- phosphorylated ERM
- EIPA
- ethyl-N-isopropylamiloride
- 4-MU
- 4-methylumbelliferone
- PBS
- phosphate-buffered saline
- PI3K
- phosphoinositide 3-kinase
- NHE1
- sodium-proton exchanger-1.
REFERENCES
- 1.Fraser J. R., Laurent T. C., Laurent U. B. (1997) J. Intern. Med. 242, 27–33 [DOI] [PubMed] [Google Scholar]
- 2.Kreil G. (1995) Protein Sci. 4, 1666–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Csoka A. B., Frost G. I., Stern R. (2001) Matrix Biol. 20, 499–508 [DOI] [PubMed] [Google Scholar]
- 4.Csóka A. B., Scherer S. W., Stern R. (1999) Genomics 60, 356–361 [DOI] [PubMed] [Google Scholar]
- 5.Atmuri V., Martin D. C., Hemming R., Gutsol A., Byers S., Sahebjam S., Thliveris J. A., Mort J. S., Carmona E., Anderson J. E., Dakshinamurti S., Triggs-Raine B. (2008) Matrix Biol 27, 653–660 [DOI] [PubMed] [Google Scholar]
- 6.Martin D. C., Atmuri V., Hemming R. J., Farley J., Mort J. S., Byers S., Hombach-Klonisch S., Csoka A. B., Stern R., Triggs-Raine B. L. (2008) Hum. Mol. Genet. 17, 1904–1915 [DOI] [PubMed] [Google Scholar]
- 7.Jadin L., Wu X., Ding H., Frost G. I., Onclinx C., Triggs-Raine B., Flamion B. (2008) FASEB J. 22, 4316–4326 [DOI] [PubMed] [Google Scholar]
- 8.Vigdorovich V., Miller A. D., Strong R. K. (2007) J. Virol. 81, 3124–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lepperdinger G., Strobl B., Kreil G. (1998) J. Biol. Chem. 273, 22466–22470 [DOI] [PubMed] [Google Scholar]
- 10.Harada H., Takahashi M. (2007) J. Biol. Chem. 282, 5597–5607 [DOI] [PubMed] [Google Scholar]
- 11.Stern R., Asari A. A., Sugahara K. N. (2006) Eur. J. Cell Biol. 85, 699–715 [DOI] [PubMed] [Google Scholar]
- 12.Jiang D., Liang J., Noble P. W. (2007) Annu. Rev. Cell Dev. Biol. 23, 435–461 [DOI] [PubMed] [Google Scholar]
- 13.Rai S. K., Duh F. M., Vigdorovich V., Danilkovitch-Miagkova A., Lerman M. I., Miller A. D. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 4443–4448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang F., Grigorieva E. V., Li J., Senchenko V. N., Pavlova T. V., Anedchenko E. A., Kudryavtseva A. V., Tsimanis A., Angeloni D., Lerman M. I., Kashuba V. I., Klein G., Zabarovsky E. R. (2008) PLoS ONE 3, e3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsu L. J., Schultz L., Hong Q., Van Moer K., Heath J., Li M. Y., Lai F. J., Lin S. R., Lee M. H., Lo C. P., Lin Y. S., Chen S. T., Chang N. S. (2009) J. Biol. Chem. 284, 16049–16059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novak U., Stylli S. S., Kaye A. H., Lepperdinger G. (1999) Cancer Res. 59, 6246–6250 [PubMed] [Google Scholar]
- 17.Bourguignon L. Y., Singleton P. A., Diedrich F., Stern R., Gilad E. (2004) J. Biol. Chem. 279, 26991–27007 [DOI] [PubMed] [Google Scholar]
- 18.Udabage L., Brownlee G. R., Nilsson S. K., Brown T. J. (2005) Exp. Cell Res. 310, 205–217 [DOI] [PubMed] [Google Scholar]
- 19.Conn E. M., Madsen M. A., Cravatt B. F., Ruf W., Deryugina E. I., Quigley J. P. (2008) J. Biol. Chem. 283, 26518–26527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clarris B. J., Fraser J. R. (1968) Exp. Cell Res. 49, 181–193 [DOI] [PubMed] [Google Scholar]
- 21.Knudson W., Bartnik E., Knudson C. B. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 4003–4007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evanko S. P., Angello J. C., Wight T. N. (1999) Arterioscler. Thromb. Vasc. Biol. 19, 1004–1013 [DOI] [PubMed] [Google Scholar]
- 23.Veithen A., Cupers P., Baudhuin P., Courtoy P. J. (1996) J. Cell Sci. 109, 2005–2012 [DOI] [PubMed] [Google Scholar]
- 24.Miranda M., Sorkina T., Grammatopoulos T. N., Zawada W. M., Sorkin A. (2004) J. Biol. Chem. 279, 30760–30770 [DOI] [PubMed] [Google Scholar]
- 25.Goldberg R. L., Toole B. P. (1984) Exp. Cell Res. 151, 258–263 [DOI] [PubMed] [Google Scholar]
- 26.Lee H. G., Cowman M. K. (1994) Anal. Biochem. 219, 278–287 [DOI] [PubMed] [Google Scholar]
- 27.Underhill C. B., Chi-Rosso G., Toole B. P. (1983) J. Biol. Chem. 258, 8086–8091 [PubMed] [Google Scholar]
- 28.de Belder A. N., Wik K. O. (1975) Carbohydr. Res. 44, 251–257 [DOI] [PubMed] [Google Scholar]
- 29.André F., Rigot V., Remacle-Bonnet M., Luis J., Pommier G., Marvaldi J. (1999) Gastroenterology 116, 64–77 [DOI] [PubMed] [Google Scholar]
- 30.Platek A., Mettlen M., Camby I., Kiss R., Amyere M., Courtoy P. J. (2004) J. Cell Sci. 117, 4849–4861 [DOI] [PubMed] [Google Scholar]
- 31.Chow G., Knudson C. B., Knudson W. (2006) Osteoarthritis Cartilage 14, 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang N. S. (2002) BMC Cell Biol. 3, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu Q., Banerjee S. D., Toole B. P. (1992) Dev. Dyn. 193, 145–151 [DOI] [PubMed] [Google Scholar]
- 34.Sleeman J., Rudy W., Hofmann M., Moll J., Herrlich P., Ponta H. (1996) J. Cell Biol. 135, 1139–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lokeshwar V. B., Bourguignon L. Y. (1991) J. Biol. Chem. 266, 17983–17989 [PubMed] [Google Scholar]
- 36.Skelton T. P., Zeng C., Nocks A., Stamenkovic I. (1998) J. Cell Biol. 140, 431–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maiti A., Maki G., Johnson P. (1998) Science 282, 941–943 [DOI] [PubMed] [Google Scholar]
- 38.Cichy J., Pure E. (2000) J. Biol. Chem. 275, 18061–18069 [DOI] [PubMed] [Google Scholar]
- 39.Sugahara K. N., Murai T., Nishinakamura H., Kawashima H., Saya H., Miyasaka M. (2003) J. Biol. Chem. 278, 32259–32265 [DOI] [PubMed] [Google Scholar]
- 40.Tsukita S., Oishi K., Sato N., Sagara J., Kawai A., Tsukita S. (1994) J. Cell Biol. 126, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown K. L., Birkenhead D., Lai J. C., Li L., Li R., Johnson P. (2005) Exp. Cell Res. 303, 400–414 [DOI] [PubMed] [Google Scholar]
- 42.Selbi W., Day A. J., Rugg M. S., Fülöp C., de la Motte C. A., Bowen T., Hascall V. C., Phillips A. O. (2006) J. Am. Soc. Nephrol. 17, 1553–1567 [DOI] [PubMed] [Google Scholar]
- 43.Ricciardelli C., Russell D. L., Ween M. P., Mayne K., Suwiwat S., Byers S., Marshall V. R., Tilley W. D., Horsfall D. J. (2007) J. Biol. Chem. 282, 10814–10825 [DOI] [PubMed] [Google Scholar]
- 44.Lee J. H., Katakai T., Hara T., Gonda H., Sugai M., Shimizu A. (2004) J. Cell Biol. 167, 327–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsuda M., Makino Y., Iwahara T., Nishihara H., Sawa H., Nagashima K., Hanafusa H., Tanaka S. (2004) J. Biol. Chem. 279, 46843–46850 [DOI] [PubMed] [Google Scholar]
- 46.Culty M., Nguyen H. A., Underhill C. B. (1992) J. Cell Biol. 116, 1055–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Webber J., Meran S., Steadman R., Phillips A. (2009) J. Biol. Chem. 284, 9083–9092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Knudson C. B., Knudson W. (1993) FASEB J. 7, 1233–1241 [PubMed] [Google Scholar]
- 49.Evanko S. P., Tammi M. I., Tammi R. H., Wight T. N. (2007) Adv. Drug Deliv. Rev. 59, 1351–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mori T., Kitano K., Terawaki S., Maesaki R., Fukami Y., Hakoshima T. (2008) J. Biol. Chem. 283, 29602–29612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yonemura S., Tsukita S., Tsukita S. (1999) J. Cell Biol. 145, 1497–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Okamoto I., Kawano Y., Tsuiki H., Sasaki J., Nakao M., Matsumoto M., Suga M., Ando M., Nakajima M., Saya H. (1999) Oncogene 18, 1435–1446 [DOI] [PubMed] [Google Scholar]
- 53.Shi M., Dennis K., Peschon J. J., Chandrasekaran R., Mikecz K. (2001) J. Immunol. 167, 123–131 [DOI] [PubMed] [Google Scholar]
- 54.Nagano O., Saya H. (2004) Cancer Sci. 95, 930–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stern R. (2003) Glycobiology 13, 105R–115R [DOI] [PubMed] [Google Scholar]
- 56.Lesley J., Kincade P. W., Hyman R. (1993) Eur. J. Immunol. 23, 1902–1909 [DOI] [PubMed] [Google Scholar]
- 57.Liu D., Liu T., Li R., Sy M. S. (1998) Front. Biosci. 3, d631–d636 [DOI] [PubMed] [Google Scholar]
- 58.Simpson M. A., Wilson C. M., Furcht L. T., Spicer A. P., Oegema T. R., Jr., McCarthy J. B. (2002) J. Biol. Chem. 277, 10050–10057 [DOI] [PubMed] [Google Scholar]
- 59.Draffin J. E., McFarlane S., Hill A., Johnston P. G., Waugh D. J. (2004) Cancer Res. 64, 5702–5711 [DOI] [PubMed] [Google Scholar]
- 60.Hosono K., Nishida Y., Knudson W., Knudson C. B., Naruse T., Suzuki Y., Ishiguro N. (2007) Am. J. Pathol. 171, 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Monz K., Mass-Kück K., Schumacher U., Schulz T., Hallmann R., Schnäker E. M., Schneider S. W., Prehm P. (2008) J. Cell. Biochem. 105, 1260–1266 [DOI] [PubMed] [Google Scholar]
- 62.Lokeshwar V. B., Schroeder G. L., Carey R. I., Soloway M. S., Iida N. (2002) J. Biol. Chem. 277, 33654–33663 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.