Abstract
CTP:phosphocholine cytidylyltransferase (CCT) is the key regulatory enzyme in the synthesis of phosphatidylcholine, the most abundant phospholipid in eukaryotic cell membranes. The CCT-catalyzed transfer of a cytidylyl group from CTP to phosphocholine to form CDP-choline is regulated by a membrane lipid-dependent mechanism imparted by its C-terminal membrane binding domain. We present the first analysis of a crystal structure of a eukaryotic CCT. A deletion construct of rat CCTα spanning residues 1–236 (CCT236) lacks the regulatory domain and as a result displays constitutive activity. The 2.2-Å structure reveals a CCT236 homodimer in complex with the reaction product, CDP-choline. Each chain is composed of a complete catalytic domain with an intimately associated N-terminal extension, which together with the catalytic domain contributes to the dimer interface. Although the CCT236 structure reveals elements involved in binding cytidine that are conserved with other members of the cytidylyltransferase superfamily, it also features nonconserved active site residues, His-168 and Tyr-173, that make key interactions with the β-phosphate of CDP-choline. Mutagenesis and kinetic analyses confirmed their role in phosphocholine binding and catalysis. These results demonstrate structural and mechanistic differences in a broadly conserved protein fold across the cytidylyltransferase family. Comparison of the CCT236 structure with those of other nucleotidyltransferases provides evidence for substrate-induced active site loop movements and a disorder-to-order transition of a loop element in the catalytic mechanism.
Introduction
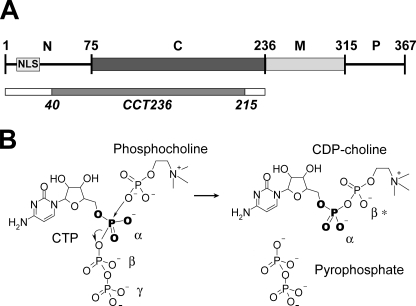
A key rate-limiting step in the synthesis of phosphatidylcholine in animal cells is the formation of the headgroup donor, CDP-choline, by transfer of a cytidylyl group from CTP to phosphocholine. This reaction is catalyzed by CTP:phosphocholine cytidylyltransferase (CCT4; EC 2.7.7.15), an enzyme subject to many layers of regulation (1–4). The ubiquitous and best studied isoform of mammalian CCT (CCTα, 367 residues) has been described as having four domains (Fig. 1A). An N-terminal domain (∼75 residues) housing its nuclear localization signal (NLS) sequence is followed by an ∼150-residue catalytic domain, an ∼60-residue membrane binding domain (domain M), and an unstructured phosphorylated tail (∼50 residues) (2, 4). CCT functions as a homodimer (5).
FIGURE 1.
Domains/regions of full-length CCTα and the CCT-catalyzed reaction. A, CCTα isoform consists of four domains or regions as follows: the N-terminal region (N; 1–75) with the NLS, the catalytic domain (C; 76–236), the membrane binding domain (M; 237–314), and the phosphorylation region (P; 315–367). The domain/region boundaries are approximate. The CCT236 construct is shown as a bar, and the region revealed in the crystal structure (40–215, chain A) is in gray. B, CCT-catalyzed mechanism for CDP-choline synthesis from CTP and phosphocholine. The nucleophilic phosphocholine phosphate becomes the β*-phosphate of the product.
CCT activation requires transformation of the enzyme from a soluble form to a membrane lipid-bound form. When the full-length soluble CCT interacts with anionic membrane surfaces, domain M transforms from a mixture of structural elements into an amphipathic α-helix (6–8). Domain M appears to act as an autoinhibitory device, whose interaction with phosphatidylcholine-deficient membranes releases an inhibitory constraint at the active site to enhance kcat by 2 orders of magnitude (9). The primary evidence for this model is the constitutive activity of a construct lacking domain M, CCT236 (9).
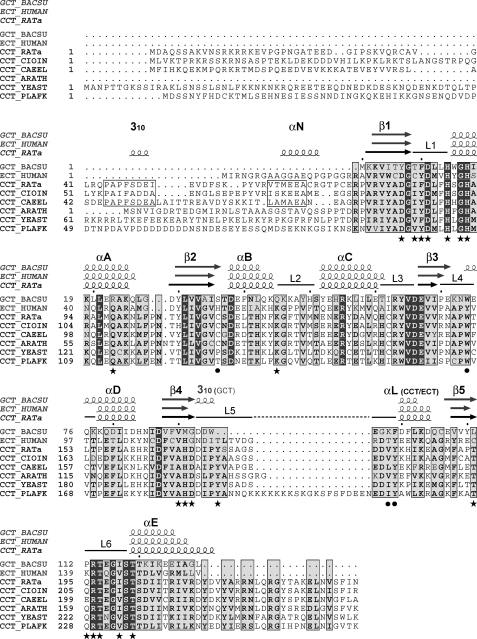
To elucidate the mechanism whereby membrane binding activates this important regulatory enzyme, we must first understand how the active form of the enzyme accomplishes catalysis. The cytidylyl transfer reaction catalyzed by CCT involves direct attack at the α-phosphate of CTP by a phosphoryl oxygen of phosphocholine acting as the nucleophile to displace the β,γ-phosphates (pyrophosphate) of CTP and form the product CDP-choline (Fig. 1B). Many aspects of this reaction mechanism resemble those of other adenylyl- and cytidylyltransferases, in which the transition state involves conversion of tetrahedral geometry at the α-phosphate to a planar bipyrimidal penta-coordinate geometry (10–12). One well characterized cytidylyltransferase with a solved structure is glycerol-phosphate cytidylyltransferase (GCT), which catalyzes the synthesis of CDP-glycerol, an intermediate in techoic acid synthesis for bacterial cell walls. Both GCT from Bacillus subtilis and CCTα from rat show random order (ternary) kinetics and have similar kcat and Km values (9, 13, 14). The catalytic domain of CCT has 34% sequence identity to GCT overall and very strong homology in several regions (Fig. 2). Crystal structures of GCT with bound CTP (PDB code 1COZ) (15) or with bound CDP-glycerol (PDB code 1N1D) (16) reveal a dimer of an α/β nucleotide binding fold composed of a twisted five-stranded parallel β-sheet flanked by five helices. The ligands are found in a pocket at the base of the β-sheet, in contact with the conserved HXGH and RTEGISTT motifs that are signatures of the GCT family of nucleotidyltransferases (15). This family includes ethanolamine-phosphate cytidylyltransferases (ECT), phosphopantetheine adenylyltransferase (PPAT), and the class I aminoacyl tRNA synthetases (aa-tRS) (10, 15–17). Multiple solved structures of PPAT and aa-tRS enzymes have been analyzed, and one as yet unanalyzed structure of human ECT was recently deposited in the Protein Data Bank (code 3ELB). The sequence similarities between GCT, ECT, and CCT imply a highly conserved catalytic domain structure. However, the presence of additional domains in CCT, the fact that the catalytic domain of CCT is regulated whereas GCT and ECT are not, and the wide variability in specific folding elements between members of this family (17) gave reasons to suspect important structural differences between them.
FIGURE 2.
Alignment of sequences and secondary structure elements of the cytidylyltransferase family. The aligned sequences are as follows: GCT from B. subtilis (P27623); domains N and C of CCT from Rattus norvegicus (P19836), Ciona intestinalis (XP 002130773) (This sequence is a translation from the Ciona genome. The others are from cloned cDNAs.), Caenorhabditis elegans (P49583), Arabidopsis thaliana (Q42555), Saccharomyces cerevisiae (P13259), and Plasmodium falciparum (P49587); ECT (residues 1–156) from Homo sapiens (Q99447). The alignment was generated by ClustalW2 (62) and then manually adjusted. Residues within a pale gray box are conserved 6/8 times, and residues within a dark gray box are identical. Open boxes surround two sequence elements in segment N of the animal CCTs that are conserved. The secondary structure elements observed in GCT (PDB code 1N1D, gray) and CCT236 (this paper; black) are indicated above the sequence alignment. The layout with secondary structure elements was generated with Espript 2.2 (63). The active site residues involved in CDP coordination are marked with stars, and residues surrounding the choline are marked with circles.
As a first step in unraveling the mechanism of autoregulation of CCT, we solved the structure of a soluble CCT fragment (CCT236) containing the N-terminal and catalytic domains (domains N and C) by x-ray crystallography at 2.2 Å resolution. This structure shows an active conformation of domain C in complex with its reaction product, CDP-choline. The C-terminal region of domain N is intimately associated with domain C and makes contacts across the dimer interface. Although the fold of domain C is closely related to that of GCT, the structure reveals two novel residues, His-168 and Tyr-173, at the active site that coordinate with the phosphocholine phosphate. These residues were not suspected to be involved in catalysis from the sequence alignment alone. Analyses of alanine substitutions at these sites confirmed their role in phosphocholine binding and efficient catalysis.
EXPERIMENTAL PROCEDURES
Baculovirus Expression and Purification of CCT236
An untagged version of residues 1–236 of rat CCTα (CCT236) was expressed in T.ni cells using baculovirus as described (8, 9). After CM-Sepharose and Blue-Sepharose chromatography, the purified protein was concentrated to 10 mg/ml using an Amicon Ultra-Spin filter, spun at 2800 × g. The protein was then subjected to gel filtration using a Sephacryl S-200 column (∼76-ml bead volume) equilibrated with 10 mm Tris-HCl (pH 7.4), 1 mm EDTA, 150 mm NaCl, 2 mm DTT. Fractions containing the purified CCT236 were pooled and concentrated to 12 mg/ml (0.45 mm) using an Amicon Ultra-Spin filter. CDP-choline was added to a final concentration of 20 mm. Approximately 2 mg of pure CCT236 was obtained per liter of cell culture suspension. The calculated molecular mass of this construct is 26,755 Da, and its sequence is the first 236 residues of UniProt accession number P19836.
E. coli Expression and Purification of Selenomethionine-incorporated CCT236
The cDNA encoding CCT236 was transferred from the baculovirus shuttle vector into pGEX6p1 via NcoI and BamHI cloning sites, to create a fusion protein with a PreScission protease-cleavable glutathione S-transferase tag. The pGEX6p1-CCT236 plasmid was transformed into the BL21-derived Rosetta cell strain (Novagen) for protein expression (18). The overexpression protocol for selenomethionine-incorporated protein was based on the method of Doublié (19). Minimal media (1 liter) were inoculated with Rosetta cells from a 10-ml overnight culture and grown at 37 °C for 17 h. A mixture of amino acids was added to reach final concentrations of selenomethionine, isoleucine, leucine, and valine (each 60 mg/liter) and lysine, phenylalanine, and threonine (each 100 mg/liter). Protein expression was induced with 0.5 mm isopropylthiogalactoside for ∼20 h at 28 °C. The cells were harvested by centrifugation at 2800 × g for 15 min and stored at −80 °C. Thawed cells were resuspended in 40 ml of phosphate-buffered saline (pH 7.4), 1 mm phenylmethylsulfonyl fluoride, 0.2 mg/ml lysozyme, 0.1 mg/ml DNase, and 1 mm DTT and were incubated for 1 h on ice. The lysed cells were cycled through a high pressure homogenizer (Avestin). The homogenate was clarified by centrifugation at 27,000 × g for 30 min.
The selenomethionine-substituted GST-CCT236 was purified by glutathione-agarose affinity and size-exclusion chromatography. The glutathione beads (0.8 ml) were equilibrated in phosphate-buffered saline with 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, 1 mm DTT. The clarified lysate was bound to beads with gentle mixing for 3 h at 4 °C. The bound fusion protein was collected by centrifugation at 460 × g for 3 min. The beads were washed three times with the equilibration buffer. The beads were resuspended in cleavage buffer (50 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1 mm DTT), and CCT236 was released from the matrix by incubation with the PreScission protease (Amersham Biosciences) overnight at 4 °C with gentle mixing. The cleaved CCT236 was applied to a Superdex 200 HR 10/30 column (GE Healthcare), equilibrated, and eluted with 10 mm Tris-HCl (pH 7.4), 1 mm EDTA, 150 mm NaCl, 2 mm DTT. The pure fractions were pooled and concentrated using an Amicon Ultra-Spin Filter at 2800 × g, and a 10-fold molar excess of CDP-choline was added. The final protein concentration used for crystallization was 13 mg/ml (0.5 mm) with 5 mm CDP-choline. This purified CCT contains residues 1–236 of rat CCTα with an N-terminal linker (GPLGSA). The specific activities of untagged CCT236 versus CCT236 with the N-terminal linker were the same (2583 ± 377 units/mg for untagged CCT236 (n = 10) and 2783 ± 260 units/mg (n = 4) for CCT236 with the linker). Matrix-assisted laser desorption ionization time-of-flight mass spectrometry gave a mass of 27,396 Da, consistent with selenomethionine incorporation into four of the five possible sites, on average. Purified CCTs were stored at −80 °C.
CCT236 Crystallization and Diffraction Data Collection
The native CCT236 (0.45 mm CCT with 20 mm CDP-choline) was crystallized using the sitting drop vapor-diffusion method with a reservoir condition that contained 0.1 m sodium acetate (pH 4.6) and 1.4 m sodium formate. The protein sample (2 μl) was mixed with 1 μl of the reservoir solution and the drop was incubated over 1 ml of the reservoir. After 7 weeks, the drop was replenished with an additional 1 μl of the same protein sample to extend crystal growth for an additional 5 months. After transfer to a cryo-solution (0.1 m sodium acetate (pH 4.6), 1.4 m sodium formate, 30% glycerol), the crystal was looped and cryo-cooled in liquid nitrogen. A highly redundant diffraction dataset was collected at the Advanced Light Source (ALS) beamline 8.2.2 using an ADSC Q315 detector. 360 images were collected with a 1° oscillation angle and a 5-s exposure time. The initial 300 images were used for data processing and refinement (Table 1).
TABLE 1.
Crystallographic data collection, phasing, and refinement statistics for the catalytic domain of Rat CCTα
The data collection statistics in parentheses are the values for the highest resolution shell. Se-Met is selenomethionine.
| Native | Se-Met incorporated | |
|---|---|---|
| Crystal parameters | ||
| Space group | P21212 | P2 |
| a, b, c | 89.0, 129.4, 43.6 Å | 129.2, 43.8, 88.3 Å |
| α, β, γ | 90.0, 90.0, 90.0° | 90.0, 90.02, 90.0° |
| Data collection statistics | ||
| Wavelength | 1.00 Å | 0.9795 Å |
| Resolution | 50.0-2.2 Å (2.28-2.20 Å) | 50.0-3.5 Å (3.63-3.50 Å) |
| Total reflections | 294,978 | 88,104 |
| Unique reflections | 26,316 | 12,964 |
| Rmergea | 0.105 (0.326) | 0.163 (0.269) |
| Mean (I)/σI | 32.7 (6.6) | 14.1 (6.8) |
| Completeness | 100.0% (100.0%) | 99.8% (99.8%) |
| Redundancy | 11.2 (10.6) | 6.4 (6.8) |
| Phasing statistics | ||
| No. of sites | 17 (out of possible 20) | |
| Overall FOMb | 0.24 | |
| Overall FOM (after density modification) | 0.63 | |
| Refinement statistics | ||
| Protein molecules in asymmetric units | 2 | |
| Residues | 353 | |
| Water molecules | 249 | |
| Total no. of atoms | 3176 | |
| Rcrystc/Rfreed | 0.22/0.27 | |
| Average B-factor (all atoms) | 27.7 Å2 | |
| Average B-factor (CDP-choline) | 19.1 Å2 | |
| r.m.s.d. on angles | 2.08° | |
| r.m.s.d. on bonds | 0.03 Å | |
a Rmerge = ΣhklΣi|Ii(hkl)− I¯(h¯k¯l¯)|/ΣhklΣiIi(hkl), where Ii(hkl) is the observed intensity, and I¯(h¯k¯l¯) is the average intensity obtained from multiple observations of symmetry-related reflections after rejections.
b FOM is figure of merit; 〈|ΣP(α)eiα/ΣP(α)|〉, where α is the phase angle and P(α) is the phase probability distribution.
c Rcryst = Σ‖Fo| − |Fc‖/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively.
d Rfree is calculated using 5% of the reflections randomly excluded from refinement.
The selenomethionine incorporated CCT236 (0.5 mm CCT; 5 mm CDP-choline) was crystallized using the vapor-diffusion hanging drop method with a reservoir consisting of 20 mm sodium citrate (pH 5.6), 0.4 m lithium sulfate, 15% PEG8000. The protein sample (1 μl) was mixed with 1 μl of the reservoir condition and incubated over a 0.5-ml volume of the reservoir. The crystals appeared within 1 week. The cryo-protectant condition was 20 mm sodium citrate (pH 5.6), 20% PEG 8000, 0.4 m lithium sulfate, and 30% glycerol. The crystal was looped and cryo-cooled in liquid nitrogen prior data collection. A complete SAD dataset was collected at the ALS beamline 8.2.1 using an ADSC Q315R detector as described above for the native crystal (Table 1).
Structure Determination and Refinement
The diffraction data were processed with the program HKL2000 (20). The programs HKL2MAP version 0.2 (21) and AUTOSOL within PHENIX version 1.3 (22) were used to solve the P2 crystal structure of selenomethionine-substituted CCT236 using single anomalous diffraction. The program AUTOBUILD within PHENIX version 1.3 automatically performed density modification and partially constructed the polypeptide chain. The rest of the molecule was built and fitted using the program COOT (23), and restrained refinements were carried out within the program REFMAC5 (24). The P21212 native crystal structure was solved by molecular replacement using the partially refined selenomethionine P2 dimer with the program MOLREP (25). The model was manually adjusted and refined using Coot (23). The data collection, phasing, and refinement statistics are summarized in Table 1. The P2 crystals have four protein chains in the asymmetric unit with a Matthews coefficient (Vm) and solvent content of 2.3 Å3 Da−1 and 47.3% respectively. The P21212 crystals have two protein chains in the asymmetric unit with a Vm and solvent content of 2.3 Å3 Da−1 and 46.4%, respectively.
Structure Analysis
The secondary structure elements were analyzed with programs DSSP (26) and PROMTIF (27). The program CONTACT within the program suite CCP4 (28) was used to identify interacting residues and measure their distances. The Protein-Protein Interaction Server (29, 30) and PISA web service (31) were used to analyze the dimeric interface, and the SSM Superpose (32) in Coot (23) was used to align structures. The CASTp server (33) was used to identify and analyze pockets on the surface, and SURFACE RACER 1.2 (34) was used to calculate the protein surface area with a probe radius of 1.4 Å. The DALI server (35) was used to find proteins with a similar protein fold. The stereochemistry of the final model was analyzed with PROCHECK (36).
Site-directed Mutagenesis and Purification of H168A and Y173A Mutant CCTs
Site-directed mutagenesis was carried out using QuickChange mutagenesis (Stratagene), following company protocols. The template was the pGEX6p1-CCT236 plasmid described above. The primers used for the H168A mutation were 5′-gattgatttcgtcgccgctgacgatatcccctactc-3′ (sense) and 5′-ctaactaaagcagcggcpgactgctataggggatgag-3′ (antisense). The primers used for Y173A mutation were 5′-catgacgatatccccgcctcttcggcagggag-3′ (sense) and 5′-ctccctgccgaagaggcggggatatcgtcatg-3′ (antisense). Annealing temperatures of 78.5 and 80 °C were used during respective PCRs. The H168A,Y173A double mutant was created by introducing the Y173A mutation into the H168A template plasmid, using QuickChange and the same mutagenic primer detailed above. The GST-CCT mutant proteins were expressed in BL21 Rosetta cells (18), purified via glutathione affinity chromatography, and cleaved with PreScission protease, as described above. The protein concentrations in the pooled elution fractions were measured by the Bradford assay (37).
CCT Enzyme Kinetics
CCT activity was measured by monitoring the conversion of 14C-labeled phosphocholine (Amersham Biosciences) into labeled CDP-choline (38). The reaction volume was 40 μl, and each reaction contained 1 μg of CCT236, 20 mm Tris (pH 7.4), 12 mm magnesium chloride, 89 mm NaCl, 10 mm DTT, and variable concentrations of the substrates, phosphocholine and CTP. The reactions were carried out for 10 min at 37 °C, with agitation, in the absence of added lipid vesicles. The phosphocholine dependence was monitored over a range of 0–5 mm with the CTP concentration held constant at 8.8 mm. The CTP dependence was monitored over a range of 0–27 mm with the phosphocholine concentration held constant at 1.5 mm.
The kinetic data were analyzed by nonlinear regression analysis using GraphPad Prism software. The initial velocity (nmol/mg/min) versus substrate concentration data were fit to the sigmoidal Hill equation, v = (Vmax [S]n)/(K*n + [S]n), where v = reaction rate; Vmax = maximum reaction rate; [S] = substrate concentration; K* = the Hill constant, i.e. the substrate concentration producing half-maximal velocity; and n = Hill coefficient. We observed substrate inhibition at high phosphocholine concentrations for CCT236 wild type and the Y173A mutant. Thus, the activity data used to calculate the K*(P-chol) for these constructs was limited to phosphocholine concentrations from 0 to 2 mm (wild type) and 0 to 1.4 mm (Y173A). CTP substrate inhibition was observed for all constructs at very high concentrations; thus the activity data used to calculate the K*[CTP] was limited to CTP concentrations of 0–20 mm (wild type), 0–5 mm (Y173A) and 0–18 mm, (H168A and H168A,Y173A).
RESULTS
Crystallization and Structure Solution
An untagged version of CCT236 yielded native crystals grown in a condition using sodium formate as a precipitant. These crystals belong to the orthorhombic space group P21212 and diffracted to 2.2 Å (Table 1). No solution to this diffraction set emerged using molecular replacement methods with GCT structures as search models. Crystals of a selenomethionine-incorporated version of CCT236 could not be reproduced in the same condition but were generated in a lithium sulfate/PEG 8000 condition. With this crystal a complete SAD dataset scaled at 3.5 Å resolution yielded the experimental phases when scaled as monoclinic P2. The partially refined model of CCT236 in the space group P2 was used as a search model to obtain phases by molecular replacement methods for the higher resolution native P21212 dataset.
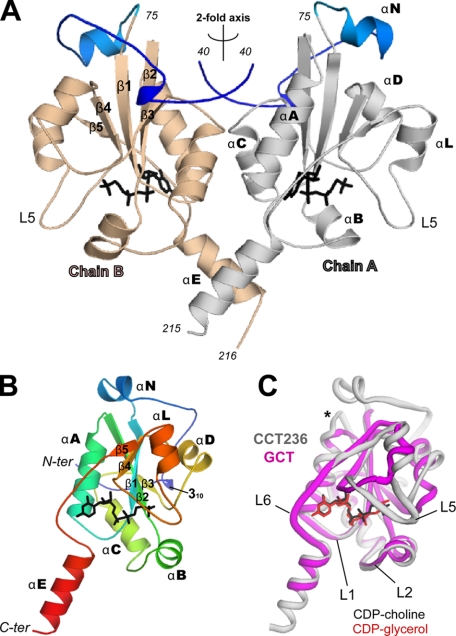
The asymmetric units of both crystal forms consist of identical homodimers. In the orthorhombic unit cell, the asymmetric unit consists of a single dimer, whereas in the monoclinic unit cell, the asymmetric unit consists of two dimers. Both crystals display almost identical crystal packing arrangements. The refined structure at 2.2 Å resolution shows electron density for 353 residues of the possible 472 residues comprising the homodimer, with two bound CDP-choline molecules and 249 solvent molecules (Fig. 3A). Electron density is lacking for the N and C termini of both chains. Chain A accounts for residues 40–215 and chain B for residues 40–216. The crystal packing contacts allow sufficient space between adjacent stacked dimers to accommodate both N- and C-terminal regions as a disordered array.
FIGURE 3.
Protein fold of CCT236 and comparison to GCT. The structures in this and all other figures were prepared using PyMOL. A, CCT236 homodimer. The asymmetric unit consisting of chain A (residues 40–215) and chain B (residues 40–216) is shown in ribbon representation. Segment N (40–75) is shown in blue in both chains, and domain C of chain A (76–215) and chain B (76–216) are shown in gray and wheat, respectively. B, ribbon diagram of the CCT236 monomer. The coloring of the chain transitions from N terminus (blue) to C terminus (red). The strands are numbered β1–5 and seven α-helices as well as a 310 helix are labeled. The active site bound reaction product, CDP-choline, is shown in stick representation (black). C, comparison of CCT236 and GCT structures. A single chain (chain A; PDB code 1N1D) of GCT (magenta; residues 1–126) is shown superimposed on chain A of CCT236 (gray; residues 40–215). The CCT bound CDP-choline (black) and the GCT bound CDP-glycerol (red) are shown in stick representation. The loops in CCT236 that form the active site are labeled L1, L2, L5, and L6. * indicates the novel extension of the β2-strand in CCT.
CCT236 Structure Displays an α/β-Fold with a Novel N-terminal Region
The visible portion of each monomer of the CCT236 dimer (Fig. 3A) contains the catalytic domain (C) plus 35 residues of the N-terminal region (N). Domain C (residues 75–215) conforms to the Rossmann fold, commonly found in nucleotide-binding proteins (39). Domain C is an α/β protein fold composed of five β-strands (β1–β5), and six α-helices (αA, αB, αC, αD, αL, and αE). This nomenclature strives to be compatible with the nomenclature of structurally analogous helices and strands in the previously solved structures of glycerol-phosphate CT (GCT) from B. subtilis (15, 16) and Staphylococcus aureus (40). The αL-helix in CCT236 replaces the 310 segments in GCT. The five β-strands assemble into a parallel, twisted β-sheet surrounded by the α-helices. Two α-helices (αA and αC) pack against one side of the β-sheet and two other helices (αD and αL) against the opposite side (Fig. 3B and supplemental Fig. S1). The αE-helix extends away from the fold and contacts its partner in the opposite subunit. The region of domain N visible in our structure is not an independent folding unit; rather, it is an intimate part of the CCT catalytic domain fold that crowns the top of domain C (Fig. 3A). In the description that follows, we refer to the residues N-terminal to the catalytic domain as segment N. Residues 40–75 of segment N loop around the entire top of domain C (Fig. 3A). This segment contains a short 310 helix (residues 48–50) and an α-helix (αN; residues 63–69), positioned near the N terminus of the β1-strand of domain C. Segment N has no counterpart in GCT (Fig. 2 and Fig. 3C).
A 2-fold rotational axis runs vertically through the center of the dimer (Fig. 3A). The visible N and C termini from each chain meet their counterparts near this rotational axis. Each chain in the dimer is virtually identical in tertiary structure, and their superimposition yields a root mean square deviation (r.m.s.d.) of 0.24 Å over 174 aligned residues. The largest deviation between chain A and B occurs in a region near the N terminus (residues 50–57 in segment N), where lack of direct contact with domain C creates inherent flexibility.
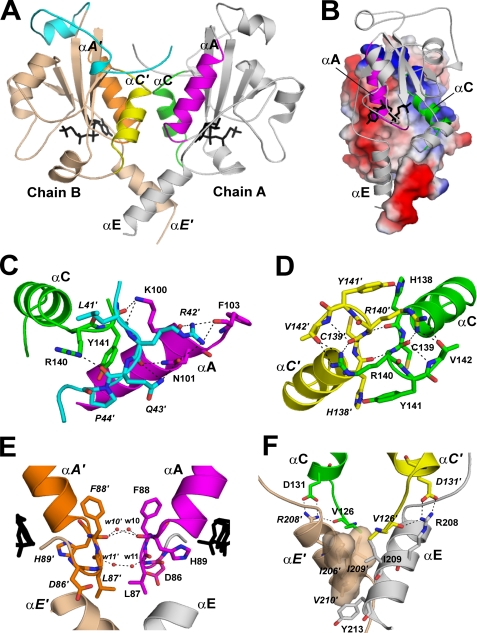
Both Segment N and Domain C Contribute to the CCT Dimerization Interface
Each CCT236 monomer has a surface-accessible area of ∼10,500 Å2. The dimerization interface buries 1,900 Å2 or 18% of the surface area of each monomer with nonpolar residues constituting 51% of the contact surface. Four structural elements contribute predominantly to the dimer interactions (Fig. 4) as follows: (i) residues 41–44 in segment N; (ii) helix A, the L1 loop preceding it, and the turn following it (residues 87–105); (iii) helix C and the L2 and L3 loops preceding and following it (residues 124–143); and (iv) helix αE (residues 206–213). Fig. 4B illustrates the charge asymmetry and resulting charge complementarity of the dimer interface. Although there is strong subunit interaction, cross-linking studies suggest that the dimer interface undergoes rearrangement upon transition from the soluble to membrane-bound form (41). It is not known whether this effect contributes to CCT activation. Segment N contributes to dimer stabilization by forging contacts with the αA-helix and L3 loop of domain C in the opposite chain. The side chains and main chain at Leu-41, Arg-42, and Gln-43 make extensive hydrogen bonding interactions with Lys-100, Asn-101, and Phe-103 at the C terminus of helix αA in the opposite chain (Fig. 4C). In addition, backbone atoms at Gln-43 and Pro-44 make polar contacts with the side chains of Arg-140 and Tyr-141 in loop L3 in the opposite chain. Further details of the segment N-domain C contacts are found in supplemental Table 1.
FIGURE 4.
Inter-subunit contacts at the CCT236 dimer interface. A, CCT dimer interface. In chain A, the structural elements of the dimer interface are labeled αA (residues 86–103; magenta), αC (residues 126–142; green), and αE (residues 203–215; gray). In chain B, the elements are labeled αA′ (residues 86–103; orange), αC′ (residues 126–142; yellow), and αE′(residues 203–216; wheat). Segment N (residues 40–75) of chain B is highlighted in cyan. These dimer interface elements are color-coded for reference to the detailed views shown in C–F, where the residues involved in the inter-chain interaction are shown in stick representation, and interactions (<3.5 Å) are indicated by dashed lines. B, surface electrostatic potential of chain B showing charge complementarity at the dimer interface. Chain A is shown in ribbon format. C, interactions between segment N (cyan) of chain B (residues 41–44) with αA (magenta) and the L3 loop following αC (green) in chain A. D, direct interactions between the L3 loops that harbor the conserved 140RYVD motif. Chain A is in green and chain B in yellow. E, interactions between the L1 loops involving ordered waters and main chain atoms at Leu-87 and Phe-88. Chain A is in magenta and chain B in orange. A portion of each CDP-choline proximal to the L1 loop is shown in black stick representation. The water molecules are labeled by the last two digits of their ID numbers in the PDB entry. F, interaction between αE and αE′ is mediated by hydrophobic residues that form complementary binding surfaces. The Arg-208 side chain in αE is anchored to αC′ by interactions with Val-126 and Asp-131.
The 140RYVD143 motif, located in loop L3 following helix C is a key contact point in the dimer interface. This motif is a signature sequence of the CT family (15) and likely serves the role of dimer stabilization in all CT folds. The two monomer chains come within 3.6 Å at the Cα of Arg-140. There are multiple inter-chain contacts in this loop, with Arg-140 as the key player. Fig. 4D illustrates a key inter-chain contact between the Arg-140 N-η1 and N-η2 atoms of one chain with the carbonyl oxygens at Cys-139 and Val-142 of the other chain. The side chains of Tyr-141 and His-138 also make inter-subunit contacts (Fig. 4D; details in supplemental Table 3). In addition to the RYVD motif, there are two other conserved elements in domain C that contribute to dimerization. In the conserved 85FDLFHXGH92 motif, Leu-87 and Phe-88 side chains directly face each other across the dimer interface, but are too distant for van der Waals contact. Rather, their main chain carbonyls interact via water molecules trapped in the dimer interface (Fig. 4E). The carbonyls at Phe-88 coordinate to w10 and w10′, and the carbonyls at Leu-87 couple to w11 and w11′. Similar water molecules linking the analogous carbonyls are found in the GCT-CDP-glycerol structure (w725 and w777 in PDB code 1N1D) (16) and in ECT (PDB code 3ELB). As well, there are other ordered water molecules trapped in cavities in the dimer interface of CCT that participate in the network of inter-chain hydrogen bonds between elements of the L1 loops and between L1 and helix αC in the partnering subunit.
At the C terminus of domain C, the two αE-helices cross over at the conserved hydrophobic residues, Ile-209 and Val-210. The interacting face of helix E is lined with the hydrophobic residues Ile-206, Ile-209, and Val-210. These three residues create a sterically complementary hydrophobic pocket for the Ile-209 side chain from the opposite helix to pack against (Fig. 4F). Tyr-213 stabilizes this interaction by further packing interactions with these hydrophobic side chains. The Ile-206 side chain of chain A is packed against the aromatic ring of Tyr-213 from chain B, whereas Tyr-213 from chain A packs against Val-210 in the opposite chain. The two Tyr-213 residues occupy different rotamers at the Cα–Cβ bond and may sample these alternate conformations.
Inter-chain contacts at helices A and C constitute important subunit interaction surfaces in both CCT236 and GCT. However, the residues involved are not conserved. In CCT236, a key inter-chain helix C-helix A interaction centers on the side chain of Asp-134 in αC, which couples to the sides chains of Ser-90 and Arg-94 in αA. Another dimer contact unique to CCT is a helix E-helix C interaction involving the side chains of Arg-208, Glu-131, and the main chain carbonyl at Val-126 (Fig. 4F). This contact also serves to anchor helix αE to the main body of the domain C fold.
Domain C Fold of Mammalian CCT Closely Aligns with Bacterial GCT and Human ECT with Several Key Differences
The sequence alignment between CCT236 and other CT enzymes is shown in Fig. 2. Of these CTs, the best characterized structurally and mechanistically is glycerol-3-phosphate cytidylyltransferase (GCT) from B. subtilis. The full GCT sequence is only 53% of the CCT236 sequence and corresponds to domain C. GCT (B. subtilis) and CCT (rat) share 34% sequence identity and 55% sequence similarity over 129 residues. The alignment of chain A of our CCT236;CDP-choline structure with chain A of the GCT;CDP-glycerol structure (PDB code 1N1D) reveals close superimposition of C-α atoms, with an r.m.s.d. of 1.55 Å over 118 aligned residues (Fig. 3C). The best superimposition is with the main chains of the β-sheets and helices A, C, and E. Segments that contain conserved motifs that forge multiple contacts with the cytidine nucleotide are closely aligned between the two CTs as follows: the L1 loop/αA-helix that houses the 89HXGH92 motif, and helix E and the preceding RTEGIST motif. Although single chains overlap well, superimposition of GCT and CCT dimers resulted in less accurate alignment (1.82 Å deviation of C-α atoms over 237 residues corresponding to the domain C region).
Although there is good conservation of the CCT domain C and GCT folds, there are three notable differences (Fig. 3C). The most apparent deviation is in the peripheral region between the β4- and β5-strands (residues 171–191). In GCT, the analogous region (residues 93–106) forms a short β-strand followed by two short 310 helices (15, 16). In CCT, there is a six-residue insertion (173YSSAGS178) that results in a longer extended loop (L5) followed by an α-helix (αL) rather than a 310 helix. This L5 loop is the least conserved region in the CT family (Fig. 2). In addition, near the N-terminal end of the β2-strand CCT has a 2-residue insertion (Pro-104 and Asn-105) that is conserved in CCTs (Fig. 2). These two residues contribute to stable intra-chain interactions between segment N and C, as described above. The conformation of loop L2 (referred to as the “40s flap” in GCT; see Ref. 16) also differs substantially between CCT and GCT, both with bound product.
The cytidylyltransferase with the greatest sequence similarity to the CCT catalytic domain is ECT. ECTs identified to date differ from CCT and GCT in that they are monomeric with two nonidentical catalytic domains. The human ECT N-terminal catalytic domain shares 41% identity (60% similarity) with rat CCT236 over 134 residues. The C-terminal catalytic domain lacks key residues in the RTEGISTS motif and may be nonfunctional. It shares 37% identity (55% similarity) with rat CCT236 over 134 residues. After we had analyzed the structure of CCT236 in relation to GCT, the coordinates for the crystal structure of human ECT were released (PDB code 3ELB). As of this writing, no paper describing this ECT structure has been published. Alignment of CCT236 chain A with the two catalytic domains of human ECT revealed excellent superimposition (supplemental Fig. S2, C and D), with an r.m.s.d. of 1.34 Å over 126 residues for the N-terminal catalytic domain and 1.53 Å over 137 residues for the C-terminal catalytic domain. The ECT N-terminal active site is unligated, and the C-terminal active site is complexed with CMP. Like GCT, ECT lacks the two-residue insertion at the beginning of the β2-strand but has a longer L5 loop like CCT. The most interesting differences to emerge from the comparison with our CCT236 structure involve the positions of the L2, L5, and L6 loops. Their position varies depending on ligand occupancy (supplemental Fig. 2B), in support of a role for mobile flaps in the catalytic mechanism (see “Discussion”).
Product CDP-choline Is Buried in a Deep Pocket at the Base of the β-Sheet
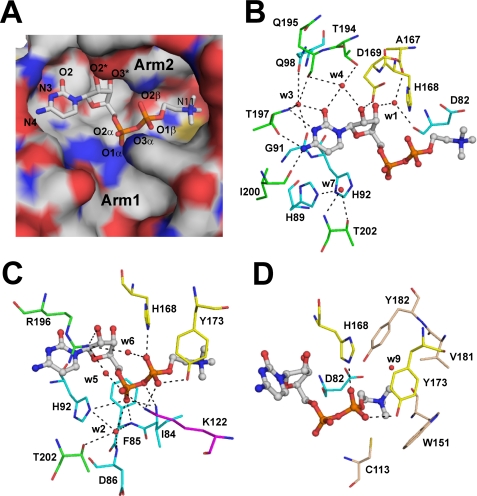
The two active sites within the CCT dimer were readily identified by CASTp (33) as the largest pockets on the surface. Each binding pocket has a solvent-accessible surface area of 770–800 Å2 and has two arms (Fig. 5A). One arm extends from the N-terminal end of helix E up to the C-terminal base of the β-sheet, and the other arm extends along the base of the β-sheet toward a pocket bounded by the L4 and L5 loops. Each arm of the pocket is about 16 Å long, and although the first arm is exposed to solvent, the second arm appears buried.
FIGURE 5.
CDP-choline in the active site of the CCT236. A, electrostatic surface representation of the active site pocket. Arg-196 and Tyr-173 were omitted from the surface representation to avoid obscuring the view of bound product. Arg-196 and Tyr-173 side chains extend over the cytidine base and trimethylammonium group, respectively (see C). B, interaction of cytidine in the active site. The hydrogen bonding interactions with the residues in the conserved RTEGISTS motif (green), HXGH motif/L1 loop (cyan), L5 loop (yellow), and solvent molecules are shown by dashed lines. The water molecules are labeled by the last digit of their ID numbers in the PDB entry. C, interaction of the phosphate groups in the active site. Lys-122 from loop L2 is shown in magenta; other elements are color-coded as in B. D, residues surrounding the choline group. Aromatic side chains as well as electrostatic interaction with Asp-82 stabilize the positive charge on the trimethylammonium group.
Within each active site pocket, there was a strong electron density for a bound ligand (supplemental Fig. S3A). A model for CDP-choline was fit easily into the difference density and refined. The B-factors for the CDP-choline are similar to those of the surrounding protein, suggesting a high occupancy for the CDP-choline. The bound product adopts a zig-zag conformation that extends across the binding cavity with the choline moiety occupying arm 2 (Fig. 5A). The cavity we refer to as arm 1 is largely empty, and it likely constitutes the binding pocket for the β- and γ-phosphates of CTP. The size and the electrostatic potential of the binding cavity are complementary to the bound ligand (Fig. 5A).
Active Site Interactions with the CDP Group Involve Many Conserved Residues in the L1, L2, L5, and L6 Loops and Several Ordered Water Molecules
The specific binding of the cytosine base is achieved primarily by direct contacts with residues from the 196RTEGIST motif in the L6 loop. The backbone carbonyls of Thr-197 and Ile-200 directly coordinate the cytosine N-4 and N-3 atoms (Fig. 5B) to establish selectivity for cytosine. These interactions are analogous to the backbone contacts at Thr-114 and Ile-117 in GCT (15, 16) and Ser-338 and Leu-340 in ECT (PDB code 3ELB). The ribose 2′-hydroxyl makes two direct contacts to active site side chains. Asp-169 at the base of β4 coordinates via its side chain (Fig. 5B), and the side chain of Arg-196 in L6 extends over the cytidine moiety to make contact with both the Asp-169 O-δ2 and the ribose 2′-hydroxyl (Fig. 5C). In GCT, these interactions are conserved in analogous residues Asp-94 and Arg-113. These residues are important for GCT activity as substitution of Asp-94 to glutamate or Arg-113 to lysine reduces kcat/Km by a factor of ∼1000 (42). In CCT, the R196K mutation reduces kcat/Km(CTP) by a factor of ∼100 (43).
In addition to direct interactions, the cytidine moiety is anchored by well coordinated water molecules. Water3 links the cytosine carbonyl oxygen to elements in αA and L6. The αA contacts involve the backbone carbonyl of Gly-91 in the 89HSGH motif and the side chain carbonyl of Gln-98, whereas the L6 loop contacts involve Thr-197 side chain O-γ and Gln-195 backbone carbonyl. Water4 bridges the cytosine carbonyl to the ribose 2′-hydroxyl and also coordinates to the L6 loop by Thr-194 side chain O-γ and Gln-195 backbone carbonyl. Finally, water1 links the ribose 3′-hydroxyl to backbone carbonyls in loop L1/β1 (at Asp-82) and β4 (at Ala-167). Analogous waters are found in ECT for w1, w3, and w4, and in GCT there are water analogs for w1 and w3.
Compared with the cytosine and ribose groups, the α- and β*-phosphates5 engage many more residues via direct hydrogen bonds with multiple structural elements in helix αA and the L1, L2, and L5 loops. The O-1 exo-oxygen of the α-phosphate directly H-bonds to the N-ϵ2 atom of His-92 in αA and the backbone NH group at Phe-85 in L1, whereas the NH of Ile-84 coordinates both exo-oxygen atoms. The Lys-122 side chain N-ζ in L2 bridges the α- and β*-phosphates (Fig. 5D). Analogous direct links to the α- and β*-phosphates occur in GCT-CDP-glycerol (PDB code 1N1D) at Lys-46 and at Thr-9 and Phe-10 (16), and in the ECT-CMP structure there are also direct links to the α-phosphate involving residues analogous to Phe-85, His-92, and Lys-122 in L1, αA, and L2. Finally, there are contacts with the β*-phosphate unique to CCT236 involving the side chains of His-168 and Tyr-173 in the L5 loop, as discussed below.
Binding of the α- and β*-phosphates also utilize three ordered water molecules, w2, w5, and w6, which engage and link elements of the L1 and L6 loops. Tables 2 and 3, supplemental Table 1, and supplemental Fig. S3B provide details of the contacts with these waters as well as further details of ligand interactions.
TABLE 2.
Hydrogen bonding interactions between the CDP portion of CDP-choline and the active site residues of CCTα
The listed distances are averages of the values measured from each active site within the homodimer. The water molecules are labeled by the last digit of their ID numbers in the PDB file.
| CDP-choline | CCTα | Residue location | Distance | GCT analog |
|---|---|---|---|---|
| Å | ||||
| Cytosine 4-NH2 | Thr-197 O | L6 | 2.9 | Thr-114 |
| Ile-200 O | L6 | 2.8 | Ile-117 | |
| Cytosine N-3 | Thr-197 N | L6 | 3.1 | Thr-114 |
| Cytosine O-2 | Water3 | 2.9 | ||
| Water4 | 2.8 | |||
| Ribose 2* OH | Water4 | 3.0 | ||
| Asp-169 O-δ2 | β4/L5 | 2.6 | Asp-94 | |
| Arg-196 N-η1 | L6 | 3.4 | Arg-113 | |
| Ribose 3* OH | Water1 | 2.8 | ||
| α-Phosphate O-1 | His-92 N-ϵ2 | αA | 3.4 | His-17 |
| Ile-84 N | L1 | 3.0 | Thr-9 | |
| Phe-85 N | L1 | 2.8 | Phe-10 | |
| Water2 | 3.2 | |||
| α-Phosphate O-2 | Water5 | 2.6 | ||
| Lys-122 N-ζ | L2 | 3.2 | Lys-46, Lys-44 | |
| α-Phosphate O-3 | Ile-84 N | 3.3 | Thr-9 | |
| β-Phosphate O-1 | Lys-122 N-ζ | L2 | 2.7 | Lys-46 |
| Tyr-173 O-η | L5 | 2.7 | ||
| β-Phosphate O-2 | His-168 N-ϵ2 | β4 | 2.6 | Trp-95 |
| Water6 | 2.6 |
TABLE 3.
Residues proximal to choline moiety
Distances from the atom closest to the trimethylamine nitrogen are reported. Values for the two active sites of the homodimer were averaged.
| CDP-choline | CCTα | Distance |
|---|---|---|
| Å | ||
| Trimethylamine N-11 | Tyr-173 O-η | 4.5 |
| Tyr-182 O-η | 4.0 | |
| Trp-151 O | 4.3 | |
| Cys-113 S-γ | 4.3 | |
| Asp-82 O-δ2 | 4.1 | |
| Val-181 C-γ2 | 5.3 | |
| Water9 | 4.3 |
Phosphocholine Binding Interactions Are Unique to CCT
There are three major differences between the CCT and GCT active sites that relate to the interactions with the phosphocholine versus phosphoglycerol groups. First, the N-ζ of the lone and critical Lys-122 interacts with the O-1 exo-oxygen of the β*-phosphate of CDP-choline (Fig. 5, C and D), whereas in GCT-CDP-glycerol, two lysines (Lys-44 and Lys-46) mediate the analogous interaction (16). Substitution of Lys-122 with arginine reduced kcat by 1000-fold and increased Km(Pchol) by 100-fold (43). Second, in the β4/L5 segment His-168 (N-ϵ2) and Tyr-173 O-η side chain atoms hydrogen bond with exo-oxygens of the β*-phosphate (Fig. 5, D and E). In GCT, Trp-95 alone carries out this function. Thus, in CCT it appears that novel active site residues, His-168 and Tyr-173, together with Lys-122 generate the hydrogen bonding interactions with the choline-phosphate that are responsible for the precise positioning of this substrate for nucleophilic attack on the α-phosphate of CTP. ECTs have histidine at a position analogous to His-168 in CCT and an adjacent threonine. In the human ECT-CMP structure, His-308 and Thr-310 interact with the ribose 3′-OH rather than the β*-phosphate. Unlike the GCT-CDP-glycerol structure, the ECT-CMP structure does not provide information on contacts with the nucleophilic substrate.
The third and most significant difference is found in the binding pocket for the choline moiety (Fig. 5E). The choline binding pocket in CCT is much more hydrophobic than the glycerol binding pocket in GCT. In GCT, specificity for glycerol is provided by a smaller pocket and by hydrogen bonding between Glu-71 and Lys-77 side chains and the two hydroxyl groups in the glycerol moiety. In CCT, specificity for choline is established by hydrophobic interactions contributed from the β4-strand and L5 loop. These include contacts with the aromatic rings of Trp-151, Tyr-173, and Tyr-182 and the side chain of Val-181. The shape and dimension of the resulting cavity (∼5 Å diameter) are ideal fits for choline. In addition, the carboxyl oxygens of Asp-82, positioned at 4.0 and 4.6 Å from the choline nitrogen atom, may provide weak ionic interactions with the quaternary amine. Interestingly, Asp-82 is conserved in cytidylyltransferases with amino alcohol substrates (CCT and ECT) but not in GCT (Fig. 2). There is also a cation-π interaction between the choline amine and the indole ring of Trp-151 in loop L4.
Other enzymes with binding pockets for choline display similar features (hydrophobicity and ion pairing with the choline amine). For example, the unrelated CCT from Streptococcus pneumoniae (PDB code 1JYK) (44) features hydrophobic residues Tyr-190, Trp-136, and Leu-159, an ionic interaction of the amine nitrogen with Asp-192, and a cation-π interaction with Trp-136. The choline binding pocket of human choline kinase (PDB code 2CKQ) (45) also consists of five hydrophobic residues and an electrostatic interaction provided by the carboxyl group of Glu-349. Mutations of the hydrophobic residues in the choline binding pocket of phospholipase C from B. cereus (PDB codes 1AH7 and 1P6D) affect substrate binding and kcat (46).
Mutational Analysis Confirms the Role of His-168 and Tyr-173 in Catalysis and Phosphocholine Binding
The ligand contacts to His-168 and Tyr-173 observed in the CCT236 structure and described above are novel to CCT. These residues are not conserved in GCTs (Fig. 2), and their role in CCT has not been tested previously. His-168 and Tyr-173 make hydrogen bonding contacts with the β*-phosphate of CDP-choline, which is the nucleophilic phosphate of the phosphocholine substrate (Fig. 1B). They are also part of the pocket complementary to the choline group. To test the hypothesis that these two residues participate in substrate binding and/or catalysis, we mutated each to alanine singly and also as a double mutation. These mutations had negligible consequence on global tertiary structure, as assessed by lack of change in the proteolytic accessibility. A time course of limited proteolysis using chymotrypsin shows essentially identical cleavage pattern for wild-type CCT236 and the three mutant proteins (supplemental Fig. S4).
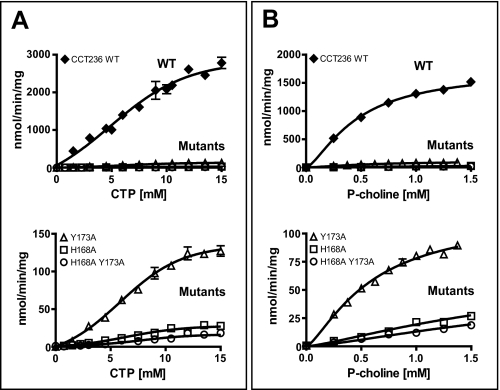
The role of His-168 and Tyr-173 in catalysis is clearly seen in the plots of substrate dependence versus reaction velocity (Fig. 6). Both the phosphocholine and CTP dependences were analyzed by a sigmoidal curve fit, as the Hill coefficients were greater than 1.0 (Table 4). Positive cooperativity toward CTP has been reported previously for both C-terminal truncated mutants (47) and full-length CCTα (43). For the Tyr-173 mutant, the Vmax value (determined from the CTP dependence curves) was reduced by a factor of 20, but the K* value for phosphocholine was unaffected. For the His-168 mutant, the Vmax value was reduced by a factor of 100 and the K* value for phosphocholine increased by ∼8-fold. The double mutant revealed a 162-fold reduction in Vmax and a 10-fold increase in K* (phosphocholine), translating into a reduced catalytic efficiency (Vmax/K*) of ∼300-fold. None of the mutations significantly affect the K*(CTP) value. These results indicate that both His-168 and Tyr-173 side chains are required for optimal catalysis, whereas His-168 appears crucial for efficient phosphocholine binding.
FIGURE 6.
Kinetic analysis of the CCT236 wild-type and active site mutant constructs. A, plots of initial velocity versus [CTP]. The P-choline concentration was held at 1.5 mm. B, plots of initial velocity versus [P-choline]. The CTP concentration was held at 8.8 mm. Data are means ± S.E. of four independent determinations (error bars are within symbol for many points). The data were fit to an equation for an allosteric sigmoidal curve (Y = Vmax Xn/K* + Xn; n = Hill coefficient) using GraphPad Prism 4. R2 values for the fits ranged from 0.88 to 0.98. The top panels show results for all constructs; the bottom panels show results for the three mutants only. Note the different y axis scales.
TABLE 4.
Kinetic parameters for wild-type CCT236 and active site mutants of CCT236
Parameters were obtained from the plots of initial velocity versus substrate concentration shown in Fig. 6, using GraphPad Prism 4 software. The Km and Vmax values are derived from the best fit to the Hill equation (see Fig. 6, legend) ± S.E. of the fit. The wild-type Vmax value from the CTP curve is more accurate, because the CTP concentration was subsaturating in the phosphocholine curves. Units for Vmax are nanomoles of CDP-choline formed (min−1) (mg CCT−1). Units for K* are in millimolar.
| CCT construct | CTP dependence |
Phosphocholine dependence |
||||
|---|---|---|---|---|---|---|
| Vmax | K* | Hill coefficient | Vmax | K* | Hill coefficient | |
| CCT236 wild type | 3399 ± 365 | 7.3 ± 1.1 | 1.6 ± 0.3 | 1724 ± 108 | 0.33 ± 0.09 | 1.4 ± 0.2 |
| Y173A | 167 ± 21 | 7.7 ± 1.2 | 1.9 ± 0.3 | 116 ± 11 | 0.48 ± 0.15 | 1.4 ± 0.2 |
| H168A | 34 ± 3 | 7.7 ± 1.0 | 2.0 ± 0.3 | 69 ± 12 | 2.5 ± 0.6 | 1.3 ± 0.3 |
| H168A,Y173A | 21 ± 6 | 8.8 ± 2.9 | 2.0 ± 0.6 | 55 ± 16 | 3.2 ± 1.2 | 1.4 ± 0.4 |
DISCUSSION
This study presents the first structural analysis of a eukaryotic cytidylyltransferase, providing a view into the workings of the active site of the enzyme that controls the rate of phosphatidylcholine synthesis and helps maintain a balanced membrane phospholipid composition (38, 48, 49). The CCT236 structure, which provides a complete catalytic domain as well as an intimately associated N-terminal extension, expands the repertoire of folds within the nucleotidyltransferase family and reveals unique features that have evolved to bind and position the phosphocholine group.
Residues 40–75 of Segment N Are an Integral Portion of the Catalytic Domain
The N-terminal region (residues 1–75) was previously regarded as a distinct domain comparable with other domains with specialized functions. However, our crystal structure of CCT236 reveals that the C-terminal half of the N “domain” (residues 40–75) wraps around the “top” of the catalytic domain, making specific and intimate contacts with it within and between individual subunits. Although it appears to be part of the catalytic domain fold of CCT, this N-terminal extension is absent from other cytidylyltransferases, including GCT and ECT, but is present in CCTs that have regulatory tail domains, from yeast to humans (see Fig. 2). GCT and CCT have similar kcat/Km values (9, 14). This argues against a participation of segment N in catalysis, and it hints at cooperation with regulatory domains. Although the N-terminal 39 residues of animal CCTs are not well conserved, sequences are conserved in the C-terminal portion corresponding to the 310 (44PAPFSDE50) and αN-helices (62VTMEEA67; mammalian CCTα numbering) that contribute to segment N-domain C contacts (Fig. 2 and supplemental Table 2).
A role for segment N in stabilization of folding and dimerization is also supported by several observations in addition to the structural data provided here. First, N-terminal truncation mutants of CCTα missing more than 39 amino acids do not express well, and the expressed protein is insoluble.6 Second, a disulfide bridge readily forms across the dimer interface at Cys-37 (41), showing that this portion of segment N contacts the corresponding region in its partner subunit. Third, a yeast two-hybrid probe of dimerization domains indicated that segment N was required for stable dimer formation (41).
Residues 1–39 are not visible in the structure. Several secondary structure predictive algorithms predict no ordered structure for this region, while correctly predicting the αN-helix at residues 63–70. Other evidence in support of a flexible N terminus is that the arginine residues within the NLS are highly accessible to the protease ArgC (50). CCTβ3, an enzymatically active isoform of CCT, initiates at a residue corresponding to Asp-28 of CCTα (51), and truncation of the first 28 residues of CCTβ1 produces a functional CCT (52). These data emphasize the lack of a structural requirement for the beginning section of the N-region. The function of this region in CCTα may be limited to nuclear localization (53) and, under some circumstances, membrane cross-bridging (5). Both roles involve the NLS sequence.
CCT Active Site Architecture Shares Key Elements with Other Nucleotidyltransferases
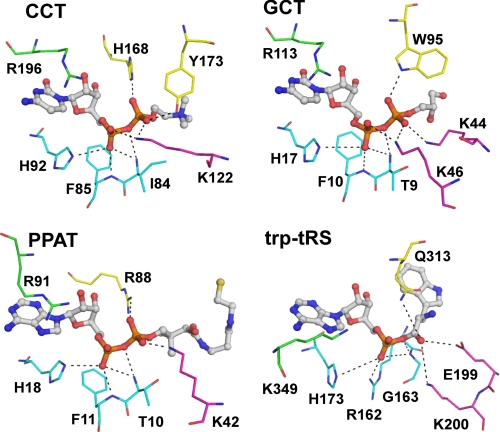
Similarities have previously been noted between the active site folds of GCT and other nucleotidyltransferases, including phosphopantetheine adenylyltransferase (PPAT) and the class 1 aminoacyl tRNA synthetases (aa-tRS) (15, 16). These enzymes, like CCT, all catalyze direct attack of a nucleophilic carboxyl or phosphoryl at the α-phosphate of CTP or ATP to displace pyrophosphate and create a nucleotidyl phosphoester. They all have some variation of a Rossmann fold but share little sequence identity with the CTs. However, there are three shared features that carry out analogous functions in the catalytic cycle. Fig. 7 shows in parallel how key analogous elements align to contact the reaction product in CCT, GCT, PPAT, or Trp-tRS as a well studied example of a class Ic aa-tRS.7
FIGURE 7.
Comparison of active site residues between nucleotidylyltransferases. CCT, residues shown to directly coordinate the α- and β-phosphates of the bound product in CCT236 are shown with CDP-choline in stick format. Residues belonging to loop L1 (conserved FDLFHXGH motif) is colored in cyan, Lys-122 from loop L2 in magenta, and both His-168 and Tyr-173 from loop L5 in yellow. Arg-196 from loop L6 (conserved RTEGISTS) that contacts the α-phosphate via a water molecule is also shown in green. Analogous residues present in other nucleotidyltransferases are shown with the same color coding schemes. GCT, glycerol-phosphate cytidylyltransferase with CDP-glycerol (PDB code 1N1D); PPAT, phosphopantetheine adenylyltransferase with dephospho-coenzyme A (PDB code 1B6T); Trp-tRS, human tryptophanyl-tRNA synthetase with Trp-AMP (PDB code 2QUJ). Note the similar ligand conformations in all four active sites.
The first element is found in the first helix of the Rossmann fold and the loop leading to it, containing the signature (T/H)XGH motif. This is the only sequence element conserved across the entire superfamily. The last histidine in this motif contacts an exo-oxygen of the α-phosphate of the bound nucleotide and thus may participate in transition state stabilization. In all four enzymes the α-phosphate also coordinates with backbone atoms in the loop preceding the HXGH motif (L1 in the CT nomenclature). The second element resides in the loop following the β5-strand of the fold. It is a poorly conserved loop that houses the RTEGIST motif in CTs, a ΦXXXXKMS(K/A)S motif in the class I aa-tRS enzymes, and WSFISSS in PPAT. The boldface positions are sites of main chain direct hydrogen bonds to the cytosine or adenine base. The arginine in the CTs or the first lysine in the Trp-tRS in this motif contacts the α-phosphate of the product, and it may assist in transition state stabilization. In PPAT, a functionally analogous arginine (Arg-91) is contributed from a separate site at the end of the β4-strand. In the third feature, three amine-containing side chains coordinate to phosphate or carboxylate oxygens of the nucleophile (illustrated with different colors in Fig. 7) as follows: a backbone NH from the L1 loop (cyan), a lysine ϵ-amino at the C-terminal end of the second helix of the fold (magenta), and a side chain NH from a residue in the β4-strand of the fold (yellow). Finally, all four enzymes share another common feature, the participation of moving loops during a catalytic cycle, as discussed below.
Novel Features of CCT Operate within a Conserved Architecture to Specify Recognition for Phosphocholine
The major differences between the CCT and GCT active site architecture relate to loops L2 and L5, which provide contacts with the nucleophilic substrate. In CCT, the L2 loop backbone positions the single Lys-122 ϵ-amino group for contact with the nucleophilic phosphate, whereas in GCT two lysines (44 and 46) engage in this interaction. The importance of Lys-122 in CCT (and Lys-46 and -44 in GCT) is well established by mutagenesis (16, 43).
The L5 loop has an extension in CCT, found also in ECT but not in GCT (Fig. 3C and supplemental Fig. S2). The side chains in the middle of this loop 174SSAGSDD180 do not contact CDP-choline. Rather, hydrophobic side chains on either side of it (Tyr-173, Val-181, and Tyr-182) make van der Waals interactions with the choline methyl groups. The L5 loop also positions the Tyr-173 hydroxyl for coordination with the nucleophilic phosphate in concert with His-168 at the C terminus of β4. The tyrosine functions as an analog to Lys-44 in GCT, although part of a separate loop; and His-168 is the functional counterpart for Trp-95 in GCT. Our kinetic analysis confirmed roles for His-168 and Tyr-173 in phosphocholine binding and/or catalysis. The histidine residue is buried deeper in the active site cleft and appears crucial for binding (Table 4), whereas the tyrosine likely assists in positioning the nucleophilic phosphate for attack. Thus, although the specific residues differ, both CTs feature three contact points from the L2 and L5 loops with the nucleophilic phosphate (see Fig. 7).
Proposal for the CCT Catalytic Mechanism
Standard kinetic analysis shows that the CCT catalytic mechanism is via a random ordered ternary complex (13), just like that of the GCT from B. subtilis (14), but different from mammalian ECT, which operates via a sequential mechanism (54). Mutational analysis in CCT and/or GCT reveals important catalytic roles for the two histidines in the HXGH site, Arg-196 (Arg-113) in L6, Lys-122 (Lys-44/46) in L2, and Asp-94 in GCT, corresponding to Asp-169 in β4 (13, 16, 42, 43) and His-168 and Tyr-173 in β4/L5 (this work). Together with the insights from the GCT and CCT236 solved structures, we propose the following catalytic mechanism.
Substrates bind in random order. CTP binds to Arm 1 of the active site cleft (Fig. 5A) via multiple contacts in the L1 and L6 loop and Asp-169 at the start of the L5 loop. Precise contacts with the β- and γ-phosphates of CTP cannot be discerned from our structure. We speculate from its position in our CCT236 structure that ordered w7 occupies a portion of the active site designated for an exo-oxygen of the CTP β-phosphate, and from analogous interactions in GCT (15) we speculate that His-89 and Thr-202 function to bind the β- and γ-phosphates, respectively. In a further analogy to GCT, Arg-196 in L6, as well as the αE-helix dipole, may contribute electrostatic stabilization of the β- and γ-phosphates (15). Phosphocholine binds within Arm 2 of the pocket. We propose, based on analyses of human Trp-tRS (55), that the same contacts with the phosphocholine in the CCT236-CDP-choline complex are used to bind the phosphocholine substrate. Those contacts are with Asp-82, Lys-122, Trp-151, and His-168 and a complementary hydrophobic pocket for the three methyl groups contributed by Cys-113 at the base of β2 and hydrophobic side chains from L4 and L5.
The trigonal bipyrimidyl transition state at the α-phosphate is generated by surrounding the electronegative oxygens with NH groups (Fig. 5). The N-ϵ2 of His-92 of the HXGH motif and the backbone NH at Ile-84 and Phe-85 pull on one of the exo-oxygens of the α-phosphate. Nitrogen atoms in the side chains of Arg-196 in L6 (via bound water w5) and Lys-122 in L2 polarize the other exo-oxygen of the α-phosphate. Orientation of the nucleophilic phosphate is accomplished by three simultaneous contacts with His-168 N-ϵ2, Lys-122 N-ζ, and Tyr-173 O-η. By analogy to the aa-tRS enzymes (55–58) and to GCT, this catalytic cycle may feature open and closed active site conformations created by loop movements of L2, L5, and L6 so that these binding elements can converge on their target atoms in the ligand.
Moving Loops during a Catalytic Cycle
A comparison of the crystal structures of unliganded GCT, GCT with CTP, and GCT with CDP-glycerol/sulfate (15, 16, 40) point to two major mobile elements in the GCT fold that make adjustments during a catalytic cycle. First, the L6/αE segment containing the RTEGISTT motif is ordered upon binding CTP as it engages the cytosine, ribose, and phosphates at multiple points (40, 59). Second, the αB/L2 segment housing Lys-44/46 that helps orient the nucleophilic phosphate moves like a flap to close up the active site. The residues that anchor the L2 flap to the main body in GCT (Arg-55 and Tyr-52) are critical for efficient catalysis (15). The αB/L2 moving flap likely operates in CCT as well, because the opening to the choline binding pocket in CCT236 is too small for efficient entry or exit of the substrate and product. Also, the hinge-forming residues and their anchors are closely preserved in CCT and GCT. In CCT (GCT) Cys-113 (Ser-36) and Val-126 (Tyr-49) act as the hinges and are anchored to Arg-132 (Arg-55) and Asp-86 (Asp-11). An alignment of the ECT active sites with and without CMP suggests a similar movement of L2 about a hinge (supplemental Fig. S2.) This alignment also provides evidence for L5 as a moving flap. In the CMP-occupied active site, the flap is drawn closer toward the ligand to enable three residues at the N-terminal side of L5 (His-307, Gly-308, and Thr-310) to forge contacts with the ribose hydroxyls. As with the L2 loop, the movement of L5 resembles a rotation about a hinge anchored on either side of the loop. We envision that in the catalytic cycles of CCT and ECT, the L2 and L5 flap movements function to allow access to the active site, forge specific contacts with the phosphocholine, exclude water, and allow product release.
The various elements of the active site share a web of hydrogen-bonding interactions, including ordered water molecules that likely coordinate movements during a reaction cycle to generate cooperative reaction kinetics. This web of interactions is charted in supplemental Fig. S3B. Coupling of the movements of the two active sites of the CCT dimer could be attributed to the location of key active site residues in L1 and in L6 that are near or at the dimer interface, as described under “Results.”
Proposed Model for Regulation of CCT Catalysis by the M Domain
A prominent model for autoinhibition by domain M proposes direct inhibitory contacts with some part of the active site (2, 4, 9). Such a mechanism was recently elucidated for the E. coli amphitropic membrane protein, pyruvate oxidase (60). Its C terminus forms an ∼23-residue amphipathic helix for membrane binding and activation, and the same segment forms an ordered structure blocking the active site and causing misalignment of a key catalytic residue in the inactive form. However, there are at least three observations that do not easily fit with this contact-inhibition model for CCT regulation. First, M domains from diverse species that exhibit lipid regulation show very little sequence conservation, an expected feature for a domain that forges specific interactions with a conserved catalytic domain. Second, the accessibility of over 30 protease sites distributed throughout domain C was not altered when comparing soluble and membrane-bound CCT (50), incompatible with a reversible docking of domain M onto C. Finally, circular dichroism analysis of the secondary structure of whole CCT and CCT fragments suggested that in the absence of lipid vesicles domain M takes on a mix of conformations, with none dominating, i.e. it appears flexible (8).
We suggest an alternative to the direct contact model, which posits that the flexibility of the domain M in the soluble form of CCT may contribute to inactivation of the enzyme by creating too much disorder in its active site. The crystallographic B-factors for the CCT236 structure (supplemental Fig. S5) suggest that although the β-sheet and helices A, C, D, and L are rigid, there is some disorder in αN, αB/L2, L5, the C terminus of L6, and especially the αE-helix. Several observations indicate that in GCT L6 and αE become more ordered commensurate with CTP binding. NMR analysis of GCT indicate that CTP binding is accompanied by a loss of conformational freedom, including at the C-terminal segment (59). In the GCT·CDP-glycerol·sulfate complex, one of the monomers of the four chain unit lacked the sulfate (mimic of pyrophosphate), and this resulted in disordering of helix E (16). In the structure of unligated GCT from S. aureus, part of the L6 loop and all of the αE-helix are not visible (40). These findings suggest that CTP binding orders this region and that L6/αE may be a suitable structural element for regulation.
As suggested by Fong et al. (40), high flexibility of helix E could prevent binding or transition state positioning of CTP. It is worth noting that CCT236, although having lipid-independent activity, has a very high CTP K*, compared with full-length lipid-bound CCT or GCT (9). The apparent weaker affinity for CTP could be a reflection of disorder at the C terminus of the catalytic domain due to lack of tethering (e.g. to a membrane surface via domain M). Interestingly, the N-terminal catalytic domain of ECT has a very highly ordered helix E (supplemental Fig. S2), due to extensive interactions with the linker segment between the two catalytic domains. This enzyme is also constitutively active (61), which supports the idea that helix αE ordering could regulate catalytic function in CCT.
Domain M of CCT, which is thought to begin at residue ∼234 (7, 50), is linked to helix αE via a linker region of ∼20 residues. This linker region may be quite flexible as there are highly accessible primary protease sites at residues 223 and 225 (50), and because it was unresolved in the CCT236 crystal. The turnover time for fully active CCT is on the order of 50–100 ms (5, 9); thus the active site residues require 50–100 ms to carry out their ordered dance, uninterrupted by the excursions of a chaotic tail domain that likely has rotational frequencies orders of magnitude higher. Binding of the M domains to membranes reduces their conformational entropy. The ordering of the M domain would facilitate the ordering of the linker segment, helix αE and loop L6. The entropic model for CCT regulation presented here is novel, yet it can be incorporated into a model invoking inhibitory contacts, if those contacts are transient and perhaps confined to a small sub-region. Our model can be tested by spectroscopic examination of the mobility of the αE-helix and the succeeding linker segment in soluble, inactive versus membrane-bound, active forms of CCT.
Supplementary Material
Acknowledgments
We thank Dr. Corie Ralston at beamline 8.2.2 of the Advanced Light Source, Lawrence Berkeley Laboratory, University of California, Berkeley. We also thank Dr. Trevor F. Moraes for help with data collection. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, United States Department of Energy under Contract DE-AC02-05CH11231.
This work was supported in part by Grant 12134 from the Canadian Institutes of Health Research.
This paper is dedicated to Dr. Claudia Kent for her pioneering work over 3 decades that cultivated a deeper understanding of the catalytic and regulatory mechanisms and the structure of cytidylyltransferases.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–3 and Figs. S1–S5.
The atomic coordinates and structure factors (code 3HL4) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
β*-Phosphate refers to the phosphoryl group of CDP-choline that originates from phosphocholine. We also sometimes refer to it as the nucleophilic phosphate. This nomenclature distinguishes this phosphate from the β-phosphate of CTP, the other substrate.
Z. Ding and J. Lee, unpublished observations.
There is considerable variation in the contacts with ligands in the numerous class I aa-tRS enzymes with solved structures. We chose to illustrate the human Trp-tRS as an example of this class that is most similar to the CTs in the organization of the active site elements.
- CCT
- α isoform of CTP:phosphocholine cytidylyltransferase
- GCT
- glycerol-phosphate cytidylyltransferase
- ECT
- ethanolamine-phosphate cytidylyltransferase
- aa-tRS
- amino acyl tRNA synthetase
- PPAT
- phosphopantetheine adenylyltransferase
- NLS
- nuclear localization sequence
- r.m.s.d.
- root mean square deviation
- PDB
- Protein Data Bank
- DTT
- dithiothreitol.
REFERENCES
- 1.Sugimoto H., Banchio C., Vance D. E. (2008) Prog. Lipid. Res. 47, 204–220 [DOI] [PubMed] [Google Scholar]
- 2.Kent C. (2005) Biochim. Biophys. Acta 1733, 53–66 [DOI] [PubMed] [Google Scholar]
- 3.Jackowski S., Fagone P. (2005) J. Biol. Chem. 280, 853–856 [DOI] [PubMed] [Google Scholar]
- 4.Cornell R. B., Northwood I. C. (2000) Trends Biochem. Sci. 25, 441–447 [DOI] [PubMed] [Google Scholar]
- 5.Taneva S., Dennis M. K., Ding Z., Smith J. L., Cornell R. B. (2008) J. Biol. Chem. 283, 28137–28148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson J. E., Rao N. M., Hui S. W., Cornell R. B. (1998) Biochemistry 37, 9509–9519 [DOI] [PubMed] [Google Scholar]
- 7.Johnson J. E., Xie M., Singh L. M., Edge R., Cornell R. B. (2003) J. Biol. Chem. 278, 514–522 [DOI] [PubMed] [Google Scholar]
- 8.Taneva S., Johnson J. E., Cornell R. B. (2003) Biochemistry 42, 11768–11776 [DOI] [PubMed] [Google Scholar]
- 9.Friesen J. A., Campbell H. A., Kent C. (1999) J. Biol. Chem. 274, 13384–13389 [DOI] [PubMed] [Google Scholar]
- 10.Izard T. (2002) J. Mol. Biol. 315, 487–495 [DOI] [PubMed] [Google Scholar]
- 11.Leatherbarrow R. J., Fersht A. R., Winter G. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 7840–7844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fersht A. R., Knill-Jones J. W., Bedouelle H., Winter G. (1988) Biochemistry 27, 1581–1587 [DOI] [PubMed] [Google Scholar]
- 13.Veitch D. P., Gilham D., Cornell R. B. (1998) Eur. J. Biochem. 255, 227–234 [DOI] [PubMed] [Google Scholar]
- 14.Park Y. S., Sweitzer T. D., Dixon J. E., Kent C. (1993) J. Biol. Chem. 268, 16648–16654 [PubMed] [Google Scholar]
- 15.Weber C. H., Park Y. S., Sanker S., Kent C., Ludwig M. L. (1999) Structure 7, 1113–1124 [DOI] [PubMed] [Google Scholar]
- 16.Pattridge K. A., Weber C. H., Friesen J. A., Sanker S., Kent C., Ludwig M. L. (2003) J. Biol. Chem. 278, 51863–51871 [DOI] [PubMed] [Google Scholar]
- 17.Bork P., Holm L., Koonin E. V., Sander C. (1995) Proteins 22, 259–266 [DOI] [PubMed] [Google Scholar]
- 18.Johnson J. E., Goulding R. E., Ding Z., Partovi A., Anthony K. V., Beaulieu N., Tazmini G., Cornell R. B., Kay R. J. (2007) Biochem. J. 406, 223–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doublié S. (1997) Methods Enzymol. 276, 523–530 [PubMed] [Google Scholar]
- 20.Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 21.Pape T., Schneider T. R. (2004) J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 22.Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 23.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24.Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 25.Vagin A., Teplyakov A. (1997) J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 26.Kabsch W., Sander C. (1983) Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 27.Hutchinson E. G., Thornton J. M. (1996) Protein Sci. 5, 212–220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collaborative Computational Project, Number 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 29.Jones S., Thornton J. M. (1995) Prog. Biophys. Mol. Biol. 63, 31–65 [DOI] [PubMed] [Google Scholar]
- 30.Jones S., Thornton J. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 13–208552589 [Google Scholar]
- 31.Krissinel E., Henrick K. (2007) J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 32.Krissinel E., Henrick K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 [DOI] [PubMed] [Google Scholar]
- 33.Liang J., Edelsbrunner H., Woodward C. (1998) Protein Sci. 7, 1884–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsodikov O. V., Record M. T., Jr., Sergeev Y. V. (2002) J. Comput. Chem. 23, 600–609 [DOI] [PubMed] [Google Scholar]
- 35.Holm L., Sander C. (1998) Nucleic Acids Res. 26, 316–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Laskowski R. A., Moss D. S., Thornton J. M. (1993) J. Mol. Biol. 231, 1049–1067 [DOI] [PubMed] [Google Scholar]
- 37.Bradford M. M. (1976) Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 38.Davies S. M., Epand R. M., Kraayenhof R., Cornell R. B. (2001) Biochemistry 40, 10522–10531 [DOI] [PubMed] [Google Scholar]
- 39.Rao S. T., Rossmann M. G. (1973) J. Mol. Biol. 76, 241–256 [DOI] [PubMed] [Google Scholar]
- 40.Fong D. H., Yim V. C., D'Elia M. A., Brown E. D., Berghuis A. M. (2006) Biochim. Biophys. Acta 1764, 63–69 [DOI] [PubMed] [Google Scholar]
- 41.Xie M., Smith J. L., Ding Z., Zhang D., Cornell R. B. (2004) J. Biol. Chem. 279, 28817–28825 [DOI] [PubMed] [Google Scholar]
- 42.Park Y. S., Gee P., Sanker S., Schurter E. J., Zuiderweg E. R., Kent C. (1997) J. Biol. Chem. 272, 15161–15166 [DOI] [PubMed] [Google Scholar]
- 43.Helmink B. A., Braker J. D., Kent C., Friesen J. A. (2003) Biochemistry 42, 5043–5051 [DOI] [PubMed] [Google Scholar]
- 44.Kwak B. Y., Zhang Y. M., Yun M., Heath R. J., Rock C. O., Jackowski S., Park H. W. (2002) J. Biol. Chem. 277, 4343–4350 [DOI] [PubMed] [Google Scholar]
- 45.Malito E., Sekulic N., Too W. C., Konrad M., Lavie A. (2006) J. Mol. Biol. 364, 136–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Martin S. F., Follows B. C., Hergenrother P. J., Trotter B. K. (2000) Biochemistry 39, 3410–3415 [DOI] [PubMed] [Google Scholar]
- 47.Yang W., Boggs K. P., Jackowski S. (1995) J. Biol. Chem. 270, 23951–23957 [DOI] [PubMed] [Google Scholar]
- 48.Attard G. S., Templer R. H., Smith W. S., Hunt A. N., Jackowski S. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9032–9036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Z., Agellon L. B., Allen T. M., Umeda M., Jewell L., Mason A., Vance D. E. (2006) Cell Metab. 3, 321–331 [DOI] [PubMed] [Google Scholar]
- 50.Bogan M. J., Agnes G. R., Pio F., Cornell R. B. (2005) J. Biol. Chem. 280, 19613–19624 [DOI] [PubMed] [Google Scholar]
- 51.Karim M., Jackson P., Jackowski S. (2003) Biochim. Biophys. Acta 1633, 1–12 [DOI] [PubMed] [Google Scholar]
- 52.Lykidis A., Murti K. G., Jackowski S. (1998) J. Biol. Chem. 273, 14022–14029 [DOI] [PubMed] [Google Scholar]
- 53.Wang Y., MacDonald J. I., Kent C. (1995) J. Biol. Chem. 270, 354–360 [DOI] [PubMed] [Google Scholar]
- 54.Tie A., Bakovic M. (2007) J. Lipid Res. 48, 2172–2181 [DOI] [PubMed] [Google Scholar]
- 55.Shen N., Zhou M., Yang B., Yu Y., Dong X., Ding J. (2008) Nucleic Acids Res. 36, 1288–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kapustina M., Weinreb V., Li L., Kuhlman B., Carter C. W., Jr. (2007) Structure 15, 1272–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Retailleau P., Huang X., Yin Y., Hu M., Weinreb V., Vachette P., Vonrhein C., Bricogne G., Roversi P., Ilyin V., Carter C. W., Jr. (2003) J. Mol. Biol. 325, 39–63 [DOI] [PubMed] [Google Scholar]
- 58.Kobayashi T., Takimura T., Sekine R., Kelly V. P., Vincent K., Kamata K., Sakamoto K., Nishimura S., Yokoyama S. (2005) J. Mol. Biol. 346, 105–117 [DOI] [PubMed] [Google Scholar]
- 59.Stevens S. Y., Sanker S., Kent C., Zuiderweg E. R. (2001) Nat. Struct. Biol. 8, 947–952 [DOI] [PubMed] [Google Scholar]
- 60.Neumann P., Weidner A., Pech A., Stubbs M. T., Tittmann K. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17390–17395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vermeulen P. S., Tijburg L. B., Geelen M. J., van Golde L. M. (1993) J. Biol. Chem. 268, 7458–7464 [PubMed] [Google Scholar]
- 62.Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 63.Gouet P., Robert X., Courcelle E. (2003) Nucleic Acids Res. 31, 3320–3323 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.