Abstract
(Dihydro)ceramide synthase 2 (cers2, formerly called lass2) is the most abundantly expressed member of the ceramide synthase gene family, which includes six isoforms in mice. CERS2 activity has been reported to be specific toward very long fatty acid residues (C22–C24). In order to study the biological role of CERS2, we have inactivated its coding region in transgenic mice using gene-trapped embryonic stem cells that express lacZ reporter DNA under control of the cers2 promoter. The resulting mice lack ceramide synthase activity toward C24:1 in the brain as well as the liver and show only very low activity toward C18:0–C22:0 in liver and reduced activity toward C22:0 residues in the brain. In addition, these mice exhibit strongly reduced levels of ceramide species with very long fatty acid residues (≥C22) in the liver, kidney, and brain. From early adulthood on, myelin stainability is progressively lost, biochemically accompanied by about 50% loss of compacted myelin and 80% loss of myelin basic protein. Starting around 9 months, both the medullary tree and the internal granular layer of the cerebellum show significant signs of degeneration associated with the formation of microcysts. Predominantly in the peripheral nervous system, we observed vesiculation and multifocal detachment of the inner myelin lamellae in about 20% of the axons. Beyond 7 months, the CERS2-deficient mice developed hepatocarcinomas with local destruction of tissue architecture and discrete gaps in renal parenchyma. Our results indicate that CERS2 activity supports different biological functions: maintenance of myelin, stabilization of the cerebellar as well as renal histological architecture, and protection against hepatocarcinomas.
Introduction
Ceramides consist of a long chain sphingoid base to which fatty acid residues of different chain length are attached via an amide bond. In addition, the fatty acid residues can differ with regard to their extent of desaturation and hydroxylation (1). Ceramides can be provided by three different biochemical pathways (i.e. de novo biosynthesis, sphingomyelinase pathway, and salvage pathway) (2). The de novo synthesis of ceramides is initiated at the cytoplasmic face of the endoplasmic reticulum by condensation of serine with palmitoyl-CoA to 3-ketosphinganine, which is subsequently reduced to sphinganine (2–5). This compound can be acylated to dihydroceramide and desaturated to yield ceramide (6, 7). Sphingomyelin, when located in the outer leaflet of the plasma membrane and corresponding endosomal membranes can be hydrolyzed by the acid sphingomyelinase (2), whereas sphingomyelin in the inner leaflet of the plasma membrane can be hydrolyzed by the neutral sphingomyelinase (8). In the salvage pathway, glycosphingolipids and sphingomyelin are degraded in lysosomes by glycosidases, acid sphingomyelinase, and acid ceramidase (2) to yield sphingosine, which, like dihydrosphingosine, can be acylated in the endoplasmic reticulum by ceramide synthases (9–10).
Ceramide synthases are transmembrane proteins located in the endoplasmic reticulum. These proteins are coded in the mouse genome by six ceramide synthase genes (cers1 to -6), previously called lass (longevity assurance) genes. Their nucleotide sequences are largely conserved in eukaryotes (11–13). CERS1 to -6 proteins carry the TLC (Tram-Lag1-CLN8) domain, which includes the catalytic LAG1 domain. In addition, each of the CERS2 to -6 proteins contain an N-terminal HOX domain whose exact function is not known (14). Recently, it has been shown that the expression of the CERS2 protein can be regulated by sphingosine 1-phosphate via an sphingosine 1-phosphate receptor-like motif (15).
Each cers gene exhibits a characteristic tissue-specific expression pattern with partially overlapping specificity. Northern blot hybridization and real time PCR analyses indicated that cerS1 is highly expressed in the brain, muscle, and testis and that cers3 is expressed in testis as well as skin, whereas cers2, -4, -5, and -6 are expressed in several tissues. cers2 has recently been reported to exhibit the highest transcript level of all cers genes. It is also strongly expressed in liver and kidney of adult mice (15–17). cers2 mRNA has also been found in peripheral and central myelinating cells (18), where its expression increased postnatally in parallel to the formation of myelin. After myelin formation is complete, cers2 mRNA expression in the brain is down-regulated (18).
In addition to its tissue expression pattern, each ceramide synthase isoform shows a characteristic substrate specificity for fatty acid residues. This has been demonstrated by overexpression of individual ceramide synthase genes or their down-regulation via transfection of small interfering RNA. CERS2 and CERS4 show a preference for very long chain fatty acyl-CoAs (C22:0–C24:0 and C20:0, respectively), whereas CERS5 and CERS6 enzymes prefer shorter fatty acyl-CoAs (C14:0–C16:0). CERS1 catalyzes synthesis of C18:0-(dihydro)ceramide mainly in neurons. CERS3 shows a rather broad fatty acyl-CoA specificity (C18:0–C24:0) (15–17, 19, 20).
Several years ago, the activity of the LASS protein in yeast (i.e. the orthologous gene of ceramide synthases in mammals) was ablated. The LASS enzyme is composed of two catalytic subunit proteins (LAC1 and LAG1) and one regulatory subunit protein (LIP1) (21). When only one catalytic component was ablated, no phenotypic abnormality and no changes in sphingolipid synthesis were noticed. The ablation of both catalytic subunits led to lethality, decreased growth, or defects in the wall of yeast cells, depending on the different genetic backgrounds of the yeast strains used (22, 23).
In this study, we have used a commercial ES4 cell clone, which carries an insertion (gene trap) in the cers2 gene to generate transgenic mice that lack cers2 mRNA. We have investigated phenotypic abnormalities in these mutant mice relative to wild type mice. In order to provide a better basis for understanding of these phenotypic abnormalities, we have complemented our analyses by a more detailed expression study of the lacZ reporter gene, which codes for β-galactosidase and is driven by the cers2 promoter.
MATERIALS AND METHODS
Generation of cers2-deficient Mice
The cers2 gene trap (gt) ES cell clone (S16–4B1) was bought from the Soriano laboratory (Mutant Mouse Regional Resource Center, Davis, CA) and analyzed by Southern blot hybridization for single integration of the gene trap vector ROSAFARY, which contains a lacZ reporter DNA fused to neomycin resistance DNA. The exact position of integration was determined by using PCRs and sequencing. The ES cells were injected into C57BL/6 mouse blastocysts following standard conditions (24). Chimeras were mated with C57BL/6 mice and the agouti-colored offspring were analyzed for integration by PCR of tail tip DNA using a cers2 intron-specific sense primer (geno_for, 5′-GGA GTC TCA GAA TTT CCA GGC ATG C-3′), which was combined with a cers2 intron-specific antisense primer (geno_rev, 5′- CAC AAT GAG CAT CAG ACA CCT GCA C-3′) and lacZ-specific antisense primer (βgeo, 5′-CAC GAC GTT GTA AAA CGA CGG GAT-3′). The heterozygous offspring (designated as cers2+/gt with 50% C57BL/6 genetic background) were mated among each other, in order to obtain homozygous cers2gt/gt mice. Mice were kept under standard housing conditions with a 12-h/12-h dark/light cycle, and with food and water ad libitum. All mice were raised in accordance with the instructions of local and state authorities regarding animal welfare.
Southern Blot Hybridization
For Southern blot hybridization of wild type (cers2+/+), heterozygous (cers2+/gt), and homozygous mice (cers2gt/gt), genomic DNA was prepared from adult liver and digested with HindIII. Genomic DNA was fractionated on 0.7% agarose gels and transferred onto nylon membrane (Hybond™-N+; Amersham Biosciences). The hybridization was performed using Quick Hyb hybridization solution (Stratagene, La Jolla, CA) at 68 °C and a 32P-labeled probe for 2 h. The probe was a 424-bp DNA fragment spanning exons 5 and 6 of the cers2 gene. Filters were first washed twice for 15 min each in 2× SSC, 0.1% SDS at 68 °C and 0.1× SSC, 0.1% SDS at 68 °C and then exposed to XAR x-ray film (Eastman Kodak Co.) at −70° using an intensifying screen.
Northern Blot Hybridization
Total RNA from adult tissues was collected using TRIzol (Invitrogen) according to the manufacturer's protocol, and 20 μg were separated by electrophoresis (25) and transferred onto HybondN nylon membrane (Amersham Biosciences) by capillary diffusion in 20× SSC. The hybridization was performed with Quick Hyb using a 624-bp PstI/NcoI DNA fragment of a cers2 expressed sequence tag clone (bought from Deutsches Ressourcenzentrum für Genomforschung, Berlin, Germany), including part of the coding region and the 3′-untranslated region of cers2. After stripping, the membrane was hybridized with a BstXI 617-bp fragment of a pSV β-galactosidase vector (Promega, Madison, WI), which contained part of the coding region of the lacZ gene.
β-Galactosidase Staining of Adult Tissues
Tissues of adult mice were frozen at −20 °C or on dry ice and embedded in Tissue-Tec (Sakura, Zoeterwoude, The Netherlands). Afterward, they were sectioned (10–20 μm) using a cryostat (MICROM HM 500 OM, Walldorf, Germany), transferred onto Superfrost Plus slides (Menzel, Braunschweig, Germany), and air-dried. Then the sections were fixed with 0.2% glutaraldehyde in 0.1 m phosphate-buffered saline, rinsed three times in wash buffer (100 mm sodium phosphate buffer (pH 7.4), 1.25 mm MgCl2, 2 mm EGTA, 0.1% Triton X-100), and stained in β-galactosidase substrate buffer (same buffer as before, supplemented with 5 mm K3(Fe(CN)6), 5 mm K4(Fe(CN)6), and 0.2% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) overnight at 37 °C. Finally, sections were washed in phosphate-buffered saline, rinsed in water, and, after air drying, mounted in Entellan (Merck). The preparations were photographed with a Nikon 90i photomicroscope equipped with a 12 MP Nikon RI camera.
Ceramide Synthase Assay
Tissue samples of 10-week-old mice were homogenized in 20 mm HEPES-KOH (pH 7.4), 25 mm KCl, 250 mm sucrose, 2 mm MgCl2, 2 mm EDTA, protease inhibitors and centrifuged at 960 × g for 10 min. The supernatant was centrifuged at 100,000 × g for 1 h to obtain the total membrane fraction. Aliquots of the membrane fraction (dissolved in 20 mm HEPES-KOH (pH 7.4), 25 mm KCl, 250 mm sucrose, 2 mm MgCl2) were frozen in liquid nitrogen and stored at −80 °C. Protein concentrations were determined using the Bio-Rad DC protein assay. Ceramide synthase activity was measured as described (11, 26), with minor modifications. Membranes were incubated with 20 μm d-erythrodihydrosphingosine (Sigma), [4,5-3H]dihydrosphingosine (0.25 μCi; Biotrend, Cologne, Germany), 20 μm defatted bovine serum albumin (Sigma), 50 μm acyl-coenzyme A (Avanti Polar Lipids) for 20 min at 37 °C. Different amounts of proteins (20–150 μg) were used, depending on the tissue and acyl-CoA, to ensure linearity of the reaction with respect to protein concentration. Reactions were stopped by adding 3 volumes of chloroform/methanol (1/2; v/v), and lipids were extracted according to Ref. 27. Lipids were separated by thin layer chromatography on HPTLC silica gel 60 plates (Merck) using chloroform/methanol/acetic acid (190/9/1; v/v/v). Radioactivity was visualized using a Bioimager screen (Fujifilm). Reaction products were scraped into scintillation counter vials, and radioactivity was determined by liquid scintillation counting.
Lipid Extraction
Brains, livers, and kidneys from mice were weighed and homogenized using an Ultra-turrax homogenizer (IKA, Staufen, Germany). Lipids from 100 mg of tissue were extracted in 4 ml of methanol, 2 ml of chloroform, and 1.6 ml of water. The extraction took place in screw cap tubes (Barloworld Scientific Ltd., Stone, UK), which were set in a shaking water bath (40 °C) overnight. Afterward, the tissue residues were separated by centrifugation, and the solvents evaporated under a stream of nitrogen. This procedure was repeated three times to ensure extensive lipid extraction. The pooled extracts were then resolved in 2 ml of chloroform/methanol (1:1; v/v). Phospholipids, which might disturb the analysis of sphingolipids, were decomposed by mild alkaline hydrolysis for 2 h at 40 °C using 150 μl of 1 m KOH. After cooling, the samples were neutralized utilizing glacial acidic acid. Upon changing the solvent ratio to methanol/chloroform/water (2:2:1.8; v/v/v), a phase separation took place with chloroform in the lower phase. For further analysis, the chloroform phase was evaporated, and the residue was solubilized in chloroform/methanol (2:1; v/v).
Nanoelectrospray Ionization-Tandem Mass Spectrometry
Analysis was performed with a triple quadrupole instrument (VG Micromass (Cheshire, UK) model Quattro II) equipped with a nanoelectrospray source and gold-sputtered capillaries as described (28). Parameters for cone voltage and collision energy of the different scan modes were used, and quantification of sphingolipids was performed according to previous publications (29, 30).
Myelin Preparation and HPTLC Analysis of Sphingolipids in Myelin, Brain, and Sciatic Nerve
Myelin was purified according to Ref. 31. Briefly, brain samples were homogenized in 10.5% sucrose using an Ultra-turrax homogenizer (IKA, Staufen, Germany). The homogenate was subjected to gradient centrifugation on a 10.5–30% sucrose step gradient. Myelin isolated from the interphase was again washed with water (three times) to remove sucrose and lyophilized.
For lipid analysis by means of HPTLC, myelin and brain samples from the three genotyped, 10-week-old mice (cers2+/+, cers2+/gt, and cers2gt/gt) were suspended in water and homogenized using a Potter-Elvehjem homogenizer (B. Braun, Melsungen, Germany), ultrasound as well as vortexing. Aliquots of the aqueous suspension were taken for protein determination, which was performed according to the BCA assay (Sigma). Lipid isolation and the removal of glycerolipids by mild alkaline hydrolysis were carried out as described above. After neutralization and phase separation (see above), the chloroform phase was separated, and chloroform was evaporated. The lipid extract was stored at −20 °C until use.
For HPTLC analysis, the extract was dissolved in chloroform/methanol (1:1; v/v). The application on HPTLC plates (20 × 10-cm silica gel 60 plates, Merck) was performed automatically using a Linomat 4 system (CAMAG, Berlin, Germany) in order to apply equal protein amounts per lane (200 μg for brain and 20 μg for myelin samples). The plates were developed in a horizontal chamber (CAMAG) using chloroform/methanol/water (70:30:4; v/v/v). After drying, the separated lipid bands were visualized by treatment of the plates with a solution of 10% CuSO4 and 8% H3PO4 (w/v) and heating them to 180 °C for 15 min. Lipid extraction from sciatic nerves and TLC was done as described (32).
Immunoblot Analyses
Brains of wild type and cers2gt/gt mice were collected in liquid nitrogen and homogenized by sonification in 1.5 ml of homogenization buffer (60 mm Tris, pH 6.8, 3% SDS) supplemented with a protease inhibitor mixture (Roche Applied Science). Homogenates were centrifuged for 5 min, and protein concentrations were determined using the BCA assay (Sigma). 100 μg of each protein lysate were separated by SDS-PAGE in a 15% gel (for myelin-associated glycoprotein (MAG) in a 12% gel). Proteins were blotted at 100 V for 120 min in transfer buffer (20 mm Tris and 150 mm glycine, pH 8.3) onto Hybond-C extra nitrocellulose (GE Healthcare). Membranes were blocked with 1× Roti (Roth, Karlsruhe, Germany) for 1 h and incubated overnight at 4 °C with a 1:2,000 dilution of rabbit anti-myelin basic protein (MBP) (AbD Serotec, Oxford, UK) or a 1:2,000 dilution of rat anti-MAG (Abcam, Cambridge, UK). After washing (three times for 10 min) in washing buffer (20 mm Tris, pH 8.0, 150 mm NaCl, and 0.2% Tween 20), membranes were incubated for 1 h at room temperature with a 1:2,500 dilution of horseradish peroxidase-conjugated goat anti-rat (Dianova, Hamburg, Germany) or a 1:2,500 dilution of horseradish peroxidase-conjugated goat anti-rabbit (Dianova) in 1× Roti, respectively, and washed again three times for 10 min in washing buffer. For detection of the bound antibodies, the SuperSignal West Pico Chemiluminescent detection kit (Pierce) was used. Standardization of immunoblots was performed by using a 1:10,000 dilution of mouse monoclonal β-tubulin antibodies (Sigma) in washing buffer containing 5% milk powder for 1 h at room temperature and a 1:10,000 diluted horseradish peroxidase-conjugated goat anti-mouse secondary antibodies (Dianova) under the same conditions.
Histological Investigations
For high resolution light microscopy based upon semithin sections as well as transmission electron microscopy, mice were deeply anesthetized with chloroform and then fixed via transcardiac perfusion with either 6% glutaraldehyde or a Karnovsky-type fixative consisting of 2% glutaraldehyde and 2% paraformaldehyde, both in 0.1 m Sörensen phosphate buffer (pH 7.4). Tissues of interest were dissected and postfixed by immersion in the same fixative at least overnight.
After thorough rinsing in phosphate buffer, regions of interest of at most 2 × 3 × 1-mm volume were either directly or after presectioning with a Leica 1200 vibratome dissected from the tissues and postfixed in 2% OsO4, dehydrated in a graded series of ethanol, and embedded in Epon 812/glycid ether (Serva). Semithin sections were cut at a thickness of 1 μm and heat-mounted on aminosilane-coated slides, either double-stained with toluidine blue and pyronine G or with p-phenylenediamine alone. Light microscopic preparations were photographed with a Nikon 90i photomicroscope equipped with a 12-megapixel Nikon RI camera.
After evaluation by light microscopy, blocks were trimmed down to regions of interest of about 1 mm2 and resectioned at 70 nm for routine TEM. Sections were mounted on 300- or 100-mesh copper grids, respectively, contrasted with uranyl acetate and lead citrate, and examined in either a 120-kV Zeiss electron microscope 910 or a 200-kV Zeiss electron microscope 924, respectively.
RESULTS
Generation of cers2 Gene Trap Mice
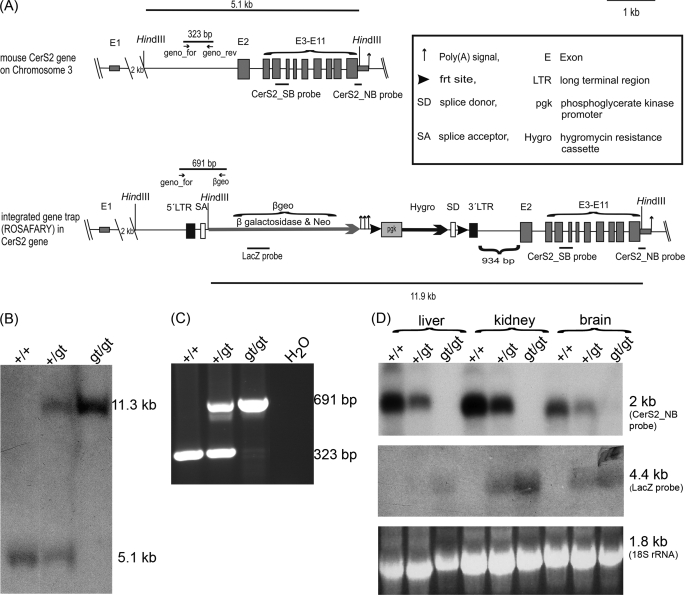
In order to generate CERS2-deficient mice, we used a commercial gene trap ES cell clone as described under “Materials and Methods.” In contrast to information provided by the company that the integration site of the gene trap vector ROSAFARY was located in exon 2 of the cers2 gene, we verified by Southern blot hybridization, PCR analyses, and sequencing that the integration site was located in intron 1, 934 bp upstream of the translational start codon of cers2 in exon 2. Exon 1 of cers2 is not translated. The gene trap vector consists of a splice acceptor, followed by DNA coding for the fusion protein βgeo (β-galactosidase and neomycin resistance protein), three polyadenylation signals, and a PGK promoter-driven hygromycin resistance cassette, which is flanked by frt sites. Between these frt sites and upstream of the hygromycin resistance DNA, a splice donor site is located. The total insertion construct is flanked by long terminal repeats (LTR) (Fig. 1A).
FIGURE 1.
Generation of cers2 gene trap mice. A, scheme of wild type and gene-trapped cers2 gene locus in ES cells for the generation of CERS2-deficient mice. The gene trap vector ROSAFARY (arrow) is integrated in intron1 of the cers2 gene. The ROSAFARY sequence consists (from 5′ to 3′) of a long terminal repeat (5′LTR), splice acceptor (SA) sequence, the coding region for the fusion protein β-galactosidase/neomycin (βgeo), polyadenylation signals, 5′-frt site, phosphoglycerate kinase (pgk) promoter, the coding region of the hygromycin resistance cassette (Hygro), splice donor sequence (SD), 3′-frt site, and 3′-long terminal repeat. B, Southern blot analysis of HindIII-digested wild type cers2+/+, cers2+/gt, and cers2gt/gt genomic DNA from liver using the external cers2_SB probe. The 5.1-kb fragment indicates the cers2 wild type allele (+), and the 11.3-kb fragment indicates the cers2 gene trap allele (gt). C, PCR analysis of cers2+/+, cers2+/gt, and cers2gt/gt tail tip genomic DNA. The cers2-specific primer combination resulted in a 323-bp amplicon for the cers2 wild type allele and a 691-bp amplicon for the cers2 gene trap allele. D, Northern blot analysis of cers2+/+, cers2+/gt, and cers2gt/gt mRNA isolated from liver, kidney, and brain. The cers2_NB probe detected a transcript of 2048-bp theoretical length. The lacZ probe detected a transcript of 4.4-kb length. The 18 S rRNA was used as a loading control.
Gene-trapped ES cells were injected into blastocysts to obtain chimeras, which were back-crossed to obtain agouti-colored offspring. Interbreeding of heterozygous cers2+/gt mice resulted in viable and fertile homozygous cers2gt/gt mice, as proven by Southern blot hybridization (Fig. 1B), PCR genotyping (Fig. 1C), and Northern blot analysis (Fig. 1D).
Ceramide Synthase Activity
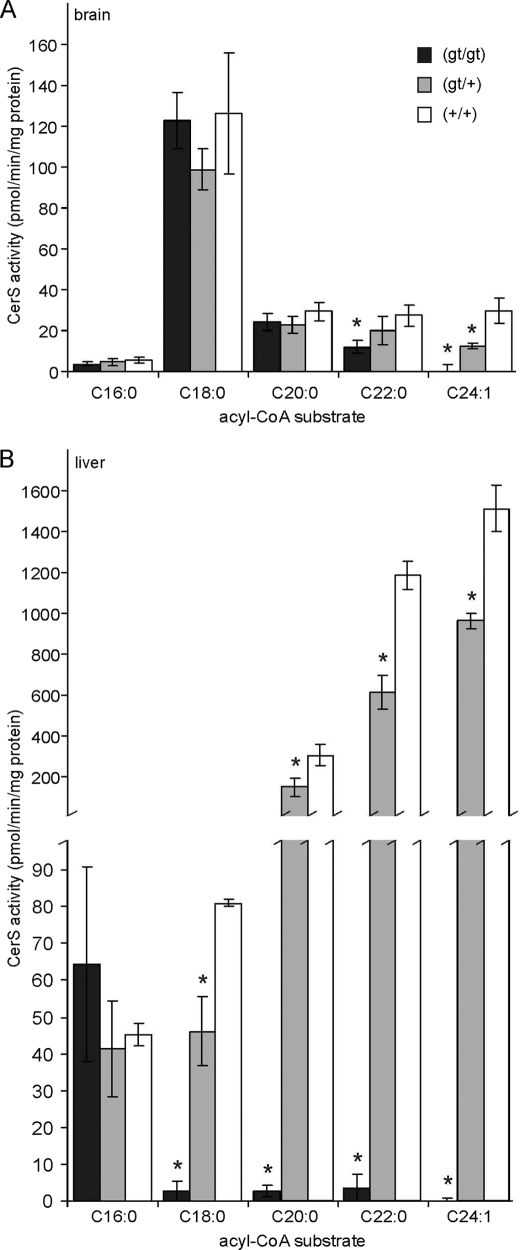
Ceramide synthase activity was determined in the brain and liver of 10-week-old mice representing all three genotypes (Fig. 2). Acyl-CoAs ranging from C16:0 to C24:1 were used as substrates. In brain samples of cers2gt/gt mice, activity toward C24:1 was undetectable, whereas activity was reduced by around 50% in heterozygous cers2+/gt mice (Fig. 2A). With C22:0-CoA, ceramide synthase activity was reduced to 44% in cers2gt/gt mice relative to wild type controls. No significant differences in ceramide synthase activity were found with the substrates C16:0-, C18:0-, and C20:0-CoA.
FIGURE 2.
Ceramide synthase activity. Shown is ceramide synthase activity in the brain (A) and liver (B) of 10-week-old cers2gt/gt, cers2+/gt, and cers2+/+ mice, using the indicated acyl-CoAs as substrates. Activity toward C24:1-CoA was undetectable in cers2gt/gt mice. Data shown are means ± S.D. (n = 3). *, statistically significant difference (p < 0.05; t test) when compared with wild type controls cers2+/+.
In brain lysates, the highest activity, irrespective of the genotype, was measured with C18:0-CoA, the specific substrate of CERS1, in agreement with its high expression level in the brain. Because activity toward C24:1-CoA was undetectable in cers2gt/gt mice in both liver and brain, incorporation of C24:1-CoA can be regarded as a specific measurement of CERS2 activity in both organs, which was 50-fold higher in the liver (1,512 ± 113 pmol/min/mg (mean ± S.D.; n = 3)) relative to the brain (30 ± 6 pmol/min/mg).
In contrast to the brain, ceramide synthase activity in the liver (Fig. 2B) was significantly reduced in cers2gt/gt and cers2+/gt mice with all substrates tested, except C16:0-CoA. With C18:0-, C20:0-, and C22:0-CoA, ceramide synthase activity was reduced in cers2gt/gt mice to 3% (C18:0) and less than 1% (C20:0, C22:0), respectively. Activity toward C24:1-CoA was undetectable in the liver of cers2gt/gt mice.
These data demonstrate that CERS2 has a relatively broad substrate specificity for acyl chain length, ranging from (at least) C18:0 to C24:1, although the longer fatty acyl-CoAs are the preferred substrates. Our in vivo data are in line with the in vitro data on CERS2 substrate specificity reported in Refs. 15 and 16.
Lipid Analysis
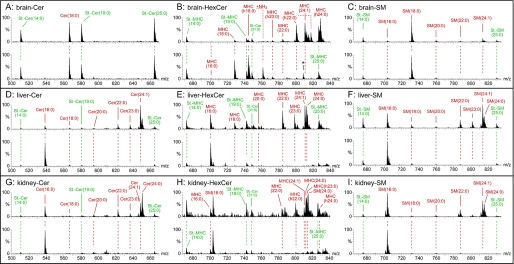
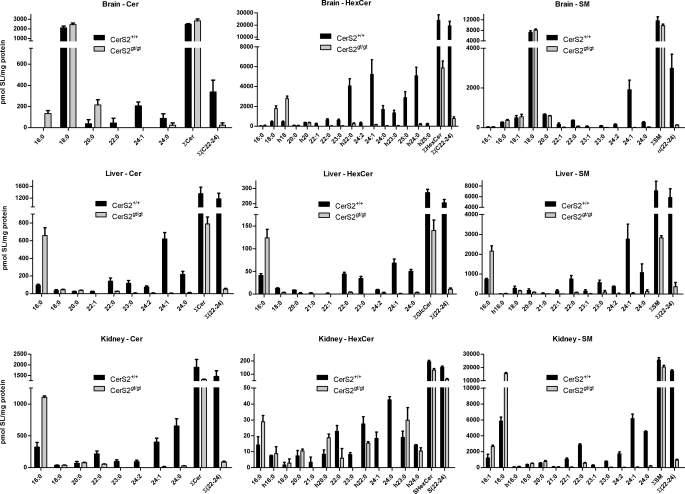
The levels of ceramides, monohexosylceramides, and sphingomyelin were determined in lipid extracts of the brain (Fig. 3, A–C), liver (Fig. 3, D–F), and kidney (Fig. 3, G–I), respectively, of wild type cers2+/+ and cers2gt/gt mice by nanoelectrospray ionization-tandem mass spectrometry. Representative mass spectra are shown in Fig. 3, and quantitative evaluations are presented in Fig. 4. Ceramide species with very long fatty acids (≥C22) were strongly reduced to very low levels or were undetectable in samples derived from all three tissues of cers2gt/gt mice. In contrast, C16:0-ceramide was significantly up-regulated in all three tissues of cers2gt/gt mice. In addition, C20:0-ceramide levels were 5-fold increased in cers2gt/gt brains.
FIGURE 3.
Precursor ion mass spectra of sphingolipids from control (upper spectrum) and Cers2gt/gt (lower spectrum) mice. Ceramides (Cer) (A, D, and G) and monohexosylceramides (MHC) (B, E, and F) were detected with the precursor ion scan m/z +264 specific for sphingolipids with d18:1-sphingosine. Sphingomyelins (SM) (C, F, and I) were analyzed with the precursor ion scan m/z +184 specific for the phosphorylcholine headgroup. Fatty acid chain lengths of SM were interpreted with the main sphingoid base sphingosine (d18:1) present in the analyzed tissues. A–C, brain; D–F, liver; G–I, kidney. Internal standards are in green, and endogenous sphingolipids are in red. m/z 808.5 marked with an asterisk in the lower spectrum of B is no hexosylceramide (HexCer), as verified with a second hexosylceramide-specific scan, neutral loss +180 atomic mass units. Quantitative evaluations of the mass spectra include also the standards Cer and hexosylceramide as well as SM standards with C31:0 acyl moieties and are shown in Fig. 4.
FIGURE 4.
Quantitative evaluation of ceramide, hexosylceramide, and sphingomyelin levels in the brain, liver, and kidney. The mass spectrometric peaks of Fig. 3, measured with three animals (n = 3), except for kidney with 2 cers2+/+ mice (n = 2), are graphically displayed as columns, indicating S.E. The very strong decrease of the different sphingolipids with C22–24 acyl chain length in cers2gt/gt mice relative to cers2+/+ mice is obvious in all cases. Instead, we found a strong increase of either C16 (liver, kidney, and brain ceramide) or C18 (brain hexosylceramide) containing sphingolipids in cers2gt/gt mice.
In accordance with the observed changes in the fatty acid composition of free ceramides, hexosylceramides and sphingomyelin with very long chain fatty acids (≥C22) were reduced to very low or undetectable levels in the brain, liver, and kidney of cers2gt/gt mice (Figs. 3 and 4). This was partially compensated by up-regulation of C16-sphingomyelin (liver and kidney), C16-hexosylceramide (liver), or C18-hexosylceramide (brain).
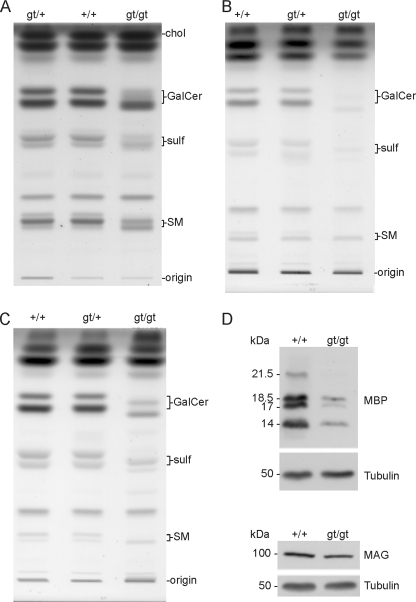
Thin layer chromatography of lipids from sciatic nerve (Fig. 5A), brain (Fig. 5B), and purified myelin (Fig. 5C) from 10-week-old mice showed a massive decrease in the amount of the fast migrating galactosylceramide and sulfatide, major components of the myelin membrane, and a concurrent increase in slower migrating sphingolipids in CerSgt/gt mice, indicating the corresponding lipids with short acyl chain ceramide (Fig. 5), which is in line with the changes observed by mass spectrometry of hexosylceramides (Fig. 4). In addition, in the brain and sciatic nerve, the total amount of galactosylceramide and sulfatide is reduced, probably as a consequence of myelin pathology (see below).
FIGURE 5.
Lipid analyses of sciatic nerve, brain, and myelin and immunoblot of myelin proteins. A–C, total lipids isolated from sciatic nerves (A), brain (B), and myelin (C) of 10-week-old wild type (+/+), heterozygous (gt/+), and cers2-deficient (gt/gt) mice were subjected to alkaline methanolysis and separated by TLC. Lipids per lane were applied according to equal protein contents (brain, 200 μg; myelin, 20 μg) or dry weight (sciatic nerves, 35 μg). Positions of lipid standards are indicated. chol, cholesterol; GalCer, galactosylceramide, sulf, sulfatide; SM, sphingomyelin. D, immunoblot analyses of MBP and MAG. These proteins were analyzed after SDS gel electrophoresis and immunoblot of brain extracts of wild type (+/+) and cers2gt/gt mice. In addition, blots were performed with anti-tubulin in order to control equal loading of the gels. In cers2gt/gt mice, MBP is decreased by about 80% and MAG by nearly 20%.
Analyses of Myelin Proteins
Since lipid and phenotypic analyses (see below) indicated abnormalities of central and peripheral myelin in cers2gt/gt mice, we analyzed the amount of myelin proteins in the brain. Fig. 5D indicates that MBP is reduced by about 80% in cers2gt/gt relative to wild type mice. In contrast, immunoblot analyses of MAG indicate that this protein appears to be decreased by only about 20% in cers2gt/gt brain. In accordance with the low level of MBP and myelin-specific sphingolipids (galactosylceramide and sulfatide) in the brain (Fig. 5B), the amount of compacted myelin isolated from 10-week-old mice was significantly reduced in cers2gt/gt mice (mg of myelin dry weight/g of brain wet weight): cers2gt/gt, 7.0 ± 1.1; cers2gt/gt, 15.4 ± 1.0; cers2+/+, 15.5 ± 1.5 (mean ± S.D.; n = 3).
Correlation between lacZ Expression and Phenotype of CERS2-deficient Mice
Since the lacZ reporter DNA of the gene trap ES cell clone is expressed under control of the cers2 promoter, we used β-galactosidase activity for indirect analysis of the expression of CERS2. Specific β-galactosidase activity (as controlled by the absence of staining in wild type tissue relative to cers2+/gt tissue) was found in virtually all organs of heterozygous cers2+/gt or homozygous cers2gt/gt mice investigated, like brain, eye, liver, heart, kidney, skin, pancreas, testis, and suprarenal glands, and was often restricted to specific cell types or subcompartments of a given tissue. We want to restrict a more detailed description of the expression to those tissues that featured an obvious pathology in cers2gt/gt mice. As an exception to this general overview, β-galactosidase activity in liver was surprisingly low when compared with the relative level of cers2 mRNA in liver and thus prohibited detailed analysis of its distribution, whereas in the kidney and brain, expression of the lacZ reporter DNA and cers2 mRNA appeared to correlate to each other (cf. Fig. 1D with Figs. 5 and 7).
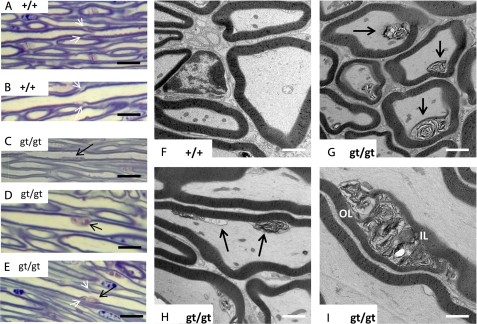
FIGURE 7.
Morphology of the trigeminal nerve in CERS2-deficient mice. A and B, morphology of trigeminal nerve axons in wild type mice. Note the regular delineation between axons and myelin sheaths and the morphology of Schmidt-Lantermann incisures in axons of different caliber (white arrows). C–E, in cers2gt/gt axons, toluidine blue-stained material was found in irregular patches attached to the inner myelin surface of a fraction of the axons (black arrows), in some cases affecting or being in the direct vicinity of Schmidt-Lantermann incisures (white arrows in E). F–I, ultrastructurally, these patches correspond to accumulations of whorled material (black arrows in G and H), apparently being derived from focally delaminated inner myelin sheath wrappings (compare cross-sections of cers2 wild type and cers2gt/gt mice in F and G). The axon membrane, therefore, is focally indented. In obliquely or longitudinally cut axons, it becomes apparent that multiple focal lesions exist (black arrows in H), between which the axon membrane readapts to the inner myelin surface. In some cases, stacks of disarranged myelin obliquely traverse the entire myelin sheath thickness in a course reminiscent of a Schmidt-Lantermann incisure with the characteristic outer and inner leaflets of compact myelin (OL and IL in I). Calibration bars, 10 μm in A–E and 1 μm in F–I.
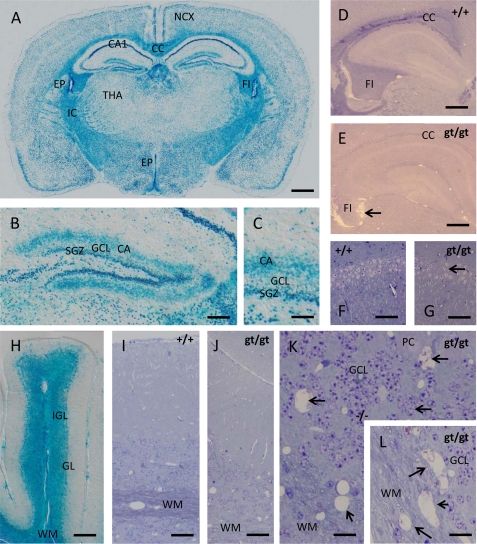
In both diencephalon and cerebral hemispheres (Fig. 6, A–C), β-galactosidase staining revealed three main sites of CERS2 expression. First, lacZ activity is abundantly present within white matter tracts, such as corpus callosum or internal capsule, whereas density and distribution of β-galactosidase-stained cells indicate the main expression in oligodendrocytes. Second, lacZ activity shows a widespread neuronal expression, whereas the labeling intensity strikingly varies with the types of neurons. lacZ reporter activity seems to be abundantly expressed in neocortical neurons of all layers, whereas its expression in the hippocampal formation is highly regionalized. Specifically, CA1 pyramidal cells feature the most intense lacZ staining seen anywhere in the CNS, but this intensive stain drops at the sharply defined CA1/CA2 border to lower levels maintained throughout the CA3 pyramidal cells. Within the dentate gyrus, the strongest lacZ reporter expression was seen in the subgranular layer (Fig. 6, B and C), the life-long active proliferative matrix of the granule cell layer, and, somewhat surprisingly, within a well defined supragranular lamina of the dentate molecular zone corresponding to the likewise laminated termination zone of the commissural and associational afferents to the granule neurons. The reasons for this strikingly compartmentalized β-galactosidase activity remain unclear at present. Third, lacZ activity was found in both choroid plexus epithelial cells and ependymal cells of the brain ventricles (cf. Fig. 6A).
FIGURE 6.
lacZ reporter gene expression and phenotypic abnormalities in the nervous system of cers2gt/gt mice. A, within the brain, β-galactosidase is expressed in all neuronal strata of the neocortex (NCX) and the white matter of the internal capsule (IC), corpus callosum (CC), and fimbria hippocampi (FI). In contrast, neuronal stain in the thalamus (THA) appears much weaker but is still present. Note the especially strong β-galactosidase staining within the CA1 field of the cornu ammonis and the ventricular ependyma (EP). B and C, within the dentate gyrus, β-galactosidase staining is especially intense within the subgranular zone (SGZ) containing the life-long active neuronal stem cell reservoir for the granule cell layer (GCL). Interestingly, the β-galactosidase signal also occupies the sublayer for commissural and associational afferents (CA) within the dentate molecular zone, an effect to be ascribed to an as yet not understood compartmentalization of β-galactosidase. D and E, when glycid ether-embedded and toluidine blue/pyronin G-stained samples of the telencephalon are compared with wild type mice (D), it is striking that the stainability of the white matter (corpus callosum and fimbria hippocampi) for basic dyes like toluidine blue is lost in cers2gt/gt mice (E), pointing toward severe biochemical alterations in myelin sheaths encompassing the loss of acidic compounds. Note the shrunken and ragged appearance of the fimbria in cers2gt/gt mice pointing toward severe axon loss (arrow in E). F and G, despite the high expression of the lacZ reporter in the hippocampal field CA1, only limited irregularities in the cell density of the pyramidal cell layer could be found (arrow in G). H, in the cerebellum, β-galactosidase is equally highly expressed in both the white matter (WM) of the medullary tree as well as in the internal granular layer (IGL), whereas the signal in the ganglionar layer (GL) is relatively weak and may reside rather in the Bergmann glia cell bodies than in the Purkinje cells. I and J, when cerebella of wild type (I) and cers2gt/gt (J) mice are compared, it is apparent that, similar to the telencephalon, the stainability of the white matter is largely lost. Again, this is not due to the loss of myelin sheath because they can be still discerned at low contrast but appears to be due to an alteration of their composition. K and L, an additional effect of CERS2 deficiency in the cerebellum, not seen in the telencephalon, is the formation of numerous small cysts (arrows) both within the gray matter of the internal granular layer (K) and the white matter (L), indicating the loss of both myelinated axons and granular layer interneurons. Calibration bars, 1 mm in A, 200 μm in B, 50 μm in C, 500 μm in D and E, 75 μm in F and G, 150 μm in H, 50 μm in I and J, and 20 μm in K and L.
In cers2gt/gt mice, we observed reduced stainability of CNS myelin by routine stains, such as toluidine blue or p-phenylenediamine, from about 3 months on, most clearly seen within the central nervous system fiber tracts, such as corpus callosum, alveus, and fimbria (Fig. 6, D and E). It is noteworthy that the fimbria in cers2gt/gt mice was smaller and presented with ragged contours (arrow in Fig. 6E) at an age of 9 months that could point to shrinkage of this fiber tract.
It is striking that despite the very high expression of β-galactosidase in the hippocampal CA1 region of cers2gt/gt mice, this is not paralleled by degenerative events of a similar magnitude in this brain region, where only scattered individual cells or small cell groups were apparently destroyed, as evidenced by the occurrence of small gaps in the CA1 pyramidal cell layer (Fig. 6, F and G).
In the cerebellum (Fig. 6H), β-galactosidase activity was again found both in white and gray matter. Strong staining was noticed not only in the medullary tree of the different folia but also at about the same level in the internal granular layer, whereas Purkinje neurons stained rather weakly if at all. β-Galactosidase staining in the Purkinje cell layer mainly resided between Purkinje cells, most probably due to Bergmann glia cells (data not shown). Similar to the situation in the cerebral hemispheres, stainability of myelin by routine stains is virtually lost in the cerebellar medullary tree (Fig. 6, I and J). However, cerebellar lesions go clearly beyond those in the telencephalon at the same stage. Both within the white matter and the internal granular layer, multiple “empty” cysts were visible in 9-month-old cers2gt/gt mice, indicative of a slowly progressing destruction of both white and gray matter during later adulthood (Fig. 6, K and L). In line with the widespread expression of cers2 in brain parenchyma and the associated neural lesions of cers2gt/gt mice, we consistently observed problems in the initiation of motor activity with these mutant mice (supplemental Fig. S1 (video)).
A more detailed analysis of the trigeminal nerve by semithin section light microscopy and transmission electron microscopy revealed subtle defects at the inner surface of myelin sheaths on about 20% of all axons investigated. By light microscopy, toluidine blue-stained material was seen in an irregular distribution attached to the inner surface of various myelin sheaths, sometimes being associated with Schmidt-Lantermann incisures (Fig. 7, compare A and B with C–E). Transmission electron microscopy revealed focal detachments of individual or groups of myelin lamellae, protruding into the periaxonal space and indenting the axons (Fig. 7, F–H), whereas longitudinal and oblique sections indicate that several such detachments may serially occur in one axon (Fig. 7H). In single cases, such myelin lesions could be seen to obliquely traverse the entire width of a myelin sheath, in a course similar to a Schmidt-Lantermann incisure (Fig. 7I).
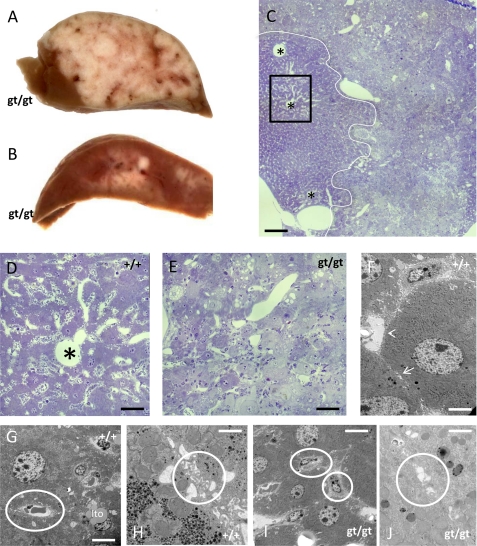
Between 7 and 9 months, striking changes were observed in the liver of CERS2-deficient mice as compared with controls. In 2 of 3 mice, multiple malignant tumors of about 1–2 mm diameter were seen, formed by unusually lipid- and glycogen-rich hepatocytes (Fig. 8, A–C). In the worst affected mouse at 9 months, these tumors took up a major part of the organ's volume (Fig. 8A). In these regions, the characteristic histoarchitecture of liver tissue was lost, with tumor cells forming compact clusters of pale cells without discernible liver lobule architecture. Ultrastrucurally, neither Disse spaces nor bile canaliculi could be found here (Fig. 8, D–J). In another animal, we could not detect overt malignant cell groups but noticed multiple liver adenomata, partially disturbing the lobular architecture of the organ.
FIGURE 8.
Phenotypic abnormalities in the liver of CERS2gt/gt mice. A and B, beyond an age of 7 months, cers2-deficient mice regularly featured multiple hepatic tumors, macroscopically appearing as gray nodules, throughout the liver parenchyma, in extreme cases (A) extending through a major part of the otherwise dark brown organ. The histological appearance of the tumor cells is that of pale hepatocytes, often filled with large lipid droplets (C). The white line indicates the separation of tumor tissue (right) and normal hepatocytes (left). Note that the normal architecture of liver lobules with central veins (asterisks) and radial sinusoids is lost in tumor tissue (C–E). The key ultrastructural hallmarks of liver tissue, Disse spaces, and bile canaliculi (arrowhead and arrow in F) are no longer discernible in tumor tissue (Disse space and bile canaliculi shown by white circles in G and H for wild type tissue; endothelia-like cells directly attached to hepatocytes and disfigured vacuolar structures in place of bile canaliculi shown by white circles in I, J for gt/gt tissue). Calibration bars, 500 μm in C, 100 μm in D and E, 5 μm in F, 10 μm in G, 1 μm in H, 10 μm in I, and 3 μm in J.
In the kidneys, most intense expression of lacZ is clearly seen in the collecting ducts, especially strong in the renal papilla (supplemental Fig. S2, A and B). Additional staining was seen in subcapsular tubuli that probably represent proximal tubules of nephrons, a finding corroborated by the visualization of strong β-galactosidase staining of tubules directly originating from individual Bowman capsules. Other tubules exhibited a weak to intermediate staining intensity, whereas lacZ expression in glomeruli was very low (supplemental Fig. S2B). Somewhat in contrast to this clear cut expression pattern of the lacZ reporter gene, discrete changes were only visible at an age of 7–9 months, when several gaps of 100-μm width were seen within the parenchyma, mainly in the vicinity of blood vessels and glomeruli (supplemental Fig. S2, C–F), thus not overtly coinciding with the highly β-galactosidase-positive collecting duct system.
DISCUSSION
cers2gt/gt mice were generated using gene-trapped ES cells in which the transgene was inserted into intron 1 of the cers2 gene. This integration apparently led to a loss of expression of the cers2 gene, because no cers2 mRNA could be detected in the resulting homozygous transgenic mice. Instead of the cers2 transcript driven by the endogenous cers2 promoter and its regulatory elements, a fusion transcript of cers2 exon 1 and the gene trap coded reporter gene cassette (βgeo) is expressed, leading to blue-stained cells by histochemical detection of β-galactosidase activity. Unexpectedly, no transcript of the hygromycin resistance gene fused to the cers2 exon 2 to exon 11 sequences was found. This may be due to a mutation in the phosphoglycerate kinase promoter, which should have driven expression of the hygromycin resistance gene independently of the cers2 promoter activity. Nevertheless, the homozygous cers2gt/gt mouse represents a functional inactivation of the cers2 gene.
In the brains of 10-week-old wild type mice, we found the enzymatic activity of CERS2 to be about 50-fold lower than in the liver. Accordingly, in cers2gt/gt brain tissue, only CERS2 activity toward the C24:1-CoA was reduced below the detection level, whereas the activity with the C22:0 component was reduced to about half, and activity with the C20:0 residues was not significantly decreased at all. Thus, any effects on phenotypic abnormalities found in the brains of cers2gt/gt mice are probably due to lack of very long chain fatty acid residues in sphingolipids.
Ceramide synthase activity and mass spectrometric analyses of ceramides in the liver, kidney, and brain of cers2gt/gt mice confirmed the in vitro data (15, 16) on fatty acid specificity of cers2. In the liver and kidney, cers2 is known to be highly expressed (15). Consequently, the relative abundance of ceramide species with very long chain fatty acid residues (≥C22) in wild type tissues was predictable and could be confirmed by mass spectrometry. Due to ablation of CERS2 activity, these ceramide species and ceramide moieties of hexosylceramides and sphingomyelins with C22–24 acyl residues were in sum strongly reduced to, on average, 4–6% of C22–24 wild type level in both organs as well as in the brain.
In the brain, the main fraction of C22:0 to C24:0 sphingolipids is present in myelin, where C22:0/C24:1-galactoside and sulfatide (2-hydroxylated and non-hydroxylated) constitute the majority of sphingolipids. During the period of myelin formation, cers2 is strongly up-regulated in myelinating cells, suggesting that CERS2 expression might limit synthesis of myelin sphingolipids (18). Consequently, we found a strong reduction in the amount of galactosylceramide and sulfatide in total brain and purified myelin of cers2gt/gt mice.
In central and peripheral myelin sheaths of cers2gt/gt mice, we noticed abnormalities in the inner lamella, which was focally detached in about 20% of all myelinated axons analyzed. This might indicate changes in the expression or localization of adhesion molecules involved in interaction between or within lamellae. Possibly in line with this, our results indicate that the level of MBP is reduced to about 20% in cers2gt/gt brain relative to its level in wild type mice. This may also affect the morphological abnormalities seen in myelin and the loss of stainability of myelin by toluidine blue in cers2gt/gt mice. The decreased level of MBP could contribute to the significant reduction in the amount of compact myelin isolated by density gradient centrifugation. This speculation may be supported by recent data underscoring the role of lipid environment for MBP positioning in myelin membranes (33).
In the brain and kidney of wild type mice, the level of CERS2 expression appears to correlate with the level of lacZ reporter gene expression in cers2+/gt mice, whereas only weak staining for β-galactosidase was found in the liver of these mice. Possibly the insertion of the gene trap vector into intron 1 of the cers2 gene destroyed a liver-specific enhancer sequence for transcriptional regulation. Previously, expression of cers2 mRNA has been demonstrated by in situ hybridization in the white matter of brain, presumably in oligodendrocytes (18). Our data show that the lacZ reporter gene is not only expressed in both neurons and (oligodendro)glia of the brain but also in the glial lineage-derived ependymal cells and epithelial cells of the choroid plexus. Degeneration of both gray and white matter in the cerebellum supports the concept that CERS2 activity is crucial not only for myelin sheaths but also for neurons. Interestingly the cers2gt/gt mice exhibit difficulties in the initiation of motor activity (cf. supplemental Fig. S1). This may be due to a neural defect.
Furthermore, it is striking that at least according to the β-galactosidase staining, CERS2 expression may vary greatly between different types of neurons. Next to the high expression in CA1 pyramidal neurons, lacZ appears to be highly expressed in the dentate subgranular zone, which stands out against the rest of the brain because it contains neuronal stem cells that proliferate and add granule neurons to the dentate gyrus throughout life. This link to cell proliferation is surprising, because brain development in general appears undisturbed. Similar to the brain lesions, liver carcinogenesis becomes obvious only during late adult life, although it may initiate much earlier. Because we could not yet precisely identify lacZ expression in distinct liver cells, we have so far not studied the initiation of hepatocarcinogenesis in cers2gt/gt mice.
Finally, in kidney lacZ expression in the renal collecting ducts and to a lesser degree in proximal tubules might point to an association with cell specializations linked to the more aggressive concentrated urine in the collecting ducts and possibly the high transport activity in the proximal tubules. Because the discrete loss of renal parenchyma observed here is linked to tubules rather than to collecting ducts, this latter notion should be tested in future experiments.
Because so far only cers2gt/gt mice have been studied that were up to 9 months old. It is possible that older animals, if they survive, will develop further complications in the liver and brain as well as additional phenotypic abnormalities (e.g. in the kidney). We conclude from our results that the loss of CERS2 activity cannot be functionally compensated by other ceramide synthase isoforms, at least in liver, myelinated cells, and perhaps also in other tissues. Further studies need to clarify the molecular mechanisms of how myelin stability in different parts of the brain as well as in peripheral nerves, liver homeostasis, and the parenchymal architecture in the kidney are affected by the loss of CERS2 activity.
Very recently, Pewzner-Jung et al. (34) have reported that cers2 is essential for normal liver homeostasis, based on the preliminary characterization of a gene trap mouse mutant.
Supplementary Material
Acknowledgments
We are grateful to B. Rau, M. Lindemann, A. Zoons, and C. Siegmund for expert technical assistance.
This work was supported by several grants from the Bonn Collaborative Research Center (SFB 645) “Regulation and Manipulation of Information Flow within Dynamic Protein and Lipid Environments” sponsored by the German Research Association (to K. W. (B2), K. S. (B3), and M. E. and V. G. (B5)).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 (video) and S2.
- ES
- embryonic stem
- gt
- gene trap
- HPTLC
- high performance thin layer chromatography
- MAG
- myelin-associated glycoprotein
- MBP
- myelin basic protein
- frt
- flippase recognition target.
REFERENCES
- 1.Kolesnick R. N., Goñi F. M., Alonso A. (2000) J. Cell. Physiol. 184, 285–300 [DOI] [PubMed] [Google Scholar]
- 2.Kolter T., Sandhoff K. (1999) Angew. Chem. Int. Ed. 38, 1532–1568 [DOI] [PubMed] [Google Scholar]
- 3.Braun P. E., Snell E. E. (1968) J. Biol. Chem. 243, 3775–3783 [PubMed] [Google Scholar]
- 4.Mandon E. C., Ehses I., Rother J., van Echten G., Sandhoff K. (1992) J. Biol. Chem. 267, 11144–11148 [PubMed] [Google Scholar]
- 5.Stoffel W., LeKim D., Sticht G. (1968) Hoppe-Seylers Z. Physiol. Chem. 349, 1637–1644 [DOI] [PubMed] [Google Scholar]
- 6.Sribney M. (1966) Biochim. Biophys. Acta 125, 542–547 [DOI] [PubMed] [Google Scholar]
- 7.Rother J., van Echten G., Schwarzmann G., Sandhoff K. (1992) Biochem. Biophys. Res. Commun. 189, 14–20 [DOI] [PubMed] [Google Scholar]
- 8.Ohanian J., Ohanian V. (2001) Cell Mol. Life Sci. 58, 2053–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tettamanti G., Bassi R., Viani P., Riboni L. (2003) Biochimie 85, 423–437 [DOI] [PubMed] [Google Scholar]
- 10.Kihara A., Mitsutake S., Mizutani Y., Igarashi Y. (2007) Prog. Lipid Res. 46, 126–144 [DOI] [PubMed] [Google Scholar]
- 11.Hirschberg K., Rodger J., Futerman A. H. (1993) Biochem. J. 290, 751–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Merrill A. H., Jr. (2002) J. Biol. Chem. 277, 25843–25846 [DOI] [PubMed] [Google Scholar]
- 13.Wang B., Shi G., Fu Y., Xu X. (2007) DNA Seq. 18, 92–103 [DOI] [PubMed] [Google Scholar]
- 14.Pewzner-Jung Y., Ben-Dor S., Futerman A. H. (2006) J. Biol. Chem. 281, 25001–25005 [DOI] [PubMed] [Google Scholar]
- 15.Laviad E. L., Albee L., Pankova-Kholmyansky I., Epstein S., Park H., Merrill A. H., Jr., Futerman A. H. (2008) J. Biol. Chem. 283, 5677–5684 [DOI] [PubMed] [Google Scholar]
- 16.Mizutani Y., Kihara A., Igarashi Y. (2006) Biochem. J. 398, 531–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizutani Y., Kihara A., Igarashi Y. (2005) Biochem. J. 390, 263–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker I., Wang-Eckhardt L., Yaghootfam A., Gieselmann V., Eckhardt M. (2008) Histochem. Cell Biol. 129, 233–241 [DOI] [PubMed] [Google Scholar]
- 19.Riebeling C., Allegood J. C., Wang E., Merrill A. H., Jr., Futerman A. H. (2003) J. Biol. Chem. 278, 43452–43459 [DOI] [PubMed] [Google Scholar]
- 20.Venkataraman K., Riebeling C., Bodennec J., Riezman H., Allegood J. C., Sullards M. C., Merrill A. H., Jr., Futerman A. H. (2002) J. Biol. Chem. 277, 35642–35649 [DOI] [PubMed] [Google Scholar]
- 21.Vallée B., Riezman H. (2005) EMBO J. 24, 730–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guillas I., Kirchman P. A., Chuard R., Pfefferli M., Jiang J. C., Jazwinski S. M., Conzelmann A. (2001) EMBO J. 20, 2655–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schorling S., Vallee B., Barz W. P., Riezman H., Oesterhelt D. (2001) Mol. Biol. Cell 12, 3417–3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagy A., Gertsenstein M., Vintersten K., Behringer R. (2003) Manipulating the Mouse Embryo, pp. 476–480, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 25.Sambrook J., Russel D. W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Vol. 1, pp. 7.31–7.34, Cold Spring Harbor, NY [Google Scholar]
- 26.Lahiri S., Lee H., Mesicek J., Fuks Z., Haimovitz-Friedman A., Kolesnick R. N., Futerman A. H. (2007) FEBS Lett. 581, 5289–5294 [DOI] [PubMed] [Google Scholar]
- 27.Bligh E. G., Dyer W. J. (1959) Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 28.Sandhoff R., Hepbildikler S. T., Jennemann R., Geyer R., Gieselmann V., Proia R. L., Wiegandt H., Grone H. J. (2002) J. Biol. Chem. 277, 20386–20398 [DOI] [PubMed] [Google Scholar]
- 29.Jennemann R., Sandhoff R., Langbein L., Kaden S., Rothermel U., Gallala H., Sandhoff K., Wiegandt H., Gröne H. J. (2007) J. Biol. Chem. 282, 3083–3094 [DOI] [PubMed] [Google Scholar]
- 30.Rabionet M., van der Spoel A. C., Chuang C. C., von Tümpling-Radosta B., Litjens M., Bouwmeester D., Hellbusch C. C., Körner C., Wiegandt H., Gorgas K., Platt F. M., Gröne H. J., Sandhoff R. (2008) J. Biol. Chem. 283, 13357–13369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Norton W. T., Poduslo S. E. (1973) J. Neurochem. 21, 759–773 [DOI] [PubMed] [Google Scholar]
- 32.Yaghootfam A., Gieselmann V., Eckhardt M. (2005) Eur. J. Neurosci. 21, 711–720 [DOI] [PubMed] [Google Scholar]
- 33.Min Y., Kristiansen K., Boggs J. M., Husted C., Zasadzinski J. A., Israelachvili J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 3154–3159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pewzner-Jung Y., Brenner O., Park H., Laviad E. L., Braun S., Stiban J., Levy M., Lahiri S., Merrill A. H., Jr., Futerman A. H. (2009) Charleston Ceramide Conference, Charleston, SC, March 11–14, 2009, Abstr, p. 29 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.