Abstract
The Aurora kinases are a family of serine/threonine protein kinases that perform important functions during the cell cycle. Recently, it was shown that Drosophila Aurora A also regulates the asymmetric localization of Numb to the basal and the partitioning-defective (Par) complex to the apical cortex of neuroblasts by phosphorylating Par6. Here, we show that Aurora A is required for neuronal polarity. Suppression of Aurora A by RNA interference results in the loss of neuronal polarity. Aurora A interacts directly with the atypical protein kinase C binding domain of Par3 and phosphorylates it at serine 962. The phosphorylation of Par3 at serine 962 contributes to its function in the establishment of neuronal polarity.
Introduction
The Aurora kinases are a family of serine/threonine protein kinases that perform essential functions during the cell cycle (1). They participate in the regulation of centrosome duplication and chromosome segregation but are also present in synapses of postmitotic hippocampal neurons (1, 2). The regulation of Aurora kinases is complex and requires their phosphorylation and interaction with specific factors (1, 3–6). Recently, it was shown that Drosophila Aurora A regulates the asymmetric localization of Numb to the basal and the Par complex to the apical cortex of neuroblasts (7–10). The highly conserved Par polarity complex is essential for the polarity of many cell types and consists of Par33 (Bazooka in Drosophila), Par6, and aPKC (11, 12). Aurora A phosphorylates Par6 and relieves its inhibition of aPKC. Active aPKC phosphorylates the Lethal (2) giant larvae protein Lgl and triggers the formation of the Par3·Par6·aPKC complex by promoting the dissociation of Lgl and recruitment of Bazooka·Par3. As a consequence, aPKC phosphorylates Numb that is released from the apical cortex and restricted to the opposite side of the neuroblast (8–10).
The Par polarity complex is required not only in Drosophila neuroblasts but also in rodent hippocampal neurons for the establishment of polarity, where it acts downstream of phosphoinositide 3-kinase and the GTPases Rap1B and Cdc42 (13–15). Cultured hippocampal neurons initially extend several short neurites (stage 2) (16). Within 24 h, one of these neurites is specified as the axon and begins to elongate rapidly (stage 3). Par3 and Par6 are present in all neurites of unpolarized neurons but become restricted to the axon in polarized stage 3 neurons, whereas active aPKC is found only at the tip of the axon (11, 13, 15). Par3 overexpression or suppression and aPKC inhibition result in defects in neuronal polarity and the loss of axons (14, 15, 17–19). Par3 performs multiple function in neurons and mediates the transport of the Par complex and Smurf2 and the activation of Rac through Stef. In neurons, aPKC phosphorylates and inactivates glycogen synthase kinase 3β and Mark2 to promote the normal extension of axons (11, 20–22).
Here, we show that Aurora A serves as a regulator of Par3 in mammalian neurons. Suppression of Aurora A by RNAi results in the loss of neuronal polarity. Aurora A interacts directly with the aPKC binding domain of Par3 and phosphorylates it at Ser962. The phosphorylation of Par3 at Ser962 is required for its function in the establishment of neuronal polarity.
EXPERIMENTAL PROCEDURES
Plasmids
Expression vectors for human Aurora A and B (23) and Par3 (24) were kindly provided by Drs. Nigg (Max-Planck-Institute for Biochemistry, Martinsried, Germany) and Macara (University of Virginia, Charlottesville), respectively. Aurora A and Aurora B deletion constructs containing the kinase and N-terminal domains were generated by PCR using the following primers: Aurora A kinase domain, AGG ATC CTT TGA AAT TGG TCG CCC TCTG and AGT CGA CAG ACT GTT TGC TAG CTG ATT CTT T; Aurora A N-terminal domain, AGG ATC CAT GGA CCG ATC TAA AGA AAA CT and AGT CGA CGT CTT CCA AAG CCC ACT GCC; Aurora B kinase domain, AGG ATC CGG CAA AGG CAA GTT TGG AAA and AGT CGA CTC AGG CGA CAG ATT GAA GGG. The amplified sequences were cloned into pMALc2X (New England Biotechnology), pGEX-4T2, and pEGFP-C1 (Clontech). Expression vectors for rat Aurora A (BC005425) and B (BC104676) were obtained from imaGenes (Berlin). Par3 deletion constructs comprising the CR1, PDZ1, PDZ2, PDZ3, and aPKC binding domain (aPKCBD) have been described before (14). The shRNA vector for the knockdown of Par3 was described previously (14). For the knockdown of Aurora A and Aurora B, the following target sequences were selected for the generation of shRNA vectors based on the pSHAG-1 or pSHAG-M2 plasmid (25, 26): TAC TTG GCG AGG GAG AAG CAA AGC AAG TTC AT (Aurora A RNAi1), CTC ATT TCA AGA CTG TTA A (Aurora A RNAi2), TGC GGA CTT TGG CTG GTC TGT GCA TGC CCC AT (Aurora B RNAi1), CAG CCT TTC ACC ATT GAC A (Aurora B RNAi2), and GTA ATT CAC AGA GAC ATA A (Aurora B RNAi3).
Neuronal Cultures and Transfection
Cultures of dissociated hippocampal neurons were prepared and transfected as described previously (27). Briefly, the hippocampus from E18 rat embryos was dissected and dissociated, and neurons were plated onto glass coverslips coated with polyornithine (Sigma) at a density of 800,000 cells/coverslip in 24-well plates. Neurons were transfected 2 h after plating, using calcium phosphate coprecipitation (14). The cells were detached after the transfection and replated onto new coverslips at a lower density (40,000–60,000 cells/coverslip in a 24-well plate). Neurons were fixed at 3 d.i.v. with 4% paraformaldehyde and 15% sucrose in phosphate-buffered saline for 20 min at 4 °C. To analyze the establishment of neuronal polarity, neurons were stained with the Tau-1 (as a marker for axons) and an anti-MAP2 antibody (minor neurites) (14). Neurites longer than 100 μm and positive for the characteristic Tau-1 staining pattern with a proximal to distal increase in staining intensity were counted as axons (14, 28). Processes shorter than 50 μm and positive for MAP2 staining were categorized as minor neurites. Neurons were regarded as unpolarized when none of the neurites showed the properties of an axon.
Immunofluorescence and Image Analysis
The following antibodies were used: Tau-1 (mAb3420, dilution 1:1000; Chemicon) and anti-MAP2 (Ab5622, 1:200; Chemicon), rabbit anti-Aurora A (1:500; Cell Signaling), mouse anti-Aurora A (Aurora A kinase/IAK-1, 1:500; BD Biosciences), anti-Aurora B (1:500, Sigma), anti-phospho-Aurora A/B/C (1:500; Cell Signaling), and Alexa Fluor-conjugated secondary antibodies (1:1000; Molecular Probes). Neuronal morphology was analyzed using the SPOT Advanced Imaging software (Diagnostic Instruments), ImageJ 1.33u (National Institutes of Health), and Adobe Photoshop. The length of axons and dendrites was determined using the SPOT software. The parametric Student's t test was used to test statistical significance of differences between two values if justified by one-way ANOVA.
Immunoprecipitation, Pulldown Assays, and Western Blotting
HEK 293T cells were grown in minimum Eagle's medium supplemented with 10% fetal bovine serum, 2 mm l-glutamine, and antibiotics. HEK 293T cells were transfected using calcium phosphate coprecipitation. GST or MBP pulldown assays were performed as described (14). To study the direct interaction of Par3 and Aurora A, bacterially expressed MBP fusion proteins were immobilized on amylose beads (New England Biolabs). After four washes with binding buffer (200 mm Tris, pH 7.4, 250 mm NaCl, 1 mm dithiothreitol), the beads were incubated with bacterial lysates containing GST fusion proteins. After five washes with pulldown buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1 mm dithiothreitol, 1.5 mm MgCl2, 5 mm EDTA, 10% glycerol, 0.1% Triton X-100, Complete protease inhibitor mixture (Roche Applied Science) in phosphate-buffered saline), bound proteins were eluted with sample buffer. For immunoprecipitation, brains from 5–10 E18 rats were lysed in 2% Triton X-100, 137 mm NaCl, 2.7 mm KCl, 8.1 mm Na2HPO4, 1.5 mm KH2PO4, and phenylmethylsulfonyl fluoride for 1 h at 4 °C, and insoluble debris was removed by centrifugation. For immunoprecipitation and Western blotting, the following antibodies were used: anti-myc (2272, 1:1000; Cell Signaling), anti-Aurora A (4718, 1:1000; Cell Signaling), anti-Aurora B (A5102, 1:1000; Sigma), anti-phospho-Aurora A/B/C (2914, 1:1000; Cell Signaling), anti-Par3 (07-330, 1:500; Upstate), anti-GST (27-457-01, 1:2000; GE Healthcare), anti-MBP (E80305, 1:2000; New England Biolabs), anti-EGFP (MMS-11P, 1:1000; BabCo), anti- α-tubulin (T9026, 1:3000; Sigma), and horseradish peroxidase-coupled secondary antibodies (1:10,000 MoBiTec, Göttingen).
In Vitro Kinase Assay
Proteins expressed in bacteria or HEK 293T cells were purified by pulldown or immunoprecipitation, respectively. Kinases were incubated with substrate proteins in 20 μl of kinase buffer containing 10 μCi of 1 μm [γ-32P]ATP at 30 °C for 30 min. The reaction was terminated by addition of sample buffer and boiling for 5 min. After separating the samples by SDS-PAGE, gels were fixed and dried and analyzed by autoradiography using the Fujifilm Image reader BAS-180. Alternatively, 1 μm ATP (Sigma) was used for kinase assays, and phosphorylated proteins were detected by Western blotting using an anti-phosphoserine antibody (Ab1603, 1:2000; Chemicon). Proteins expressed in HEK 293T cells were dephosphorylated by calf intestinal alkaline phosphatase (CIP) using 10 units of CIP in 50 μl of reaction buffer at 37 °C for 60 min prior to their use in kinase assay.
RESULTS
Aurora A and B Are Expressed in Neurons
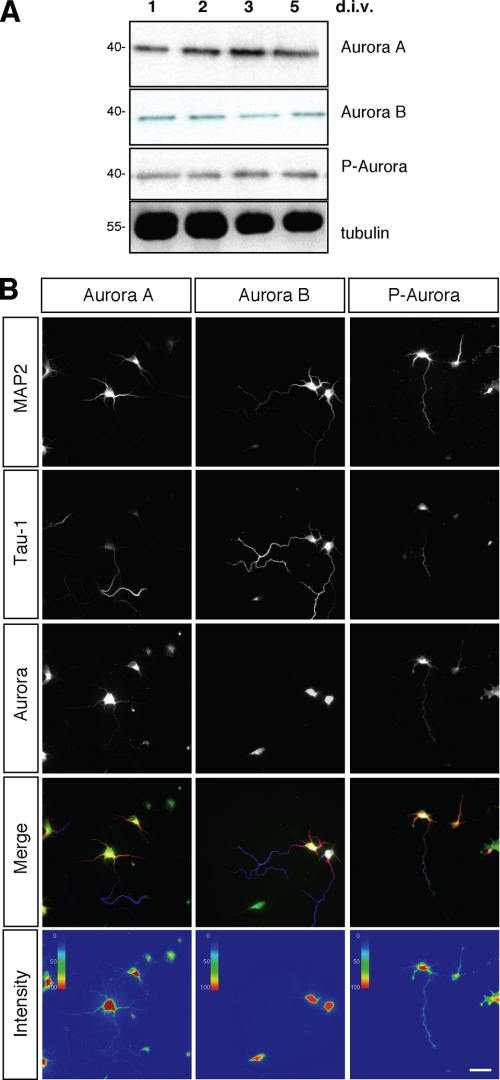
We first analyzed the expression of Aurora kinases in postmitotic neurons by Western blotting and immunofluorescence. Aurora A and B could be detected in cultured cortical neurons prepared from E18 rat embryos between 1 and 5 d.i.v. (Fig. 1A). Dividing cells were removed by treatment with arabinoside cytosine to ensure that only postmitotic neurons are present. Hippocampal neurons from E18 rat embryos were fixed at 3 d.i.v. and analyzed by immunofluorescence to determine the subcellular localization of Aurora kinases (Fig. 1B). Although both kinases were present in the soma, only Aurora A was detected in all neurites and showed a punctate staining pattern. No enrichment in axons or minor neurites was apparent. Phosphorylation at Thr288 and Thr232 in the activation loop is essential for the activity of Aurora A and B, respectively (29). Active Aurora kinases could be detected in cortical (Fig. 1A) and hippocampal neurons (Fig. 1B) using an antibody specific for active Aurora kinases phosphorylated at Thr288 or Thr232. Active Aurora kinases were present in the soma and all neurites. The subcellular localization of phosphorylated Aurora kinases was similar to that of Aurora A, suggesting that the signal mainly reflects phosphorylated Aurora A.
FIGURE 1.
Expression of Aurora A and B in neurons. A, cortical neurons prepared from E18 rat embryos were cultured in the presence of arabinoside cytosine to kill dividing cells. At 1, 2, 3, and 5 d.i.v., the expression of Aurora A, Aurora B, and phosphorylated Aurora was analyzed by Western blotting using anti-Aurora A, anti-Aurora B, and anti-phospho-Aurora antibodies. An anti-tubulin antibody was used to confirm that comparable amounts of protein were loaded. Numbers indicate molecular mass in kDa. B, hippocampal neurons from E18 rat embryos were fixed at 3 d.i.v. and stained with antibodies specific for Aurora A, Aurora B, and phosphorylated Aurora (P-Aurora) kinases, MAP2, and the Tau-1 antibody. Intensity representations of the fluorescence signals for Aurora A, Aurora B, and phosphorylated Aurora A and B were obtained using ImageJ software with the plug-in Intensity Analysis. Scale bar, 40 μm.
Aurora A Regulates the Formation of Axons
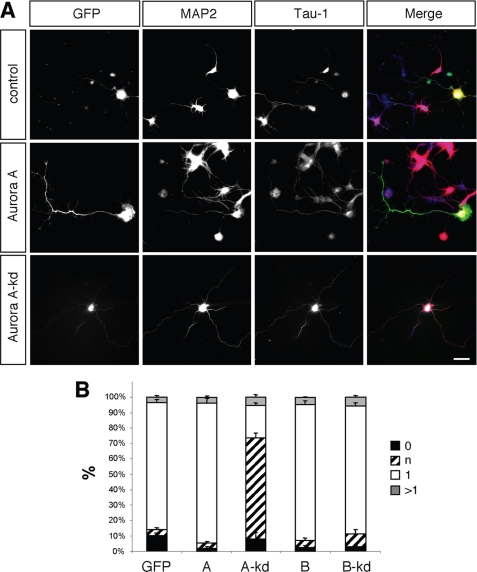
To investigate the function of Aurora kinases, hippocampal neurons were transfected with expression vectors for wild type or kinase-dead Aurora A or B at 0 d.i.v. and analyzed at 3 d.i.v. (Fig. 2). All constructs were expressed at comparable levels (supplemental Fig. S1). The majority of control neurons transfected with a vector for GFP formed a single Tau-1-positive axon and several MAP2-positive minor neurites (means ± S.E.: 82 ± 2%, n = 3 experiments, 43 neurons in total). Expression of wild type Aurora A or B did not significantly affect the number of axons/cell (Aurora A: 1 ± 0.02 axons per cell, n = 3 experiments, 51 neurons; Aurora B: 1 ± 0.03, 32 neurons; GFP: 1.0 ± 0.02, 43 neurons). However, the length of axons was significantly increased (GFP: 187 ± 6 μm, n = 3 experiments, 43 neurons; Aurora A: 225 ± 10 μm, 52 neurons; Aurora B: 229 ± 15 μm, 32 neurons). Expression of a kinase-dead Aurora A disrupted the formation of axons. Instead of a single long axon (longer than 100 μm and showing Tau-1 immunoreactivity in their distal segments) and several short minor neurites (neurites shorter than 50 μm), neurons extended multiple neurites with an average length of 78 ± 4 μm (GFP: 4 + 1%; Aurora A kinase-dead: 66 ± 3%, 32 neurons) that were only weakly positive for Tau-1 staining. Although these neurites were significantly longer than minor neurites, they lacked the characteristics of normal axons and were categorized as indeterminate neurites. In addition, a significant reduction in the length of the remaining axons could be observed (Fig. 2; 140 ± 9 μm, 32 neurons). Kinase-dead Aurora B did not show any significant effect. These results suggest that kinase-dead Aurora A has a dominant negative effect.
FIGURE 2.
Effect of Aurora expression on neuronal differentiation. A, hippocampal neurons were transfected at 0 d.i.v. with expression vectors for GFP (control), wild type, and kinase-dead (kd) mutants of GFP-Aurora A or B (green). Transfected cells were analyzed at 3 d.i.v. by staining with an anti-MAP2 (red) and the Tau-1 antibody (blue). Scale bar, 40 μm. B, the development of neuronal polarity was analyzed by determining the number of unpolarized neurons without an axon (0, black bars), unpolarized neurons with one or more neurites of undefined identity (indeterminate neurites longer than 50 and weak of absent Tau-1 staining; n, hatched bars), polarized neurons with a single axon (1, white bars), and neurons with multiple axons (>1, gray bars) (t test: multiple indeterminate neurites: p < 0.01, A kinase-dead compared with GFP; means ± S.E.). ANOVA (p < 0.0001) and a parametric Student's t test were used to test statistical significance.
Aurora A Is Necessary for the Formation of Axons
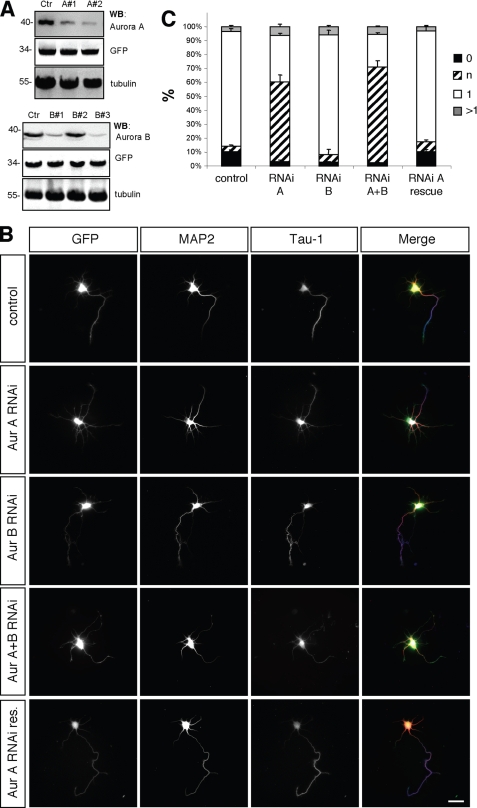
To investigate whether Aurora kinases are required for the normal development of hippocampal neurons, endogenous Aurora A or B was suppressed by RNAi. The efficiency of the shRNA expression vectors was tested by Western blotting after coexpression with Aurora A or B in HEK 293T cells and by immunofluorescence after expression in hippocampal neurons (Fig. 3A and supplemental Fig. S2). Two of the constructs tested showed efficient suppression of Aurora A (shRNA-A1 and A2) or Aurora B (shRNA-B1 and B3). Human Aurora A was resistant to shRNA-A2 and was used for rescue experiments (Fig. 3).
FIGURE 3.
Aurora A is required for neuronal differentiation. A, HEK 293T cells were transfected with vectors for EGFP, rat Aurora A (upper panel) or rat Aurora B (lower panel), and pSHAG-1 or vectors for shRNAs directed against Aurora A (A1, A2) or Aurora B (B1, B2, or B3) as indicated. Aurora A and B expression was analyzed by Western blotting (WB) using antibodies specific for Aurora A or Aurora B, GFP (transfection control), and α-tubulin (loading control). Numbers indicate molecular mass in kDa. B, hippocampal neurons were transfected at 0 d.i.v. with vectors for GFP, pSM2 (control), shRNAs directed against Aurora A (Aur A RNAi: sh-Aurora A2), Aurora B (Aur B RNAi: sh-Aurora B3), Aurora A and B (Aur A+B RNAi: sh-Aurora A2 + B3), or human GFP-Aurora A (Aur A RNAi res.) as indicated. Transfected cells were analyzed at 3 d.i.v. by staining with an anti-MAP2 (red) and the Tau-1 (blue) antibody. Scale bar, 40 μm. C, the development of neuronal polarity was analyzed by determining the number of unpolarized neurons without an axon (0, black bars), unpolarized neurons with one or more neurites of undefined identity (indeterminate neurites longer than 50 and weak of absent Tau-1 staining; n, hatched bars), polarized neurons with a single axon (1, white bars), or neurons with multiple axons (>1, gray bars) (t test: multiple indeterminate neurites: p < 0.01, Aur A RNAi, Aur A+B RNAi compared with GFP; means ± S.E.). ANOVA (p < 0.0001) and a parametric Student's t test were used to test statistical significance.
Hippocampal neurons were transfected with the shRNA expression vectors at 0 d.i.v. and analyzed at 3 d.i.v. (Fig. 3). Suppression of Aurora A by RNAi resulted in the loss of neuronal polarity. In controls, the majority of the neurons (GFP: 83 ± 2%, n = 3 experiments, 45 neurons) extended a single axon with an average length of 187 ± 6 μm (45 neurons). After knockdown of Aurora A, 58 ± 2% of the neurons lost their polarity and extended one or more MAP2-positive neurites with an increased length of 77 ± 2 μm (33 neurons). These neurites were longer than normal minor neurites but shorter than axons and showed a weak or absent Tau-1 staining. In addition, the length of the remaining axons was significantly reduced (sh-Aurora A: 139 ± 7 μm, 33 neurons). Suppression of Aurora B did not have a significant effect on axons or minor neurites. Knockdown of both Aurora kinases did not have a more severe effect than that of Aurora A alone. Coexpression of human Aurora A, which is resistant to shRNA-A2 (supplemental Fig. S2), together with the Aurora A shRNA rescued the loss of endogenous Aurora to a large extent. 76 ± 3% (23 neurons) of the neurons transfected with the vector for human Aurora A formed a single axon (control (Aurora shRNA): 33 ± 1%, 32 neurons). Thus, Aurora A but not B is required for axon formation, and the loss of Aurora A results in a loss of neuronal polarity.
Aurora Kinases Interact Directly with Par3
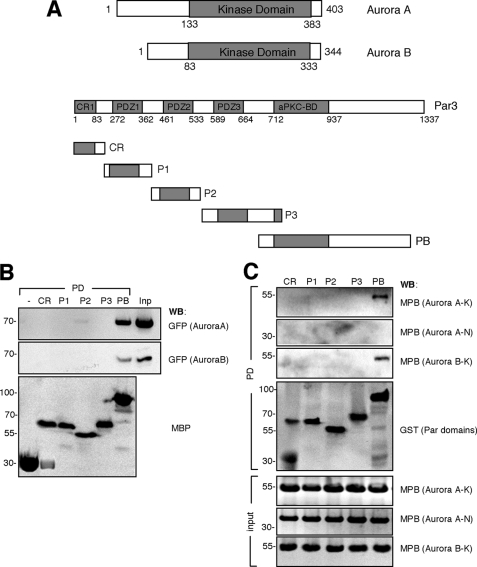
The knockdown of Aurora A showed that it is required for the formation of axons. The phenotype of suppressing Aurora A by RNAi is similar to that of a loss of Par3, which also results in the loss of axons (14). In Drosophila neuroblasts, Aurora A regulates the Par polarity complex (10). To investigate whether Aurora kinases interacts with the mammalian Par complex in neurons, a pulldown assay was performed using MBP fusion proteins of individual Par3 domains and GFP-tagged Aurora A. These experiments showed that Aurora A and B interact with Par3 and identified a Par3 fragment comprising the aPKC binding domain (Par3-aPKCBD) as the domain that mediates interaction with Aurora A and B (Fig. 4). To confirm that this interaction is direct, bacterially expressed MBP fusion proteins for different domains from Aurora A or B were used in a pulldown assay with bacterially expressed GST fusion proteins for different Par3 domains. These experiments confirmed a direct interaction between Par3-aPKCBD and the kinase domain of Aurora A and B (Fig. 4C).
FIGURE 4.
Interaction of Aurora kinases with Par3. A, schematic representations of Aurora A and B, Par3, and individual domains of Par3 (CR, CR1, conserved region 1, amino acids 1–200; P1–3, PDZ domains 1–3, P1, amino acids 200–400, P2, 400–560; P3, 560–780; PB, aPKCBD, 700–1337) are shown. The numbers refer to the positions in the amino acid sequence. B, bacterially expressed MBP fusion proteins of different Par3 domains were coupled to amylose beads as indicated and incubated with lysates of HEK 293T cells transfected with the expression for GFP-tagged Aurora A or Aurora B (Inp, input). The expression of comparable amounts of protein was confirmed by Western blotting (WB) using anti-MBP and anti-GFP antibodies. Numbers indicate molecular mass in kDa. C, bacterially expressed GST fusion proteins of different Par3 domains (CR, P1, P2, P3, and PB) were coupled to glutathione-Sepharose beads as indicated and incubated with bacterially expressed MBP fusion proteins of the N-terminal (Aurora A–N, amino acids 1–133) or kinase domains of Aurora A (Aurora A–K, amino acids 133–403) or B (Aurora B–K, amino acids 83–344). The expression of comparable amounts of protein was confirmed by Western blotting using anti-MBP and anti-GST antibodies. Numbers indicate molecular mass in kDa.
Aurora A Phosphorylates Par3 at Ser962
To determine whether Aurora A phosphorylates Par3, wild type and kinase-dead mutants for Aurora A and B were coexpressed with Par3-aPKCBD in HEK 293T cells. Phosphorylated proteins were immunoprecipitated using an anti-phosphoserine antibody and analyzed by Western blotting (Fig. 5A). Two proteins were detected at 100 and 70 kDa that correspond to phosphorylated Par3 and autophosphorylated Aurora A. Coexpression of inactive Aurora A did not result in detectable phosphorylation of Par3. Aurora B phosphorylated Par3 only weakly. To confirm that Aurora A directly phosphorylates Par3, an in vitro kinase assay was performed using [γ-32P]ATP and the purified MBP fusion proteins for wild type or a kinase-dead Aurora A or B together with a purified MBP fusion protein of the Par3-aPKCBD. The kinase assay showed that the Par3-aPKCBD was phosphorylated by wild type Aurora A, whereas no signal was obtained for inactive Aurora A (kinase-dead) or Aurora B (Fig. 5B). In addition, signals for autophosphorylated Aurora A and Aurora B were detectable.
FIGURE 5.
Aurora A kinase phosphorylates Par3. A, HEK 293T cells were transfected with expression vectors for GFP-Aurora and GFP-Par3-aPKCBD. Phosphorylated proteins were immunoprecipitated (IP) with an anti-phosphoserine antibody. The expression of comparable amounts of GFP-tagged protein was analyzed by Western blotting (WB) using an anti-GFP antibody. Numbers indicate molecular mass in kDa. B, bacterially expressed Aurora A (AuA), Aurora B (AuB), and kinase dead-Aurora A (Aukd) were purified and incubated with purified MBP fusion proteins of Par3-aPKCBD in kinase buffer containing [γ-32P]ATP. Proteins were resolved by autoradiography (32P, top) and staining with Coomassie Blue (CB, bottom).
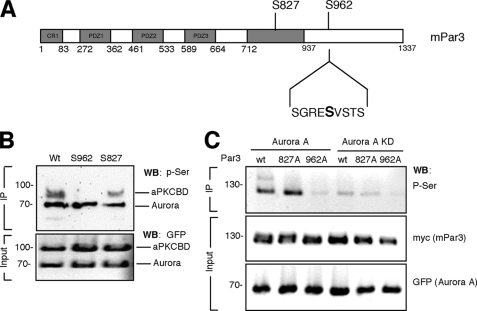
Par3 contains two sequences that match the consensus RX(ST)(LV) for Aurora A phosphorylation (30, 31) at positions 219–222 (RSSL) and 960–963 (RESV) located within the aPKCBD. To test whether Aurora A phosphorylates Par3 at Ser962, wild type Par3-aPKCBD and the nonphosphorylatable S962A and S827A mutants were expressed in HEK 293T cells, dephosphorylated, purified by immunoprecipitation, and used for an in vitro kinase assay with Aurora A. The nonphosphorylatable mutant aPKCBD S827A for the aPKC phosphorylation site was still phosphorylated by Aurora A (Fig. 6B). Only wild type Par3-aPKCBD and the S827A mutant could be phosphorylated by Aurora A, but no signal indicating phosphorylation was detectable for the S962A mutant.
FIGURE 6.
Aurora A kinase phosphorylates Par3 at Ser962. A, a schematic representation of Par3 and the sequence surrounding Ser962 (amino acids 958–966) is shown. The numbers refer to the positions in the amino acid sequence. B, wild type (Wt) and mutant GFP-Par3-aPKCBD (Wt, S827A, S962A) and GFP-tagged Aurora A were expressed separately in HEK 293T cells and purified by immunoprecipitation (IP) with an anti-GFP antibody. Purified kinases and GFP-Par3-PKCBD were mixed and incubated in kinase buffer. Phosphorylated Par3-aPKCBD was detected by Western blotting (WB) using an anti-phosphoserine (p-Ser) antibody C, Myc-tagged wild type (wt) or mutant (S827A, S962A) full-length Par3 were immunoprecipitated with an anti-myc antibody from lysates of transfected HEK 293T cells after coexpression with GFP-tagged wild type (Aurora A) or kinase-dead (KD) Aurora A. Phosphorylated proteins and the expression of comparable amounts of myc-tagged and GFP-tagged proteins in lysates were analyzed by Western blotting using anti-phosphoserine, anti-myc and anti-GFP antibodies, respectively. Numbers indicate molecular mass in kDa.
To confirm that Aurora A phosphorylate Par3 at Ser962, HEK 293T cells were cotransfected with vectors for wild type or the nonphosphorylatable full-length Par3 S827A (aPKC phosphorylation site) and S962A mutants and wild type or kinase-dead Aurora A (Fig. 6C). Proteins were immunoprecipitated and phosphorylated proteins detected by Western blotting using an anti-phosphoserine antibody. A strong signal was detected for wild type and S827A Par3 but not for Par3-S962A or when kinase-dead Aurora A were expressed. These results show that Aurora A phosphorylates Par3 at Ser962.
Phosphorylation of Par3 at Ser962 Is Required for Par3 Function in Neurons
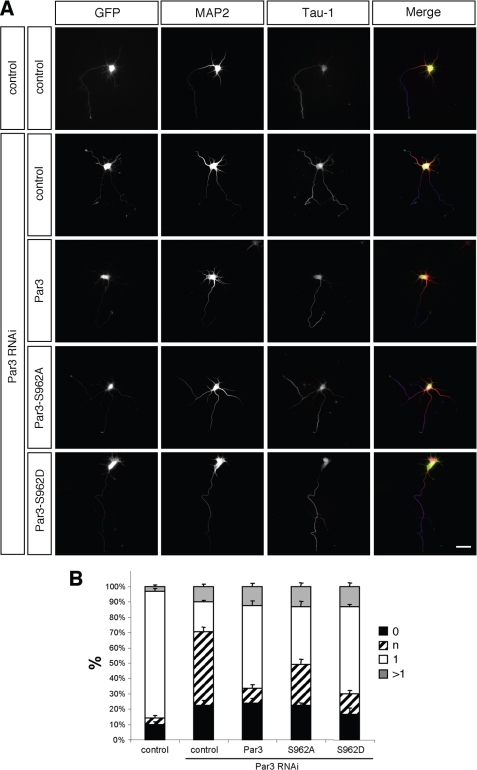
To investigate whether phosphorylation of Par3 at Ser962 is required for axon formation, we tested the ability of the phosphomimic Par3-S962D and the nonphosphorylatable Par3-S962A mutants to rescue the loss of endogenous Par3 (Fig. 7). Hippocampal neurons were transfected with expression vectors for a Par3 shRNA and wild type or mutant Par3 that are resistant to the RNAi vector. Suppression of Par3 blocked axon formation and resulted in the formation of multiple indeterminate neurites (Fig. 7; 48 ± 4%, n = 3 experiments, 37 neurons) as described previously (14). The expression of wild type Par3 or Par3-S962D rescued the loss of endogenous Par3 to a large extent (RNAi + Par3: 10 ± 1% neurons with indeterminate neurites, 40 neurons; RNAi + Par3-S962D: 13 ± 1%; 31 neurons, p < 0.001 compared with Par3-RNAi). There was no significant difference in the rescue by wild type Par3 and Par3-S962D. By contrast, Par3-S962A showed a reduced ability to rescue the loss of endogenous Par3 (27 ± 1% neurons with indeterminate neurites, 44 neurons, p < 0.001, control + Par3 RNAi compared with S962A + Par3 RNAi). The phenotype of neurons expressing Par3-S962A instead of the endogenous Par3 after its knockdown is similar to that of neurons after suppression of Aurora A. This indicates that phosphorylation of Par3 at Ser962 promotes its function during axon formation.
FIGURE 7.
Role of Par3 phosphorylation for neuronal polarity. A and B, hippocampal neurons were transfected at 0 d.i.v. with vectors for GFP (control), myc-tagged Par3, Par3-S962A (S962A), or Par3-S962D (S962D), and pSM2 (control) or a vector for an shRNA directed against rat Par3 (RNAi), as indicated. Transfected cells were analyzed at 3 d.i.v. by staining with an anti-MAP2 (red) and the Tau-1 (blue) antibody. Scale bar, 40 μm. B, the development of neuronal polarity was analyzed by determining the number of unpolarized neurons without an axon (0, black bars), unpolarized neurons with one or more neurites of undefined identity (indeterminate neurites longer than 50 and weak of absent Tau-1 staining; n, hatched bars), polarized neurons with a single axon (1, white bars), and neurons with multiple axons (>1, gray bars) (t test: multiple indeterminate neurites: p < 0.001, control + Par3 RNAi compared with control, wild type Par3 + Par3 RNAi, S962D + Par3 RNAi, S962A + Par3 RNAi, and S962A + Par3 RNAi compared with wild type Par3 + Par3 RNAi, S962D + Par3 RNAi; means ± S.E.). ANOVA (p < 0.0001) and a parametric Student's t test were used to test statistical significance.
DISCUSSION
In addition to its function during the cell cycle, Aurora A also regulates asymmetric cell division in Drosophila neuroblasts by phosphorylating Par6 (10). Here, we show that rodent Aurora A phosphorylates Par3 at Ser962 and that Par3 Ser962 phosphorylation is required in hippocampal neurons for the formation of axons.
Aurora A and B are expressed in hippocampal neurons during the initial stages of their differentiation. Suppression of Aurora A by RNAi results in the loss of neuronal polarity. After knockdown of Aurora A, neurons formed multiple neurites that were longer than minor neurites but lacked the characteristics of axons. These neurites were shorter than axons and showed no or only weak staining for the axonal marker Tau-1. Expression of inactive Aurora A had a similar phenotype, indicating that it has a dominant negative effect. Overexpression of Aurora A did not affect axon formation, probably because it has to be activated by upstream kinases or by association with other proteins (1, 3–6). Alternatively, Aurora kinases might not be sufficient by themselves to have an effect on axon formation. Although Aurora B is also expressed in hippocampal neurons, expression of inactive Aurora B or its suppression by RNAi did not affect axon formation. These results demonstrate a novel function for Aurora A, which belongs to a growing list of proteins with important functions in cell cycle regulation and in postmitotic neurons (32).
The phenotype of suppressing endogenous Aurora A in neurons is similar to that of Par3 (14). We could show that Aurora A interacts directly with the Par3-aPKCBD and phosphorylates it. The phosphorylation of Par3 at Ser962 is important for axon formation. Wild type Par3 and the phosphomimic mutant Par3-S962D rescued the loss of endogenous Par3. By contrast, the nonphosphorylatable mutant Par3-S962A could not efficiently compensate for the knockdown of Par3. These results suggests that phosphorylation of Par3 at Ser962 by Aurora A promotes its function in axon formation.
The ability to rescue the loss of endogenous Par3 by Par3-S962D but not Par3-S962A indicates that Par3 is one of the targets for Aurora A in hippocampal neurons. Unlike Par6, Par3-Ser962 phosphorylation does not appear to be involved in aPKC activation because the inhibition of aPKC (15) and suppression of Aurora A result in different phenotypes. Par3 phosphorylation at Ser962 could change the affinity for its interaction partners, as it has been reported for other kinases such as aPKC, ROK, and Src family kinases (33–35). Phosphorylation of Par3 at Tyr1127 reduces the association of Par3 with LIM kinase 2 (35, 36). aPKC phosphorylates Par3 at Ser827, which decreases its affinity for aPKC (33). The Aurora A phosphorylation site is located close to the Par3-aPKCBC. This region of Par3 contains the binding sites for aPKC, Kif3A, Tiam-1, Stef, Mark2, and Numb (17, 18, 33, 37–41). However, preliminary results show that Aurora A phosphorylation does not affect the binding of these proteins. Future experiments will have to identify additional binding partners of Par3 that might be affected by Aurora A.
Aurora A is necessary for the polarization of Drosophila neuroblasts and regulates aPKC recruitment to the Par complex (10). Aurora A phosphorylates Par6 and releases its inhibition of aPKC. Although Aurora A also phosphorylates mammalian Par6, the phosphorylation of Par3 does not appear to be conserved. Sequence comparison shows that the Aurora A phosphorylation site in Par3 is conserved in vertebrates but is not present in Drosophila Bazooka.
In summary, our results show that Aurora A is required for the establishment of neuronal polarity. It binds to and phosphorylates Par3. The phosphorylation of Par3 at Ser962 contributes to its function in the establishment of neuronal polarity. The loss of Aurora A or a failure to phosphorylate Par3 at Ser962 leads to the loss of axons and the formation of multiple indeterminate neurites.
Supplementary Material
Acknowledgments
We thank Drs. Nigg and Macara for plasmids.
This work was supported by Deutsche Forschungsgemeinschaft Grants SFB 629 and GRK 1050 (to A. W. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
- Par
- partitioning-defective
- Par6
- mammalian partitioning-defective 6
- ANOVA
- analysis of variance
- aPKC
- atypical protein kinase C
- aPKCBD
- aPKC binding domain
- d.i.v.
- days in vitro
- E18
- embryonic day 18
- GFP
- green fluorescent protein
- GST
- glutathione S-transferase
- MAP
- microtubule-associated protein
- Mark2
- microtubule affinity-regulating kinase 2
- MBP
- maltose-binding protein
- RNAi
- RNA interference
- shRNA
- short hairpin RNA
- Smurf2
- Smad ubiquitination regulatory factor 2.
REFERENCES
- 1.Carmena M., Earnshaw W. C. (2003) Nat. Rev. Mol. Cell Biol. 4, 842–854 [DOI] [PubMed] [Google Scholar]
- 2.Huang Y. S., Jung M. Y., Sarkissian M., Richter J. D. (2002) EMBO J. 21, 2139–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eyers P. A., Erikson E., Chen L. G., Maller J. L. (2003) Curr. Biol. 13, 691–697 [DOI] [PubMed] [Google Scholar]
- 4.Hirota T., Kunitoku N., Sasayama T., Marumoto T., Zhang D., Nitta M., Hatakeyama K., Saya H. (2003) Cell 114, 585–598 [DOI] [PubMed] [Google Scholar]
- 5.Hutterer A., Berdnik D., Wirtz-Peitz F., Zigman M., Schleiffer A., Knoblich J. A. (2006) Dev. Cell 11, 147–157 [DOI] [PubMed] [Google Scholar]
- 6.Tsai M. Y., Wiese C., Cao K., Martin O., Donovan P., Ruderman J., Prigent C., Zheng Y. (2003) Nat. Cell Biol. 5, 242–248 [DOI] [PubMed] [Google Scholar]
- 7.Berdnik D., Knoblich J. A. (2002) Curr. Biol. 12, 640–647 [DOI] [PubMed] [Google Scholar]
- 8.Lee C. Y., Andersen R. O., Cabernard C., Manning L., Tran K. D., Lanskey M. J., Bashirullah A., Doe C. Q. (2006) Genes Dev. 20, 3464–3474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang H., Somers G. W., Bashirullah A., Heberlein U., Yu F., Chia W. (2006) Genes Dev. 20, 3453–3463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wirtz-Peitz F., Nishimura T., Knoblich J. A. (2008) Cell 135, 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arimura N., Kaibuchi K. (2007) Nat. Rev. Neurosci. 8, 194–205 [DOI] [PubMed] [Google Scholar]
- 12.Suzuki A., Ohno S. (2006) J. Cell Sci. 119, 979–987 [DOI] [PubMed] [Google Scholar]
- 13.Schwamborn J. C., Püschel A. W. (2004) Nat. Neurosci. 7, 923–929 [DOI] [PubMed] [Google Scholar]
- 14.Schwamborn J. C., Khazaei M. R., Püschel A. W. (2007) J. Biol. Chem. 282, 35259–35268 [DOI] [PubMed] [Google Scholar]
- 15.Shi S. H., Jan L. Y., Jan Y. N. (2003) Cell 112, 63–75 [DOI] [PubMed] [Google Scholar]
- 16.Dotti C. G., Sullivan C. A., Banker G. A. (1988) J. Neurosci. 8, 1454–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nishimura T., Kato K., Yamaguchi T., Fukata Y., Ohno S., Kaibuchi K. (2004) Nat. Cell Biol. 6, 328–334 [DOI] [PubMed] [Google Scholar]
- 18.Nishimura T., Yamaguchi T., Kato K., Yoshizawa M., Nabeshima Y., Ohno S., Hoshino M., Kaibuchi K. (2005) Nat. Cell Biol. 7, 270–277 [DOI] [PubMed] [Google Scholar]
- 19.Schwamborn J. C., Müller M., Becker A. H., Püschel A. W. (2007) EMBO J. 26, 1410–1422 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Chen Y. M., Wang Q. J., Hu H. S., Yu P. C., Zhu J., Drewes G., Piwnica-Worms H., Luo Z. G. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 8534–8539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang H., Guo W., Liang X., Rao Y. (2005) Cell 120, 123–135 [DOI] [PubMed] [Google Scholar]
- 22.Yoshimura T., Kawano Y., Arimura N., Kawabata S., Kikuchi A., Kaibuchi K. (2005) Cell 120, 137–149 [DOI] [PubMed] [Google Scholar]
- 23.Honda R., Körner R., Nigg E. A. (2003) Mol. Biol. Cell 14, 3325–3341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joberty G., Petersen C., Gao L., Macara I. G. (2000) Nat. Cell Biol. 2, 531–539 [DOI] [PubMed] [Google Scholar]
- 25.Paddison P. J., Caudy A. A., Bernstein E., Hannon G. J., Conklin D. S. (2002) Genes Dev. 16, 948–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paddison P. J., Caudy A. A., Hannon G. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1443–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwamborn J. C., Li Y., Püschel A. W. (2006) Methods Enzymol. 406, 715–727 [DOI] [PubMed] [Google Scholar]
- 28.Mandell J. W., Banker G. A. (1996) J. Neurosci. 16, 5727–5740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walter A. O., Seghezzi W., Korver W., Sheung J., Lees E. (2000) Oncogene 19, 4906–4916 [DOI] [PubMed] [Google Scholar]
- 30.Cheeseman I. M., Anderson S., Jwa M., Green E. M., Kang J., Yates J. R., 3rd, Chan C. S., Drubin D. G., Barnes G. (2002) Cell 111, 163–172 [DOI] [PubMed] [Google Scholar]
- 31.Ferrari S., Marin O., Pagano M. A., Meggio F., Hess D., El-Shemerly M., Krystyniak A., Pinna L. A. (2005) Biochem. J. 390, 293–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank C. L., Tsai L. H. (2009) Neuron 62, 312–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagai-Tamai Y., Mizuno K., Hirose T., Suzuki A., Ohno S. (2002) Genes Cells 7, 1161–1171 [DOI] [PubMed] [Google Scholar]
- 34.Nakayama M., Goto T. M., Sugimoto M., Nishimura T., Shinagawa T., Ohno S., Amano M., Kaibuchi K. (2008) Dev. Cell 14, 205–215 [DOI] [PubMed] [Google Scholar]
- 35.Wang Y., Du D., Fang L., Yang G., Zhang C., Zeng R., Ullrich A., Lottspeich F., Chen Z. (2006) EMBO J. 25, 5058–5070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X., Macara I. G. (2006) J. Cell Biol. 172, 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benton R., St. Johnston D. (2003) Cell 115, 691–704 [DOI] [PubMed] [Google Scholar]
- 38.Chen X., Macara I. G. (2005) Nat. Cell Biol. 7, 262–269 [DOI] [PubMed] [Google Scholar]
- 39.Nishimura T., Kaibuchi K. (2007) Dev. Cell 13, 15–28 [DOI] [PubMed] [Google Scholar]
- 40.Traweger A., Wiggin G., Taylor L., Tate S. A., Metalnikov P., Pawson T. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10402–10407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang H., Macara I. G. (2006) Nat. Cell Biol. 8, 227–237 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.