Abstract
Afflicted neurons in Alzheimer disease have been shown to display an imbalance in the expression of TrkA and p75NTR at the cell surface, and administration of nerve growth factor (NGF) has been considered and attempted for treatment. However, wild-type NGF causes extensive elaboration of neurites while providing survival support. This study was aimed at developing recombinant NGF muteins that did not support neuritogenesis while maintaining the survival response. Critical residues were identified at the ligand-receptor interface by point mutagenesis that played a greater importance in neuritogenesis versus survival. By combining point mutations, two survival-selective recombinant NGF muteins, i.e./7-84-103 and KKE/7-84-103, were generated. Both muteins reduced neuritogenesis in PC12 (TrkA+/p75NTR+) cells by >90%, while concurrently retaining near wild-type survival activity in MG139 (TrkA+ only) and PCNA fibroblast (p75NTR+-only) cells. Additionally, survival in both naive and terminally differentiated PC12 cells was shown to be intermediate between NGF and negative controls. Dose-response curves with 7-84-103 showed that the differentiation curve was shifted by about 100-fold, whereas the EC50 for survival was only increased by 3.3-fold. Surface plasmon resonance analysis revealed a 200-fold decrease in binding of 7-84-103 to TrkA. The retention of cell survival was attributed to maintenance of signaling through the Akt survival pathway with reduced MAPK signaling for differentiation. The effect of key mutations along the NGF receptor interface are transmitted inside the cell to enable the generation of survival-selective recombinant NGF muteins that may represent novel pharmacologic lead agents for the amelioration of Alzheimer disease.
Introduction
Neurotrophins are a family of closely related proteins that have diverse functions ranging from neuronal survival and differentiation to regulation of axonal and dendritic outgrowth, synapse formation, cell migration, and cellular proliferation (1–5). The family of neurotrophins was established with the discovery of its founding member, nerve growth factor (NGF)3 (3, 6, 7). Brain-derived neurotrophic factor (BDNF), neurotrophin 3 (NT-3), and neurotrophin 4/5 (NT-4/5) possess a high sequence homology (∼50%) and adopt similar tertiary structures (7–12). Neurotrophins bind selectively to a family of 140-kDa Tropomyosin-receptor-kinases (Trk), i.e. NGF to TrkA, brain-derived neurotrophic factor and NT-4 to TrkB, and NT-3 to TrkC, via interactions with a nanomolar affinity (Kd ∼10−9 m) (13–16). All neurotrophins can also bind a 75-kDa common neurotrophin receptor, p75NTR, via similar affinity interactions (17, 18).
NGF is a target-derived polypeptide synthesized by the hippocampus and retrogradely transported within axons of cholinergic interneurons (striatum), magnocellular cholinergic neurons (basal forebrain), and Purkinje cells (cerebellum) (19–22). NGF is secreted as a propeptide of 305 residues (∼35 kDa), pro-NGF, and subsequently cleaved at the N terminus by serine proteases to generate a smaller 120-amino acid mature polypeptide (∼13 kDa) (23, 24). The folded monomer is stabilized by three disulfide bridges that form the core cystine-knot motif, characteristic of all neurotrophins (25, 26). Each monomer is composed of two pairs of antiparallel β-strands connected by four β-hairpin loops (25). NGF possesses highly conserved hydrophobic residues along the elongated central axis of the molecule, which facilitates its tight association into a noncovalent homodimer (Kd <10−13 m) (25, 27). The solvent-exposed β-hairpin loops (I–V), along with the N and C termini, are highly variable across neurotrophins and play a functional role in establishing specificity in receptor activation (12, 17, 26, 28–31).
The crystal structure of a symmetric complex of dimeric NGF bound to a pair of TrkA-d5 domains identified two critical patches of similar sizes along the ligand-receptor interface (25). The conserved patch consists of the central β-sheet forming the core of homodimeric NGF and the C-terminal loops of TrkA-d5. This patch is highly conserved among all the neurotrophins suggesting an evolutionarily conserved binding motif (25, 30). The most strictly conserved residue is Arg103, which is implicated to be the most important binding determinant, especially for the interaction between NT-3 and TrkC (25, 32). Most of the residues that mediate high affinity interactions between NGF and TrkA lie in the specificity patch that consists of the solvent-accessible N terminus of NGF and the ABED sheet of TrkA. Upon interacting with the hydrophobic pocket of the ABED sheet, residues 6–9 adopt a helical conformation with their side chains buried within the interface. Deletion of residues in the N terminus greatly decreases activity (33, 34). Moreover, examination of the Δ9/13 NGF mutein4 (deletion of residues 9–13) demonstrated a greater than 100-fold decrease in affinity to TrkA while maintaining binding to p75NTR (33). An extended binding surface was described for TrkA binding along NGF variable regions I, II, and V of the NGF dimer (12, 23, 29–31, 35, 36), and a mutein selective for TrkA binding over p75NTR by mutation of Lys34, Lys36, and Glu35 to alanine was developed (29). Moreover, a histidine in the conserved patch, His84, was shown to be important for activity (37). However, many of these early studies did not focus on discriminating between various cellular activities such as survival and differentiation.
NGF binding induces TrkA receptor dimerization, trans-autophosphorylation of a specific set of tyrosine residues, and intracellular tyrosine kinase domain activation (6, 38–40). Three major signaling pathways are subsequently activated. The PI 3-kinase pathway activates the pro-survival factor Akt and accounts for over 80% of neurotrophin-mediated survival in neurons (14, 41, 42). The MAPK pathway has been implicated in both survival and differentiation (14, 17). Finally, the phospholipase Cγ pathway is involved primarily in Ca2+ regulation (38). Signaling via the p75NTR has been thought to be involved in the induction of negative signals like apoptosis and growth arrest through its death domain (14, 43). However, the presence of Trk in proximity to p75NTR has been shown to silence pro-apoptotic pathways and activate the pro-survival transcription factor, NF-κB, which in turn activates Akt (6, 14, 44, 45). Thus, both receptors converge in the mediation of survival by synergistically activating Akt in cells co-expressing TrkA and p75NTR. Experimental evidence has suggested that the affinity of neurotrophin binding as well as the efficiency of signaling are augmented with the expression of both receptors. Thus, “high affinity receptor” binding appears to include contributions from both p75NTR and Trk receptors (17, 46–49), although other viewpoints exist (50).
The importance of these receptor interactions is well demonstrated in neurodegenerative conditions such as Alzheimer disease. Protein immunohistochemistry and mRNA hybridization studies led to the discovery that protein levels and mRNA levels for both receptors are reduced in early Alzheimer disease even in the presence of stable NGF expression (51–54). A shift from mature NGF to pro-NGF, which binds p75NTR and induces apoptosis (55), was also noted (54–56). Thus, the unopposed neurodegeneration in Alzheimer disease is exacerbated by a reduction in signaling from TrkA receptors coupled with apoptotic signal enhancement by pro-NGF binding p75NTR (17, 51, 54, 57). A small phase I clinical trial of ex vivo NGF gene delivery showed promise in curtailing neuronal apoptosis in eight patients with mild Alzheimer disease (58). At the 22-month follow up, Mini-Mental State Exam scores demonstrated a reduction in rates of cognitive decline, whereas positron emission tomography scans demonstrated significantly greater cortical glucose uptake. Although these observations were highly encouraging, at autopsy the cholinergic basal forebrain in these patients demonstrated a trophic response of immature neuritic processes (59). Although this response is necessary for target innervation during development, we suggest that adult brains afflicted with Alzheimer disease possessing an established axonal infrastructure may further suffer from this type of new and incomplete neuritogenesis.
Therefore, our study was directed at the generation of a survival-specific recombinant NGF mutein with lessened neuritogenesis. By using a limited set of rationally selected mutations within the specificity, conserved, and variable regions (implicated as the three most important receptor binding epitopes on NGF), we proposed to identify key residues along the ligand-receptor interface that played a more discriminatory role in signal transduction. By combining mutations to these key residues, we were able to generate muteins capable of selectively activating a signal transduction pathway for survival.
MATERIALS AND METHODS
Site-directed Mutagenesis
Single residue point mutations were made to wild-type mβ-NGF in the pBlueBac vector using the QuikChange® site-directed mutagenesis kit (Stratagene). Site-specific oligonucleotide primers, harboring the point mutations of interest, were designed using Scitools™ and Biotools™. A single set of forward and reverse primers was used per reaction to generate the desired point mutations. Briefly, insert-containing vectors were PCR-amplified with mutagenic primers using 2.5 units of the ultra-efficient, heat-stable PfuUltra™ DNA polymerase (200 mm Tris-HCl, pH 8.8, 20 mm MgSO4, 100 mm KCl, 100 mm (NH4)2SO4, 1% Triton X-100, 1 μg/ml bovine serum albumin). Incubation with 10 units of DpnI (50 mm potassium acetate, 20 mm Tris acetate, 10 mm magnesium acetate, 1 mm dithioerythritol, pH 7.9) at 37 °C for 60 min completely digested methylated parental cDNA. Escherichia coli were transformed with the PCR products and grown at 37 °C overnight. Isolated colonies were picked for a small scale amplification and sequence verification. Positive clones, harboring the point mutations of interest, subsequently underwent a large scale amplification and sequence verification.
Multisite-directed Mutagenesis
Multiple point mutations were made to wild-type mβ-NGF or to the KKE mβNGF mutant (K32A/K34A/E35A) in the pBlueBac vector using QuikChange® multisite-directed mutagenesis kit (Stratagene). Up to three mutant unidirectional primers, oriented in either the 5′ → 3′ or 3′ → 5′ direction, were used to generate triple mutations in wild-type mβNGF or KKE mβNGF; primers were borrowed from the previous site-directed mutagenesis protocol. Briefly, insert-containing vectors were PCR-amplified with 2.5 units of enzyme blend with PfuTurbo™ DNA polymerase (200 mm Tris-HCl, pH 8.8, 20 mm MgSO4, 100 mm KCl, 100 mm (NH4)2SO4, 1% Triton X-100, 1 μg/ml bovine serum albumin). Parental strand digestion and transformations into E. coli were performed as described previously. Isolated colonies were picked, amplified, sequenced, and further decontaminated (of RNA and protein contaminants). Positive clones were concentrated to 1 μg/μl.
Cell Lines and Culturing Conditions
Sf21 cells, from the fall armyworm, Spodoptera frugiperda, were grown in Grace's insect cell medium (catalog number 11605, Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum at 27 °C and subcultured every 2 days (60). For expression assays, Sf21 cells were grown in serum-free Excell-401 medium at 27 °C for 5 days. Cells were visualized using phase contrast at 20–200-fold magnification with a Nikon Diaphot microscope. PC12 cells (TrkA+, p75NTR+) (a kind gift from L. A. Greene, New York University), PCNA cells (TrkA−, p75NTR+) (a kind gift from E. M. Shooter, Stanford University), and MG139 cells (TrkA+, p75NTR−) (a kind gift from P. A. Baker, McGill University) were grown in complete Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% heat-inactivated horse serum, 5% heat-inactivated fetal bovine serum, 4.5 mg/ml glucose, 4 mm glutamine, 100 units penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin B at 37 °C in a humid environment with 6.5% CO2, and subcultured every 3 days (61–63). For the neuritogenesis assay, DMEM-N1, supplemented with 20 nm progesterone, 100 μm putrescine, 5 μg/ml insulin, 5 ng/ml selenium, 5 μg/ml transferrin, 4 mm glutamine, 100 units penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin B, was used for growth of PC12 cells at 37 °C in a humid environment with 6.5% CO2 for 3 days (64). For survival assays, serum-free DMEM, supplemented with 4 mm glutamine, 100 units of penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin B, was used for growth of PCNA and MG139 cells at 37 °C in a humid environment with 6.5% CO2 for up to 120 h. For immunoprecipitation studies, cells were first grown in complete DMEM for 72 h at 37 °C in a humid environment with 6.5% CO2 and then grown for an additional 24 h with complete DMEM diluted 1:30.
Antibodies
Rabbit polyclonal NGF antibody (M-20) raised against the N terminus of mouse βNGF was from Santa Cruz Biotechnology, Inc. Rabbit polyclonal NGF antibody (MC-51) raised against intact mouse βNGF was from Cedarlane Labs (CLMCNET-051, Ontario, Canada). Rabbit polyclonal TrkA (C-14) antibody raised against a peptide within the extracellular domain of human TrkA was from Santa Cruz Biotechnology, Inc. Monoclonal anti-phosphotyrosine antibody, generated against mouse hybridoma 4G10 cells, was from Upstate Cell Signaling Solutions. Rabbit polyclonal antibodies, p44/42 MAPK, phospho-p44/42 MAPK (Thr202/Tyr204), Akt, and phospho-Akt (Ser473), were from Cell Signaling Technology, Inc. Horseradish peroxidase-conjugated goat anti-rabbit IgG was from Kirkegaard & Perry Laboratories, Inc. Horseradish peroxidase-conjugated donkey anti-goat IgG was from Santa Cruz Biotechnology, Inc. Horseradish peroxidase-conjugated goat anti-mouse IgG was from Bio-Rad.
Transfection into Sf21 Cells
Baculoviral transfer vector (pBlueBac 4.5), containing the gene of insert (wild-type or mutated mβNGF), was co-transfected with triple cut, linearized Bac-N-Blue® viral DNA (Invitrogen) into Sf21 insect cells. The transfer vector and viral DNA were incubated with Cellfectin® reagent for 15 min before incubation with Sf21 cells (2 × 106 cells) in a 60-mm dish supplemented with serum-free Grace's insect cell medium. Transfected Sf21 cells were subsequently maintained at 27 °C in Grace's insect cell media supplemented with 10% heat-inactivated fetal bovine serum in a sealed plastic bag. At 72 h post-transfection, medium was aspirated for a subsequent plaque assay. High titer baculoviral stocks were then generated and ranged between 1.4 × 107 and 2.7 × 108 plaque-forming units/ml.
Recombinant NGF Mutein Expression
Expression of recombinant wild-type or mutant NGF (rwtNGF or recombinant mutein NGF) was performed according to the protocol from Invitrogen with certain modifications. Briefly, Sf21 insect cells were adapted to serum-free Excel-401 medium and grown in 25-cm2 flasks to 80% confluency (4 × 106 cells). Cells were then infected with rwtNGF or recombinant mutein NGF baculovirus at a high multiplicity of infection (5–10). At 120 h post-infection, cultures were aspirated and centrifuged for 5 min at 3,000 × g to remove cellular debris. Expression levels were quantified via Western blot analysis following SDS-PAGE separation using the MC-51 antibody. Yields in crude medium ranged from 0.25 to 2.0 mg/liter and were used without further purification after careful standardization by the immunoblots (65).
Survival Assay (Trypan Blue)
Exponentially dividing MG139, PCNA, and PC12 cells were grown in serum-free DMEM and seeded at a concentration of 3.0 × 103 cells/well in collagen-coated 96-well plates or at a concentration of 5.0 × 105 cells/well in poly-l-ornithine coated 24-well plates. Cells were treated with 2 nm rwtNGF, 2 nm recombinant mutein NGF, or vector alone and grown at 37 °C, 6.5% CO2 for 24, 48, 72, or 96 h in serum-free DMEM. At 24-h blocks, ∼50 μl of 0.4% trypan blue dye solution was added to each well and incubated for 1 min at room temperature. The entire volume of the well was then discarded, and the fraction of surviving cells was determined in triplicate by counting at least 200 single cells/well.
Survival Assay (XTT)
Exponentially dividing MG139, PCNA, and PC12 cells were grown in serum-free DMEM and seeded at a concentration of 5 × 103 cells/well (MG139) or 1 × 104 cells/well (PCNA and PC12) in uncoated 96-well plates. Cells were treated with 2 nm rwtNGF, 2 nm recombinant mutein NGF, or vector alone and grown at 37 °C, 6.5% CO2 for 72 h (MG139 and PC12) or 96 h (PCNA) in serum-free DMEM. To determine the percentage of surviving, metabolically active cells in each well, 50 μl of XTT dye solution was added to each well at the appropriate time point, i.e. 72 or 96 h, and incubated at 37 °C, 6.5% CO2 for an additional 4–6 h. The number of cells actively converting the dye to the formazan metabolite was determined in triplicate using a scanning multiwell spectrophotometer (TECAN) measuring absorbance at 492 nm and subtracting background at 650 nm.
PC12 Terminal Differentiation
This procedure was performed as described by Vaghefi et al. (66). Exponentially dividing PC12 cells were grown in complete DMEM. Terminal differentiation was performed by placing PC12 cells in poly-l-ornithine-coated 96-well plates in the presence of 15% serum and 2 nm mβNGF for 14 days. To ensure maximum viability of the terminally differentiated PC12 cells, NGF and serum were replaced every 3 days.
PC12 Neurite Outgrowth Assay
This bioassay was performed as described by Reinhold and Neet (64). Exponentially dividing PC12 cells were grown in defined medium. Cells were seeded at a concentration of 3.0 × 103 cells/well in collagen-coated 96-well plates. Cells were incubated for 24 h at 37 °C, 6.5% CO2 in complete DMEM before treatment with rwtNGF, recombinant mutein NGF, or vector alone. Cells were incubated at 37 °C, 6.5% CO2 for an additional 48 h. Percent neuritogenesis was determined in triplicate by counting at least 200 single cells per well and calculating the proportion of cells possessing processes longer than 1.5 times their cell body in length.
Immunoprecipitation and Immunoblotting
These assays were performed as described in the protocol of Upstate Cell Signaling Solutions with slight modifications (67). Exponentially dividing PC12 cells were grown in complete medium until 90% confluency, washed, and re-incubated overnight with complete medium diluted 1:30 (0.5% serum). Cells were subsequently incubated with 2 nm mβNGF, 2 nm recombinant mutein NGF, or buffer alone at 37 °C for 20 min. Cells were washed three times with phosphate-buffered saline to remove excess media and growth factors. RIPA buffer (protease inhibitor mixture, 1 mm phenylmethanesulfonyl fluoride, 1 mm activated sodium orthovanadate, 1 mm sodium fluoride) was then added to induce cell lysis. Cells were rocked at 4 °C for 15–20 min and centrifuged to remove debris. Cell lysates were used for MAPK and Akt phosphorylation studies and immunoprecipitation studies of TrkA phosphorylation profiles.
For SDS-PAGE and immunoblotting, the NuPAGE Novex BisTris gel electrophoresis system (Invitrogen) was used. Gels were blotted onto nitrocellulose membranes in the NuPAGE Transfer Buffer, washed in PBS-T, incubated in 10% nonfat milk, and then washed with PBS-T. Membranes were incubated in primary antibody in 5% nonfat milk overnight, washed, visualized, with horseradish peroxidase-conjugated secondary antibody, and detected by chemiluminescence using the ECL Plus detection reagents (GE Healthcare).
Mutein Purification Protocol
Selected recombinant NGF muteins were expressed in Sf21 cells and purified to greater than 95% purity using the following two-step protocol: cation exchange chromatography followed by immunoaffinity chromatography. The protein was concentrated and partially purified in the first step from the expression media using a weak cation exchange column, carboxymethyl fast flow (catalog number 17-5091-01, GE Healthcare). The column was equilibrated and washed with 20 mm Tris-HCl buffer, pH 8.0, and the bound proteins were eluted isocratically using 50 mm Tris-HCl, pH 9.0, 0.5 m NaCl buffer. The semi-purified protein was further purified using an immunoaffinity column. The immunoaffinity column was prepared using the N60 monoclonal antibody (68) against NGF that had been cross-linked with cyanogen bromide to Sepharose beads. The column was washed with 50 mm Tris-HCl, pH 9.0, 0.5 m NaCl followed by 10 mm Tris-HCl, pH 8.0, and finally with double distilled water. The bound protein was eluted using 0.2 m glycine-HCl buffer, pH 2.3, and the elution aliquots were immediately neutralized using 2.0 m Tris solution for storage.
Surface Plasmon Resonance (SPR) Spectroscopy
The TrkA ECD was prepared at 5 μg/ml in 10 mm sodium acetate coupling buffer, pH 4.5, and immobilized to a CM4 sensor chip using N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide/N-hydroxysuccinimide coupling chemistry on a Biacore 3000 instrument (Biacore, Piscataway, NJ) as described previously (69). All reagents were automatically introduced over the sensor chip in 10 mm HEPES, pH 7.4, 0.15 m NaCl, and 0.005% v/v surfactant P20 (HBS-P) at a flow rate of 30 μl/min with a blank chip subtracted as control for nonspecific surface binding. Binding isotherms were determined at 25 °C. The sensor chip surface was regenerated by treating with 10 mm glycine-HCl, pH 2, at a flow rate of 10 μl/min. The sensorgram data were evaluated with the BIAevaluation software (version 3.2, Biacore).
RESULTS
Point Muteins That Lie in the Ligand-Receptor Interface Demonstrate the Largest Decline in Neuritogenesis while Preserving Survival
Twenty recombinant NGF point muteins whose residues lie within the specificity, conserved, or variable regions were selected for study (Fig. 1). Cell lines expressing either TrkA only (MG139), p75NTR only (PCNA), or both receptors (PC12) were tested for survival induction (or apoptosis in PCNA cells) with these muteins. Individual point mutations were found to have no effect on cellular survival in MG139 or PC12 cells or on apoptosis in PCNA cells (65). Cellular differentiation was studied by assessing the ability of recombinant NGF point muteins to induce neuritogenesis in PC12 cells. Point muteins showed a statistically significant decline in neurite outgrowth (12–20%), which was not thought to be biologically significant (65). However, muteins could be divided into two groups uncovering a subset of residues that accomplished ∼18–20% reduction and hence warranted further investigation. These residues were found in the ligand-receptor interface between NGF and TrkA, either within the specificity patch or within the conserved patch.
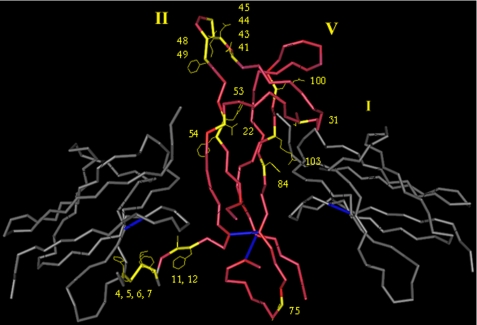
FIGURE 1.
Schematic representation of selected residues thought to play an important role in ligand-receptor interactions between NGF and TrkA-d5. For illustrative purposes, an NGF monomer is shown bound to a dimer of the TrkA-d5 domains (Protein Data Bank code 1WWW). The residues chosen for this study are highlighted. Loop regions I, II, and V are indicated. Red, NGF backbone; yellow, selected residues; gray, TrkA-d5 domains; blue, disulfide bonds. Rendered in Cn3D (NCBI).
Four Triple Muteins Were Generated to Study the Combined Effect of Mutations within Spatially Distinct Regions along the NGF-TrkA Interface
We reasoned that combining the small effects of point mutations in different portions of the molecule could lead to greater effects on overall function. Residues were chosen based on a combination of established structural data and analysis of our point mutein functional data on biological activity. Recombinant triple mutein F7A/H84A/R103A incorporated mutations within the specificity patch, F7A, and the conserved patch, H84A and R103A. Focus was also placed on mutations within the variable regions. Therefore, a second triple mutein was generated that incorporated simultaneous mutations within the variable, specificity, and conserved regions of NGF, i.e. F7A/N45A/R103A.
Because the intent of this investigation was to generate a survival-specific mutein that can function as a pharmacologic agent in the treatment of neurodegenerative diseases, the implications of binding to neurons, like oligodendrocytes and glial cells, exclusively expressing the pro-apoptotic receptor, p75NTR, were considered. Ibanez et al. (29) synthesized an NGF molecule (KKE) wherein simultaneous mutations of three key residues, i.e. Lys32, Lys34, and Glu35 that formed a positively charged patch at one end of the neurotrophin molecule, successfully abrogated interaction with p75NTR (26). By combining these mutations with the original triple mutations, two recombinant KKE hextuple muteins were also generated, i.e. KKE F7A/N45A/R103A and KKE F7A/H84A/R103A. For ease in subsequent discussions, muteins were designated as recombinant NGF triple muteins, 7-84-103 (SS-1) and 7-45-103 (SS-2), or recombinant KKE hextuple muteins, KKE/7-84-103 (SS-3) and KKE/7-45-103 (SS-4), where the SS designation stands for “survival-selective” or “signal-selective.” To thoroughly delineate the functional properties of recombinant NGF and KKE triple muteins, additional negative controls were incorporated into this study. These included Δ9/13 (induces 100-fold reduction in TrkA binding), KKE (abolishes p75NTR binding), and a new combined mutein KKE/Δ9/13 (Δ9/13 + KKE) (29, 33).
Recombinant Muteins Containing the KKE Triple Mutation Show Lower Levels of Expression than Recombinant NGF Triple Muteins
Analysis of expression by Western blot demonstrated that, in general, muteins generated on an NGF backbone expressed at higher levels than muteins generated on a KKE backbone. As a result, KKE, KKE/Δ9/13, KKE/7-45-103, and KKE/7-84-103 achieved significantly lower expression levels (∼3–6.3-fold) than Δ9/13, 7-45-103, and 7-84-103 (∼1.1–1.6-fold) compared with rwtNGF (65).
Our results differ somewhat from previous studies, because our approach incorporated key differences, such as the use of an insect cell expression system, routine expression quantification, and usage of crude cell lysate for cellular studies to allow direct comparison of the various mutants. Woo et al. (33) expressed Δ9/13 in 2.5% serum-supplemented medium to augment expression levels of Sf21 cells and subsequently purified the mutein for biochemical analysis. An analysis of stability via equilibrium denaturation was carried out to demonstrate that stability of Δ9/13 was similar to rwtNGF. Also, KKE was synthesized by Ibanez et al. (29) via transfection of COS cells with mutant plasmid DNA, leading to relative expression levels as high as 65% (compared with ∼19% in our study). Therefore, results obtained in this part of the study must be interpreted with consideration of these differences.
Recombinant NGF and KKE Triple Muteins Supported Survival in MG139 Cells
The interaction of recombinant NGF and KKE triple muteins with TrkA in mediating survival was studied in the MG139 cell line (TrkA+/p75NTR−). With two separate survival assays, i.e. trypan blue and XTT, we found that the recombinant triple muteins generally supported high levels of survival comparable with rwtNGF (Fig. 2). Key differences did exist between the assays.
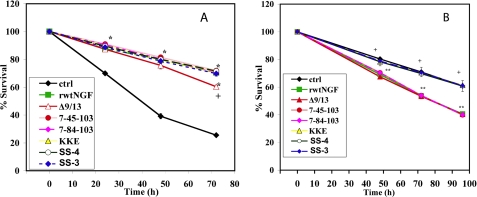
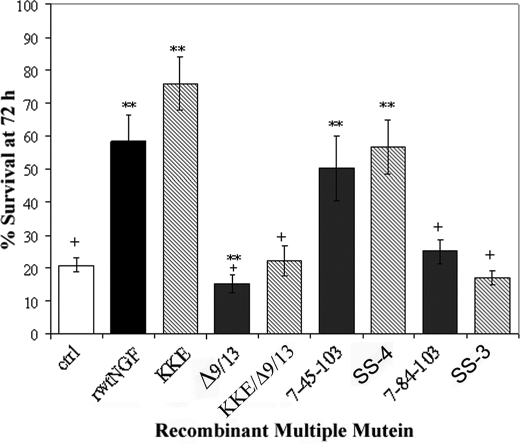
FIGURE 2.
Cellular survival determined by the trypan blue assay. MG139 cells (A) or PCNA cells (B) received insect cell supernatant containing 0.7 nm rwtNGF, Δ9/13, 7-45-103 (SS-2), 7-84-103 (SS-1), KKE/7-45-103 (SS-4), KKE/7-84-103 (SS-3), or vector-treated controls (ctrl). Cells were grown in serum-free DMEM for 72 h (MG139) or 96 h (PCNA) at 37 °C, 6.5% CO2. At 24-h time points, 0.4% trypan blue was added to each plate for assessment of survival. Percent survival was determined by comparing the number of surviving cells excluding blue dye to total number of cells counted. At least 200 cells were counted per well. Data are presented as mean ± S.D. with n = 3–4 independent expressions per data point, **, p < 0.01 significantly different from vector-treated controls (♦) for that time point; +, p < 0.01 significantly different from rwtNGF (■) for that time point.
With the trypan blue assay, for instance, Δ9/13 demonstrated a slightly different survival curve compared with the other recombinant triple muteins which demonstrated survival at or close to wild-type levels (Fig. 2A). At 24 h, Δ9/13 showed survival comparable with rwtNGF, i.e. 87%. Over the next 48 h, however, a progressive decline in survival was noted (Fig. 2A, Δ). Using cold chase binding studies to measure the affinity of Δ9/13 for TrkA, Woo et al. (33) previously showed that even 10 nm Δ9/13 (versus 0.7 nm used here) was inadequate to significantly inhibit binding of 100 pm 125I-NGF. However, cellular studies of survival were never conducted with this recombinant mutein in TrkA-only cells like MG139. The data presented here show only an ∼10% reduction in survival at 72 h from rwtNGF with Δ9/13. Thus, in MG139 cells, trypan blue staining showed that Δ9/13 supported survival reasonably well, whereas the other muteins were indistinguishable from NGF.
With the XTT assay, recombinant triple muteins harboring the N45A mutation showed levels of survival comparable with rwtNGF, whereas muteins harboring the H84A mutation differed significantly from both negative and positive controls (Fig. 3A). The margin of increase over negative controls was comparable with the margin of decrease from rwtNGF, i.e. an ∼1.3-fold difference in both cases. Thus, in MG139 cells, XTT quantification showed that 7-84-103 and KKE/7-84-103 supported an intermediate level of survival. Although Δ9/13 demonstrated a similar trend in the previous assay, it induced no survival by the XTT assay (Fig. 3A).
FIGURE 3.
Cellular survival determined by XTT assay. MG139 cells (A) or PCNA cells (B) received insect cell supernatant containing 0.7 nm rwtNGF, Δ9/13, KKE, 7-45-103 (SS-2), KKE/7-45-103 (SS-4), 7-84-103 (SS-1), KKE/7-84-103 (SS-3), or vector-treated controls (ctrl). Cells were grown in serum-free DMEM for 72 h (MG139) or 96 h (PCNA) at 37 °C, 6.5% CO2. Cells were treated with XTT reagent at 72 h and incubated for an additional 12 h. The number of cells actively converting the dye to the formazan metabolite was determined in triplicate using a scanning multiwell spectrophotometer (TECAN) measuring absorbance at 492 nm and subtracting background at 650 nm. Data are presented as mean ± S.D. with n = 3 independent expressions per data point. **, p < 0.05 significantly different from vector-treated controls; +, p < 0.01 significantly different from rwtNGF.
Recombinant NGF Triple Muteins Promoted Cell Death and Recombinant KKE Triple Muteins Supported Survival in PCNA Cells
To study the interaction of recombinant NGF and KKE triple muteins with p75NTR in inducing cell death, we chose to study the PCNA cell line (TrkA−/p75NTR+) using the same two survival assays as before. Based on trypan blue data, triple muteins were divided into inducers of survival and inducers of cell death. Recombinant muteins that harbored the KKE mutations, knocking out p75NTR binding, did not induce death of PCNA cells and maintained a similar level of survival to vector-treated controls, as expected. Conversely, recombinant NGF triple muteins caused PCNA cell death similar to rwtNGF (Fig. 2B).
Survival measurements obtained by XTT assay confirmed trypan blue data for survival in PCNA cells for recombinant triple muteins, i.e. recombinant NGF triple muteins induced cell death whereas recombinant KKE triple muteins induced survival in PCNA cells (Fig. 3B). However, recombinant NGF triple mutein 7-45-103 differed significantly from both positive and negative controls (+ and ** in Fig. 3B). In this experiment, the 7-45-103 mutein demonstrated an intermediate survival response between vector-treated controls and rwtNGF. This result was believed to be an inaccurate measure of the true survival potential of this mutein. The difference could be explained by the sensitivity of the XTT assay to changes in temperature, CO2 saturation, and cell number per well. Multiple XTT assays have been conducted that conclusively showed that the true survival potential of 7-45-103 was closer to rwtNGF and significantly different from negative controls (data not shown). Those data have not been reported in Fig. 3B because they were done in separate experiments and did not cover the full range of muteins currently being represented.
Recombinant NGF and KKE Triple Muteins Achieved a Statistically and Biologically Relevant Reduction in Differentiation of PC12 Cells
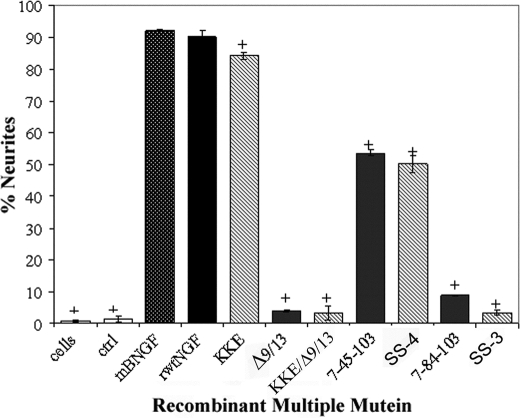
PC12 cells were chosen to study the interaction of recombinant triple muteins with TrkA and p75NTR in the induction of neuritogenesis. Recombinant triple muteins produced significantly lower neuritogenesis in PC12 cells (Fig. 4). More specifically, N45A-harboring muteins, i.e. 7-45-103 and KKE/7-45-103, attained ∼1.7–1.8-fold reduction in neuritogenesis, achieving a maximum of 59 and 55% neurite outgrowth, respectively. H84A-harboring muteins, i.e. 7-84-103 and KKE/7-84-103, attained an even greater reduction in neurite outgrowth that ranged from ∼10-fold for the former to >25-fold for the latter compared with rwtNGF and mβNGF. These values corresponded to an ∼10% neurite outgrowth response with 7-84-103 and a mere ∼4% response with KKE/7-84-103. Recombinant muteins Δ9/13 and KKE/Δ9/13 showed similar levels of neurite outgrowth and further confirmed observations by Woo et al. (33) on the ∼3% biological activity of Δ9/13 at a concentration of 1 nm (compared with ∼4% biological activity at 2 nm in our study).
FIGURE 4.
Neurite outgrowth in PC12 cells. PC12 cells grown in defined medium for 24 h received insect cell supernatant containing 0.7 nm purified mβNGF, rwtNGF, KKE, Δ9/13, KKE/Δ9/13, 7-45-103 (SS-2), KKE/7-45-103 (SS-4), 7-84-103 (SS-1), KKE/7-84-103 (SS-3), vector-treated controls (ctrl), or no treatment at all (cells). They were grown for an additional 48 h at 37 °C, 6.5% CO2. Neurite outgrowth was assessed by comparing cells bearing processes >1.5 time their cell body in length to total cells counted. At least 200 cells were counted per well. Data are presented as mean ± S.D. with n = 3–4 independent expressions per data point. +, p < 0.01 significantly different from recombinant wild-type NGF.
To display these results, PC12 cells grown with 7-84-103 and KKE/7-84-103 were photographed (Fig. 5, E and F). For a proper comparison, mβNGF was incorporated to demonstrate the extent of neuritogenesis achievable at 72 h (Fig. 5B). Additionally, muteins that demonstrated similar trends to 7-84-103 and KKE/7-84-103, i.e. a near-abrogation of differentiation in PC12 cells, were incorporated and include Δ9/13 and KKE/Δ9/13 (Fig. 5, C and D, respectively). Cells bearing neurites have been distinguished from cells considered negative for neurites (Fig. 5, B, arrow versus C, arrowhead, respectively). The assay was carried on for an additional 4 days and showed that at 7 days post-treatment, cells treated with 7-84-103 or KKE/7-84-103 were still deficient in extending neurites.
FIGURE 5.
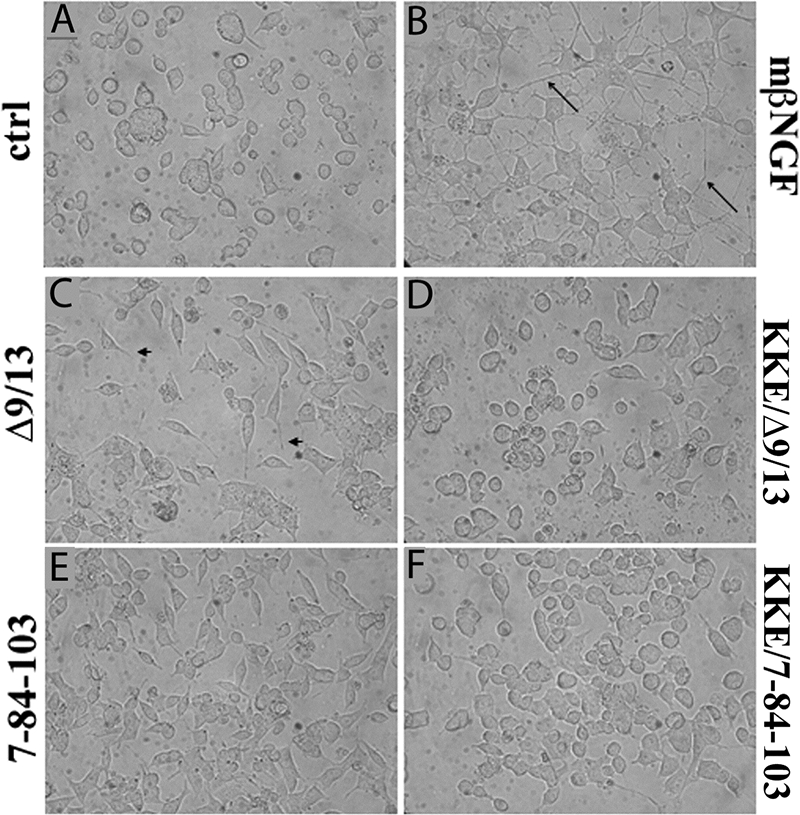
Photographic representation of neuritogenesis in PC12 cells. PC12 cells grown in defined medium for 24 h received insect cell supernatant containing 0.7 nm mβNGF, Δ9/13, KKE/Δ9/13, 7-84-103 (SS-1), KKE/7-84-103 (SS-3), or vector-treated controls (ctrl). Representative photographs at 72 h from six independent experiments are shown. Arrow, mature processes >1.5 times cell body in length. Arrowhead, immature processes <1.5 times cell body in length.
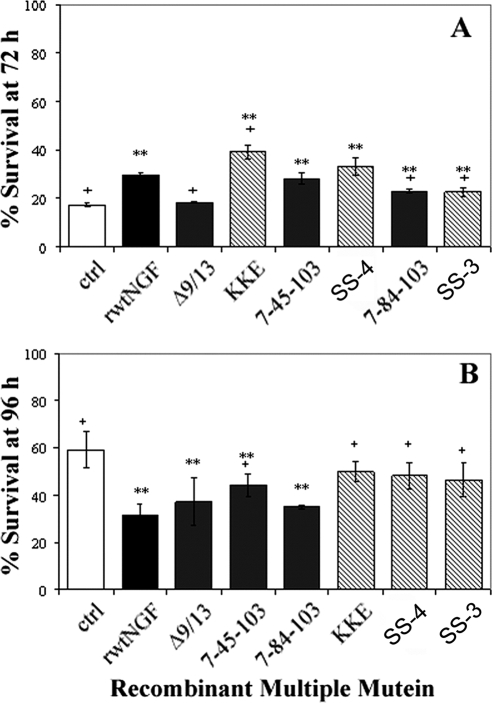
Recombinant Muteins 7-84-103 (SS-1) and KKE/7-84-103 (SS-3) Did Not Support Survival of PC12 Cells by the XTT Assay but Showed Intermediate Survival of PC12 Cells by the Trypan Blue Assay
The additional assessment of survival in PC12 cells co-expressing TrkA and p75NTR was necessary to substantiate those findings of putative survival-specific muteins. These data with the XTT assay showed that N45A-harboring recombinant triple muteins, 7-45-103 and KKE/7-45-103, induced a 50–57% survival of PC12 cells at 72 h, comparable with rwtNGF (58%), whereas H84A-harboring recombinant triple muteins 7-84-103 and KKE/7-84-103 attained a 17–25% survival in PC12 cells at 72 h comparable with negative controls (21%) (Fig. 6). The Δ9/13 mutein was shown to be significantly lower in survival than both vector-treated controls and rwtNGF, demonstrating a true lack of survival-inducing potential (Fig. 6, + and **), not previously shown with this mutein.
FIGURE 6.
Survival of naive PC12 cells by XTT assay. PC12 cells received insect cell supernatant containing 0.7 nm rwtNGF, KKE, Δ9/13, KKE/Δ9/13, 7-45-103 (SS-2), KKE/7-45-103 (SS-4), 7-84-103 (SS-1), KKE/7-84-103 (SS-3), or vector-treated controls (ctrl). Cells were grown in serum-free DMEM for 72 h at 37 °C, 6.5% CO2. Cells were treated with XTT reagent at 72 h and assayed as described in Fig. 3. Data are presented as mean ± S.D. with n = 3 independent expressions per data point, **, p < 0.05 significantly different from vector-treated controls; +, p < 0.01 significantly different from rwtNGF.
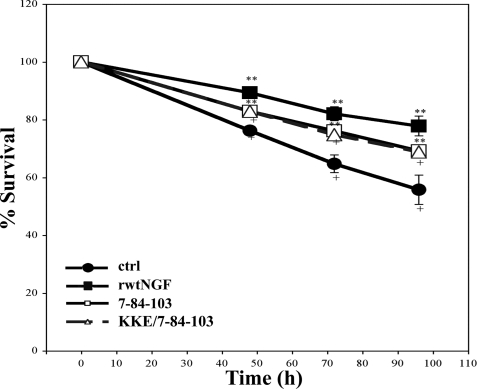
To further investigate the survival-inducing properties of 7-84-103 and KKE/7-84-103 in PC12 cells, we also performed the trypan blue assay. By comparing survival measurements at 24-h intervals to vector-treated controls and rwtNGF, we found that an intermediate level of survival was produced by 7-84-103 and KKE/7-84-103 in PC12 cells (Fig. 7). By 72 h, the two recombinant triple muteins were shown to be intermediate in survival, achieving an ∼1.2-fold difference from each control. These data recapitulate and complement the differences obtained earlier in MG139 cells.
FIGURE 7.
Survival of naive PC12 cells by trypan blue assay. PC12 cells received insect cell supernatant containing 0.7 nm rwtNGF, 7-84-103 (SS-1), KKE/7-84-103 (SS-3), or vector-treated controls (ctrl). Cells were grown in serum-free DMEM for 72 h at 37 °C, 6.5% CO2. At 24-h time points, 0.4% trypan blue was added and assayed as described in Fig. 2. Data are presented as mean ± S.D. with n = 3 independent expressions per data point. **, p < 0.05 significantly different from vector-treated controls (●) for that time point; +. p < 0.05 significantly different from vector-treated controls (■) for that time point.
Purified 7-84-103 (SS-1) Showed Similar Survival and Differentiation Properties
To confirm the results of the findings with the most interesting NGF triple mutein, i.e. SS-1 (7-84-103), this protein, as well as Δ9/13 (33), was expressed in Sf21 insect cells in batch amounts and purified by ion exchange and immunoaffinity chromatography (see “Materials and Methods”) to a single band. The availability of purified mutein also allowed the examination of higher concentration treatments. Purified SS-1 gave similar results to those at a somewhat lower concentration in the crude Sf21 cell supernatant (cf. Figs. 4 and 7) with about 30% of mβNGF PC12 cell differentiation and 75% of mβNGF survival by trypan blue assay (Fig. 8A).
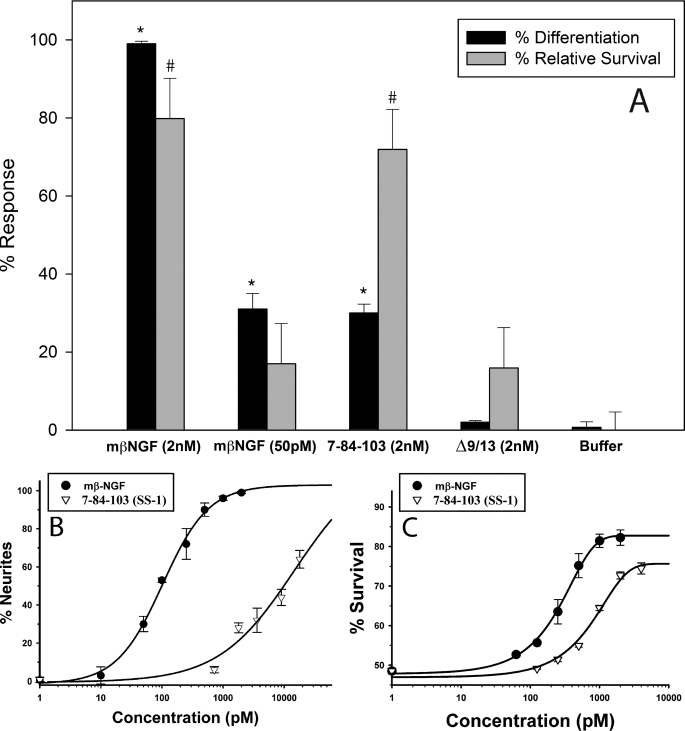
FIGURE 8.
Survival and neuritogenesis of PC12 cells treated with purified NGF or muteins. A, survival (trypan blue) and differentiation were determined (as in Fig. 4 and Fig. 7) after treatment with purified mβNGF, 7-84-103 (SS-1), and Δ9/13 at the concentrations indicated. Cells bearing neurites were counted after 48 h of treatment in defined medium. Relative survival at 72 h was calculated as a percentage of the fold increase in survival with respect to buffer alone. Calculated relative survival = {(actual % survival for sample/actual % survival for buffer) − 1} × 100%. The actual survival observed for buffer alone was 45.2% and that for wtNGF (2 nm) was 81.3%. Data are presented as mean ± S.D. with n = 3 independent assays per data point. *, unadjusted p < 0.001 significantly different for % differentiation with respect to buffer; #, unadjusted p < 0.001 significantly different for % relative survival with respect to buffer. Statistical significance was calculated by one-way analysis of variance using the Holm-Sidak method in SigmaPlot. B, dose-response curve for neurite outgrowth. C, dose response for survival (trypan blue). These data were obtained as described in A at varying concentrations.
Discrimination between Differentiation and Survival by the SS-1 (7-84-103) Mutein Is Largely Due to a Greater Shift in the Differentiation Dose-Response Curve
To determine whether the distinction between wtNGF and SS-1 survival and differentiation assays at 0.7 or 2 nm mutein was due to an altered concentration dependence, purified SS-1 was examined at different concentrations in the neuritogenesis assay (Fig. 8B) and in the trypan blue survival assay (Fig. 8C). In the PC12 cell differentiation assay, the EC50 for wtNGF was found to be about 100 pm, in good agreement with the literature, whereas the EC50 for SS-1 was about 10 nm, an increase of 100-fold. In the PC12 survival assay, the EC50 for wtNGF was found to be about 290 pm, whereas the EC50 for SS-1 was about 950 pm, an increase of only 3.3-fold. A discrimination ratio can be defined as the ratio of the change in EC50 for differentiation to the change in EC50 for survival, both relative to wild-type NGF. A discrimination ratio calculated from these EC50 values relative to wtNGF (Fig. 8, B and C) is about 30 (100-fold increase divided by a 3.3-fold increase).
SS-1 (7-84-103) Has Greatly Reduced Affinity for TrkA as Measured by SPR Analysis
The difference in dose-response curves for SS-1 relative to wtNGF suggests that the affinity for TrkA may be decreased with this mutein. To directly measure this interaction, we chose to utilize SPR. The TrkA ECD was immobilized, and various concentrations of purified SS-1 were passed over the chip (supplemental Fig. 1). Multiple experiments were done with three levels of response units of TrkA immobilized. The average values of the equilibrium dissociation constants are given in Table 1. The Kd value for SS-1 (7-84-103) binding to TrkA is about 175 to 240 times larger than the Kd value for wtNGF binding. At this concentration range, the SPR data are less precise, and therefore, the standard deviations are high. Nevertheless, it is clear that the affinity is greatly reduced by these three mutations.
TABLE 1.
Affinity of SS-1 (7-84-103) for TrkA determined by Biacore SPR
Kd values were obtained using BIAevaluation 3.2 by a Langmuir fit to the constants or by a steady state fit to the plateau values at different concentrations. The wtNGF data are from two separate chips at low (172) and medium (246) response unit values immobilized with the same result and agree well with previous results (69). The SS-1 data are from five runs over 2 days on three separate chips at low (172), medium (246), and high (432) response unit values immobilized. ND means not determined.
| NGF/mutein | Kd ± S.D. (Langmuir global fit) | Kd ± S.D. (steady state fit) |
|---|---|---|
| nm | nm | |
| NGF wild type | 2.1 | ND |
| SS-1 (7-84-103) | 370 ± 210 | 480 ± 120 |
Recombinant NGF Triple Mutein 7-84-103 (SS-1) Supported Survival in Terminally Differentiated PC12 Cells
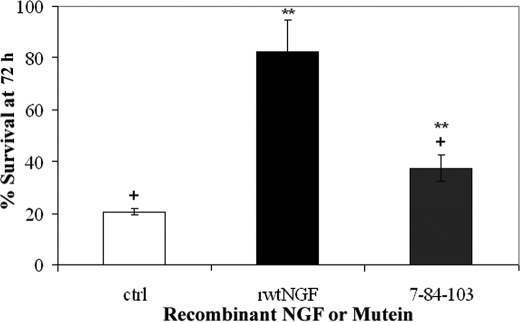
As a further test of the survival-inducing potential of H84A-harboring muteins, survival of terminally differentiated PC12 cells was studied by the XTT assay. PC12 cells treated with NGF for 7–10 days terminally differentiate into a neuronal phenotype, possessing many of the markers of mature neurons, including becoming NGF-dependent for survival, even in the presence of serum (66). SS-1 (7-84-103) induced an intermediate level of survival between negative control and rwtNGF (Fig. 9), similar to that previously seen with the XTT assay in MG139 cells and the trypan blue assay in naive PC12 cells. This mutein supported an ∼1.8-fold increase in survival over negative control and a similar ∼2.2-fold reduction from rwtNGF, which represents an overall survival of ∼40% at 72 h of NGF withdrawal. The KKE/Δ9/13 mutein showed no survival activity in this assay, as expected (data not shown).
FIGURE 9.
Survival of terminally differentiated PC12 cells by XTT assay. Terminally differentiated PC12 cells received purified 2 nm rwtNGF, 7-84-103 (SS-1), or vector-treated controls (ctrl). Cells were grown in 15% serum-supplemented DMEM containing 2 nm mβNGF for 14 days at 37 °C, 10% CO2. Media were removed, and cells were treated with purified rwtNGF or 7-84-103 muteins for 72 h and then incubated with XTT reagent for an additional 6 h and assayed as described in Fig. 3. Data are presented as mean ± S.D. with n = 3 independent wells per data point. **, p < 0.01 significantly different from vector-treated controls; +, p < 0.01 significantly different from mβNGF.
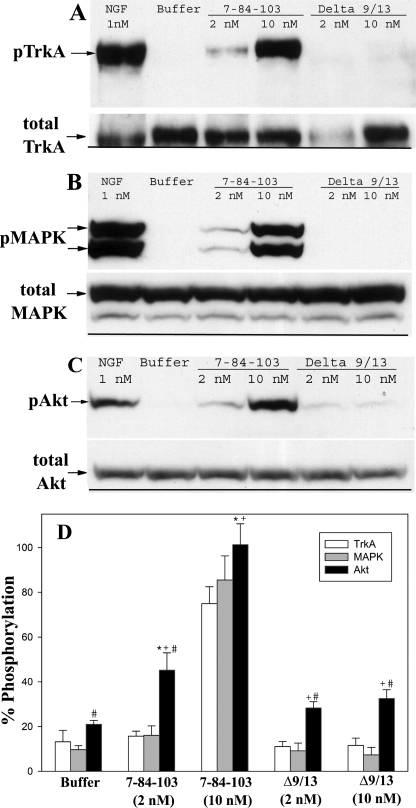
Survival-selective Response of SS-1 (7-84-103) Is Accompanied by Increased Activation of Akt Compared with MAPK
In PC12 and neuronal cells, Akt and MAPK phosphorylation and activation have both been shown to be part of the survival response, whereas sustained MAPK activation is needed for a differentiation response (70–72). Phosphorylation of TrkA, MAPK, and Akt was examined to understand the survival/differentiation signaling properties of 7-84-103. TrkA phosphorylation was examined by immunoprecipitating TrkA with an anti-Trk antibody, and the phosphorylation levels were determined by blotting with a pan-anti-phosphotyrosine antibody. Phosphorylation of MAPK and Akt was determined directly in cell lysates with specific antibodies. We focused on the 7-84-103 mutant, because it expressed well and was the best mutein or triple mutein in terms of minimal differentiation along with good survival. NGFΔ9/13 was used as a negative/minimum response control and mβNGF as the positive control. Purified proteins (mβNGF, Δ9/13, and 7-84-103) were used for these studies.
TrkA phosphorylation data showed lower levels of phospho-TrkA by 2 and 10 nm 7-84-103 compared with mβNGF, but 7-84-103 was significantly higher than that induced by 10 nm Δ9/13 (Fig. 10A). Hence, activation of TrkA by 7-84-103 was observed, which increased with higher concentrations. Activation of TrkA by 7-84-103 was dose-dependent. A similar trend was noted with respect to MAPK phosphorylation (Fig. 10B). Akt phosphorylation levels, on the other hand, were higher for 10 nm 7-84-103 than that observed for 1 nm mβNGF (Fig. 10C). Comparable with our other cellular responses, Akt phosphorylation induced by Δ9/13 was much lower than mβNGF and 7-84-103. The results overall indicate activation of TrkA by 7-84-103, which leads to comparatively low activation of the MAPK pathway, but nearly as good activation of the Akt pathway as that seen with mβNGF. Thus, the survival signal, Akt, correlates with the cellular survival shown earlier when PC12 cells are treated with the survival-selective mutein 7-84-103.
FIGURE 10.
Phosphorylation of TrkA, MAPK, and Akt (protein kinase B) in PC12 cells. Cells were treated with the indicated concentration of mβNGF, mutein, or control and assayed for phosphorylated proteins as described under “Materials and Methods.” A, Western blot of pTrkA and total TrkA (lower panel); arrows show position of phospho-TrkA band. B, Western blot of pMAPK and total MAPK (lower panel); arrows show position of phospho-MAPK doublet band. C, Western blot of pAKT and total AKT (lower panel); arrow shows position of phospho-Akt band. D, quantification of TrkA, MAPK, and Akt phosphorylation. Data are presented as mean ± S.D. of three separate experiments. *, unadjusted p < 0.001 significantly different for Akt phosphorylation with respect to buffer; #, unadjusted p < 0.001 significantly different for Akt phosphorylation with respect to MAPK phosphorylation for the same treatment; +, unadjusted p < 0.001 significantly different for Akt phosphorylation with respect to TrkA phosphorylation for the same treatment. Statistical significance calculated by one-way analysis of variance using the Holm-Sidak method in SigmaPlot.
DISCUSSION
General Aspects of Intracellular Signaling; EGFR Example to Suggest a Signal-selective Ligand
Signaling from tyrosine kinase receptors is a more complicated process than might be expected from the relatively simple requirement for a receptor dimer to trans-autophosphorylate tyrosine residues on its ICD kinase domain. Several distinct mechanisms have been observed for receptor dimerization, or alteration of an existing receptor dimer, to cause activation subsequent to ligand binding to the ECD (73, 74). Phosphorylation of a tyrosine in a loop in the active site typically, but not always, follows to autoactivate the ICD kinase by moving the loop out of its inhibitory position in the active site. Finally, the kinase continues to phosphorylate tyrosine residues in the C-terminal tail of the adjacent ICD of the receptor tyrosine kinase that provide a docking site for Src homology 2 domain adaptor proteins or enzymes (73, 75, 76). Evidence with the EGF receptor has suggested an allosteric mechanism between two asymmetrically interacting EGFR ICDs that explains how this signal may be transduced across the cell membrane (77). In this case, the activation loop does not have to be phosphorylated, and the monomeric form of the EGFR ICD was shown to be auto-inhibited. Their model suggests that the two ICDs interact in an asymmetric manner via a 3-fold screw axis in which the C-lobe of one kinase ICD pushes the N-lobe of the other ICD toward the active site to form a critical Lys721–Glu738 ionic bond to activate the kinase activity. Indeed, a cytosolic protein inhibitor of EGFR, Mig6, has been shown to bind to this ICD interface (78). The details of how the C-terminal tail tyrosines subsequently become phosphorylated are still unknown. However, kinetic studies of EGFR kinase activity have shown that the preference for Gab1- and Shc-docking proteins is different between the unliganded and EGF-liganded states (79). We hypothesized that the complex process of the formation of the signal transduction complex would depend upon the intricate relationship of the ICDs with docking proteins and thus be susceptible to manipulation by the manner in which the ligand, NGF in our case, brought two TrkA ECDs (and hence the ICDs and their docking partners) together, thereby altering the signal to the cell.
Survival-selective NGF Muteins
Recombinant NGF and KKE triple muteins were generated to study survival in MG139, PCNA, and PC12 (naive and terminally differentiated) cells via two survival assays, i.e. trypan blue and XTT, and to study differentiation in PC12 cells via the neurite outgrowth assay (summarized in Table 2). Data highlighted a difference between triple muteins containing the N45A mutation and triple muteins, including the H84A mutation. Although the former maintained survival in MG139 and PC12 cells and showed ∼50% less neuritogenesis in PC12 cells than rwtNGF, the latter induced slightly lower levels of survival in MG139 and PC12 cells, while greatly reducing neuritogenesis in PC12 cells. The similarity in measurements between a recombinant NGF triple mutein and its KKE counterpart also suggested that loss of p75NTR binding was inconsequential to the induction of survival or differentiation in these cell lines. Therefore, the modulatory role of p75NTR within the high affinity receptor complex was not critical in these cases. Assays in PCNA cells confirmed that incorporation of the KKE mutation limited apoptosis through p75NTR. The results of 7-84-103 and KKE/7-84-103 with MG139 cells (TrkA+, p75NTR−) showed that these muteins still possess the ability to interact with the TrkA receptor. However, Δ9/13 showed a consistent reduction in survival assays, usually at levels comparable with negative controls, consistent with a lack of interaction with TrkA (33).
TABLE 2.
Summary of intracellular signaling properties of a triple mutein (SS-1) and a hextuple mutein (SS-3)
Comparison of crude Sf21 cell supernatant at 0.7 nm NGF mutein, based upon semiquantitative Western immunoblotting to determine mutein concentration as follows: wild type = levels of survival attained comparable to rwtNGF; intermediate = levels of survival attained between rwtNGF and vector-treated controls; none = control levels; ND = not determined; <10% = less than 10% of rwtNGF neurite production.
| Cell line/test | Assay | 7-84-103 | KKE/7-84-103 |
|---|---|---|---|
| MG139, survival | Trypan blue | Wild type | Wild type |
| XTT | Intermediate | Intermediate | |
| PCNA, apoptosis | Trypan blue | Wild type | None |
| XTT | Wild type | None | |
| PC12 naive, survival | Trypan blue | Intermediate | Intermediate |
| XTT | None | None | |
| PC12 differentiated, survival | XTT | Intermediate | ND |
| PC12 naive, differentiation | Neuritogenesis | <10% | <10% |
Molecular Surface Areas of NGF for Neuritogenesis/ Differentiation
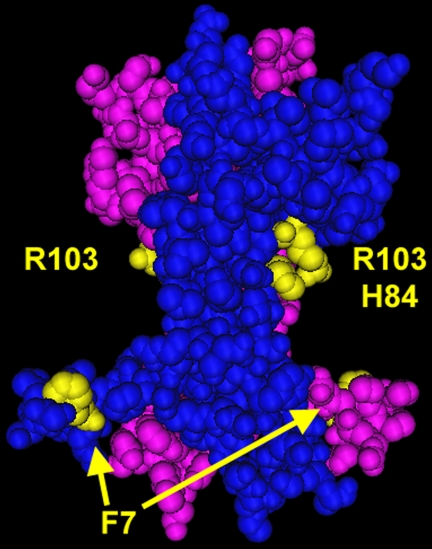
Data on neuritogenesis more specifically pointed toward the likelihood of residues Phe7, His84, and Arg103 playing a critical role in biological activity. These residues are shown in Fig. 11. The calculated neuritogenic potential of each recombinant triple mutein suggested that His84 (conserved patch) played a more discriminatory role than Asn45 (variable loop II) in neurite outgrowth. When the former residue was mutated to alanine, in 7-84-103 and KKE/7-84-103, the maximum neuritic response was almost 40% lower than when His84 was left unaltered in 7-45-103 and KKE/7-45-103. Earlier data on recombinant NGF point muteins had shown that each of these residues when mutated individually produced an ∼20% reduction in neuritogenesis in PC12 cells (65). With 7-84-103 and KKE/7-84-103, a cumulative effect was shown when combining mutations to these residues. In 7-45-103 and KKE/7-45-103, the level of neuritogenesis reflected the presence of two of these key residues with no additional effect with the presence of Asn45. Taken together, these data suggested that limited mutations within the specificity and conserved patches were additive and led to a reduction of neuritogenesis.
FIGURE 11.
Structural location of key residues to make SS-1. The dimer of NGF is shown with each subunit colored separately in cyan and magenta. Residues Phe7, His84, and Arg103 are highlighted in yellow, as indicated. Rendered in Cn3D (NCBI).
Survival Measurements
Considering observations of survival in only MG139 and PC12 cells (without regard to differentiation effects) clearly distinguished mutations at Phe7, Asn45, and Arg103 from mutations, including His84. Mutations to the former three residues were shown to be ineffective in reducing survival from wild-type levels in MG139 and PC12 cells. However, incorporation of a mutation to His84 led to an appreciable decline in survival support from wild-type NGF in both these cell lines. This apparent difference was clarified when the location of residues being mutated was addressed. With 7-45-103 and KKE/7-45-103, point mutations were made to separate regions of NGF, i.e. the specificity patch, variable loop II, and the conserved patch. Data from recombinant point muteins have already suggested that single mutations to any of these residues were not efficient in affecting survival (65). In 7-84-103 and KKE/7-84-103, however, two point mutations were made in close proximity within the same region, i.e. His84 and Arg103, within the conserved patch. Based on the survival data, it appeared that combining mutations in close proximity may have an additive effect on ligand-receptor interactions and ultimately reduce the efficiency of at least some aspects of intracellular signaling. This cumulative effect on survival and differentiation would explain the reduction in both survival and differentiation signals with the triple and hextuple muteins, 7-84-103 and KKE/7-84-103. These comparative analyses showed that independent point mutations do not achieve a biologically significant reduction in signaling; however, when mutations are incorporated in close proximity within critical regions at the ligand-receptor interface, survival and differentiation are both affected, with a more pointed effect seen with the latter. The greater decrease in differentiation and the relative maintenance of survival suggest the possibility of multiple binding epitopes for the mediation of survival, while concurrently highlighting more limited patches needed for the induction of differentiation.
Although consistent with our conclusions, notable differences were nevertheless found in measurements obtained by the trypan blue assay versus the XTT assays. In all tested cases, the trypan blue assay recorded higher levels of survival than the XTT assay. When the XTT assay returned measurements of survival that were comparable with negative controls, the trypan blue assay demonstrated intermediate levels of survival (as was the case with Δ9/13 in MG139 cells). Similarly, when the XTT assay returned intermediate levels of survival, the trypan blue assay demonstrated wild-type levels of survival (as was the case with 7-84-103 and KKE/7-84-103 in MG139 cells). To explain this trend, it is important to understand that both assays measure a slightly different aspect of survival, i.e. total cell counts by trypan blue versus mitochondrial activity by XTT. Therefore, the inherent difference between these two assays is in the measure of the metabolic activity of cells, which is a more appropriate measure of survival in actively dividing cells. The direct comparison in Fig. 8 substantiates this conclusion because the same percentage of surviving cells (75–80% by trypan blue) between 7-84-103 and mβNGF have different amounts of neuritogenesis (99 versus 31%). The greater apparent level of survival with XTT assay for wild type over 7-84-103 (Figs. 6 and 9) may reflect this greater metabolic activity of differentiating cells. Alternatively, cells that have undergone cell cycle arrest are viable cells that may retain a lower level of metabolic activity than actively dividing cells. An interesting possibility then is the potential for recombinant muteins, such as 7-84-103 and KKE/7-84-103, to cause cell cycle arrest and prevent cell division while simultaneously maintaining survival.
To explore these phenomena further, additional studies were undertaken in cells that have permanently undergone cell cycle arrest, i.e. terminally differentiated PC12 cells, and in the process have become dependent on NGF for survival. Confirmation of the survival-maintaining properties of the tested muteins was obtained in these cells wherein both His84-incorporating triple muteins supported intermediate survival levels compared with controls. Therefore, our survival analyses with differentiated PC12 cells confirm that 7-84-103 retained viability-supporting properties. This result highlights the survival-inducing properties of H84A-harboring muteins, especially in a neuron-like population of cells that would be pertinent to the search for a pharmacologic lead agent in the treatment of neurodegenerative disorders.
Molecular Surface Areas of NGF for Survival
The maintenance of survival may be understood by focusing on the variable regions that were left virtually untouched by this study. Previously, regions I, II, and V have been implicated in forming a continuous binding surface for TrkA, most probably at the C-terminal linker region (Fig. 1) (25, 30, 31, 35, 36, 50). The data reported here suggest that the variable regions may be necessary for the induction of the appropriate conformation of the receptor for survival signals. When coupled to the different downstream survival pathways, these multiple extracellular interactions may enable NGF to maintain crucial receptor interactions that maintain viability while allowing differentiation to be much more affected. Thus, survival may be controlled by multiple interactions of TrkA with the distinct and discontinuous variable regions of NGF, whereas differentiation may be activated by limited, highly specific interactions of TrkA with the specificity and conserved patches of NGF.
The survival-promoting properties observed with whole cell assays were verified upon looking at the intracellular signals in differentiation (MAPK) and survival (MAPK and Akt) pathways. Because the MAPK pathway contributes both to differentiation and survival (14, 17), activation of this kinase does not distinguish between cellular outcomes. However, the clear positive signal from phosphorylated Akt clearly indicates that the 7-84-103 mutein is functional in maintaining the viability of the cells. Whether the intermediate level of MAPK activation denotes the decreased level of differentiation of PC12 cells by 7-84-103 is problematic at this stage.
Mechanism of Distinction between Survival and Differentiation Signal Transduction
As with other tyrosine kinases, NGF binding induces TrkA receptor dimerization, trans-autophosphorylation, and intracellular tyrosine kinase domain activation (6, 38–40). These ICDs are maintained in an inactive state by the interaction of a pseudosubstrate tyrosine within an activation loop (Tyr674) from nearby residues. Phosphorylation of a specific set of tyrosine residues (Tyr670, Tyr674, and Tyr675) within the activation loop of the TrkA kinase domain keeps the kinase ICD active (6, 38–40, 80). Two tyrosines located in the juxtamembrane domain (Tyr490) and C terminus (Tyr785) of Trk serve as docking sites for adaptor proteins. The three major signaling pathways include the PI 3-kinase/Akt pathway (through Tyr(P)490), the MAPK pathway (through Tyr(P)490), and the phospholipase Cγ pathway (through Tyr(P)785) (6, 14, 17, 38–40, 70, 81–83). The phosphorylated Tyr(P)490 site of TrkA binds Shc via a phosphotyrosine binding (PTB) domain and initiates formation of a signal transduction particle that includes FRS2, SOS, Grb2, Gab-1, activated PI 3-kinase, and activated Ras (59, 84). PI 3-kinase acts indirectly via phosphatidylinositol 3,4,5-trisphosphate to activate Akt, which is a serine/threonine kinase that phosphorylates and inhibits apoptotic proteins, such as Bad, Forkhead, and pro-caspase-9 (14, 17). PI 3-kinase also activates the inhibitor of apoptosis, a family of caspase inhibitors. The PI 3-kinase/Akt pathway accounts for over 80% of neurotrophin-mediated survival (14, 41, 42). Ras activates the MAPK pathway, which is implicated in both survival and differentiation. MAPK is a serine/threonine protein kinase that has cytosolic and nuclear targets (17, 72, 85). Nuclear MAPK influences gene expression by phosphorylating several transcription factors, such as Elk-1 and CREB, which ultimately maintain a robust and sustained MAPK activation that supports differentiation and neuritogenesis (70–72). Cytoplasmic MAPK activates MAPK/extracellular signal-regulated kinase kinase (MEK), which promotes survival by augmenting the expression of anti-apoptotic proteins, such as Bcl-2 and cAMP-response element-binding protein (14). Therefore, depending on the upstream activators and downstream effectors of MAPK, survival, differentiation, or both signals can be enhanced.
At 2 nm, the SS-1 mutein (7-84-103) shows a 75% retention of survival support and only a 30% retention of differentiation support in PC12 cells (Fig. 8A). The ratio of these activities is 2.5 (0.75:0.3). However, the concentration curves demonstrate that this discrimination is in part due to a higher EC50 for 7-84-103, compared with wtNGF (Fig. 8, B and C). The discrimination ratio, as defined under “Results,” is analogous to a specificity ratio for mutation of enzymes based on their change in Km values. A discrimination ratio of 30 based on the EC50 values (Fig. 8, B and C) highlights a remarkably large divergent effect on the two measured activities.
Another comparison that may be made is between the dose-response curves and the binding affinity determined by SPR analysis. Only a 3.3-fold change in EC50 for survival (Fig. 8C) was observed, but the change in EC50 for differentiation (100-fold, Fig. 8B) and the change in affinity for TrkA by SPR (175–225-fold, Table 1) were comparable and much larger. This comparison suggests that differentiation tracks more closely to affinity than does survival. However, it should be well noted that the SPR method measures binding to “naked” TrkA, whereas in the cellular studies TrkA is modulated by the presence of p75NTR and possibly other cellular or membrane components. Moreover, both survival and differentiation involve signaling cascades and therefore may have quite different saturation characteristics than simple equilibrium binding to a cell surface receptor.
The 7-84-103 mutein clearly supports activation of the PI 3-kinase/Akt pathway, which leads to survival, over the Ras/MAPK pathway, from which differentiation is supported. These differences in signaling pathways (Akt and MAPK, Fig. 10) indicate that these are truly qualitative differences that result from a different mode of binding to the receptor and not simply an affinity change that affects all cellular responses equally. We propose that the arrangement of the signal transduction complex of at least six proteins centered on Tyr490 is sensitive to the interaction of the ICDs of TrkA itself, which in turn is determined by how the ECDs of the receptor are brought together. This interpretation is supported by the decreased level of TrkA phosphorylation with 7-84-103 treatment. Thus, wild-type NGF juxtaposes the two ICDs in a signal transduction complex that generates a “normal” ratio of MAPK to Akt signaling. In contrast, the survival-selective muteins 7-84-103 and KKE/7-84-103 have shifted this output ratio in favor of Akt over MAPK signaling and hence survival over neuritogenesis. Whether the design of the 7-84-103 mutein is optimal at this stage, or whether improvements in discrimination between survival and differentiation pathways is possible, is not yet known.
Conclusions
The design and characterization of two mechanism-selective recombinant NGF muteins, i.e. 7-84-103 (SS-1) and KKE/7-84-103 (SS-3), have illuminated the possibility of designer growth factors. Based on multiple cellular survival assays, a notable difference in the ratio of survival to neuritogenesis was observed, as compared with rwtNGF, which was reflected in the corresponding EC50 values. Importantly, a difference in signaling through Akt, a survival pathway, has been found. The careful, stepwise mutagenesis of NGF has thus led to the generation of two survival-selective muteins that are potentially therapeutic lead candidates for Alzheimer disease or other neurodegenerative disorders. Furthermore, these results show the proof of principle that one can, in fact, dissect intracellular pathways for a given receptor via modifications to its ligand. Further improvement and characterization of these reagents will be enlightening.
Supplementary Material
Acknowledgments
We thank Promila Pagadala for helpful discussions of these experiments. We also thankfully acknowledge the use of the Biacore 3000 SPR instrument in the Department. of Biochemistry, Medical College of Wisconsin, and the help of Drs. Nancy Dahms and Richard Bohnsack in obtaining and interpreting the SPR data.
This work was supported, in whole or in part, by National Institutes of Health Grant NS24380 from the USPHS.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
Mutant NGF (mutein) names are as follows: (a) recombinant muteins from the literature are KKE = K32A/K34A/E35A (abolishes p75NTR binding (29)) and Δ9/13 = deletion of sequence from 9 to 13 (100-fold reduction in TrkA binding (33)); (b) recombinant triple muteins (survival-selective) studied herein 7-84-103 = (SS-1) = F7A/H84A/R103A; and 7-45-103 = (SS-2) = F7A/N45A/R103A; (c) recombinant KKE hextuple muteins: KKE/7-84-103 = (SS-3) = KKE combined with 7-84-103 and KKE/7-45-103 = (SS-4) = KKE combined with 7-45-103; (d) recombinant combined mutein KKE/Δ9/13 = (Δ9/13 combined with KKE).
- NGF or mβNGF
- nerve growth factor (β-subunit)
- DMEM
- Dulbecco's modified Eagle's medium
- EGF
- epidermal growth factor
- EGFR
- EGF receptor
- ECD
- extracellular domain
- ICD
- intracellular domain
- NT
- neurotrophin
- p75NTR
- common neurotrophin receptor
- PC12
- pheochromocytoma 12 cell line
- rwtNGF
- recombinant wild-type NGF
- SPR
- surface plasmon resonance
- Trk
- tropomyosin related kinase
- XTT
- sodium 3′-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis(4-methoxy-6-nitro) benzene sulfonic acid
- PCNA
- fibroblast cell line ectopically expressing p75NTR
- MAPK
- mitogen-activated protein kinase
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- PI
- phosphatidylinositol.
REFERENCES
- 1.Segal R. A. (2003) Annu. Rev. Neurosci. 26, 299–330 [DOI] [PubMed] [Google Scholar]
- 2.Lindsay R. M. (1988) J. Neurosci. 8, 2394–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levi-Montalcini R. (1987) Science 237, 1154–1162 [DOI] [PubMed] [Google Scholar]
- 4.Katz D. M., Erb M., Lillis R., Neet K. (1990) Exp. Neurol. 110, 1–10 [DOI] [PubMed] [Google Scholar]
- 5.Diamond J., Foerster A., Holmes M., Coughlin M. (1992) J. Neurosci. 12, 1467–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bibel M., Barde Y. A. (2000) Genes Dev. 14, 2919–2937 [DOI] [PubMed] [Google Scholar]
- 7.Petruska J. C., Mendell L. M. (2004) Neurosci Lett. 361, 168–171 [DOI] [PubMed] [Google Scholar]
- 8.Barde Y. A., Edgar D., Thoenen H. (1982) EMBO J. 1, 549–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ernfors P., Ibáñez C. F., Ebendal T., Olson L., Persson H. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 5454–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hallböök F., Ibáñez C. F., Persson H. (1991) Neuron 6, 845–858 [DOI] [PubMed] [Google Scholar]
- 11.Timm D. E., de Haseth P. L., Neet K. E. (1994) Biochemistry 33, 4667–4676 [DOI] [PubMed] [Google Scholar]
- 12.Ibáñez C. F. (1994) J. Neurobiol. 25, 1349–1361 [DOI] [PubMed] [Google Scholar]
- 13.Neet K. E., Campenot R. B. (2001) Cell. Mol. Life Sci. 58, 1021–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaplan D. R., Miller F. D. (2000) Curr. Opin. Neurobiol. 10, 381–391 [DOI] [PubMed] [Google Scholar]
- 15.Cordon-Cardo C., Tapley P., Jing S. Q., Nanduri V., O'Rourke E., Lamballe F., Kovary K., Klein R., Jones K. R., Reichardt L. F. (1991) Cell 66, 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soppet D., Escandon E., Maragos J., Middlemas D. S., Reid S. W., Blair J., Burton L. E., Stanton B. R., Kaplan D. R., Hunter T., Nikolics K., Parade L. F. (1991) Cell 65, 895–903 [DOI] [PubMed] [Google Scholar]
- 17.Lad S. P., Neet K. E., Mufson E. J. (2003) Curr. Drug Targets CNS Neurol. Disord. 2, 315–334 [DOI] [PubMed] [Google Scholar]
- 18.Dechant G., Barde Y. A. (2002) Nat. Neurosci. 5, 1131–1136 [DOI] [PubMed] [Google Scholar]
- 19.Mobley W. C., Woo J. E., Edwards R. H., Riopelle R. J., Longo F. M., Weskamp G., Otten U., Valletta J. S., Johnston M. V. (1989) Neuron 3, 655–664 [DOI] [PubMed] [Google Scholar]
- 20.Cohen-Cory S., Dreyfus C. F., Black I. B. (1991) J. Neurosci. 11, 462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Korsching S. (1993) J. Neurosci. 13, 2739–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrup K., Sunter K. (1987) J Neurosci. 7, 829–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ibáñez C. F., Hallböök F., Ebendal T., Persson H. (1990) EMBO J. 9, 1477–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott J., Selby M., Urdea M., Quiroga M., Bell G. I., Rutter W. J. (1983) Nature 302, 538–540 [DOI] [PubMed] [Google Scholar]
- 25.Wiesmann C., Ultsch M. H., Bass S. H., de Vos A. M. (1999) Nature 401, 184–188 [DOI] [PubMed] [Google Scholar]
- 26.McDonald N. Q., Lapatto R., Murray-Rust J., Gunning J., Wlodawer A., Blundell T. L. (1991) Nature 354, 411–414 [DOI] [PubMed] [Google Scholar]
- 27.Bothwell M. A., Shooter E. M. (1977) J. Biol. Chem. 252, 8532–8536 [PubMed] [Google Scholar]
- 28.Ibáñez C. F., Ebendal T., Persson H. (1991) EMBO J. 10, 2105–2110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ibáñez C. F., Ebendal T., Barbany G., Murray-Rust J., Blundell T. L., Persson H. (1992) Cell 69, 329–341 [DOI] [PubMed] [Google Scholar]
- 30.Ibáñez C. F., Ilag L. L., Murray-Rust J., Persson H. (1993) EMBO J. 12, 2281–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ilag L. L., Lönnerberg P., Persson H., Ibáñez C. F. (1994) J. Biol. Chem. 269, 19941–19946 [PubMed] [Google Scholar]
- 32.Urfer R., Tsoulfas P., Soppet D., Escandón E., Parada L. F., Presta L. G. (1994) EMBO J. 13, 5896–5909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woo S. B., Timm D. E., Neet K. E. (1995) J. Biol. Chem. 270, 6278–6285 [DOI] [PubMed] [Google Scholar]
- 34.Kahle P., Burton L. E., Schmelzer C. H., Hertel C. (1992) J. Biol. Chem. 267, 22707–22710 [PubMed] [Google Scholar]
- 35.Kullander K., Kaplan D., Ebendal T. (1997) J. Biol. Chem. 272, 9300–9307 [DOI] [PubMed] [Google Scholar]
- 36.Kullander K., Ebendal T. (1994) J. Neurosci. Res. 39, 195–210 [DOI] [PubMed] [Google Scholar]
- 37.Woo S. B., Neet K. E. (1996) J. Biol. Chem. 271, 24433–24441 [DOI] [PubMed] [Google Scholar]
- 38.Cunningham M. E., Stephens R. M., Kaplan D. R., Greene L. A. (1997) J. Biol. Chem. 272, 10957–10967 [DOI] [PubMed] [Google Scholar]
- 39.Friedman W. J., Greene L. A. (1999) Exp. Cell Res. 253, 131–142 [DOI] [PubMed] [Google Scholar]
- 40.Cunningham M. E., Greene L. A. (1998) EMBO J. 17, 7282–7293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crowder R. J., Freeman R. S. (1998) J. Neurosci. 18, 2933–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartlett S. E., Reynolds A. J., Weible M., Heydon K., Hendry I. A. (1997) Brain Res 761, 257–262 [DOI] [PubMed] [Google Scholar]
- 43.Rabizadeh S., Oh J., Zhong L. T., Yang J., Bitler C. M., Butcher L. L., Bredesen D. E. (1993) Science 261, 345–348 [DOI] [PubMed] [Google Scholar]
- 44.Khursigara G., Orlinick J. R., Chao M. V. (1999) J. Biol. Chem. 274, 2597–2600 [DOI] [PubMed] [Google Scholar]
- 45.Jiang Y., Woronicz J. D., Liu W., Goeddel D. V. (1999) Science 283, 543–546 [DOI] [PubMed] [Google Scholar]
- 46.Lee K. F., Li E., Huber L. J., Landis S. C., Sharpe A. H., Chao M. V., Jaenisch R. (1992) Cell 69, 737–749 [DOI] [PubMed] [Google Scholar]
- 47.Mahadeo D., Kaplan L., Chao M. V., Hempstead B. L. (1994) J. Biol. Chem. 269, 6884–6891 [PubMed] [Google Scholar]
- 48.Bibel M., Hoppe E., Barde Y. A. (1999) EMBO J. 18, 616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barker P. A. (2007) Neuron. 53, 1–4 [DOI] [PubMed] [Google Scholar]
- 50.Wehrman T., He X., Raab B., Dukipatti A., Blau H., Garcia K. C. (2007) Neuron. 53, 25–38 [DOI] [PubMed] [Google Scholar]
- 51.Counts S. E., Nadeem M., Wuu J., Ginsberg S. D., Saragovi H. U., Mufson E. J. (2004) Ann. Neurol. 56, 520–531 [DOI] [PubMed] [Google Scholar]
- 52.Salehi A., Ocampo M., Verhaagen J., Swaab D. F. (2000) Exp. Neurol. 161, 245–258 [DOI] [PubMed] [Google Scholar]
- 53.Mufson E. J., Conner J. M., Kordower J. H. (1995) Neuroreport 6, 1063–1066 [DOI] [PubMed] [Google Scholar]
- 54.Counts S. E., Mufson E. J. (2005) J. Neuropathol. Exp. Neurol. 64, 263–272 [DOI] [PubMed] [Google Scholar]
- 55.Lee R., Kermani P., Teng K. K., Hempstead B. L. (2001) Science 294, 1945–1948 [DOI] [PubMed] [Google Scholar]
- 56.Fahnestock M., Michalski B., Xu B., Coughlin M. D. (2001) Mol. Cell. Neurosci. 18, 210–220 [DOI] [PubMed] [Google Scholar]
- 57.Mufson E. J., Ma S. Y., Cochran E. J., Bennett D. A., Beckett L. A., Jaffar S., Saragovi H. U., Kordower J. H. (2000) J. Comp. Neurol. 427, 19–30 [DOI] [PubMed] [Google Scholar]
- 58.Tuszynski M. H., Thal L., Pay M., Salmon D. P., U H. S., Bakay R., Patel P., Blesch A., Vahlsing H. L., Ho G., Tong G., Potkin S. G., Fallon J., Hansen L., Mufson E. J., Kordower J. H., Gall C., Conner J. (2005) Nat. Med. 11, 551–555 [DOI] [PubMed] [Google Scholar]
- 59.Schlessinger J., Lemmon M. A. (2003) Sci. STKE 2003, RE12. [DOI] [PubMed] [Google Scholar]
- 60.Vaughn J. L., Goodwin R. H., Tompkins G. J., McCawley P. (1977) In Vitro 13, 213–217 [DOI] [PubMed] [Google Scholar]
- 61.Greene L. A., Tischler A. S. (1976) Proc. Natl. Acad. Sci. U.S.A. 73, 2424–2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Radeke M. J., Misko T. P., Hsu C., Herzenberg L. A., Shooter E. M. (1987) Nature 325, 593–597 [DOI] [PubMed] [Google Scholar]
- 63.Barker P. A., Lomen-Hoerth C., Gensch E. M., Meakin S. O., Glass D. J., Shooter E. M. (1993) J. Biol. Chem. 268, 15150–15157 [PubMed] [Google Scholar]
- 64.Reinhold D. S., Neet K. E. (1989) J. Biol. Chem. 264, 3538–3544 [PubMed] [Google Scholar]
- 65.Mahapatra S. (2008) Identification of Critical Residues within the Conserved and Specificity Patches of Nerve Growth Factor Leading to Survival or Differentiation. Ph.D. thesis, Rosalind Franklin University of Medicine and Science, North Chicago: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vaghefi H., Hughes A. L., Neet K. E. (2004) J. Biol. Chem. 279, 15604–15614 [DOI] [PubMed] [Google Scholar]
- 67.Lad S. P., Neet K. E. (2003) J. Neurosci. Res. 73, 614–626 [DOI] [PubMed] [Google Scholar]
- 68.Neet K. E., Fanger M. W., Baribault T. J. (1987) Methods Enzymol. 147, 186–194 [DOI] [PubMed] [Google Scholar]
- 69.Woo S. B., Whalen C., Neet K. E. (1998) Protein Sci. 7, 1006–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kao S., Jaiswal R. K., Kolch W., Landreth G. E. (2001) J. Biol. Chem. 276, 18169–18177 [DOI] [PubMed] [Google Scholar]
- 71.Qian X., Riccio A., Zhang Y., Ginty D. D. (1998) Neuron 21, 1017–1029 [DOI] [PubMed] [Google Scholar]
- 72.Grewal S. S., York R. D., Stork P. J. (1999) Curr. Opin. Neurobiol. 9, 544–553 [DOI] [PubMed] [Google Scholar]
- 73.Ward C. W., Lawrence M. C., Streltsov V. A., Adams T. E., McKern N. M. (2007) Trends Biochem. Sci. 32, 129–137 [DOI] [PubMed] [Google Scholar]
- 74.Stroud R. M., Wells J. A. (2004) Sci. STKE. 2004, re7. [DOI] [PubMed] [Google Scholar]
- 75.Bose R., Holbert M. A., Pickin K. A., Cole P. A. (2006) Curr. Opin. Struct. Biol. 16, 668–675 [DOI] [PubMed] [Google Scholar]
- 76.Pellicena P., Kuriyan J. (2006) Curr. Opin. Struct. Biol. 16, 702–709 [DOI] [PubMed] [Google Scholar]
- 77.Zhang X., Gureasko J., Shen K., Cole P. A., Kuriyan J. (2006) Cell 125, 1137–1149 [DOI] [PubMed] [Google Scholar]
- 78.Zhang X., Pickin K. A., Bose R., Jura N., Cole P. A., Kuriyan J. (2007) Nature 450, 741–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fan Y. X., Wong L., Deb T. B., Johnson G. R. (2004) J. Biol. Chem. 279, 38143–38150 [DOI] [PubMed] [Google Scholar]
- 80.Zhang Y., Moheban D. B., Conway B. R., Bhattacharyya A., Segal R. A. (2000) J. Neurosci. 20, 5671–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yoon S. O., Soltoff S. P., Chao M. V. (1997) J. Biol. Chem. 272, 23231–23238 [DOI] [PubMed] [Google Scholar]
- 82.Lad S. P., Peterson D. A., Bradshaw R. A., Neet K. E. (2003) J. Biol. Chem. 278, 24808–24817 [DOI] [PubMed] [Google Scholar]
- 83.Loeb D. M., Stephens R. M., Copeland T., Kaplan D. R., Greene L. A. (1994) J. Biol. Chem. 269, 8901–8910 [PubMed] [Google Scholar]
- 84.Gotoh N. (2008) Cancer Sci. 99, 1319–1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gómez N., Cohen P. (1991) Nature 353, 170–173 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.