Abstract
Sirtuins comprise a family of enzymes found in all organisms, where they play a role in diverse processes including transcriptional silencing, aging, regulation of transcription, and metabolism. The predominant reaction catalyzed by these enzymes is NAD+-dependent lysine deacetylation, although some sirtuins exhibit a weaker ADP-ribosyltransferase activity. Although the Sir2 deacetylation mechanism is well established, much less is known about the Sir2 ADP-ribosylation reaction. We have studied the ADP-ribosylation activity of a bacterial sirtuin, Sir2Tm, and show that acetylated peptides containing arginine or lysine 2 residues C-terminal to the acetyl lysine, the +2 position, are preferentially ADP-ribosylated at the +2 residue. A structure of Sir2Tm bound to the acetylated +2 arginine peptide shows how this arginine could enter the active site and react with a deacetylation reaction intermediate to yield an ADP-ribosylated peptide. The new biochemical and structural studies presented here provide mechanistic insights into the Sir2 ADP-ribosylation reaction and will aid in identifying substrates of this reaction.
Introduction
Sirtuins, also known as Sir2 enzymes, are a universally conserved class of NAD+-dependent acetyl-lysine specific deacetylases (1). Sir2 deacetylation substrates include histones (2), FoxO transcription factors (3, 4), p53 (5), and PGC-1α (6, 7). Sir2 enzymes regulate multiple biological pathways including gene silencing (8, 9), transcription (3, 4), and fat metabolism (10).
The sirtuin deacetylation reaction consumes acetyl lysine and NAD+, yielding deacetylated lysine, nicotinamide, and O-acetyl ADP-ribose (11). In the first step of the enzymatic reaction, the acetyl lysine reacts with the C1′ of the NAD+ nicotinamide-ribose, leading to the release of nicotinamide and formation of an O-alkylamidate intermediate (11, 12). Next, the nicotinamide-ribose 2′OH reacts with the O-alkylamidate intermediate to generate a 1′2′-bicyclic species and the deacetylated lysine (11). Finally, hydrolysis of the 1′2′-bicyclic species yields 2′ O-acetyl ADP-ribose (11, 13). The Sir2 deacetylation reaction is inhibited by the reaction product, nicotinamide(14, 15), which binds to the Sir2 active site and reacts with the O-alkylamidate to regenerate NAD+ and acetyl lysine.
Alhough Sir2 enzymes are known primarily as protein deacetylases, sirtuins were first identified as NAD+-dependent ADP-ribosyltransferases (16). The Escherichia coli sirtuin, CobB, ribosylates a small molecule, 5,6-dimethylbenzimidazole (17, 18). Subsequent studies demonstrated that CobB can also ADP-ribosylate protein substrates (18). Other sirtuins including human SIRT1(19), human SIRT4 (20), and a trypanosomal sirtuin (21) can ADP-ribosylate protein substrates. The Trypanosoma brucei Sir2 enzyme, TbSir2Rp1, is a particularly well characterized sirtuin with ADP-ribosyltransferase activity. TbSir2Rp1 has dual deacetylase and ADP-ribosyltransferase activities on histone substrates (21); however, its deacetylation activity is 5 orders of magnitude greater than its ADP-ribosyltransferase activity (22). The TbSir2Rp1 ribosylation activity is greatly enhanced by acetyl lysine (22). One model that could explain the increased Sir2 ADP-ribosyltransferase activity with acetylated substrates is that a deacetylation reaction intermediate, possibly the O-alkylamidate intermediate, reacts with a nucleophilic amino acid on the substrate protein to yield an ADP-ribosylated product (22). How a nucleophilic side chain enters the Sir2 active site or what amino acid side chains can be ADP-ribosylated by sirtuins is not known.
In structures of sirtuins bound to an acetylated p53 peptide and to an S-alkylamidate intermediate (23–27), a methionine located two residues C-terminal to the acetyl lysine, the +2 position, inserts into the Sir2 active site (see Fig. 1A). This suggested that substrates containing nucleophilic side chains at this position could potentially attack the O-alkylamidate intermediate and become ADP-ribosylated. To test this model, we performed a series of biochemical assays on the Thermotoga maritima sirtuin, Sir2Tm, and the mammalian sirtuin, SIRT1, using various peptides and found that these sirtuins ADP-ribosylate arginine at the +2 position in an acetylated peptide substrate. To gain further mechanistic insights, we solved a structure of Sir2Tm bound to the Arg-acetylated +2 peptide. The structural and biochemical data presented here describe a plausible mechanism for Sir2-mediated ADP-ribosylation of acetylated substrates.
FIGURE 1.
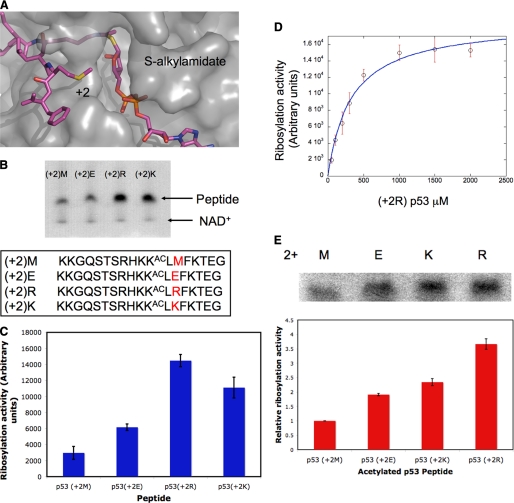
Sirtuin-mediated ribosylation of acetyl p53 peptides. A, the methionine of the acetyl p53 peptide, which is two amino acids C-terminal to the acetyl lysine, points toward the S-alkylamidate intermediate in the Sir2Tm-S-alkylamidate intermediate structure. B, acetyl p53 peptides with different amino acids two positions C-terminal to the acetyl lysine were reacted with Sir2Tm and [32P]NAD+, and the reactions were resolved by SDS-PAGE gels. (+2)M, (+2) Met; (+2)E, (+2) Glu; (+2)R, (+2) Arg; (+2)K, (+2) Lys. C, ribosylation activity for the reactions from B was quantified by densitometry. Error bars indicate S.D. D, Sir2Tm was reacted with various concentrations of acetyl p53 Arg peptide (B), and the data were fit to the Michaelis-Menten model. Error bars indicate S.D. E, SIRT1 was reacted with the p53 acetyl peptides described in B and [32P]NAD+. The reactions were separated by SDS-PAGE gels, and SIRT1 ribosylation activity was quantified by densitometry. Error bars indicate S.D.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Recombinant Sir2Tm proteins were expressed and purified as described previously (28).
ADP-ribosylation Assays
Sir2Tm (33 ng/μl) was incubated for 4 h at 37 °C with 1 μCi of [32P]NAD+, 1 mm NAD+, and peptide (0.5 mm) with a final reaction volume of 30 μl. The peptide sequences were based on the p53 sequence that was previously crystallized with Sir2Tm; however, these acetyl p53 peptides had different residues substituted two amino acids C-terminal to the acetyl lysine. The sequences of the peptides are KKGQSTSRHK(KAC)LXFKTEG, where X is Met, Lys, Arg, or Glu. The reaction buffer used in all experiments consisted of 50 mm CHES,2 pH = 9.2, 100 mm NaCl, and 1 mm dithiothreitol. To resolve the radiolabeled peptides, the reaction mixtures were separated by electrophoresis on NuPAGE 4–12% Bis-Tris SDS-PAGE gels (Invitrogen), and unincorporated [32P]NAD+ was washed out by soaking the gels in destaining solution (10% acetic acid and 5% ethanol). Gels were dried and exposed to film, and the amount of radiolabel transferred to the peptide was quantified by densitometry with the ImageJ software package.
Edman Degradation of the Acetyl p53 +2 Peptide
Sir2Tm, at a concentration of 100 μm, was incubated with 1 mm +2 Arg-acetylated p53 peptide, with the sequence KKGQSTSRHK(KAC)LRFKTEG, and 10 μCi of [32P]NAD+ in a final reaction volume of 50 μl for 2 h at 37 °C. The reaction mixture was resolved on an SDS-PAGE gel. The p53 peptide band was cut out of the gel and homogenized. The peptide was extracted by incubating the gel slice in 500 μl of 100 mm sodium acetate, 0.1% SDS, 50 mm dithiothreitol at 30 °C for 10 h followed by centrifugation to pellet the gel pieces. The peptide was then extracted from the supernatant by acetone precipitation.
Manual Edman degradation was performed on the peptide sample to identify the ADP-ribosylated residue. Briefly, 20 μl of 5% (v/v) solution of phenylisothiocyanate in pyridine was added to the peptide sample and incubated for 30 min at 45 °C followed by the addition of 200 μl of a 10:1 (v/v) heptane-ethyl acetate solution. The upper organic layer was collected, and the aqueous phase was extracted with a 2:1 (v/v) heptane-ethyl acetate solution. The aqueous phase was lyophilized in a SpeedVac evaporator and then resuspended in 50 μl of 100% trifluoroacetic acid and incubated for 10 min at 45 °C. Then, 50 μl of pyridine and 250 μl of butyl acetate were added, and the butyl acetate layer was collected because the N-terminal thiazolinone amino acid partitions into the butyl acetate. The butyl acetate fraction was lyophilized and resuspended in 20 μl of water. This reaction cycle was repeated 19 times. The collected thiazolinone amino acids from each cycle were added to 5 ml of scintillation fluid, and the radioactivity was quantified with a scintillation counter.
Peptide Binding Assays
Sir2Tm (0.4 μm) was incubated with various peptide concentrations, and the changes in intrinsic tyrosine fluorescence (excitation wave length of 280 nm) due to binding of Sir2Tm to peptide were measured on an ISS Chronos fluorescence lifetime spectrophotometer. The p53 2 + Glu, Met, and Lys peptide binding quenched the tyrosine fluorescence of Sir2Tm, whereas binding of the p53 +2 Arg peptide enhanced the fluorescence signal. Binding curves were fit with equation 1
where ΔF is the change in fluorescence signal between the baseline and highest fluorescence signal.
X-ray Crystallographic Data Collection and Processing
Crystals of Sir2Tm bound to acetylated p53 peptide, KKGQSTSRHK(KAC)LXFKTEG (where X is Arg or Glu), were produced and frozen as described previously (23). The crystals formed in the space group P212121 with unit cell dimensions a = 46.5 Å, b = 60.4 Å, and c = 107 Å.
X-ray diffraction data were recorded from frozen crystals at the BioCARS facility at Argonne National Laboratory on the 14-BM-C beamline equipped with an ADSC Quantum 315 detector. Data were reduced with HKL2000 (29)and CCP4 (30). Initial phases were calculated using Sir2Tm bound to p53 acetyl peptide (Protein Data Bank (PDB) ID 2H4F). Model building was done with Coot (31), and refinement was performed with CNS (32) and CCP4. Crystallographic statistics are summarized in Table 1.
TABLE 1.
Diffraction data and refinement statistics
| Diffraction data | |
| Space group | P212121 |
| Unit cell (Å) | 45.72, 58.38, 106.5 |
| Resolution range (Å) | 22.6–1.50 |
| Measured reflections | 165,438 |
| Completeness (outer shell) (%) | 98.5 (96.2) |
| Average I/σ I | 15.5 |
| Multiplicity | 3.8 |
| Rmerge (outer shell) | 0.09 (.34) |
| Refinement statistics | |
| Unique reflections | 43,439 |
| Test set (5%) | 2,319 |
| Total atoms | 2,135 |
| Protein | 1,853 |
| Peptide | 54 |
| Waters | 228 |
| R (%) | 20.5 |
| Rfree (%) | 22.3 |
| r.m.s.d.a | |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.3 |
a r.m.s.d., root mean square deviation.
RESULTS
The Amino Acid Two Positions C-terminal to the Acetyl Lysine Affects Ribosylation of Substrate Peptides
We wished to test the hypothesis that a side chain in the peptide substrate could react with a Sir2 deacetylation reaction intermediate, likely the O-alkylamidate intermediate, and become ADP-ribosylated. One intriguing proposal for Sir2-mediated ribosylation is that a nucleophile on the acetylated protein substrate could insert into the Sir2 active site and react with the O-alkylamidate to yield ADP-ribosylated substrate (22). In all structures of Sir2Tm bound to an acetylated p53 peptide, there is a methionine two residues C-terminal to the acetyl lysine (the +2 position) that inserts into the Sir2 active site through an opening adjacent to the acetyl lysine binding tunnel (Fig. 1A). In the structure of Sir2Tm bound to an S-alkylamidate intermediate, the +2 methionine points at the C1′ of the S-alkylamidate and is 7 Å away from the C1′ (Fig. 1A) (23), raising the possibility that a side chain nucleophile at the +2 position could react with the O-alkylamidate intermediate and become ADP-ribosylated. Although the +2 methionine is not expected to react with a reaction intermediate, nucleophilic amino acids would have a higher probability of attacking a Sir2 reaction intermediate.
To determine whether Sir2Tm could ADP-ribosylate a side chain nucleophile at the +2 position, we synthesized a series of peptides containing different amino acids at the +2 position and assayed Sir2Tm ADP-ribosylation activity for these peptides (Fig. 1B). The sequence of the peptides was based on the acetylated p53 peptide previously crystallized with Sir2Tm (25). The sequence of the peptides assayed was KKGQSTSRHK(KAC)LXFKTEG, where X was Met, Lys, Arg, or Glu (Fig. 1B). Sir2Tm, which has robust deacetylation activity, was incubated with the acetylated peptides and [32P]NAD+ (Fig. 1B). As seen by autoradiography (Fig. 1, B and C) and also confirmed by mass spectrometry (supplemental data), acetylated peptides with arginine and lysine substitutions at the +2 position had a 5- and 4-fold increase, respectively, in incorporated 32P radiolabel as compared with the methionine-containing peptide, presumably due to ADP-ribosylation. The acetylated peptide with a glutamic acid substitution had only a 2-fold increase in labeling relative to the wild type peptide (Fig. 1, B and C).
To determine whether the ADP-ribosylation of the acetylated peptide was the result of Sir2Tm enzymatic activity, and not a nonenzymatic reaction with [32P]NAD+, Sir2Tm was reacted with 1 mm NAD+ at various concentrations of the acetylated +2 Arg peptide. At high peptide concentrations, the ADP-ribosyltransferase activity is saturated, suggesting that the observed labeling is enzymatic. The data were fit to the Michaelis-Menten equation (Fig. 1D), which showed that the Km for the Sir2Tm ribosylation reaction is 350 μm as compared with a Km of 150 μm for the deacetylation reaction. These observations are consistent with two models; either the arginine or lysine two amino acids C-terminal to the acetyl lysine in the acetylated p53 substrate is ADP-ribosylated by Sir2Tm, or the lysine and arginine promote ADP-ribosylation of another residue in the p53 peptide.
To determine whether other sirtuins share the same ADP-ribosylation activity as Sir2Tm, mouse SIRT1 was assayed using the p53 peptide variants described in Fig. 1B. As shown in Fig. 1E, SIRT1 ADP-ribosylation activity is 4-fold higher for the +2 arginine peptide than for the +2 methionine peptide. The acetylated peptides with lysine and glutamic acid substitutions at the +2 position only exhibited a 2.5- and 2-fold increase in labeling, respectively, as compared with the acetylated peptide containing methionine at that position (Fig. 1E). The increase in ADP-ribosylation activity for both a mammalian and a bacterial sirtuin with an acetylated p53 peptide containing arginine at the +2 position demonstrates that ADP-ribosylation activity is conserved in at least a subset of sirtuins.
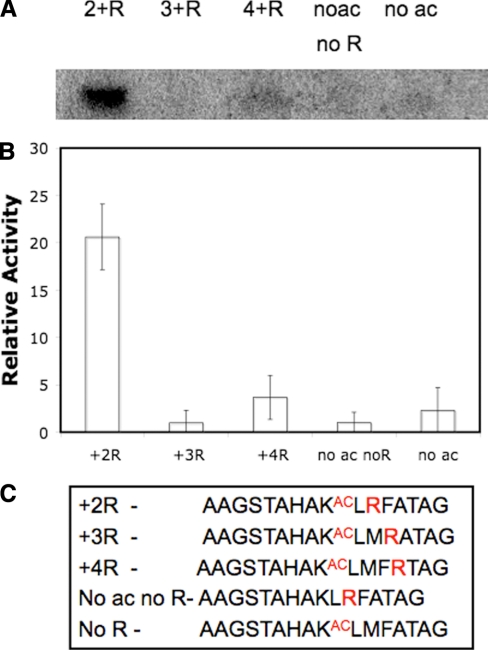
To determine whether the increased ADP-ribosylation of the arginine-substituted peptide was dependent on the precise position of the substituted arginine within the acetylated peptide, the ADP-ribosylation of peptides with arginine substituted at different positions was compared. As shown in Fig. 2, peptides with arginine at positions +3 and +4 showed negligible labeling as compared with the acetylated +2 arginine peptide. The peptides with arginine substituted at other positions have the same amount of background ADP-ribosylation as both acetylated and unacetylated peptide (Fig. 2). This demonstrates that the position of the arginine two residues C-terminal to the acetyl lysine is important for the ribosylation reaction to occur and that it is not simply the presence of arginine in the peptide that gives rise to the increased labeling. The unacetylated +2 arginine peptide was not significantly ADP-ribosylated, indicating that acetyl lysine enhances Sir2Tm ribosylation activity (Fig. 2) and supporting the previous observation that acetyl lysine is necessary for significant ADP-ribosylation (22).
FIGURE 2.
Sir2Tm ribosylation of peptides containing arginine at different positions. Sir2Tm was incubated with [32P]NAD+ and different peptides that have arginine substituted at different positions in the acetyl p53 peptide sequence. A, the autoradiograph of the reactions containing Sir2Tm-, [32P]NAD+-, and p53-based peptides. R indicates arginine; no ac, without acetyl; no R, without arginine. B, quantification of A by densitometry based on data from three replicates of the reaction. Error bars indicate average deviation C, sequences of the peptides used in A.
Sir2Tm Directly Ribosylates the +2 Arginine of the Acetyl p53 Peptide
To determine which amino acid in the +2 Arg peptide (Fig. 1B) is ADP-ribosylated by Sir2Tm, we performed manual Edman degradation on the acetylated p53 +2 arginine peptide that had been incubated with Sir2Tm and [32P]NAD+. In each cycle of Edman degradation, the N-terminal amino acid is isolated. Because the NAD+ is radiolabeled and the sequence of the peptide is known, ADP-ribosylated residues were identified by relating the radioactivity in each cycle number to the primary sequence of the acetyl p53 peptide. Cycle number 13 contained a significant amount of radiolabel, indicating that the +2 arginine is ADP-ribosylated by Sir2Tm (Fig. 3). This is the arginine that was substituted for methionine in the wild type p53 peptide and resulted in increased Sir2Tm ribosylation (Fig. 1, B and C). Radiolabel was also present in cycle number 2, indicating that an N-terminal lysine was also ADP-ribosylated by Sir2Tm (Fig. 3).
FIGURE 3.
Sir2Tm directly ribosylates the arginine two amino acids C-terminal to acetyl lysine. Manual Edman degradation was performed on the acetyl p53 Arg peptide that was incubated with Sir2Tm and [32P]NAD+. The sites of ribosylation were determined by relating the Edman degradation cycle to the known protein sequence. Error bars indicate average deviation.
Sir2Tm Peptide Binding Affinity and Ribosylation Activity Are Not Correlated
To determine whether the different levels of ADP-ribosylation activity for the acetylated +2 arginine and +2 lysine peptides were due to different binding affinities of Sir2Tm for these peptides, we measured the Kd values for Sir2Tm binding to acetylated peptides containing different amino acid substitutions at the +2 position. To measure Kd values, we developed a fluorescence-based binding assay that uses the intrinsic tyrosine fluorescence of Sir2Tm. The binding isotherms were fit to determine the Kd of the Sir2Tm-acetylated p53 peptide interaction (see “Experimental Procedures”). Sir2Tm binds more tightly to the +2 methionine (Kd of 2.7 μm) and +2 glutamic acid (Kd of 0.11 μm) peptides, as compared with the +2 lysine (Kd of 27.8 μm) and +2 arginine (Kd of 28.2 μm) peptides. Interestingly, Sir2Tm has the highest ADP-ribosylation activity for the acetylated p53 arginine and lysine peptides, yet Sir2Tm binds to these peptides with the lowest affinities. Conversely, the methionine and glutamic acid peptides show the lowest level of labeling by Sir2Tm, yet Sir2Tm binds to these peptides with the highest affinity. Given the lack of correlation between binding affinity and ADP-ribosylation of the acetylated peptides, the differences in ADP-ribosylation activity with different peptide substrates cannot be ascribed to differences in binding affinity.
Crystal Structure of a Sir2Tm-p53 Arginine Peptide Complex
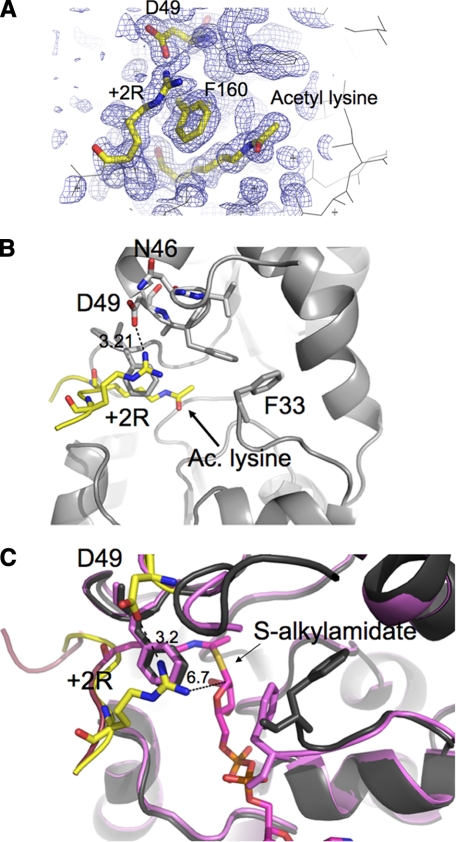
To explore how Sir2Tm uses the +2 arginine peptide as an ADP-ribosylation substrate, we determined the crystal structure of a Sir2Tm bound to acetylated p53 peptide containing arginine at the +2 position to 1.5 Å resolution (PDB code 3JR3 and Fig. 4A). The binding of the acetyl lysine residue in the active site is identical to that observed in previous structures of sirtuins bound to acetylated peptides (Fig. 4A). The arginine two amino acids C-terminal to the acetyl lysine enters the Sir2 active site through a cleft that lies adjacent to the acetyl lysine binding tunnel (Fig. 4, A and B). The +2 arginine forms a hydrogen bond with Asp-49 of Sir2Tm (3.2 Å) and stacks against Phe-160 of Sir2Tm, which is essential for binding the acetyl lysine (Fig. 4B). The Sir2 Asp-49 and Phe-160 contacts to the +2 arginine of the acetylated p53 peptide orient the arginine toward the Sir2 active site (Fig. 4B).
FIGURE 4.
Structure of the Sir2Tm acetyl +2 Arg (+2R) p53 peptide. The structure of the Sir2Tm-acetyl p53 Arg peptide complex was determined to 1.5 Å resolution. A, the 2Fo − Fc electron density map (1 σ). B, the Sir2Tm (gray) Asp-49 contact with the acetyl p53 Arg (yellow). C, superposition of the Sir2Tm-S-alkylamidate intermediate (purple) and Sir2Tm (gray)-acetyl p53 Arg (yellow) structures.
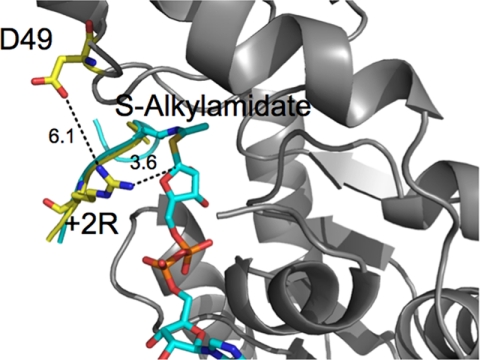
Because it has been proposed that a side chain nucleophile on a protein substrate could likely react with the O-alkylamidate to generate an ADP-ribosylated protein (22), we examined whether the +2 arginine was in a position to react with the C1′ in a complex with the O-alkylamidate intermediate. However, a superposition of the present structure with that of the Sir2Tm-S-alkylamidate structure shows that the position of the +2 arginine would place the guanidino nitrogen 6.7 Å away from the C1′ carbon of the S-alkylamidate, which appears too far for the +2 Arg to react with the O-alkylamidate intermediate (Fig. 4C). Because of the long distance between the arginine and S-alkylamidate, either the arginine or the imidate intermediate would have to move for ADP-ribosylation of arginine to occur. In the crystal structure, the arginine is not fully extended (Fig. 4A). However, by rotating the bonds in the arginine side chain, the arginine can be repositioned within van der Waals contact distance (3.6 Å) of the C1′ of the S-alkylamidate (Fig. 5). This suggests that it is possible for the arginine to move farther into the Sir2 active site and contact the imidate intermediate. It is possible that the addition of NAD+ and formation of the O-alkylamidate would promote the +2 arginine to adopt a more extended conformation that could react with the O-alkylamidate intermediate to yield the ADP-ribosylated product.
FIGURE 5.
Model of a Sir2Tm acetyl p53 Arg-S-alkylamidate complex. The Sir2Tm S-alklyl amidate-p53 +2 Arg (+2R) peptide complex was modeled to determine whether the arginine could approach the intermediate.
DISCUSSION
The work presented here provides mechanistic insights into how acetylated substrates exhibit enhanced ADP-ribosylation by Sir2Tm. In several structures of either Sir2Tm or Sir2Af2 bound to an acetylated p53 peptide, the peptide methionine located two amino acids C-terminal to the acetyl lysine inserts into the enzyme active site just adjacent to the acetyl lysine (23–26). Previously, Kowieski et al. (22) proposed that a nucleophilic side chain on an acetylated protein substrate could intercept a Sir2 deacetylation reaction intermediate. This proposal and previous Sir2-acetylated peptide structures led us to speculate that a nucleophilic amino acid substituted at the +2 position could react with the O-alkylamidate intermediate to yield an ADP-ribosylated product. Based on the dimensions and geometry of the Sir2Tm active site (26), the +2 amino acid from the substrate peptide needs to be longer than 5 Å to enter the active site, and ideally, longer than 6 Å. We found that substituting lysine or arginine at this key position of the acetylated p53 peptide indeed results in increased enzymatic ADP-ribosylation by Sir2Tm (Figs. 1, B and D, and 2). The acetyl p53 +2 Arg peptide also stimulates ADP-ribosylation activity of SIRT1 (Fig. 1E), suggesting that ADP-ribosylation activity is conserved in at least a subset of Sir2 enzymes. Peptides with arginine substitutions at positions other than two amino acids C-terminal to the acetyl lysine are far less efficiently ADP-ribosylated (Fig. 2), indicating that ADP-ribosylation by Sir2Tm is sensitive to the position of the substituted arginine. Acetyl lysine residues that are substrates for Sir2 enzymes are located in loop regions of proteins, so using peptide substrates is reasonable. However, it would be informative to analyze the Sir2 ribosylation activity on a protein substrate.
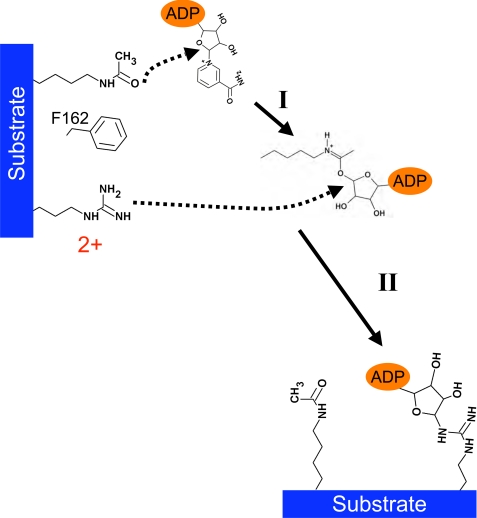
The +2 Arg of the acetylated peptide was directly ADP-ribosylated by Sir2Tm. Based on the structure of Sir2Tm bound to the acetyl p53 +2 Arg peptide, the +2 Arg enters the Sir2Tm active site adjacent to the acetyl lysine binding tunnel. In this position, the +2 arginine could react with a Sir2 deacetylation reaction intermediate to become ADP-ribosylated. Our biochemical and structural data support a mechanism in which the +2 arginine reacts with the O-alkylamidate intermediate generating ADP-ribosylated arginine, nicotinamide, and acetyl lysine (Fig. 6, step I). Next, the +2 arginine would react with the O-alkylamidate to generate ADP-ribosylated arginine, nicotinamide, and acetyl lysine (Fig. 6, step II).
FIGURE 6.
Structure-based mechanism of acetyl-dependent Sir2Tm-mediated ribosylation. In the first step of the reaction (step I), the acetyl lysine reacts with NAD+ to generate the O-alkylamidate. The O-alkylamidate can react with nicotinamide to regenerate the starting reactants, NAD+ and acetyl lysine, react with the 2′OH from the nicotinamide-ribose, leading to deacetylation products, or react with the arginine, resulting in ribosylation of the peptide substrate (step II).
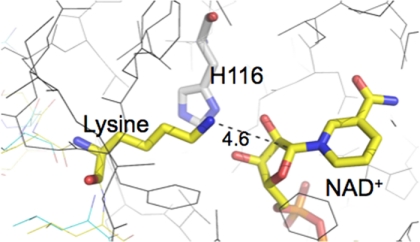
An N-terminal lysine is also ADP-ribosylated in the acetylated +2 Arg peptide, suggesting an alternate pathway for ADP-ribosylating substrates (Fig. 3). In structures of Sir2Tm bound to the acetylated p53 peptide, this N-terminal lysine cannot access the Sir2Tm active site. The N-terminal lysine is disordered in all of the Sir2Tm-acetyl p53 peptide complex structures and extends out into solvent. There are at least three models that could explain the ADP-ribosylation of this lysine. First, the reaction product O-acetyl ADP-ribose or NAD+ could react with this lysine nonenzymatically to yield a ribosylated product. There are three additional lysines in the acetyl p53 peptide that are not ribosylated (Fig. 2), arguing that this nonenzymatic mechanism is not likely. A second model would involve binding of a first acetyl peptide and formation of the O-alkylamidate intermediate. Next, another peptide could bind to Sir2Tm and insert the N-terminal lysine into the active site to react with the O-alkylamidate intermediate or another deacetylation intermediate. Based on all identified entry tunnels to the Sir2 active site, the lysine from the second peptide would not be long enough to gain access to the active site, suggesting that this mechanism is not likely. A third model involves Sir2Tm binding the N-terminal lysine through the acetyl lysine binding tunnel instead of the acetyl lysine. A crystal structure of Sir2Tm bound to an unacetylated p53 peptide demonstrates that Sir2Tm can bind unacetylated lysine peptides within the acetyl lysine binding tunnel (24). The unacetylated lysine can insert into the active site and come within 4.5 Å of the C1′ of NAD+ (Fig. 7). Next, the lysine could then directly attack the C1′ of NAD+ to yield a ribosylated lysine and nicotinamide. Further studies will be required to assess this proposed mechanism and determine whether other post-translational modifications including propionyl-lysine, which is a substrate for Sir2Tm-mediated depropionylation(33), are substrates for the Sir2 ribosylation reaction.
FIGURE 7.
Model of Sir2Tm bound to deacetylated peptide and NAD+. A model based on the Sir2Tm deacetylated peptide of Sir2Tm bound to deacetylated p53 peptide and NAD+ was built using the Quanta software package (Accelerys). The peptide lysine and NAD+ are colored in yellow, and the conserved histidine 116 is colored in gray.
The significance of the ADP-ribosylation activity of sirtuins to their physiological function remains an open question. The best established role for this activity is the regulation of the mitochondrial enzyme, glutamate dehydrogenase, by SIRT4 (20). Glutamate dehydrogenase had previously been shown to be inactivated by ADP-ribosylation, and SIRT4 was subsequently identified as the enzyme responsible for the ADP-ribosylation (20). Although it is not known whether ADP-ribosylation of glutamate dehydrogenase is dependent upon prior acetylation of that enzyme, it was recently found that glutamate dehydrogenase is acetylated at multiple sites (34), raising the possibility that ADP-ribosylation of this enzyme could proceed as described here for Sir2Tm. Although other mono-ADP-ribosylated proteins have been identified in eukaryotic cells (35), it remains to be determined which enzymes are responsible for adding this post-translational modification in vivo.
Supplementary Material
Acknowledgments
We thank Guy Macha and the staff at BioCARS beamline 14-BM-C at Advanced Photon Source (APS). We also thank Kamau Fahie (Johns Hopkins School of Medicine) for helpful discussions and technical assistance for the mass spec analysis.
This work was supported by National Science Foundation Grant MCB-0615815.
The atomic coordinates and structure factors (code 3JR3) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- CHES
- 2-(cyclohexylamino)ethanesulfonic acid
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1.Frye R. A. (2000) Biochem. Biophys. Res. Commun. 273, 793–798 [DOI] [PubMed] [Google Scholar]
- 2.Smith J. S., Brachmann C. B., Celic I., Kenna M. A., Muhammad S., Starai V. J., Avalos J. L., Escalante-Semerena J. C., Grubmeyer C., Wolberger C., Boeke J. D. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6658–6663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Daitoku H., Hatta M., Matsuzaki H., Aratani S., Ohshima T., Miyagishi M., Nakajima T., Fukamizu A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 10042–10047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motta M. C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L. (2004) Cell 116, 551–563 [DOI] [PubMed] [Google Scholar]
- 5.Luo J., Nikolaev A. Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. (2001) Cell 107, 137–148 [DOI] [PubMed] [Google Scholar]
- 6.Rodgers J. T., Lerin C., Haas W., Gygi S. P., Spiegelman B. M., Puigserver P. (2005) Nature 434, 113–118 [DOI] [PubMed] [Google Scholar]
- 7.Nemoto S., Fergusson M. M., Finkel T. (2005) J. Biol. Chem. 280, 16456–16460 [DOI] [PubMed] [Google Scholar]
- 8.Johnson L. M., Kayne P. S., Kahn E. S., Grunstein M. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 6286–6290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braunstein M., Rose A. B., Holmes S. G., Allis C. D., Broach J. R. (1993) Genes Dev. 7, 592–604 [DOI] [PubMed] [Google Scholar]
- 10.Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De Oliveira R., Leid M., McBurney M. W., Guarente L. (2004) Nature 429, 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauve A. A., Celic I., Avalos J., Deng H., Boeke J. D., Schramm V. L. (2001) Biochemistry 40, 15456–15463 [DOI] [PubMed] [Google Scholar]
- 12.Smith B. C., Denu J. M. (2006) Biochemistry 45, 272–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson M. D., Denu J. M. (2002) J. Biol. Chem. 277, 18535–18544 [DOI] [PubMed] [Google Scholar]
- 14.Jackson M. D., Schmidt M. T., Oppenheimer N. J., Denu J. M. (2003) J. Biol. Chem. 278, 50985–50998 [DOI] [PubMed] [Google Scholar]
- 15.Sauve A. A., Schramm V. L. (2003) Biochemistry 42, 9249–9256 [DOI] [PubMed] [Google Scholar]
- 16.Tanny J. C., Dowd G. J., Huang J., Hilz H., Moazed D. (1999) Cell 99, 735–745 [DOI] [PubMed] [Google Scholar]
- 17.Tsang A. W., Escalante-Semerena J. C. (1998) J. Biol. Chem. 273, 31788–31794 [DOI] [PubMed] [Google Scholar]
- 18.Frye R. A. (1999) Biochem. Biophys. Res. Commun. 260, 273–279 [DOI] [PubMed] [Google Scholar]
- 19.Liszt G., Ford E., Kurtev M., Guarente L. (2005) J. Biol. Chem. 280, 21313–21320 [DOI] [PubMed] [Google Scholar]
- 20.Haigis M. C., Mostoslavsky R., Haigis K. M., Fahie K., Christodoulou D. C., Murphy A. J., Valenzuela D. M., Yancopoulos G. D., Karow M., Blander G., Wolberger C., Prolla T. A., Weindruch R., Alt F. W., Guarente L. (2006) Cell 126, 941–954 [DOI] [PubMed] [Google Scholar]
- 21.García-Salcedo J. A., Gijón P., Nolan D. P., Tebabi P., Pays E. (2003) EMBO J. 22, 5851–5862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kowieski T. M., Lee S., Denu J. M. (2008) J. Biol. Chem. 283, 5317–5326 [DOI] [PubMed] [Google Scholar]
- 23.Hawse W. F., Hoff K. G., Fatkins D. G., Daines A., Zubkova O. V., Schramm V. L., Zheng W., Wolberger C. (2008) Structure 16, 1368–1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cosgrove M. S., Bever K., Avalos J. L., Muhammad S., Zhang X., Wolberger C. (2006) Biochemistry 45, 7511–7521 [DOI] [PubMed] [Google Scholar]
- 25.Avalos J. L., Celic I., Muhammad S., Cosgrove M. S., Boeke J. D., Wolberger C. (2002) Mol Cell 10, 523–535 [DOI] [PubMed] [Google Scholar]
- 26.Hoff K. G., Avalos J. L., Sens K., Wolberger C. (2006) Structure 14, 1231–1240 [DOI] [PubMed] [Google Scholar]
- 27.Avalos J. L., Boeke J. D., Wolberger C. (2004) Mol Cell 13, 639–648 [DOI] [PubMed] [Google Scholar]
- 28.Smith J. S., Avalos J., Celic I., Muhammad S., Wolberger C., Boeke J. D. (2002) Methods Enzymol. 353, 282–300 [DOI] [PubMed] [Google Scholar]
- 29.Otwinowski Z., Minor W. (1997) Method Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 30.Bailey S. (1994) Acta Crystallogr. D 50, 760–76315299374 [Google Scholar]
- 31.Emsley P., Cowtan K. (2004) Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 32.Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 33.Garrity J., Gardner J. G., Hawse W., Wolberger C., Escalante-Semerena J. C. (2007) J. Biol. Chem. 282, 30239–30245 [DOI] [PubMed] [Google Scholar]
- 34.Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 35.Leno G. H., Ledford B. E. (1990) FEBS Lett. 276, 29–33 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.