Abstract
Most apicomplexan parasites harbor a relict chloroplast, the apicoplast, that is critical for their survival. Whereas the apicoplast maintains a small genome, the bulk of its proteins are nuclear encoded and imported into the organelle. Several models have been proposed to explain how proteins might cross the four membranes that surround the apicoplast; however, experimental data discriminating these models are largely missing. Here we present genetic evidence that apicoplast protein import depends on elements derived from the ER-associated protein degradation (ERAD) system of the endosymbiont. We identified two sets of ERAD components in Toxoplasma gondii, one associated with the ER and cytoplasm and one localized to the membranes of the apicoplast. We engineered a conditional null mutant in apicoplast Der1, the putative pore of the apicoplast ERAD complex, and found that loss of Der1Ap results in loss of apicoplast protein import and subsequent death of the parasite.
Introduction
Apicomplexa are a phylum of obligate parasites that include the causative agents of malaria, toxoplasmosis, and cryptosporidiosis. Recent evidence suggests that apicomplexans evolved from a free-living photosynthetic ancestor (1). This ancestry is reflected in the presence of a chloroplast-like organelle, the apicoplast (2). While no longer engaged in photosynthesis, the apicoplast is essential to parasite survival and home to several critical biosynthetic pathways. Bioinformatic and experimental evidence suggests that the apicoplast is engaged in the synthesis of fatty acids, isoprenoids, and heme (3, 4). Genetic or pharmacological ablation of these pathways blocks parasite growth and the apicoplast, therefore, is currently considered a prime target for antiparasitic drug development (5–7). The organelle was derived by secondary endosymbiosis and reflects the successful union of a red alga and a heterotrophic eukaryote (8). A large number of algal genes were transferred to the host nucleus, and their products must now be routed back into the organelle. Trafficking occurs via the secretory pathway and is guided by an N-terminal bipartite targeting sequence (9). Transport from the ER to the apicoplast is believed to be vesicle mediated and to sidestep the Golgi apparatus (10, 11). Once delivered to the apicoplast, proteins have to cross three additional membranes. Recently, we demonstrated that a homolog of Tic20, a component of the translocon of the inner chloroplast membrane (Tic)4 complex in plants, is likely required for protein import across the innermost membrane of the apicoplast (12). To date bioinformatics searches have failed to identify a matching translocon of the outer chloroplast membrane in apicomplexans (13). Analysis of the genome of the remnant nucleus of the algal endosymbiont in cryptomonads showed the presence of an ER-associated protein degradation (ERAD) system and offered a candidate for a translocon across the third and potentially the second membrane (14). Classically, ERAD functions in the homeostasis of the secretory pathway by retrieving misfolded proteins from the ER that are subsequently degraded by the cytoplasmic proteasome. Der1 is a candidate for the membrane protein pore of the translocon (15). The ATPase Cdc48 together with its cofactors Ufd1 and Npl4 recognizes substrate emerging on the cytosolic side of the ER (16) (Fig. 7a). ATP hydrolysis then drives extraction into the cytoplasm followed by degradation in the proteasome (17). Recently Der1 homologs have been shown to localize to the plastids of a range of secondary plastid-containing organisms (14, 19, 20), but evidence linking these proteins to a functional role in apicoplast import is lacking. Here we use Toxoplasma as a genetic model to rigorously test the function of an ERAD-derived system in apicoplast biogenesis. We demonstrate the presence of two differentially localized systems, one in the ER/cytoplasm and one in the apicoplast. These systems are phylogenetically distinct and the apicoplast ERAD is derived from the algal endosymbiont. Importantly, we provide direct genetic and biochemical evidence for an essential function of the ERAD membrane component Der1Ap in apicoplast protein import.
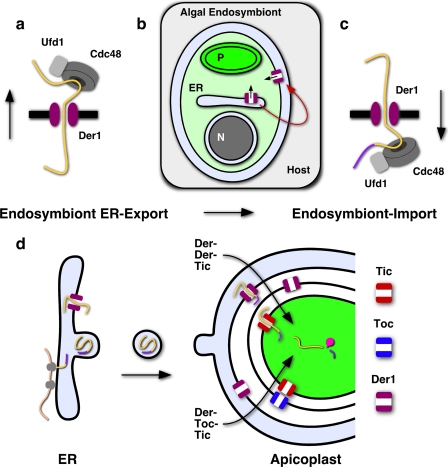
FIGURE 7.
Multiple distinct translocons act in apicoplast protein import. a, Der1, Cdc48, and Ufd1 are believed to be core components of the translocon of the ERAD pathway. b, duplication of the Der1 gene in the algal endosymbiont and relocation of its protein product to its plasma membrane enables c, protein import from the host endomembrane system. d, schematic representation of trafficking of nuclear-encoded proteins to the lumen of the apicoplast comparing a model employing subsequent Der1, Toc, and Tic translocons with a model in which the Toc translocon has been subsequently replaced by a second Der1 translocon. Cargo proteins are shown in yellow. The transit peptide (purple) is cleaved upon arrival. Note that trafficking of certain apicoplast membrane proteins appears not to require a bipartite signal sequence (26) and might occur through a different mechanism.
MATERIALS AND METHODS
Gene Identification and Gene Tagging
Yeast sequences for Der1, Cdc48, and Ufd-1 (GenBank™ IDs P38307, NP_010157, and NP_011562, respectively) were used as query sequences for BLAST searches against the T. gondii genome and GenBank™ databases. Initial RT-PCR experiments indicated that the automated gene predictions for the respective T. gondii homologs did not identify the beginning and end for all genes correctly. We performed 5′- and 3′-RACE using the SMART RACE cDNA amplification kit (BD Biosciences), and the resulting PCR products were cloned and sequenced. The sequences of primers used are listed in supplemental Table ST1. Using this approach, we experimentally verified the ends of six genes and the 3′-end of UfdAp. The 5′-end of the UfdAp gene was identified by RT-PCR taking advantage of array-based promoter predictions (21). The experimentally validated T. gondii genes described in this study are Der1-1ER (FJ976521), Der1-2ER (FJ976522), Cdc48Cy (FJ976518), Ufd1Cy (FJ976516), Der1Ap (FJ976520), Cdc48Ap (FJ976519), and Ufd1Ap (FJ976517). The full-length coding sequences were amplified from T. gondii cDNA using primers introducing flanking BglII and AvrII restriction sites, subcloned into plasmid pCR2.1 (Invitrogen) and subsequently introduced into the equivalent sites of either plasmid pCTH or pCTM35 placing them under the control of the T. gondii α-tubulin promoter and fusing a 3× HA tag (pCTH) or a 3× c-Myc tag (pCTM3), respectively, to the 3′-end. These constructs were stably introduced into RH strain T. gondii parasites using chloramphenicol selection (22).
To study the localization of UfdAp, we tagged the 3′-end of the gene by targeting the native locus with a cosmid clone of the respective locus (ToxoX83, see ToxoDB and Ref. 23) modified to contain a 3×HA tag by recombineering.6 Briefly, a modification cassette containing a 3× HA tag, a phleomycin marker for selection in T. gondii, and a gentamycin marker (24) for selection in bacterial cells was amplified from the plasmid template pH3BG. The modification cassette was flanked by 50 bp of targeting sequence introduced in the primer sequence to guide recombination into the cosmid at the appropriate sites at the 3′-end of the Ufd1 gene (see supplemental Table ST2). The resultant PCR product was electroporated into Escherichia coli EL250 cells (25) previously transfected with the ToxoX83 cosmid. Recombination was induced by heat shock and recombinant clones were isolated by double selection on kanamycin and gentamycin. The resulting modified cosmid clone was transfected in RH strain T. gondii parasites, and stable clones were obtained through phleomycin selection.
Parasite Culture and Isolation of Der1Ap Conditional Mutant
Parasites were cultured and genetically manipulated as described (22). To generate a conditional knock-out of Der1Ap, we generated a parental strain expressing an inducible copy of Der1Ap. The Der1Ap-coding sequence was introduced into the pDt7s4H vector (12), and the resulting construct was transfected into the T. gondii TATi strain, which expresses a tetracycline transactivator protein (27) using pyrimethamine selection. In this background, we disrupted the native Der1Ap locus. To engineer a targeting construct, we amplified ∼2 kb up- and downstream of the Der1Ap coding sequence (see supplemental Table ST3 for primers) and ligated the amplicon into the SalI and KpnI and SpeI and AatII sites, respectively, of vector pTCY. Linearized plasmid was transfected into the parental strain and selected on chloramphenicol. Chloramphenicol-resistant YFP-negative parasites were cloned by cell sorting as described (5). To complement the Der1Ap knock-out, we introduced a Der1Ap minigene under the control of a heterologous Tom22 promoter and a phleomycin-resistance cassette in the knock-out parasite line by phleomycin selection (28). To generate a Der1Ap knock-out cell line expressing apicoplast-targeted RFP, we digested the ptubFNR-RFP/sagCAT vector (29) with BglII and NotI and ligated the FNR-RFP containing fragment into the equivalent sites of pBTH(Der1Ap). The resultant pBTR(FNR) construct was stably transfected into the Der1Ap knock-out and parental lines using phleomycin selection. Parasite growth was measured by fluorescence and plaque assay (22) in the presence and absence of 0.5 μm anhydrotetracycline (ATc).
Microscopy
For immunofluorescence analysis, human fibroblasts were infected with the indicated parasite strain, fixed 24 h after infection with 3% paraformaldehyde, and permeabilized with 0.25% Triton X-100 in PBS. Primary antibodies used were rabbit anti-acyl-carrier protein (ACP) (1:1000 dilution), rat anti-HA (1:100 to 1:500), mouse anti-GFP (1:400), and mouse anti-c-Myc (1:200). Secondary antibodies used were goat anti-rabbit Alexa Fluor 546 (1:500), goat anti-rat Alexa Fluor 488, and rabbit anti-mouse Alexa Fluor 488 (1:200, Invitrogen). Images were collected on an Applied Precision Delta Vision or a Leica DIRBE microscope, and images were deconvolved and adjusted for contrast using Softworx and Openlab software.
For cryo-electron microscopy, infected cells were fixed in 4% paraformaldehyde/0.05% glutaraldehyde (Polysciences Inc.) in 100 mm PIPES buffer. Samples were then embedded in 10% gelatin and infiltrated overnight with 2.3 m sucrose/20% polyvinyl pyrrolidone in PIPES at 4 °C. Samples were frozen in liquid nitrogen and sectioned with a cryo-ultramicrotome. Sections were probed with the indicated primary antibodies followed by the appropriate secondary antibody conjugated to 12 or 18 nm colloidal gold, stained with uranyl acetate/methylcellulose, and analyzed by transmission EM as described previously (30).
Western Blotting
Protein samples were loaded onto precast 12% Bis-Tris and 3–8% Tris-acetate NuPAGE gels (Invitrogen). After electrophoresis, proteins were transferred to nitrocellulose membrane. Blots were probed with antibodies against ACP (1:1000–1:2000 dilution; a kind gift from Geoff McFadden, University of Melbourne (9), GRA8 (1:200,000), a kind gift from Gary Ward, University of Vermont (31), anti-HA (1:100 to 1:500; clone 3F10, Roche Applied Science), anti-c-Myc (1:50 to 1:100; clone 9E10, Roche Applied Science), GFP (1:1000; Torry Pines Biolabs), and anti-CDC48Ap (1:500; generated in this study, see supplemental methods). Horseradish peroxidase-conjugated anti-rat and anti-rabbit antibodies (Pierce) were used at 1:5000 to 1:10,000 dilutions, whereas horseradish peroxidase-conjugated anti-mouse antibodies (TrueBlot, eBioscience) were used at a 1:1000 dilution.
Pulse-chase Labeling and Immunoprecipitation
Pulse-chase experiments were carried out essentially as described previously (12). Briefly, host cell cultures were infected with 2 × 106 parasites and grown in the presence or absence of ATc. 2 days after infection, pulse-chase labeling with 100 μCi/ml [35S]cysteine and [35S]methionine (MP Biomedicals) was conducted as described under “Results.” To test parasites that were exposed to ATc 3 and 4 days, cultures were preincubated with drug in the previous passage for the appropriate time. Proteins of interest were purified by immunoprecipitation, separated by SDS-PAGE, and visualized by autoradiography. Antibodies used were anti-MIC5, a kind gift from Vern Carruthers, University of Michigan (32), anti-lipoic acid (Calbiochem), anti-RFP (Roche Applied Science), and anti-apicoplast-Cpn60 (rabbit serum raised against recombinant protein for this study).
Phylogenetic Analyses
For the phylogenetic analysis of Cdc48, we used sequences from 30 taxa (GenBank™ accession numbers for Cdc48 proteins are provided under supplemental methods) and generated a multiple sequence alignment in ClustalX. 900 unambiguously aligned amino acid positions were used for further analysis (alignments available on request). This data set was subjected to maximum likelihood phylogenetic analysis using RAxML version 7.0.4 (33). A phylogenetic tree was constructed using the GAMMA+P-Invar evolutionary model, and the model parameters were alpha: 0.981286, invar: 0.114318, and Tree-Length: 3.905532. Bootstrap analyses were conducted using 100 replicates (34).
RESULTS
Two Differentially Localized Sets of ERAD Components in T. gondii
Using the ERAD components from yeast and their recently identified cryptophyte homologs (14) as query sequences, we identified the genes for four putative Der1, two Ufd1, and two Cdc48 homologs in the T. gondii genome. Interestingly, the plastid-less apicomplexan Cryptosporidium retains only two Der1 homologs and a single Ufd1 and Cdc48. The coding sequences of seven of these candidates were amplified by PCR from T. gondii cDNA and introduced into parasite expression constructs that resulted in C-terminal fusion with an epitope tag (see “Materials and Methods” and supplemental Table ST1). The resulting constructs were transfected into parasites, and stable transgenic lines were obtained by drug selection. Ectopic expression of proteins from a non-native promoter can potentially impact their localization. To ensure that the observed localization patterns are physiological we also introduced an epitope tag directly into the genomic locus of one gene (Ufd1AP, see “Materials and Methods” for more detail), and we raised antibodies to recombinant protein for a second gene (Cdc48Ap, see supplemental methods).
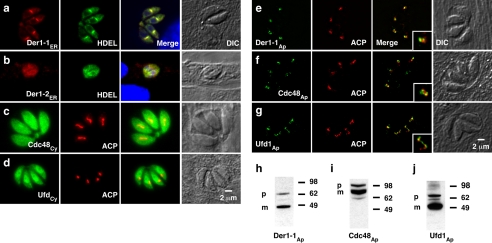
Using immunofluorescence assays we found that different T. gondii ERAD proteins localize to distinct subcellular compartments. Fig. 1, a and b shows the localization of two Der1 homologs to a perinuclear structure that colocalizes with P30-GFP-HDEL, a marker for the T. gondii ER (35). One homolog each of Cdc48 and Ufd1 is found in the cytosol. The localization of these four proteins is comparable with the localization of their homologs in yeast and mammalian cells and is consistent with their likely role in the classical ERAD pathway. A second set of ERAD components localized to the apicoplast. Fig. 1 e, f, and g shows localization of Der1Ap-HA, Cdc48Ap-c-Myc, and Ufd1Ap-HA and the apicoplast ACP for comparison. Apicoplast proteins typically contain an N-terminal signal for apicoplast targeting, which is cleaved upon arrival (9, 36). Sequence analysis predicts the presence of such a signal in the apicoplast ERAD proteins. Western blot analysis reveals two bands for each protein, suggesting N-terminal processing (Fig. 1, h–j), and the respective molecular masses are consistent with the predicted full-length precursor and the processed smaller mature form of the proteins.
FIGURE 1.
T. gondii ERAD components are associated with the ER/cytoplasm and the apicoplast. a–g, immunofluorescence analysis of parasites transfected with the coding sequences of seven putative T. gondii ERAD proteins carrying a C-terminal epitope tag (g, the Ufd1Ap epitope tag was constructed by inserting the tag directly into the genomic locus, see supplemental methods). We also expressed Cdc48Ap as a recombinant protein and raised a specific antiserum. Localization of the native protein detected with this antibody is indistinguishable from the protein derived from the tagged transgene (Fig. 2 and supplemental Fig. S1). Insets in the merge in panels e–g show a 2-fold higher magnification. h–j, Western blot analysis of apicoplast ERAD components. As for most apicoplast proteins, two bands are apparent (p, precursor; m, mature protein (9, 36)). ACP, luminal apicoplast marker, HDEL, T. gondii ER marker P30-GFP-HDEL (35).
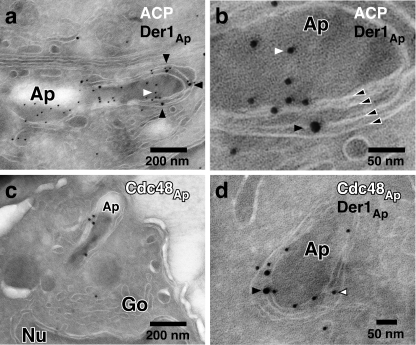
Whereas ERAD proteins localize to the apicoplast, they do not colocalize fully with the luminal marker ACP (see insets in Fig. 1, e–g). In silico modeling of the protein structure of Der1Ap suggests the presence of transmembrane domains, whereas Cdc48Ap and Ufd1Ap are predicted to be soluble. To study their intraorganellar localization, we performed immuno-electron microscopy. As shown in Fig. 2, Der1Ap and Cdc48Ap localize to an organelle that is bound by four membranes and can be labeled for the apicoplast marker ACP. Der1Ap and Cdc48Ap were confined to the periphery of the organelle, where both proteins colocalize, whereas ACP was found mainly in the lumen of the apicoplast. We conclude that a set of ERAD components is likely associated with the apicoplast membranes.
FIGURE 2.
The ERAD components Der1Ap and Cdc48Ap localize to the membrane compartment surrounding the apicoplast. Host cells infected with a transgenic T. gondii line expressing an HA-tagged version of Der1Ap were fixed, frozen, and sectioned with a cryo-ultramicrotome. Sections were incubated with a rat antibody to HA (a, b, d, black arrowheads), a rabbit serum to ACP (9) (a and b, white arrowhead), and a newly developed rabbit antibody against recombinant Cdc48Ap (c and d, white arrowhead with black outline, see also supplemental Fig. S1) followed by secondary antibody conjugated to 12 or 18 nm colloidal gold, stained with uranyl acetate/methylcellulose, and analyzed by transmission electron microscopy. Panel b shows a higher magnification of the cell shown in panel a, the four membranes of the apicoplast are indicated by black arrowheads outlined in white. Ap, apicoplast; Go, golgi; Nu, nucleus.
Der1Ap Is Essential for Parasite Growth
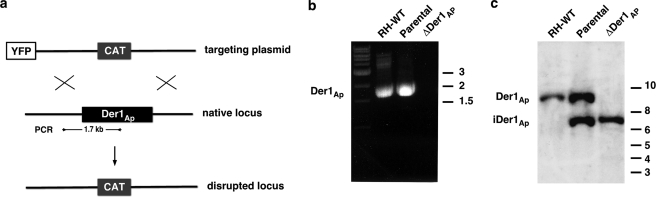
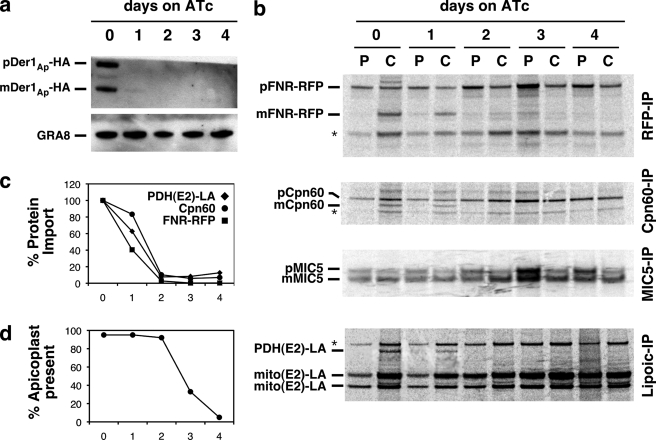
To genetically dissect the function of apicoplast ERAD components, we engineered a conditional null mutant in Der1Ap, the presumptive membrane pore of the putative translocon. We introduced an epitope-tagged Der1Ap minigene under the control of a tetracycline-regulatable promoter into a parasite strain expressing the tetracycline transactivator protein (37). In this background, we replaced the coding sequence of the native Der1Ap locus with a chloramphenicol acetyl transferase (CAT) selectable marker by double homologous recombination (5, 12). We confirmed disruption of the locus in chloramphenicol-resistant clonal parasites by PCR and Southern blot analysis (Fig. 3). In this mutant line, Der1Ap expression can be suppressed by culture in the presence of ATc. To measure inducible Der1Ap-HA expression, we grew parasites for 0–4 days on ATc, harvested parasites, and detected Der1Ap-HA protein by Western blot. We found that Der1Ap-HA levels were greatly reduced after 1 day on ATc and undetectable after 2 days (Fig. 5a).
FIGURE 3.
Disruption of the native Der1Ap locus. a, Der1Ap locus was disrupted by homologous recombination using a targeting construct carrying a CAT marker flanked by 2 kb of gene-specific upstream and downstream sequences. Recombinants were isolated using chloramphenicol to select for CAT expression, and cell sorting to select against YFP expression. Wild type (RH-WT), parental (carrying the native (Der1Ap) as well as an inducible minigene version (iDer1Ap), and ΔDer1Ap (carrying only the inducible version) were tested for the presence of the native locus by b, PCR (position of primers is indicated in a), or c, Southern blot using the radiolabeled coding sequence of Der1Ap as probe and Nde1 restriction to distinguish the native and inducible locus.
FIGURE 5.
Genetic ablation of Der1Ap results in loss of apicoplast protein import and organellar defects. a, Western blot analysis of Der1Ap-HA levels in ΔDer1Ap in response to ATc treatment (the dense granule protein GRA8 serves as loading control). b, pulse-chase (P/C) analysis of post-translational modification of apicoplast (FNR-RFP, Cpn60, PDH-E2), mitochondrial (mito(E2)), and secretory (MIC5) proteins. The data shown are representative of three independent experiments. Bands labeled with an asterisk likely represent RFP expression from an internal translation start site, mitochondrial Cpn60, and human PDH-E2 (12), respectively. Note that ATc treatment in parental parasites has no effect on apicoplast protein import (see supplemental Fig. S2 and Ref. 12) c, phosphorimager quantification of mature apicoplast-targeted protein bands shown in b. d, number of “four parasite” vacuoles in which each parasite has a clearly discernable apicoplast (n = 100 for each data point, see supplemental Fig. S3 for additional detail).
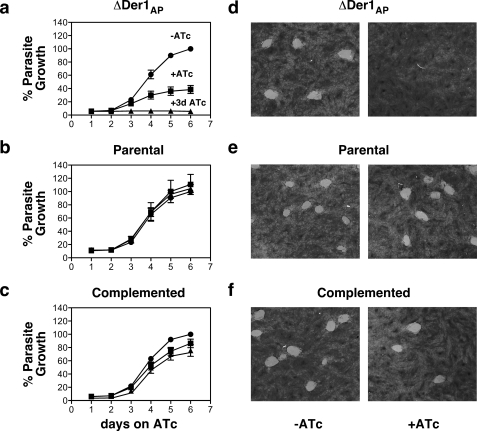
We next asked whether Der1Ap was essential for parasite growth. We introduced a red fluorescent protein into both mutant and parental lines and monitored parasite growth by measuring fluorescence intensity. In the absence of ATc, both cell lines grow at the same rate (Fig. 4, a and b). Whereas ATc appears to have no effect on the parental strain, growth of the mutant slows dramatically after 3 days of treatment. Pretreatment for 3 days prior to the assay blocks growth entirely. Growth of the mutant is restored when we reintroduce the Der1Ap coding sequence under a constitutive promoter (Fig. 4c), demonstrating the specificity of the observed phenotype. As a separate measure for parasite growth, we performed plaque assays. In this assay, confluent host cell cultures are infected with a small number of parasites and incubated for 8 days. Repeated rounds of invasion, growth, and egress result in the formation of clearings in the host cell monolayer that can be visualized by crystal violet staining. These experiments demonstrated minimal plaque formation in cultures infected with mutant parasites when these were grown in the presence of ATc, whereas parental and complemented strains showed no attenuation in growth (Fig. 4, d–f). We conclude that Der1Ap function is essential for parasite growth.
FIGURE 4.
Der1Ap is essential for parasite growth. a–c, fluorescence growth assays for the ΔDer1Ap mutant carrying the inducible copy only (a), the parental strain carrying both inducible and native Der1Ap (b), and a ΔDer1Ap clone complemented with the Der1Ap gene driven by a constitutive promoter (c, all strains were engineered to express RFP). Assays were performed in triplicate (error bars reflect S.D., note that the error bar is only shown if larger than the symbol (>3%)) in the absence (circles) and presence (squares) of 0.5 μm ATc or after 3 days of ATc preincuabtion (triangles). d–f, plaque assays measuring impact of loss of Der1AP. Confluent HFF cultures were infected with 400 parasites of the ΔDer1Ap mutant (d), parental (e), or complemented (f) strain, respectively, and cultured for 9 days in the absence (−ATc) or presence (+ATc) of anhydrous tetracycline. Cultures were fixed and stained as described under “Materials and Methods.” Note loss of growth in the mutant under ATc that is restored by gene complementation.
Genetic Knockdown of Der1Ap Ablates Apicoplast Protein Import
Whereas recent studies suggest that ERAD-derived proteins are found in several organisms harboring secondary plastids (19, 20, 38), experimental evidence for their function is largely missing. The apicoplast ERAD system could act equivalently to the ER system in quality control-associated protein export from the apicoplast, or alternatively in protein import into the organelle (14). To test the latter possibility, we measured apicoplast protein import in the Der1Ap mutant by assaying the maturation and post-translational modification of cargo proteins (12). We introduced a transgenic marker for the apicoplast lumen, FNR-RFP (29), into the mutant background. This strain was then used to perform pulse-chase labeling experiments. We labeled parasites for 1 h with 35S-labeled amino acids (pulse, P), followed by washout and incubation in nonradioactive medium for 2 h (chase, C). We next measured processing or modification of several apicoplast-targeted and control proteins by immunoprecipitation, gel electrophoresis, and autoradiography. As shown in Fig. 5b, FNR-RFP occurs as a 38-kDa precursor during the pulse and is processed during the chase to yield the mature form of 27 kDa. ATc treatment results in a significant reduction and subsequent loss of FNR-RFP maturation after 1 and 2 days of treatment, respectively (Fig. 5, b and c). Equivalent results were obtained using an antibody against Cpn60, a native luminal apicoplast chaperone, whereas treatment had no apparent effect on the maturation of MIC5, a secretory protein targeted to the micronemes (32) (Fig. 5, b and c). As an independent measure of protein import, we determined the level of lipoylation of the apicoplast pyruvate dehydrogenase E2 subunit (PDH-E2) (5, 12). Protein import is a prerequisite for lipoylation of PDH-E2 as the process requires two apicoplast resident enzymes, and the precursor molecule octanoyl-ACP that is synthesized de novo in the lumen of the organelle by the type II fatty acid synthesis system (5, 39). An antibody was used to immunoprecipitate specifically lipoylated proteins from parasite lysate. After 1 day on ATc, the level of lipoylated PDH-E2 was reduced, falling to undetectable levels after 2 days (Fig. 5, b and c). Several mitochondrial enzymes are similarly lipoylated and modification relies on successful import into the mitochondrion. This process showed no observable defect in the Der1Ap mutant. We conclude that ablation of protein import in the Der1Ap mutant is specific to the apicoplast. To ensure that the observed defects are not an artifact of ATc treatment, we performed control import experiments using the Der1Ap parental strain. In this strain, ATc treatment for 4 days had no apparent effect on the maturation of Cpn60 or the lipoylation of PDH-E2 (see supplemental Fig. S2).
Defects in apicoplast replication have been shown to affect apicoplast protein import (40). To test whether Der1Ap has a role in apicoplast replication, we performed live imaging of Der1Ap mutant parasites expressing FNR-RFP. We observed no defects for the first 2 days, a decrease in plastid numbers after 3 days, and widespread organellar loss after 4 days (Fig. 5d and supplemental Fig. S3). We conclude that Der1Ap does not directly affect apicoplast replication, and we establish the following sequence of consequences of ATc treatment in the Der1Ap mutant: Der1Ap protein levels are severely depleted after 1 day, directly coinciding with a loss of protein import, leading to subsequent defects in organellar biogenesis and parasite growth on days 3 and 4 post-treatment. Our data strongly support a direct role for Der1Ap in protein import into the apicoplast.
The Apicoplast ERAD System Is Derived from the Red Algal Endosymbiont
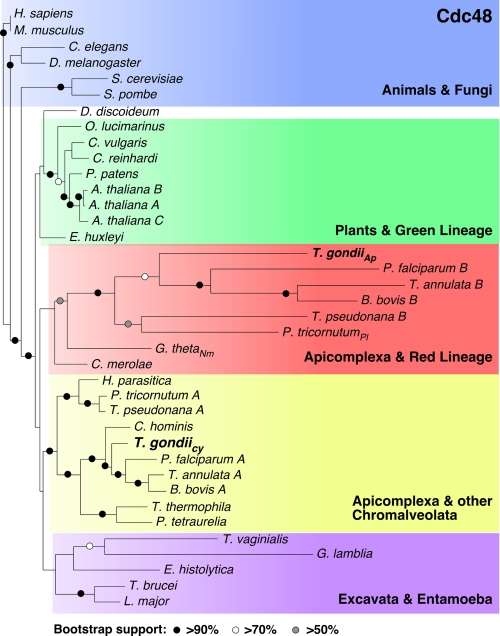
The apicoplast ERAD system could have evolved by duplication of ERAD genes of the host or alternatively could be derived by horizontal gene transfer from the algal endosymbiont. We performed phylogenetic analyses for the T. gondii ERAD components. The strong sequence divergence of Der1 proteins precluded the construction of meaningful alignments and trees. However, Cdc48 proteins are highly conserved providing 900 unambiguously aligned residues for robust analysis. We found that the two T. gondii proteins were of divergent phylogenetic origin (Fig. 6). The cytoplasmic protein forms a well-supported clade with homologs from chromalveolates (including photosynthetic and plastid-less taxa). This placement is consistent with the current model of vertical evolution of Apicomplexa as established by phylogenetic analyses using numerous protein and ribosomal RNA sequences (41, 42). In contrast, Cdc48Ap clusters with proteins from organisms harboring secondary plastids of red algal origin. Whereas analysis of Ufd1 did not provide full resolution of the tree of life because of a lower level of sequence conservation when compared with Cdc48, it fully supported divergent ancestry of the cytoplasmic and apicoplast protein (supplemental Fig. S4). We conclude that the apicoplast ERAD system is likely derived from the red algal endosymbiont, whereas the classical ER resident system was inherited vertically.
FIGURE 6.
Divergent origins of T. gondii Cdc48 proteins. RAxML maximum likelihood tree derived from an alignment of Cdc48 proteins from 30 taxa (900 unambiguously aligned amino acid characters were used after manual inspection, GenBank™ accession numbers are provided in the supplemental methods). Bootstrap analyses were conducted using 100 replicates. Nm, nucleomorph (remnant endosymbiont nucleus), Pl, plastid, Ap, apicoplast, Cy, cytoplasm.
DISCUSSION
Endosymbiosis is now well established as a mechanism that has played a crucial role in the evolution of eukaryotic cells. One organism, the symbiont, is engulfed by a second organism, the host, and a stable symbiotic relationship ensues in which the endosymbiont loses its independence and gradually evolves into an organelle that is controlled by the host and serves the host metabolic needs. A common hallmark of this process is massive horizontal gene transfer from the endosymbiont to the host (43). This gene transfer affords control to the host but also requires the establishment of mechanisms to reroute proteins that are now encoded and synthesized by the host back into the symbiont. A large body of work on mitochondria and chloroplasts has demonstrated the presence of elaborate protein translocons in the inner and outer membranes of these organelles (44, 45) that specifically recognize targeting information and deliver cargo proteins accordingly to the organellar lumen or various membrane compartments.
The apicoplast is the product of secondary endosymbiosis, the enslavement of a single celled eukaryotic alga. Compared with their primary progenitors, secondary plastids are surrounded by additional membranes that must be traversed by nuclear-encoded proteins (a total of four membranes surround the apicoplast). The evolution of mechanisms to traverse these additional membranes must have occurred early in organelle acquisition, and a simple solution would have been to use existing protein transport complexes. Our previous studies support this model and have shown that transport over the innermost apicoplast membrane is dependent on elements derived from the translocon of the inner chloroplast membrane of the endosymbionts chloroplast (12). The conservation of the Tic complex would make it appear likely that the Toc complex is equally conserved. However, so far genome searches have failed to identify Toc components in Apicomplexa or diatoms (13). The presence of the Toc complex might be masked by a high level of sequence divergence of its components or alternatively indicate that it has been replaced by a different mechanism. How proteins might cross the third or periplastid membrane is of particular interest as this membrane is thought to be a derivative of the endosymbionts plasma membrane. Based on the discovery of genes encoding elements of the ERAD system in the nucleomorph in cryptophytes, Sommer et al. (14) proposed that this complex was retooled to function in trafficking of plastid proteins (proteins of ERAD origin have also been speculated to be part of the peroxisomal proteome (46)). Most recently, this model has received support from studies on a variety of organisms bearing secondary plastids including the current study. ERAD-associated proteins are found in the membranous compartment surrounding secondary plastids in the apicomplexans Plasmodium and Toxoplasma ((19, 20) and Figs. 1 and 2 and supplemental Fig. S1), the diatom Phaeodactylum (19), and the cryptophyte Guillardia (14). Light and electron microscopy experiments indicate that these proteins are associated with the outer membranes of the plastid, but they lack resolution to tie the system to a specific membrane. Interestingly, split GFP assays in Phaeodactylum demonstrate the presence of two Der1 homologs in the third membrane. However, it is important to note that these experiments do not exclude the presence of Der1 homologs in other plastid membranes. Additional markers are needed to establish if the ERAD system is limited to the third membrane or might also be found in the second membrane and thus have replaced the Toc complex previously present in this membrane (Fig. 7d).
Whereas the presence of an ERAD translocon in the membranes of plastids was consistent with a role in plastid biology, it was unclear what this role of the ERAD complex might be. Functional data discriminating between a role in protein import versus protein export and quality control have as yet been missing. Whereas symbiont-derived Der1 proteins in Phaeodactylum have been shown to interact at a steady state level with fusion proteins targeted to the periplastid compartment, no interaction was observed with fusion proteins targeted to the plastid lumen (38). In the current study, we have used the ability to construct conditional mutants in T. gondii to devise a rigorous test of the protein import hypothesis. We isolated a T. gondii Der1Ap mutant and demonstrated that this protein is essential for apicoplast protein import and parasite survival. We have previously shown that apicoplast protein import across the innermost membrane is essential for Toxoplasma survival (12). In contrast, genetic disruption of classical ERAD in yeast or mammalian systems does not affect cell viability (18), and mutant cells have to be subjected to stressors resulting in the accumulation of misfolded proteins to produce a viability phenotype. More directly, we have developed and validated assays that track post-translational modifications of reporters that are restricted to the apicoplast lumen and thus require successful import (12). Applying these assays to the current study, we demonstrate a direct correlation between the loss of Der1Ap and a complete loss of apicoplast protein import (Fig. 5). These genetic and biochemical experiments provide the strongest evidence to date that the novel plastid ERAD system has a direct and essential role in plastid protein import.
Our phylogenetic analyses of CDC48 and Ufd1 indicate that the T. gondii ERAD systems are phylogenetically distinct and that the apicoplast system is derived from the red algal endosymbiont (Fig. 6 and supplemental Fig. S4). This is consistent with the presence of homologs in the cryptophyte nucleomorph, a remnant of the algal nucleus (14). It thus appears that the Der1 protein of the red algal symbiont was re-targeted from its original location in the ER to the symbiont to its plasma membrane, where it could now function in importing plastid-targeted proteins from the host secretory pathway (Fig. 7, a–c). This represents a remarkably simple and elegant solution for the complex problem of how to evolve protein exchange between host and endosymbiont at the beginning of their relationship. Once targeted to the endosymbionts cytoplasm proteins could take advantage of established mechanisms, namely the Toc and Tic, to gain access to the chloroplast. We hypothesize that the apicoplast employs a series of specific translocons that reflect the diverse evolutionary origin of the membranes in which they reside (Fig. 7d).
Supplementary Material
Acknowledgments
We thank Jessica Kissinger for help with phylogenetic analyses, Carrie Brooks for technical assistance, Julie Nelson for help with flow cytometry, and Vern Carruthers, Gary Ward, and Geoff McFadden for antibodies.
This work was supported, in whole or in part, by National Institutes of Health Grant AI64671 (to B. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables ST1–ST3, Methods, and Figs. S1–S4.
The nucleotide sequences reported in this paper has been submitted to the GenBank™/EBI Data Bank with accession number(s) FJ976521, FJ976522, FJ976518, FJ976516, EJ976520, FJ976519, and FJ976517.
G. G. van Dooren, unpublished data.
C. F. Brooks, G. G. van Dooren, and B. Striepen, unpublished data.
- Tic
- translocon of the inner chloroplast membrane
- ERAD
- endoplasmic reticulum-associated protein degradation
- RACE
- rapid amplification of cDNA ends
- HA
- hemagglutinin
- GFP
- green fluorescent protein
- ACP
- acyl-carrier protein
- PIPES
- 1,4-piperazinediethanesulfonic acid
- ATc
- anhydrotetracycline
- Toc
- translocon of the outer chloroplast membrane
- CAT
- chloramphenicol acetyltransferase
- RFP
- red fluorescent protein.
REFERENCES
- 1.Moore R. B., Oborník M., Janouskovec J., Chrudimský T., Vancová M., Green D. H., Wright S. W., Davies N. W., Bolch C. J., Heimann K., Slapeta J., Hoegh-Guldberg O., Logsdon J. M., Carter D. A. (2008) Nature 451, 959–963 [DOI] [PubMed] [Google Scholar]
- 2.Köhler S., Delwiche C. F., Denny P. W., Tilney L. G., Webster P., Wilson R. J., Palmer J. D., Roos D. S. (1997) Science 275, 1485–1489 [DOI] [PubMed] [Google Scholar]
- 3.Ralph S. A., van Dooren G. G., Waller R. F., Crawford M. J., Fraunholz M. J., Foth B. J., Tonkin C. J., Roos D. S., McFadden G. I. (2004) Nat. Rev. Microbiol. 2, 203–216 [DOI] [PubMed] [Google Scholar]
- 4.Fichera M. E., Roos D. S. (1997) Nature 390, 407–409 [DOI] [PubMed] [Google Scholar]
- 5.Mazumdar J., Wilson E., Masek K. A., Hunter C., Striepen B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 13192–13197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jomaa H., Wiesner J., Sanderbrand S., Altincicek B., Weidemeyer C., Hintz M., Türbachova I., Eberl M., Zeidler J., Lichtenthaler H. K., Soldati D., Beck E. (1999) Science 285, 1573–1576 [DOI] [PubMed] [Google Scholar]
- 7.Goodman C. D., McFadden G. I. (2007) Current Drug Targets 8, 15–30 [DOI] [PubMed] [Google Scholar]
- 8.Gould S. B., Waller R. F., McFadden G. I. (2008) Annu Rev. Plant Biol. 59, 491–517 [DOI] [PubMed] [Google Scholar]
- 9.Waller R. F., Keeling P. J., Donald R. G., Striepen B., Handman E., Lang-Unnasch N., Cowman A. F., Besra G. S., Roos D. S., McFadden G. I. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 12352–12357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DeRocher A., Gilbert B., Feagin J. E., Parsons M. (2005) J. Cell Sci. 118, 565–574 [DOI] [PubMed] [Google Scholar]
- 11.Tonkin C. J., Struck N. S., Mullin K. A., Stimmler L. M., McFadden G. I. (2006) Mol. Microbiol. 61, 614–630 [DOI] [PubMed] [Google Scholar]
- 12.van Dooren G. G., Tomova C., Agrawal S., Humbel B. M., Striepen B. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 13574–13579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tonkin C. J., Kalanon M., McFadden G. I. (2008) Traffic 9, 166–175 [DOI] [PubMed] [Google Scholar]
- 14.Sommer M. S., Gould S. B., Lehmann P., Gruber A., Przyborski J. M., Maier U. G. (2007) Mol. Biol. Evol. 24, 918–928 [DOI] [PubMed] [Google Scholar]
- 15.Ye Y., Shibata Y., Yun C., Ron D., Rapoport T. A. (2004) Nature 429, 841–847 [DOI] [PubMed] [Google Scholar]
- 16.Meyer H. H., Shorter J. G., Seemann J., Pappin D., Warren G. (2000) EMBO J. 19, 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flierman D., Ye Y., Dai M., Chau V., Rapoport T. A. (2003) J. Biol. Chem. 278, 34774–34782 [DOI] [PubMed] [Google Scholar]
- 18.Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., Walter P. (2000) Cell 101, 249–258 [DOI] [PubMed] [Google Scholar]
- 19.Spork S., Hiss J. A., Mandel K., Sommer M., Kooij T. W., Chu T., Schneider G., Maier U. G., Przyborski J. M. (2009) Eukaryot. Cell 8, 1134–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalanon M., Tonkin C. J., McFadden G. I. (2009) Eukaryot. Cell 8, 1146–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gissot M., Kelly K. A., Ajioka J. W., Greally J. M., Kim K. (2007) PLoS Pathog 3, e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Striepen B., Soldati D. (2007) in Toxoplasma gondii: The Model Apicomplexan - Perspective and Methods (Weiss L. M., Kim K. eds) pp. 391–415, Elsevier [Google Scholar]
- 23.Gubbels M. J., Lehmann M., Muthalagi M., Jerome M. E., Brooks C. F., Szatanek T., Flynn J., Parrot B., Radke J., Striepen B., White M. W. (2008) PLoS Pathog 4, e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poteete A. R., Rosadini C., St. Pierre C. (2006) BioTechniques 41, 261–262, 264 [DOI] [PubMed] [Google Scholar]
- 25.Lee E. C., Yu D., Martinez de Velasco J., Tessarollo L., Swing D. A., Court D. L., Jenkins N. A., Copeland N. G. (2001) Genomics 73, 56–65 [DOI] [PubMed] [Google Scholar]
- 26.Karnataki A., Derocher A., Coppens I., Nash C., Feagin J. E., Parsons M. (2007) Mol. Microbiol. 63, 1653–1668 [DOI] [PubMed] [Google Scholar]
- 27.Meissner M., Schlüter D., Soldati D. (2002) Science 298, 837–840 [DOI] [PubMed] [Google Scholar]
- 28.Messina M., Niesman I., Mercier C., Sibley L. D. (1995) Gene 165, 213–217 [DOI] [PubMed] [Google Scholar]
- 29.Striepen B., Crawford M. J., Shaw M. K., Tilney L. G., Seeber F., Roos D. S. (2000) J. Cell Biol. 151, 1423–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brossier F., Starnes G. L., Beatty W. L., Sibley L. D. (2008) Eukaryot. Cell 7, 664–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carey K. L., Donahue C. G., Ward G. E. (2000) Mol. Biochem. Parasitol. 105, 25–37 [DOI] [PubMed] [Google Scholar]
- 32.Brydges S. D., Harper J. M., Parussini F., Coppens I., Carruthers V. B. (2008) Biol. Cell 100, 253–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stamatakis A. (2006) Bioinformatics 22, 2688–2690 [DOI] [PubMed] [Google Scholar]
- 34.Stamatakis A., Hoover P., Rougemont J. (2008) Syst. Biol. 57, 758–771 [DOI] [PubMed] [Google Scholar]
- 35.Hager K. M., Striepen B., Tilney L. G., Roos D. S. (1999) J. Cell Sci. 112, 2631–2638 [DOI] [PubMed] [Google Scholar]
- 36.van Dooren G. G., Su V., D'Ombrain M. C., McFadden G. I. (2002) J. Biol. Chem. 277, 23612–23619 [DOI] [PubMed] [Google Scholar]
- 37.Meissner M., Reiss M., Viebig N., Carruthers V. B., Toursel C., Tomavo S., Ajioka J. W., Soldati D. (2002) J. Cell Sci. 115, 563–574 [DOI] [PubMed] [Google Scholar]
- 38.Hempel F., Bullmann L., Lau J., Zauner S., Maier U. G. (2009) Mol. Biol. Evol. 26, 1781–1790 [DOI] [PubMed] [Google Scholar]
- 39.Thomsen-Zieger N., Schachtner J., Seeber F. (2003) FEBS Lett. 547, 80–86 [DOI] [PubMed] [Google Scholar]
- 40.van Dooren G. G., Reiff S. B., Tomova C., Meissner M., Humbel B. M., Striepen B. (2009) Curr. Biol. 19, 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keeling P. J. (2009) J. Eukaryot Microbiol. 56, 1–8 [DOI] [PubMed] [Google Scholar]
- 42.Cavalier-Smith T. (2002) Int. J. Syst Evol. Microbiol. 52, 297–354 [DOI] [PubMed] [Google Scholar]
- 43.Kleine T., Maier U. G., Leister D. (2009) Annu Rev. Plant. Biol. 60, 115–138 [DOI] [PubMed] [Google Scholar]
- 44.Neupert W., Herrmann J. M. (2007) Annu. Rev. Biochem. 76, 723–749 [DOI] [PubMed] [Google Scholar]
- 45.Jarvis P. (2008) New Phytol. 179, 257–285 [DOI] [PubMed] [Google Scholar]
- 46.Gabaldón T., Snel B., van Zimmeren F., Hemrika W., Tabak H., Huynen M. A. (2006) Biol. Direct 1, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.