Abstract
TEM-1 β-lactamase is the most common plasmid-encoded β-lactamase in Gram-negative bacteria and is a model class A enzyme. The active site of class A β-lactamases share several conserved residues including Ser70, Glu166, and Asn170 that coordinate a hydrolytic water involved in deacylation. Unlike Ser70 and Glu166, the functional significance of residue Asn170 is not well understood even though it forms hydrogen bonds with both Glu166 and the hydrolytic water. The goal of this study was to examine the importance of Asn170 for catalysis and substrate specificity of β-lactam antibiotic hydrolysis. The codon for position 170 was randomized to create a library containing all 20 possible amino acids. The random library was introduced into Escherichia coli, and functional clones were selected on agar plates containing ampicillin. DNA sequencing of the functional clones revealed that only asparagine (wild type) and glycine at this position are consistent with wild-type function. The determination of kinetic parameters for several substrates revealed that the N170G mutant is very efficient at hydrolyzing substrates that contain a primary amine in the antibiotic R-group that would be close to the Asn170 side chain in the acyl-intermediate. In addition, the x-ray structure of the N170G enzyme indicated that the position of an active site water important for deacylation is altered compared with the wild-type enzyme. Taken together, the results suggest the N170G TEM-1 enzyme hydrolyzes ampicillin efficiently because of substrate-assisted catalysis where the primary amine of the ampicillin R-group positions the hydrolytic water and allows for efficient deacylation.
Introduction
β-Lactam antibiotics (e.g. penicillins and cephalosporins) are among the most often prescribed drugs to treat bacterial infections (1, 2). Their frequent use over several decades, however, has led to the widespread selection of β-lactam-resistant bacteria. β-Lactamases catalyze the hydrolysis of β-lactam antibiotics and represent the most common mechanism of bacterial resistance to the drugs (3). There are four different classes (A–D) of β-lactamases based on primary sequence homology. Class B β-lactamases are metallo-enzymes that use zinc ions to hydrolyze the β-lactam ring, whereas classes A, C, and D β-lactamases utilize a catalytic serine as the primary nucleophile (4).
Class A β-lactamases exhibit a broad substrate hydrolysis profile that includes penicillins, cephalosporins, and, for a few enzymes, carbapenems (5–7). TEM-1 β-lactamase is a class A enzyme and is the most common plasmid-encoded β-lactamase in Gram-negative bacteria (8). TEM-1 β-lactamase efficiently hydrolyzes penicillins and many cephalosporins, but it is not an effective catalyst for extended spectrum cephalosporin turnover (9). In addition, small molecule mechanism-based inhibitors of TEM-1 β-lactamase such as clavulanic acid are often used in conjunction with penicillins to avoid hydrolysis of the antibiotic (10). TEM-1 β-lactamase-mediated resistance has evolved over the past several years, however, because of mutations in the blaTEM-1 gene that result in amino acid substitutions that allow the TEM enzyme to hydrolyze extended spectrum cephalosporins or to avoid the action of mechanism-based inhibitors (11, 12).
The current view of the class A β-lactamase catalytic mechanism is divided into two stages: acylation and deacylation (13). During the acylation step, a proton is removed from the catalytic Ser70 residue (14). The process by which the proton is removed is currently under debate as to whether it is transferred to the side chain amine group of Lys73 directly or if it is transferred to a water molecule that is coordinated by Ser70, Glu166, and Asn170 (14–22). Currently, there is evidence for both pathways, and it has been proposed that perhaps both actions are possible and function together to remove the proton from Ser70 (23). The oxygen of Ser70 then attacks the carbonyl group to break the amide bond of the β-lactam (20). At this point, an acyl-intermediate is formed, and the same water that was coordinated by Ser70, Glu166, and Asn170 in acylation is activated to attack the covalent bond formed in the acyl-intermediate structure, which results in hydrolysis of the β-lactam antibiotic and regeneration of the enzyme (20, 22, 24, 25).
An omega loop structure within the TEM-1 active site contains residues Glu166 and Asn170 that coordinate the hydrolytic water discussed above (22). Glu166 is known to be crucial for deacylation, and changing this glutamate to other residues results in the formation of a stable acyl-enzyme intermediate (24, 26–29). For example, a TEM-1 E166N enzyme has been crystallized, and its structure has been determined in the acyl-enzyme form (20). Although the significance of Glu166 is well documented (24, 26–28), less work has been done to determine the functional role of Asn170 even though it forms hydrogen bonds to Glu166 and to the hydrolytic water that is also coordinated by Glu166 (22, 30).
The importance of Asn at position 170 for ampicillin hydrolysis was assessed by randomizing the position 170 codon to create a library containing all 20 possible amino acids. The library was sorted for mutants proficient at ampicillin hydrolysis by introducing the library into Escherichia coli and spreading the transformed cells on agar plates containing a concentration of ampicillin that selected for mutants exhibiting wild-type levels of hydrolysis. DNA sequencing of the blaTEM-1 gene from functional clones indicated only asparagine (wild type) or glycine at position 170 was consistent with high level function. The determination of kinetic parameters for several substrates revealed that the N170G enzyme is very efficient at hydrolyzing substrates that contain a primary amine in the antibiotic side chain. In addition, the x-ray structure of the N170G enzyme in the absence of substrate indicated that the position of the catalytically active water was altered in comparison with the wild-type structure. The kinetic parameters and structure determination results suggest that the N170G TEM-1 enzyme efficiently hydrolyzes substrates containing an amino group in the side chain, such as ampicillin, because of substrate-assisted catalysis, whereby the amine coordinates the hydrolytic water and facilitates an efficient deacylation reaction.
EXPERIMENTAL PROCEDURES
Construction of the Position 170 Random Library and Selection of Functional Clones
The position 170 random library was constructed by overlap extension PCR using sense and antisense strand primers complementary to the regions surrounding codon 170 (31). The oligonucleotides contained the sequence NNN, where N is any of the four nucleotides, in place of codon 170. The sense and antisense strand primers outside of the blaTEM-1 gene were PD-bla1 and TEM-XbaI-bot, which contained the SacI and XbaI sites, respectively, at the 5′-ends as follows: TEM-N170X, top, 5′-GGG AAC CGG AGC TGN NNG AAG CCA TAC CAA A-3′; TEM-N170X, bottom, 5′-CGT TTG GTA TGG CTT CNN NCA GCT CCG GTT CC-3′; PD-bla1, 5′-CGG GGA GCT CGT TTC TTA GAC GTC AGG TGG CAC-3′; and TEM-XbaI, bottom, 5′-GCG TGG TGC AAG TCT AGA TTA CCA ATG CTT AA-3′.
The pBG66 plasmid contains the wild-type blaTEM-1 gene and was used as template for the PCRs. The final PCR product was digested with SacI and XbaI restriction endonucleases and ligated into SacI-XbaI-cut pTP123 plasmid (32). The ligation mixtures were used to transform E. coli XL-1-Blue (Stratagene) by electroporation, and colonies were selected on LB agar plates containing 12.5 μg/ml chloramphenicol. Plasmid DNA was isolated from several transformants, and the region encoding the Asn170 codon was sequenced to ensure that different codons were present at position 170 among the transformants. The colonies were pooled from the LB plus 12.5 μg/ml chloramphenicol plates to create the N170X library.
The library cells were then spread on LB agar plates containing either 12.5 μg/ml chloramphenicol or 1 mg/ml ampicillin. The agar plates containing 1 mg/ml ampicillin were used to select clones from the library that provide wild-type levels of ampicillin resistance because this is the highest concentration of ampicillin on which an E. coli cell containing the TEM-1 plasmid can grow (33, 34). The plates were then incubated at 37 °C overnight. The clones that survived on the 1 mg/ml ampicillin agar plates were picked and grown in LB broth containing 12.5 μg/ml chloramphenicol, and plasmid DNA was purified using the QIAprep Spin Maniple kit. Once the plasmids were isolated, the blaTEM-1 gene was sequenced to determine the amino acid sequence of position 170 among the functional clones.
Site-directed Mutagenesis
The N170G and N170A mutants were constructed in the pET-TEM-1 plasmid using overlap extension PCR (31). The pET-TEM-1 plasmid used here encodes the wild-type mature portion of TEM-1 β-lactamase fused to the ompA signal sequence as previously described (35). The T7 forward and reverse primers were used in combination with the primers to introduce the glycine and alanine substitutions (N170G forward, 5′-GTT GGG AAC CGG AGC TGG GTG AAG CCA TAC CAA ACG-3′; N170G reverse, 5′-CGT TTG GTA TGG CTT CAC CCA GCT CCG GTT CCC AAC-3′; N170A forward, 5′-GTT GGG AAC CGG AGC TGG CTG AAG CCA TAC CAA ACG-3′; and N170A reverse, 5′-CGT TTG GTA TGG CTT CAG CCA GCT CCG GTT CCC AAC-3′). The PCR product was introduced into the pET-TEM-1 vector through a DNA ligation reaction using T4 ligase and the BamH1 and Nde1 restriction sites. The ligation reactions were then transformed into XL-1 Blue E. coli cells. DNA sequencing was used to confirm that the mutation was introduced and that no extraneous mutations occurred.
The N170G, E166A, and E166A/N170G mutants were created by overlap extension PCR with oligonucleotides encoding the desired mutations (31). The TEM-1 E166A/N170G clone is a double mutant. The sense and antisense strand primers outside of the blaTEM-1 gene were PD-bla1 and TEM-XbaI-bot, which contained the SacI and XbaI sites, respectively, as described for the library construction above. The final PCR product was digested with SacI and XbaI and ligated into SacI-XbaI cut pTP123 plasmid (32). The ligation mixtures were used to transform E. coli XL-1-Blue by electroporation, and colonies were selected on LB agar plates containing 12.5 μg/ml chloramphenicol. DNA sequencing was used to identify clones containing the correct mutant sequence.
Minimum Inhibitory Concentration Determination
The plasmids encoding the wild-type and mutant TEM-1 genes were transformed into E. coli XL1-Blue cells and grown overnight in 4 ml of LB broth containing 12.5 μg/ml chloramphenicol. The overnight culture was diluted 100-fold and grown at 37 °C for 3–4 h until the cultures reached mid-log (A600 = 0.4–0.5). A cotton swab was placed into the culture and then spread over a warm LB agar plate. An Etest strip (AB Biodisk) was placed on the plate and incubated at 37 °C overnight. The minimum inhibitory concentration (MIC)3 was determined the next day in accordance with the company standards.
The MIC was also determined by liquid broth dilution by doubling the amount of ampicillin in culture to confirm the observations above and to determine an accurate MIC of the E. coli cells containing the wild-type TEM-1 and the N170G mutant plasmids. Overnight cultures were grown at 37 °C by inoculating 4 ml of LB broth containing 12.5 μg/ml chloramphenicol with single colonies. 104 colony-forming units/ml cells were used to inoculate LB broth containing increasing amounts of ampicillin starting with a concentration of 2 μg/ml. The MIC was determined after an 18-h incubation at 37 °C.
Kinetic Analysis of the β-Lactamase Enzymes
The in vitro kinetic parameters were determined as previously described (9). The amount of wild-type TEM-1 or mutant enzyme that was used varied between the enzymes and the substrates tested. In addition, a 1-cm-pathlength quartz cuvette was used to monitor ampicillin and penicillin G hydrolysis, whereas a 0.1-cm quartz cuvette was used for cephalothin and cephalexin reactions due to the high absorption of the cephems at the concentrations relevant to the assay. The initial velocities were measured with a DU 800 spectrophotometer, and the data were fitted to the Michaelis-Menten equation, v = Vmax[S]/(Km + [S]), using GraphPad Prism5 to determine kcat and Km. When the TEM-1 mutants had high Km values that prevented determination of Vmax, the catalytic efficiency (kcat/Km) was determined using the equation, v = kcat/Km [E][S], where [S] ≪ Km (36). The initial velocities were measured in at least duplicate and averaged to determine the kinetic parameters.
TEM1 β-Lactamase Purification and Crystallization
TEM-1 β-lactamase and its mutants were purified as described previously using a zinc chelating column followed by gel filtration (9). After the gel filtration column, the N170G TEM-1 β-lactamase was dialyzed into 60 mm sodium phosphate buffer, pH 7.8. The sample was condensed to a concentration of 30 mg/ml. Hanging drops were then set up using a TTP LabTech mosquito instrument. The reservoir contained 70 μl of 0.2 m LiCl, 0.1 m Hepes, pH 7.0, and 20% polyethylene glycol 6000 (Qiagen) and a 2-μl hanging drop (1:1 ratio of protein solution:mother liquor), and the trays were maintained at 20 °C. The crystals were visible after 24 h and were harvested after 2 weeks. The crystals were cryoprotected with the mother liquor plus 20% glycerol and stored in liquid nitrogen until data collection.
Data Collection and Refinement
The x-ray crystallography data were collected using the Advanced Photon Source at Argonne National Laboratory Synchrotron. The data were integrated and scaled using HKL2000 (37). Six molecules/asymmetric unit were found using molecular replacement with the Phaser program as a part of the CCP4i suite (38, 39). The reference molecule (Protein Data Bank code 1BTL) used was the wild-type TEM-1 β-lactamase molecule (22). After molecular replacement, a total omit map was created by the program SFCHECK to minimize model bias (40). The model was then fitted to the total omit map using Coot (41). The Phenix program was used to refine the structure with TLS and noncrystallographic symmetry restraints. Ordered solvent was also added using Phenix (42). A list of the data collection and refinement statistics are provided in Table 3. The analysis of the structure was done using both the Pymol and Coot programs (43). The coordinates of the TEM-1 N170G mutant structure have been deposited in the Protein Data Bank (code 3JYI).
TABLE 3.
Data collection and refinement statistics for TEM-1 N170G β-lactamase structure (Protein Data Bank code 3JYI)
| Space group | P43212 |
| Unit cell a, b, c (Å) | 88.1, 88.1, 500.4 |
| Resolution range (Å) | 50–2.70 (2.75–2.70) |
| Molecules/asymmetric unit | 6 |
| Number of unique reflections | 55,684 |
| Completeness (%)a | 99.9 (99.2) |
| Redundancya | 6.5 (5.5) |
| I/σa | 16.1 (2.0) |
| Rmerge (%)a | 10.8 (56.0) |
| Rwork (%)/Rfree(%) | 23.0/26.0 |
| Number of protein atoms | 12242 |
| Number of waters | 174 |
| Average B-factor protein (solvent) | 53.36 (44.10) |
| Root mean square deviations | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.1 |
| Ramachandran analysis | |
| Preferred (%) | 94.1 |
| Allowed (%) | 4.5 |
| Outliers (%) | 1.4 |
a The values in parentheses correspond to the highest resolution shell.
Molecular Modeling
The TEM-1 E166N acylated intermediate structure (Protein Data Bank code 1FQG) with penicillin was used to approximate the space available in the acylated intermediates of the N170G and N170A mutants. These substitutions were introduced to the 1FQG acyl-enzyme structure in conjunction with the addition of an amine group to the penicillin substrate to create ampicillin. The appropriate distance measurements were estimated using Pymol (43). The models and distances were used to assess the potential of the primary amine from the ampicillin side chain to fill the void left from the position 170 mutants in the acyl-intermediate of catalysis.
RESULTS
Randomization of Position 170 and Selection of Functional Mutants
As a first step in understanding the role of asparagine at position 170 in the active site of TEM-1 β-lactamase, the effects of amino acid substitutions at this residue were studied by randomizing the position 170 codon from AAT to NNN, where N is any of the four nucleotides to create a library of all possible amino acid substitutions. To determine which amino acid substitutions at position 170 are consistent with wild-type levels of function, the random library was introduced into E. coli, and the transformed cells were spread on agar plates containing a concentration of ampicillin (1 mg/ml) that selects for wild-type levels of ampicillin hydrolysis by β-lactamase. The DNA sequence of the bla gene of functional mutants revealed that the spectrum of amino acid substitutions that are consistent with high level ampicillin hydrolysis includes only the wild-type asparagine and glycine (Fig. 1). This result indicates that, other than the wild-type asparagine, only a glycine substitution at this position results in an enzyme able to hydrolyze ampicillin at levels comparable with wild type. It is also interesting to note that for both asparagine and glycine, two different codons are represented among the functional mutants, suggesting that many codons were sampled from the library, but only those encoding Asn or Gly are consistent with high level function.
FIGURE 1.
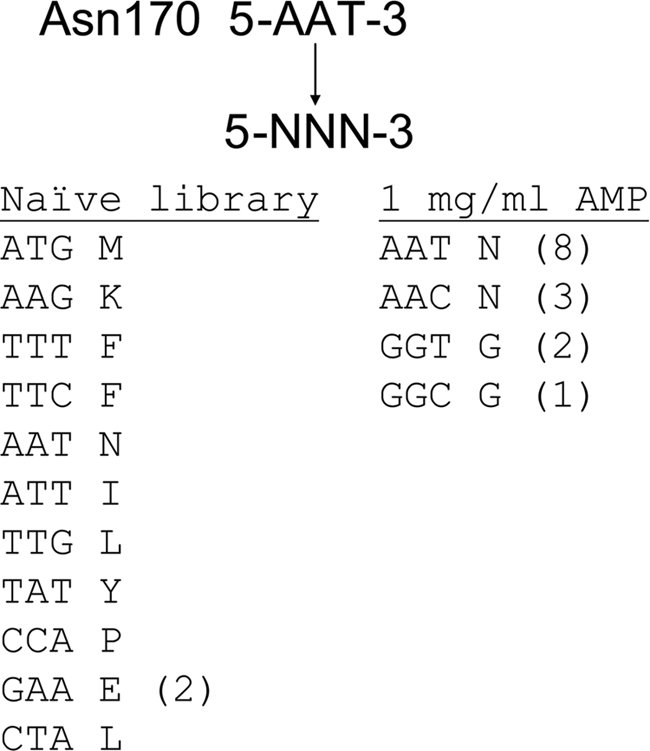
Results of DNA sequence analysis of mutants from the N170X random library. The column on the left shows sequences of N170X mutants picked from the naive library before selection on ampicillin. These mutant sequences indicate that the Asn170 position was randomized in the library and that there is no obvious bias among the sequences. The column on the right indicates sequences of mutants from the N170X library after selection for functional clones on agar plates containing 1 mg/ml ampicillin. The numbers in parentheses indicate the number of times the indicated codon was identified among the mutants sequenced.
Characterization of the TEM-1 N170G Mutant
To confirm that E. coli containing the N170G mutant exhibits resistance levels similar to wild type, the MIC of ampicillin was determined and found to be in the same range as wild type (Table 1). The mutant was further characterized by expressing N170G in E. coli and purifying the enzyme to homogeneity as judged by SDS-PAGE fractionation and visualization. Kinetic parameters were determined for ampicillin hydrolysis for wild-type TEM-1 and the N170G enzyme, and it was found that Km values were similar, whereas kcat of N170G was approximately one-half that of wild type (Table 2). Therefore, consistent with the random library selection results, the N170G enzyme proficiently hydrolyzes ampicillin.
TABLE 1.
Minimum inhibitory concentrations of ampicillin for omega loop mutants
| E. coli containing TEM-1 plasmids | Broth dilution MIC | Etest MIC |
|---|---|---|
| μg/ml | μg/ml | |
| E. coli XL-1 Blue | 2 | 1.5 |
| pTP123 | 2 | 1.5 |
| Wild-type TEM-1 | 2048 | >256 |
| N170G | 2048 | >256 |
| E166A | 4 | 2 |
| E166A/N170G | 8 | 4 |
TABLE 2.
Kinetic parameters for hydrolysis of β-lactam antibiotics by TEM-1 and mutant β-lactamases
kcat is expressed in s−1, and Km is expressed in μm. NA, not available, Km too high to determine accurately. ND, not detected.
| Enzyme | Substrates |
|||
|---|---|---|---|---|
| Ampicillin | Penicillin G | Cephalexin | Cephalothin | |
| Wild-type | ||||
| kcat | 1085 ± 137 | 950 ± 41 | 13 ± 1 | 146 ± 16 |
| Km | 38 ± 18 | 28 ± 5 | 1162 ± 250 | 182 ± 58 |
| kcat/Km | 29 | 34 | 0.01 | 0.80 |
| N170G | ||||
| kcat | 467 ± 76 | 64 ± 2 | 16 ± 3 | 31 ± 5.0 |
| Km | 56 ± 28 | 3 ± 0.5 | 2590 ± 758 | 1501 ± 449 |
| kcat/Km | 8.3 | 21 | 0.006 | 0.02 |
| N170A | ||||
| kcat | 22 ± 1 | 45 ± 2 | 1 ± 0.08 | NA |
| Km | 13 ± 3 | 8 ± 2 | 1561 ± 294 | >2000 |
| kcat/Km | 1.7 | 5.6 | 0.001 | 0.004 |
| E166A | ||||
| kcat | ND | ND | ND | ND |
| Km | ND | ND | ND | ND |
| kcat/Km | ND | ND | ND | ND |
| E166A/N170G | ||||
| kcat | 0.9 ± 0.08 | 0.8 ± 0.04 | ND | ND |
| Km | 24 ± 5.4 | 20 ± 3.5 | ND | ND |
| kcat/Km | 0.04 | 0.04 | ND | ND |
To assess the impact of the N170G substitution on substrate specificity, kinetic parameters were determined for the wild-type and mutant enzymes for penicillin G hydrolysis. The wild-type enzyme hydrolyzed penicillin G with kcat and Km values similar to those obtained for wild type with ampicillin as substrate (Table 2). In contrast, the N170G enzyme exhibited greatly reduced kcat and Km values for penicillin G hydrolysis compared with those obtained with the same enzyme for ampicillin. In addition, the N170G enzyme exhibited greatly reduced kcat and Km values for penicillin G hydrolysis compared with those observed for the wild-type enzyme with this substrate. Thus, the wild-type enzyme hydrolyzes ampicillin and penicillin G with similar kinetic parameters, whereas the N170G enzyme exhibits significantly different specificity between these substrates. The reduction in Km of the N170G enzyme for penicillin G hydrolysis served to balance the lowered kcat value so that catalytic efficiency (kcat/Km) was reduced less than 2-fold compared with wild type (Table 2). Nevertheless, the striking difference in kcat and Km values suggests that a rate-limiting step may be altered for penicillin G hydrolysis by the N170G enzyme.
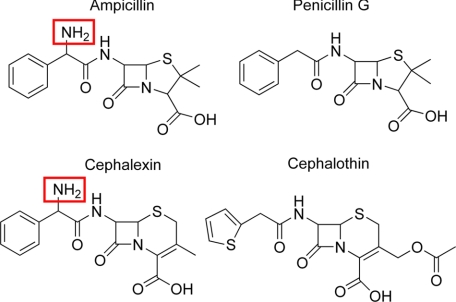
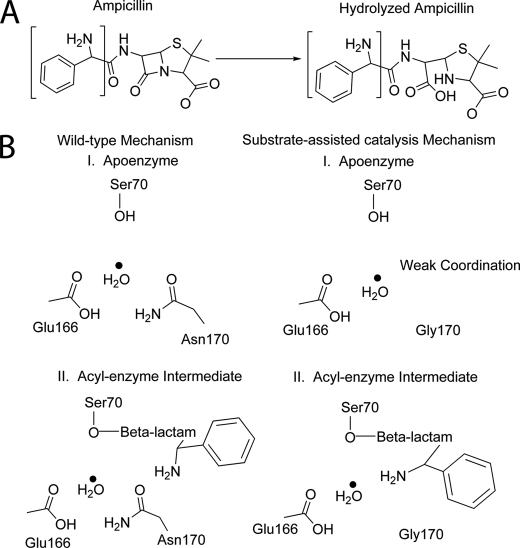
The only difference in structure between ampicillin and penicillin G is the presence of an amino group positioned after the amide group and adjacent to the benzyl group in the side chain of ampicillin (Fig. 2). This difference in substrate structure presumably results in a change in kinetic parameters of the N170G enzyme for ampicillin versus penicillin G hydrolysis and for the difference in penicillin G hydrolysis by the N170G β-lactamase versus penicillin G hydrolysis by the wild-type enzyme. In the wild-type TEM-1 enzyme structure, the Asn170 side chain forms a hydrogen bond with the catalytic water molecule (22), presumably to help position the water for interaction with Glu166 (18, 20), which serves as a general base to activate the water for deacylation (Fig. 3) (20, 24, 29).
FIGURE 2.
Schematic illustration of penam and cephem substrates used in this study.
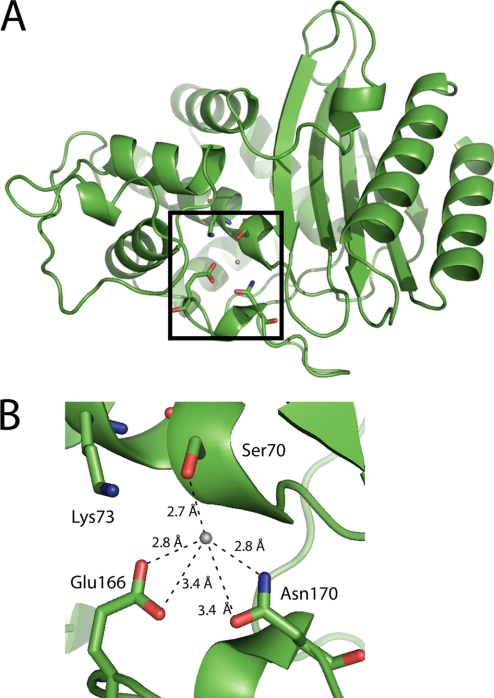
FIGURE 3.
The diagram illustrates the overall structure and the active site of the wild-type TEM-1 enzyme (22). A, a cartoon representation of the overall structure of TEM-1 β-lactamase is shown with the active site boxed. B, the active site is shown with several of the conserved residues labeled and the hydrogen bonding environment near the hydrolytic water illustrated including Asn170, which is one of the three residues that position the water (Protein Data Bank code 1BTL) (22).
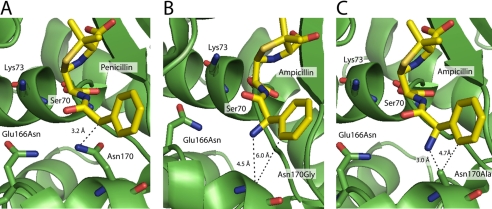
The structure of a TEM-1 E166N mutant that had been acylated by penicillin G was previously solved (20), and the carbon position to which the amino group of ampicillin is attached is only 3.2 Å away from the nitrogen in the side chain of Asn170 that hydrogen bonds to the water (Fig. 4A). When the Gly170 substitution is modeled into the acyl-intermediate structure, the distance from the carbon of interest on penicillin to the alpha carbon of glycine is 5.3 Å. When the primary amine of ampicillin is added to the N170G acyl-intermediate model, the distance from the modeled nitrogen to the alpha carbon of glycine is 4.5 Å. Furthermore, the benzene ring of the substrate is a full 6.0 Å away from the alpha carbon of glycine (Fig. 4B). Thus, there is sufficient space for the amino group from the ampicillin R substituent to be repositioned to occupy the former position of the Asn170 side chain. These findings suggest that the amino group in ampicillin may replace the Asn170 side chain nitrogen in the N170G enzyme and serve to position the catalytic water molecule for the deacylation reaction by substrate-assisted catalysis. This hypothesis would explain why the N170G enzyme hydrolyzes ampicillin with kinetic parameters similar to the wild-type enzyme but exhibits a significantly reduced kcat for penicillin G hydrolysis.
FIGURE 4.
Molecular modeling of the TEM-1 β-lactamase acyl-intermediate. A, the acyl-intermediate with TEM-1 (in green) covalently bonded to penicillin G is shown to illustrate that the benzene ring of penicillin (in yellow) is close to Asn170 (Protein Data Bank code 1FQG) (20). The carbon position that contains the amine group in ampicillin is 3.2 Å away from the side chain of Asn170. B, when ampicillin and the glycine 170 substitution are modeled into the acyl-intermediate structure, the alpha carbon of glycine is 4.5 and 6.0 Å from the primary amine and the benzene ring of the ampicillin R substituent, respectively. C, TEM-1 ampicillin acyl-enzyme model from B with the alanine substitution modeled into position 170.
Molecular modeling of the TEM-1 E166N enzyme acylated with penicillin G (20) indicates that the addition of the amino group to mimic ampicillin requires removal of the entire Asn170 side chain to allow room for the amino group to occupy a position to hydrogen bond to the catalytic water. When an N170A substitution was modeled into the acyl-intermediate structure, the carbon position to which the amino group of ampicillin would be attached is only 3.8 Å from the methyl group of Ala170. When the amino group of ampicillin is modeled into the acyl-intermediate, the distance from the modeled nitrogen to the methyl group of Ala170 is only 3.0 Å. In addition, the benzene ring of the substrate is 4.7 Å away from the methyl group of Ala170 (Fig. 4C). This suggests that the methyl group of Ala170 could create steric clashes with the amino group of ampicillin, hindering the primary amine from coordinating the hydrolytic water. Thus, the modeling results suggest that even an N170A substitution does not create sufficient space for the amino group to occupy the position of the Asn170 side chain nitrogen.
To test these modeling results, an N170A TEM-1 enzyme was created by site-directed mutagenesis and purified to determine its kinetic properties. The N170A enzyme hydrolyzed penicillin G with kinetic parameters very similar to those observed for this substrate with the N170G enzyme. However, the kinetic parameters for ampicillin hydrolysis were greatly different for the N170A enzyme compared with wild-type or the N170G enzyme in that both kcat and Km values were reduced, which is similar to the observations with the N170G enzyme with penicillin G as substrate (Table 2). These findings are consistent with the molecular modeling results and suggest that there is not sufficient space for the amino group of ampicillin to occupy a position to hydrogen bond with the catalytic water molecule in the N170A TEM-1 enzyme.
X-ray Structure Determination of the TEM-1 N170G Enzyme
The hypothesis that the TEM-1 N170G enzyme hydrolyzes ampicillin via substrate-assisted catalysis, whereby the side chain amino group helps to position the catalytic water to participate in deacylation, predicts that the catalytic water is absent or in an altered position in the N170G enzyme in the absence of substrate. This prediction was tested by determining the x-ray structure of the N170G enzyme. The N170G enzyme was crystallized exhibiting a large unit cell (a = 88.1 Å, b = 88.1 Å, c = 500.4 Å) with the space group P43212. Molecular replacement found six molecules/asymmetric unit, and the structure was refined to 2.7 Å resolution (Table 3).
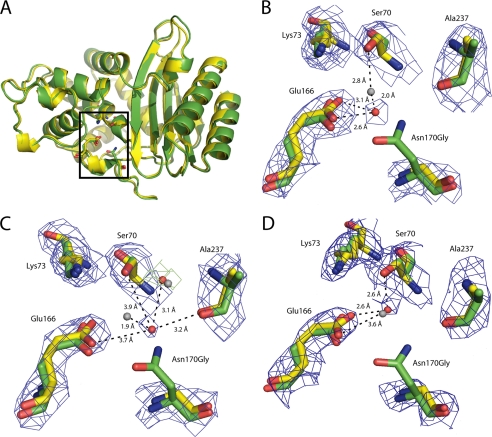
When the native TEM-1 (Protein Data Bank code 1BTL) and N170G mutant structures were aligned, the root mean square deviation was found to be ∼0.51 Å for all six molecules, indicating that the overall structure of the N170G mutant was strongly conserved (Fig. 5A). Position 170 is located at the apex of the omega loop (residues 161–179). When this region was aligned to the wild-type structure, the root mean square deviation was merely 0.23 Å for all six molecules. The low root mean square deviation values indicate that the structure of the omega loop is also strongly maintained in the N170G enzyme relative to wild type. In addition, the position of Glu166 is not significantly altered in the N170G structure, despite the fact that glutamate forms a hydrogen bond with the Asn170 side chain in the wild-type structure.
FIGURE 5.
The crystal structures shown are of the TEM-1 N170G mutant and the wild-type TEM-1 enzyme (Protein Data Bank code 1BTL) superimposed (22). A, superimposition of the molecules demonstrates that the overall structure and the omega loop of N170G (in yellow) and TEM-1 (in green) are highly conserved. However, the hydrolytic water is moved or absent in five of the six TEM-1 molecules. B, illustration of the superimposition of chain A of the N170G mutant structure onto the structure of the wild-type enzyme. In this diagram, the hydrolytic water in the mutant structure (red sphere) is ∼2.0 Å away from its location (gray sphere) in the wild-type structure. C, illustration of chain B where the hydrolytic water is moved away from both Glu166 and Ser70 and is coordinated by the water in the oxyanion hole and the main chain carbonyl group of Ala237. Hydrolytic water from mutant structure is a red sphere, and the wild-type water is a gray sphere. D, position of the hydrolytic water (red sphere) in chain F of the mutant structure. The hydrolytic water occupies a similar position as the water in the wild-type structure (gray sphere). The electron density map (2Fo − Fc) is shown in blue at a contour level of 1.0 σ and the difference map (Fo − Fc) in green at a contour level of 3.0 σ. Some of the other active site waters and peripheral side chains were removed for clarity.
The most significant observation in the N170G enzyme structure is that the catalytic water molecule that is coordinated in the wild-type structure by Ser70, Glu166, and Asn170 is partially occupied and exhibits altered coordination in the N170G structure. The density for these catalytic waters are only weakly present in the total omit map and were found only after refinement. The catalytic water is found in five of the six N170G molecules. However, they were not all present in the same locations throughout the six molecules and were coordinated by different residues throughout the active site. In chains A and E, the hydrolytic water was repositioned about 2 and 2.4 Å away from its original location, respectively. This change in position allows the water to be coordinated by only Glu166 while moving away from the catalytic Ser70 (Fig. 5B). In chains B and C, the water is coordinated by the main chain carbonyl group of Ala237 and the conserved water found in the oxyanion hole of the enzyme. The catalytic water is repositioned 1.9 and 1.6 Å away from its original location, respectively. This new coordination allows the water to move to a position where it is >3.6 Å away from both Glu166 and Ser70 (Fig. 5C). In contrast, chain F revealed the hydrolytic water in a very similar position to the wild-type structure. In this chain, the hydrolytic water has only moved 0.5 Å away from its original position and is still coordinated by Glu166 and Ser70 (Fig. 5D). This partial occupancy among the N170G molecules suggests that the hydrolytic water is still present in active site; however, it is loosely coordinated in comparison with the wild-type structure. These findings are consistent with the predictions of the substrate-assisted catalysis hypothesis in that, in the absence of the ampicillin substrate, the deacylation water is absent or moved. Thus, the rate of the deacylation reaction would be expected to be altered.
Hydrolysis of Cephalosporin Substrates with N170G Enzyme
The penam substrates ampicillin and penicillin G offer a clear test of the role of the amine group in the side chain of the antibiotic because the only difference between these molecules is the amine group. The cephem substrates cephalexin and cephalothin offer a similar comparison in that cephalexin contains a side chain structure identical to ampicillin, and cephalothin lacks the amine group in the side chain (Fig. 2). The cephalothin molecule also differs in that it contains a thiophene rather than benzyl ring and a different R-group at the C3 position of the cephalosporin ring structure (Fig. 2). The N170G enzyme hydrolyzes cephalexin with similar kinetic parameters as the wild-type enzyme, which is similar to the behavior of this enzyme with the analogous ampicillin as substrate (Table 2). In contrast, the N170G enzyme displays greatly reduced catalytic efficiency compared with the wild-type enzyme for cephalothin hydrolysis because of a reduction in kcat and a large increase in Km. Thus, hydrolysis of the cephalosporin substrates by the N170G enzyme follows the same general trends as observed for the penams with the substrates containing an amine side chain hydrolyzed with similar kinetic parameters as by the wild-type enzyme, whereas substrates lacking an amine side chain are hydrolyzed with altered kinetic parameters compared with wild type.
Role of Glu166 in the Function of the N170G Enzyme
As described above, the Glu166 residue is critical for β-lactam hydrolysis and, in particular, the deacylation reaction of the wild-type TEM-1 β-lactamase (18–21, 24, 27, 28). According to the substrate-assisted catalysis model, the Glu166 residue will also be essential for N170G hydrolysis of ampicillin in that it is hypothesized to serve the same function as in wild type as a general base in the deacylation reaction. Therefore, the impact of a E166A substitution on N170G function was assessed by constructing and analyzing the E166A single mutant and E166A/N170G double mutant. As expected the E166A mutant exhibited little activity, with an ampicillin MIC value of 2 μg/ml by the E-test method and 4 μg/ml by broth dilution (Table 1). Both of these numbers are barely above the ampicillin MIC obtained for the E. coli XL1-Blue strain in the absence of a β-lactamase (Table 1). The E166A/N170G double mutant also exhibited little activity, which is consistent with Glu166 playing an important role in the hydrolysis of ampicillin by the N170G enzyme. Surprisingly, however, the E166A/N170G double mutant exhibited a 2-fold increased ampicillin MIC value relative to the E166A mutant. It was expected that the E166A/N170G double mutant would provide less ampicillin resistance because of the removal of two residues that position the catalytic water molecule for deacylation.
The E166A and E166A/N170G enzymes were purified, and kinetic parameters for substrate hydrolysis were determined to further characterize these enzymes. It was not possible to detect ampicillin or penicillin G hydrolysis by the E166A enzyme at concentrations up to 100 μm of enzyme and 1 mm of substrate. In contrast, the E166A/N170G enzyme exhibited measurable catalytic activity at 100 nm of enzyme. The enzyme hydrolyzed ampicillin and penicillin G with similar kcat and Km values for both substrates. Although the catalytic efficiency of the E166A/N170G mutant is far below wild-type levels, the enzyme clearly exhibits increased substrate hydrolysis relative to the deacylation-deficient E166A enzyme (Table 2). Taken together, these results indicate, first, that the Glu166 residue is important for high level ampicillin hydrolysis by the N170G enzyme, and therefore the substrate-assisted catalysis mechanism is dependent on the Glu166 side chain, and, second, that the removal of the Asn170 side chain somehow enhances the catalytic activity of the E166A enzyme.
DISCUSSION
The class A β-lactamase TEM-1 presents a significant clinical antibiotic resistance problem because of mutations that alter the substrate specificity to allow hydrolysis of extended spectrum cephalosporins or provide for inhibitor resistance (3–8). The serine β-lactamase mechanism is similar to that of serine proteases (44), in that a proton is abstracted from the side chain oxygen of serine that acts a nucleophile and attacks the amide bond of the β-lactam ring (14, 15, 17–21, 24). An acyl-intermediate is then formed, and a highly ordered water molecule is activated for deacylation to hydrolyze the β-lactam and return the enzyme to its native state (20, 22, 27–29).
We previously compiled a collection of over 150 class A β-lactamase sequences, and analysis of this collection indicated that the residues Ser70, Glu166, and Asn170 are stringently conserved throughout class A enzymes (45). The catalytic Ser70 and Glu166 are invariant in class A β-lactamases because of their direct role in catalysis. Asn170 is also conserved in over 96% of class A β-lactamase sequences, indicating that the three residues that coordinate the hydrolytic water are conserved through the vast majority of class A β-lactamases.
To investigate the significance of residue 170, the codon for this position was randomized to create all 20 possible amino acids in the TEM-1 enzyme. When E. coli containing this library was selected on agar plates containing a high concentration of ampicillin, the surviving clones encoded only asparagine or glycine at position 170. Further characterization of the N170G mutant revealed that its ampicillin MIC was the same as E. coli containing the wild-type enzyme, and the steady-state kinetics for the mutant and wild-type enzymes for ampicillin are also comparable. However, when the N170G enzyme was assayed against other β-lactam antibiotics, it was observed that the substrates capable of being hydrolyzed at levels comparable with wild type were those containing a primary amine in the side chain of the antibiotic, suggesting a role for this group and the possibility of substrate-assisted catalysis by the N170G enzyme.
Substrate-assisted catalysis refers to the ability of a functional group in a substrate to contribute to the catalytic mechanism of the enzyme (46). Classical evidence of substrate-assisted catalysis is when a functional group or residue is subtracted from the enzyme with a loss of activity that is then restored by the replacement of a similar functional group on the substrate. In the case of the N170G TEM-1 mutant, the amine group of the asparagine is replaced by the primary amine of ampicillin or cephalexin. Examination of the acyl-intermediate structure of E166N TEM-1 covalently bonded to penicillin G (20) suggests that the primary amine of ampicillin could fill the void left by the N170G mutant. In support of the substrate-assisted catalysis hypothesis, it was found that removal of the amine group of ampicillin, i.e. penicillin G, resulted in kinetic parameters that are drastically altered for the N170G mutant but not wild type, suggesting that the amino group is participating in its own catalysis in the mutant enzyme. Similar methods have been used to investigate substrate-assisted catalysis of engineered serine proteases (47, 48). When a H64A substitution was made in the catalytic triad of the serine protease BPN′, it altered the substrate specificity of the enzyme to preferentially hydrolyze peptide substrates containing histidine at the proper positions. However, even the histidine-containing substrates did not restore the catalytic efficiency of the protease near wild-type levels, unlike the N170G mutant presented in this paper (47, 48).
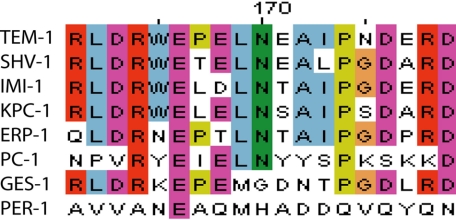
A glycine at position 170, instead of the conserved asparagine has been found in the wild-type class A β-lactamase GES-1 (49, 50), (Fig. 6). GES-1 is an extended spectrum β-lactamase that has been shown to alter its substrate specificity by altering the residue at position 170 (6, 7, 49–54). One such substitution included a mutation to asparagine at this position in the GES-2 enzyme (52). The GES-1 structure was solved at atomic resolution, 1.1 Å, and revealed that the hydrolytic water was loosely coordinated despite the coordination of Ser70 and Glu166 because the density was found only in the later stages of refinement in the Fo − Fc difference Fourier maps (53). The authors also state that other surrounding waters were better defined than this key hydrolytic water (53). The coordination of the hydrolytic water in the GES-1 enzyme is similar to that of the N170G TEM-1 structure presented here with the removal of the conserved Asn170. In our structure of N170G TEM-1, the water was found in five of the six molecules only after refinement and is improperly coordinated in four of the six molecules. This partial occupancy and improper coordination indicates that its use in enzyme catalysis would be inefficient and more difficult for the water to be properly activated by Glu166 for deacylation.
FIGURE 6.
Protein sequence alignment of the omega loop (positions 161–179) of several class A β-lactamases chosen as representative of the large enzyme family. Over 96% of class A β-lactamases contain an asparagine at position 170. GES-1 and PER-1 are shown as examples of the class A β-lactamases where the asparagine is absent. In addition, the omega loop sequence of the PER-1 enzyme is significantly different from the majority of class A enzymes. This figure was made using ClustalW2 (63).
The PER-1 enzyme is an extended spectrum class A β-lactamase that contains a histidine instead of asparagine at position 170 (Fig. 6) (55–58). Examination of its crystal structure revealed that the omega loop is altered to allow for a glutamine at position 69 to act as the third ligand of the hydrolytic water instead of asparagine (59). This structure suggests that a third ligand to coordinate the hydrolytic water is important for function in that there has been a selection for three coordination sites at least two independent times.
As discussed above, the coordination of the hydrolytic water by Asn170 is highly conserved among class A β-lactamases. The results of this study suggest that the amino group provided by the ampicillin substrate serves the role of Asn170 by coordinating the hydrolytic water in the N170G enzyme (Fig. 7). This mechanism of substrate-assisted catalysis for N170G TEM-1 is similar to that of cytochrome P450eryF, where crystallographic and kinetic analysis demonstrated that the substrate, 6-deoxyerythronolide B, stabilizes a catalytic water (60–62). The activity of this enzyme is decreased when a key hydroxyl group of the substrate is missing. Similarly, the activity of the N170G mutant is altered when the primary amine of the ampicillin side chain is removed. This change in activity in conjunction with the movement of the key hydrolytic water in the native structure suggests that the primary amine is aiding in the positioning of the hydrolytic water to facilitate its activation by Glu166.
FIGURE 7.
Schematic representation of the wild-type β-lactamase mechanism compared with the proposed substrate-assisted catalysis mechanism of the TEM-1 N170G mutant. A, hydrolysis reaction catalyzed by β-lactamase shown with ampicillin as the β-lactam substrate. The region of ampicillin implicated in substrate-assisted catalysis and shown in B of this figure is bracketed on the structure. B, the wild-type mechanism (left) and the substrate-assisted catalysis mechanism (right) are divided into two stages. In the wild-type mechanism, the hydrolytic water is coordinated by Ser70, Glu166, and Asn170, whereas the hydrolytic water in the N170G mutant is weakly coordinated in the absence of substrate. The primary amine of ampicillin is shown to act as a ligand for the water in the acyl-intermediate step of catalysis instead of the wild-type Asn170 residue side chain to position the catalytic water to be activated by Glu166 for deacylation.
The role of Glu166 in the context of the N170G substitution was assessed both within the cell and using purified protein. The double mutant E166A/N170G was found to exhibit drastically reduced activity compared with the N170G substitution alone. This marked decrease in activity demonstrates that Glu166 is important for the catalytic mechanism of the N170G enzyme, as it is for the wild-type enzyme (20, 24, 26, 27, 55). A puzzling observation, however, is the increase in activity of the E166A/N170G double mutant in comparison with the single mutation of E166A. The mechanism is unclear, but the improvement in catalytic activity is not likely to be due to substrate-assisted catalysis because the E166A/N170G enzyme is able to hydrolyze both ampicillin and penicillin at similar rates. The poor catalytic efficiency toward ampicillin and penicillin G is likely due to inefficient activation of water for deacylation because of the absence of Glu166 (18, 20, 24, 26–28, 55). The lack of both the Glu166 and Asn170 side chains may result in an enzyme that relies on nonenzyme catalyzed water attack on the acyl-intermediate for deacylation. The E166A/N170G substitutions could result in a more open active site to allow more access for water molecules to enter the active site and function in deacylation.
In conclusion, the coordination of the hydrolytic water is an important component for efficient hydrolysis of β-lactam antibiotic by class A β-lactamases. Sequence alignments and structural data suggest that two independent mechanisms for water coordination have evolved among class A enzymes, and evidence is provided here for a third, substrate-assisted mechanism where the primary amine of the substrate fulfills the role of the amine group of Asn170 in water positioning for TEM-1 β-lactamase. The presence of the antibiotic side chain amine allows for proficient catalysis at rates comparable with wild type rates. Thus, it appears that TEM-1-mediated β-lactam hydrolysis is most effective when a third ligand is present to coordinate the hydrolytic water in the active site of the enzyme.
Acknowledgments
We thank Kevin Phillips and members of the Baxter/Webb lab at the Methodist Hospital Research Institute for the use and assistance with TTP LabTech mosquito instrument. We also thank David Marciano of Baylor College of Medicine for helpful discussions. We acknowledge the SBC-CAT 19ID Beamline at the Advanced Photon Source and its staff for help during data collection.
This work was supported, in whole or in part, by National Institutes of Health Grant AI32956 (to T. P.). This work was also supported by a training fellowship from the Keck Center Pharmacoinformatics Training Program of the Gulf Coast Consortia (National Institutes of Health Grant T90 DK070109-05).
The atomic coordinates and structure factors (code 3jyi) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- MIC
- minimum inhibitory concentration.
REFERENCES
- 1.Livermore D. M., Woodford N. (2006) Trends Microbiol. 14, 413–420 [DOI] [PubMed] [Google Scholar]
- 2.Frère J. M., Joris B. (1985) Crit. Rev. Microbiol. 11, 299–396 [DOI] [PubMed] [Google Scholar]
- 3.Frère J. M. (1995) Mol. Microbiol. 16, 385–395 [DOI] [PubMed] [Google Scholar]
- 4.Paterson D. L., Bonomo R. A. (2005) Clin. Microbiol. Rev. 18, 657–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matagne A., Lamotte-Brasseur J., Frère J. M. (1998) Biochem. J. 330, 581–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh T. R. (2008) Curr. Opin. Infect. Dis. 21, 367–371 [DOI] [PubMed] [Google Scholar]
- 7.Walther-Rasmussen J., Høiby N. (2007) J. Antimicrob. Chemother. 60, 470–482 [DOI] [PubMed] [Google Scholar]
- 8.Shah A. A., Hasan F., Ahmed S., Hameed A. (2004) Res. Microbiol. 155, 409–421 [DOI] [PubMed] [Google Scholar]
- 9.Cantu C., 3rd, Huang W., Palzkill T. (1997) J. Biol. Chem. 272, 29144–29150 [DOI] [PubMed] [Google Scholar]
- 10.Georgopapadakou N. H. (2004) Expert Opin. Investig. Drugs 13, 1307–1318 [DOI] [PubMed] [Google Scholar]
- 11.Chaïbi E. B., Sirot D., Paul G., Labia R. (1999) J. Antimicrob. Chemother. 43, 447–458 [DOI] [PubMed] [Google Scholar]
- 12.Knox J. R. (1995) Antimicrob. Agents Chemother 39, 2593–2601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christensen H., Martin M. T., Waley S. G. (1990) Biochem. J. 266, 853–861 [PMC free article] [PubMed] [Google Scholar]
- 14.Chen C. C., Smith T. J., Kapadia G., Wäsch S., Zawadzke L. E., Coulson A., Herzberg O. (1996) Biochemistry 35, 12251–12258 [DOI] [PubMed] [Google Scholar]
- 15.Damblon C., Raquet X., Lian L. Y., Lamotte-Brasseur J., Fonze E., Charlier P., Roberts G. C., Frère J. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 1747–1752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamotte-Brasseur J., Dive G., Dideberg O., Charlier P., Frère J. M., Ghuysen J. M. (1991) Biochem. J. 279, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lietz E. J., Truher H., Kahn D., Hokenson M. J., Fink A. L. (2000) Biochemistry 39, 4971–4981 [DOI] [PubMed] [Google Scholar]
- 18.Minasov G., Wang X., Shoichet B. K. (2002) J. Am. Chem. Soc. 124, 5333–5340 [DOI] [PubMed] [Google Scholar]
- 19.Mustafi D., Sosa-Peinado A., Makinen M. W. (2001) Biochemistry 40, 2397–2409 [DOI] [PubMed] [Google Scholar]
- 20.Strynadka N. C., Adachi H., Jensen S. E., Johns K., Sielecki A., Betzel C., Sutoh K., James M. N. (1992) Nature 359, 700–705 [DOI] [PubMed] [Google Scholar]
- 21.Strynadka N. C., Martin R., Jensen S. E., Gold M., Jones J. B. (1996) Nat. Struct. Biol. 3, 688–695 [DOI] [PubMed] [Google Scholar]
- 22.Jelsch C., Mourey L., Masson J. M., Samama J. P. (1993) Proteins 16, 364–383 [DOI] [PubMed] [Google Scholar]
- 23.Meroueh S. O., Fisher J. F., Schlegel H. B., Mobashery S. (2005) J. Am. Chem. Soc. 127, 15397–15407 [DOI] [PubMed] [Google Scholar]
- 24.Adachi H., Ohta T., Matsuzawa H. (1991) J. Biol. Chem. 266, 3186–3191 [PubMed] [Google Scholar]
- 25.Herzberg O., Moult J. (1987) Science 236, 694–701 [DOI] [PubMed] [Google Scholar]
- 26.Escobar W. A., Tan A. K., Fink A. L. (1991) Biochemistry 30, 10783–10787 [DOI] [PubMed] [Google Scholar]
- 27.Escobar W. A., Tan A. K., Lewis E. R., Fink A. L. (1994) Biochemistry 33, 7619–7626 [DOI] [PubMed] [Google Scholar]
- 28.Guillaume G., Vanhove M., Lamotte-Brasseur J., Ledent P., Jamin M., Joris B., Frère J. M. (1997) J. Biol. Chem. 272, 5438–5444 [DOI] [PubMed] [Google Scholar]
- 29.Leung Y. C., Robinson C. V., Aplin R. T., Waley S. G. (1994) Biochem. J. 299, 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zawadzke L. E., Chen C. C., Banerjee S., Li Z., Wäsch S., Kapadia G., Moult J., Herzberg O. (1996) Biochemistry 35, 16475–16482 [DOI] [PubMed] [Google Scholar]
- 31.Ho S. N., Hunt H. D., Horton R. M., Pullen J. K., Pease L. R. (1989) Gene 77, 51–59 [DOI] [PubMed] [Google Scholar]
- 32.Petrosino J., Rudgers G., Gilbert H., Palzkill T. (1999) J. Biol. Chem. 274, 2394–2400 [DOI] [PubMed] [Google Scholar]
- 33.Huang W., Petrosino J., Hirsch M., Shenkin P. S., Palzkill T. (1996) J. Mol. Biol. 258, 688–703 [DOI] [PubMed] [Google Scholar]
- 34.Petrosino J. F., Palzkill T. (1996) J. Bacteriol. 178, 1821–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marciano D. C., Pennington J. M., Wang X., Wang J., Chen Y., Thomas V. L., Shoichet B. K., Palzkill T. (2008) J. Mol. Biol. 384, 151–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fersht A. (1985) Enzyme Structure and Function, p. 99, W. H. Freeman and Company, New York [Google Scholar]
- 37.Otwinowski Z. M., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 38.McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 40.Vaguine A. A., Richelle J., Wodak S. J. (1999) Acta Crystallogr. D Biol. Crystallogr 55, 191–205 [DOI] [PubMed] [Google Scholar]
- 41.Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 42.Zwart P. H., Afonine P. V., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., McKee E., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C., Adams P. D. (2008) Methods Mol. Biol. 426, 419–435 [DOI] [PubMed] [Google Scholar]
- 43.DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, Palo Alto, CA [Google Scholar]
- 44.Perona J. J., Craik C. S. (1995) Protein Sci. 4, 337–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marciano D. C., Brown N. G., Palzkill T. (2009) Protein Sci. 18, 2080–2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dall'Acqua W., Carter P. (2000) Protein Sci. 9, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carter P., Wells J. A. (1987) Science 237, 394–399 [DOI] [PubMed] [Google Scholar]
- 48.Carter P., Wells J. A. (1988) Nature 332, 564–568 [DOI] [PubMed] [Google Scholar]
- 49.Poirel L., Le Thomas I., Naas T., Karim A., Nordmann P. (2000) Antimicrob. Agents Chemother. 44, 622–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giakkoupi P., Tzouvelekis L. S., Tsakris A., Loukova V., Sofianou D., Tzelepi E. (2000) Antimicrob. Agents Chemother. 44, 2247–2253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bae I. K., Lee Y. N., Jeong S. H., Hong S. G., Lee J. H., Lee S. H., Kim H. J., Youn H. (2007) Diagn Microbiol. Infect. Dis. 58, 465–468 [DOI] [PubMed] [Google Scholar]
- 52.Poirel L., Weldhagen G. F., Naas T., De Champs C., Dove M. G., Nordmann P. (2001) Antimicrob. Agents Chemother. 45, 2598–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith C. A., Caccamo M., Kantardjieff K. A., Vakulenko S. (2007) Acta Crystallogr. D Biol. Crystallogr. 63, 982–992 [DOI] [PubMed] [Google Scholar]
- 54.Wang C., Cai P., Chang D., Mi Z. (2006) J. Antimicrob. Chemother. 57, 1261–1262 [DOI] [PubMed] [Google Scholar]
- 55.Bouthors A. T., Dagoneau-Blanchard N., Naas T., Nordmann P., Jarlier V., Sougakoff W. (1998) Biochem. J. 330, 1443–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bouthors A. T., Delettré J., Mugnier P., Jarlier V., Sougakoff W. (1999) Protein Eng. 12, 313–318 [DOI] [PubMed] [Google Scholar]
- 57.Nordmann P., Naas T. (1994) Antimicrob. Agents Chemother. 38, 104–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nordmann P., Ronco E., Naas T., Duport C., Michel-Briand Y., Labia R. (1993) Antimicrob. Agents Chemother 37, 962–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tranier S., Bouthors A. T., Maveyraud L., Guillet V., Sougakoff W., Samama J. P. (2000) J. Biol. Chem. 275, 28075–28082 [DOI] [PubMed] [Google Scholar]
- 60.Cupp-Vickery J. R., Han O., Hutchinson C. R., Poulos T. L. (1996) Nat. Struct. Biol. 3, 632–637 [DOI] [PubMed] [Google Scholar]
- 61.Andersen J. F., Hutchinson C. R. (1992) J. Bacteriol. 174, 725–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nebert D. W., Nelson D. R., Adesnik M., Coon M. J., Estabrook R. W., Gonzalez F. J., Guengerich F. P., Gunsalus I. C., Johnson E. F., Kemper B., et al. (1989) DNA 8, 1–13 [DOI] [PubMed] [Google Scholar]
- 63.Thompson J. D., Gibson T. J., Higgins D. G. (2002) Current Protocols in Bioinformatics, pp. 1–22, John Wiley and Sons, New York [Google Scholar]