Abstract
The flavoenzyme uridine 5’-diphosphate galactopyranose mutase (UGM or Glf) catalyzes the interconversion of UDP-galactopyranose and UDP-galactofuranose. The latter is a key building block for cell wall construction in numerous pathogens, including Mycobacterium tuberculosis. Mechanistic studies of UGM suggested a novel role for the flavin, and we previously provided evidence that the catalytic mechanism proceeds through a covalent flavin-galactose iminium. Here, we describe 2.3 and 2.5 Å resolution X-ray crystal structures of the substrate-bound enzyme in oxidized and reduced forms, respectively. In the latter, the substrate C1 is 3.6 Å from the nucleophilic flavin N5 position. This orientation is consistent with covalent catalysis by flavin.
UDP-galactopyranose mutase, which mediates the isomerization of its eponymous substrate (UDP-Galp) into UDP-Galf, plays important roles in both prokaryotic and eukaryotic pathogens (1, 2). For example, UDP-Galf is a precursor to the galactoconjugates of the cell wall in Mycobacterium tuberculosis. Thus, the gene for UGM (glf) is essential for mycobacterial viability (3), and UGM inhibitors can block the growth of mycobacterial cells (4). These findings are significant because mycobacteria cause devastating diseases, most famously tuberculosis (5). UGM function also is critical in eukaryotes, as has been observed for several Aspergillus species (6, 7). Given the absence of Galf residues in humans, UGM is an attractive therapeutic target. The development of inhibitors would be accelerated by clarification of the enzyme’s catalytic mechanism.
The identification of UGM as an FAD-containing enzyme prompted questions about the role of its cofactor. A hallmark of flavoenzymes is their ability to catalyze multiple kinds of electron transfer reactions (8). UGM must be reduced for activity (9), yet there are no candidate reducible functional groups on its substrates, UDP-Galp or UDP-Galf. It is known, however, that during turnover the UDP group is released transiently from galactose (10). To reconcile these observations, we proposed that the UGM flavin acts as a nucleophile, and, in a substitution reaction, captures the anomeric carbon position of the substrate at the reactive N5 position of flavin (Figure 1A) (11, 12). Formation of iminium ion 3 allows opening of the sugar ring, which can then close to the furanose form. The inability of 5-deaza-FAD to promote conversion (13) is consistent with the proposed nucleophilic role of N5. The intermediacy of the covalent iminium intermediate is supported by our finding that a hydride reducing agent can trap covalent adduct 3R during turnover (12) (Figure 1A). In the final reaction step, UDP serves as a nucleophile to displace the flavin, releasing product (Figure 1A).
Figure 1.
(A) Covalent flavin mechanism for UGM turnover. After nucleophilic attack, turnover proceeds through iminium intermediate 3. (B) Flavin covalent adduct as evidence of iminium intermediate: if proposed flavin-galactose iminium intermediate 3 is formed, it should be prone to reduction with cyanoborohydride. Improved isolation of covalent adduct 3R was achieved by optimizing reaction conditions. >90% of flavin can be isolated as trapped adduct. (Top curve reproduced from (12).) (C) 1H-NMR of reduced flavin and trapped adduct - shows similar chemical shifts for C6 and C9 protons in 3R.
Ensuing data from structural studies have provided little insight into the relevance of the putative covalent intermediate for the catalytic mechanism. For example, the reduced structure of Klebsiella pneumoniae UGM possesses a flavin butterfly conformation (pucker) that appeared to disfavor the covalent mechanism (14). These results, however, do not preclude the covalent catalysis mechanism because the barrier to interconversion between one puckered form and the other is low (4–5 kcal/mol) (15). The first structure of UGM with a bound ligand (the substrate mimic UDP-glucose) indicates that the UDP moiety is tightly held by conserved hydrophobic residues and the sugar moiety is proximal to the flavin as required for covalent catalysis (16). Nevertheless, the flavin and the anomeric carbon of the glucose moiety are poorly aligned for nucleophilic attack (16). Still, modeling suggests that UDP-Galp can be oriented in the active site such that the proposed covalent catalysis pathway proceeds (16). Given the intriguing features of UGM and its potential value as a therapeutic target, we focused on illuminating its catalytic mechanism.
Although there is indirect evidence that flavin-sugar conjugate 3R represents the trapped covalent adduct (12, 13), its structure had not been determined unequivocally. We therefore conducted experiments to elucidate the connectivity of this product. After optimizing our published conditions, we generated the flavin-galactose adduct in sufficiently high yield (> 90%) to isolate it for NMR structural studies (Figure 1B). These investigations indicated that the C6 and C9 protons are in chemically related environments, as their 1H NMR chemical shifts are similar (Figure 1C). This observation is consistent with the structure of 3R because the reduction of iminium 3 would yield similar open chain sugar substituents at flavin N5 and N10. NOESY experiments (Supporting Information) further support this structural assignment; both the C6 and C9 protons exhibit through-space coupling to carbohydrate protons. Together, these NMR experiments indicate that the structure of the trapped adduct is that depicted for 3R, an N5-alkylflavin.
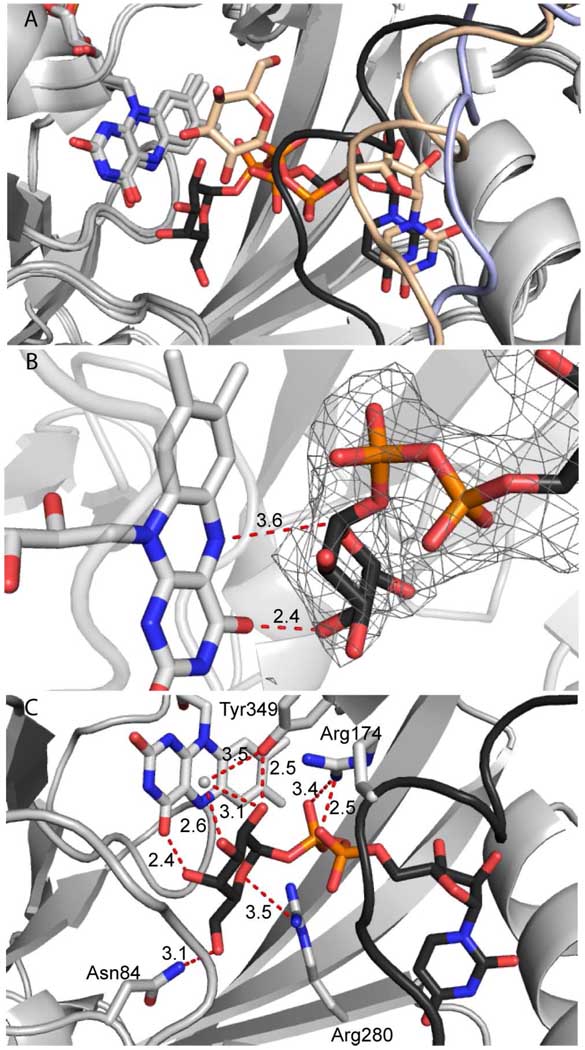
To investigate if substrate is bound in a manner consistent with covalent catalysis, we turned to X-ray crystallography. Crystals of UGM from Klebsiella pneumoniae were grown in the presence of UDP-Glc (16) and then soaked in high concentrations of the substrate UDP-Galp under aerobic conditions. The structure of UGM bound to UDP-Galp was solved by molecular replacement and refined to 2.3 Å resolution (Supplementary Table 1). The overall architecture of this dimeric enzyme is similar to that of previously reported structures of UGM. Specifically, both monomers of the UGM:UDP-Galp homodimer resemble the closed orientation observed in the UDP-Glc structure (3GF4), the only previously reported structure of a UGM-ligand complex (16). The mobile loop (residues 167–177) (14, 17) is closed over each active site (Figure 2A). The location of the uridine moiety is similar to that observed in UGM:UDP-Glc (16). In contrast, the positions of the sugar residues in the two complexes differ. Unexpectedly, in the oxidized UGM–substrate complex, the putative nucleophilic site on the flavin and the electrophilic position of the sugar are further apart than in the UDP-Glc structure. The anomeric position of the UDP-Galp is located a distant 8 Å from N5 of flavin (Figure 2A). Thus, while UDP-Galp is in the active site in the oxidized UGM complex, it is not poised for catalysis.
Figure 2.
Substrate binding and reduction create catalytically competent UGM active site. (A) Binding of UDP-Galp (wheat carbons) to oxidized UGM causes the mobile loop (light blue, from apo UGM pdb 2BI7 (14)) to close and form a helix (wheat). Reduction causes UDP-Galp (black carbons) and the loop (black) to shift closer to the flavin. (B) Nucleophilic flavin N5 and flavin C4 carbonyl approach the anomeric carbon and C4-OH, respectively, of the substrate UDP-Galp (distances in Å) in the reduced structure. The FO-FC omit map, calculated without ligand, is contoured two standard deviations above the mean (grey mesh). (C) Conserved residues as well as flavin and an ordered water molecule (grey sphere) orient the galactopyranose.
We tested whether reduction might influence the active site conformation. The electronic properties of the isoalloxazine ring in the oxidized and reduced flavin forms differ significantly (8), and the nucleophilicity of the flavin N5 should be enhanced dramatically upon reduction. Moreover, changes in flavin redox state result in local changes in conformation (18). The hypothesis that UGM might respond structurally to flavin reduction is supported by the finding that reduction of the enzyme increases its affinity for substrate (17, 19). Given these incentives, we soaked the oxidized crystals containing UDP-Galp in cryoprotectant containing 100 mM sodium dithionite. These conditions provided the means to solve the reduced structure at 2.5 Å resolution (Supplementary Table 1).
Comparison of the oxidized and reduced UGM substrate complexes is revealing, as one of the subunits (monomer B) of the dimer undergoes significant conformational changes. Specifically, flavin reduction results in a translocation of the mobile loop approximately 4 Å toward substrate (Figure 2A). Moreover, the relative orientation of the flavin and the UDP-Galp substrate shifts. The uridine portion of the ligand moves subtly toward the flavin, while the galactose moiety undergoes a more dramatic repositioning. This substrate orientation is in general similar to a minimized model proposed based on the UGM:UDP-Glc structure (16), although the Galp moiety is rotated approximately 90 degrees about the glycosidic bond. Intriguingly, in the crystal structure the C1 position of the galactose moiety is immediately adjacent to the flavin N5 (Figure 2A and 2B).
The change in substrate and cofactor proximity in the reduced, substrate bound form results in an arrangement in which UGM is poised for covalent catalysis. The flavin N5 is perched for attack at the anomeric carbon C1 of UDP-Galp with the UDP moiety positioned to serve as a leaving group. Moreover, the distance separating the nucleophilic flavin N5 and anomeric position is 3.6 Å (Figure 2B). The positioning of the sugar moiety is mediated by interactions with the flavin (see below), Tyr349, Arg280, Asn84 and an ordered water molecule (Figure 2C). The catalytically essential Arg174 (20) coordinates both the α- and β-phosphates of UDP, and its guanidinium moiety is located ~3.3 Å from the planar face of Tyr349 (Figure 2C). This conformation raises the intriguing possibility that a cation-π interaction between Arg174 and Tyr349 stabilizes the closed, substrate bound orientation (21).
An interesting feature of the complex is that the galactose C4-OH is in position to engage in a hydrogen bond with the C4 carbonyl of the reduced flavin (Figure 2B). This apparent hydrogen bond may be important for the ability of the enzyme to discriminate against UDP-Glc as a substrate (16). Specifically, the equatorial C4-OH of UDP-Glc could not engage in such an interaction. The hydrogen bond between the UDP-Galp C4-OH and the flavin C4 carbonyl also may provide a means to shuttle the proton from C4-OH to the nascent C5-OH after ring opening
The reduced UDP-Galp structure presented herein represents crystallographic evidence that directly supports a nucleophilic mechanism by the flavin cofactor for UGM catalysis. Covalent, nucleophilic catalysis is thus a weapon we are only beginning to appreciate from the chemical arsenal of the flavin cofactor. Additionally, our structures illuminate the active state of UGM and thereby provide a springboard for the design of potent inhibitors.
Supplementary Material
Footnotes
This research was supported by the NIH (NIAID 063596 to LLK and a T32 GM08349 Training Grant position to TDG) and the W.M. Keck Foundation (to KTF). We thank K. Satyshur for technical assistance and B. Fox for use of equipment. Oxidized and reduced UGM:UDP-Galp structures are in the PDB as 3INR and 3INT, respectively.
SUPPORTING INFORMATION
Experimental details and support information, 1H-1H NOESY of 3R, FO-FC omit maps for ligands, and animation of the bound ligand are available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Nassau PM, Martin SL, Brown RE, Weston A, Monsey D, McNeil MR, Duncan K. J. Bacteriol. 1996;178:1047–1052. doi: 10.1128/jb.178.4.1047-1052.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedersen LL, Turco SJ. Cell. Mol. Life Sci. 2003;60:259–266. doi: 10.1007/s000180300021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan F, Jackson M, Ma Y, McNeil M. J. Bacteriol. 2001;183:3991–3998. doi: 10.1128/JB.183.13.3991-3998.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dykhuizen EC, May JF, Tongpenyai A, Kiessling LL. J. Am. Chem. Soc. 2008;130:6706–6707. doi: 10.1021/ja8018687. [DOI] [PubMed] [Google Scholar]
- 5.World Health Organization. WHO/HTM/TB/2008. 2008;393
- 6.Schmalhorst P, Krappmann S, Vervecken W, Rohde M, Muller M, Braus G, Contreras R, Braun A, Bakker H, Routier F. Eukaryot. Cell. 2008;7:1268–1277. doi: 10.1128/EC.00109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damveld R, Franken A, Arentshorst M, Punt P, Klis F, Van Den Hondel C, Ram A. Genetics. 2008;178:873–881. doi: 10.1534/genetics.107.073148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Massey V. Biochem. Soc. Trans. 2000;28:283–296. [PubMed] [Google Scholar]
- 9.Sanders DA, Staines AG, McMahon SA, McNeil MR, Whitfield C, Naismith JH. Nat. Struct. Biol. 2001;8:858–863. doi: 10.1038/nsb1001-858. [DOI] [PubMed] [Google Scholar]
- 10.Barlow JN, Girvin ME, Blanchard JS. J. Am. Chem. Soc. 1999;121:6968–6969. [Google Scholar]
- 11.Soltero ML, Carlson EE, Kiessling LL. A Proposal for the Catalytic Mechanism of UDP-Galactopyranose Mutase. Abstracts of the 29th Steenbock Symposium: Cofactors, coenzymes and catalysis; Madison, WI. University of Wisconsin-Madison; 2003. [Google Scholar]
- 12.Soltero-Higgin M, Carlson E, Gruber TD, Kiessling LL. Nat. Struct. Mol. Biol. 2004;11:539–543. doi: 10.1038/nsmb772. [DOI] [PubMed] [Google Scholar]
- 13.Huang Z, Zhang Q, Liu HW. Bioorg. Chem. 2003;31:494–502. doi: 10.1016/j.bioorg.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 14.Beis K, Srikannathasan V, Liu H, Fullerton SW, Bamford VA, Sanders DA, Whitfield C, McNeil MR, Naismith JH. J. Mol. Biol. 2005;348:971–982. doi: 10.1016/j.jmb.2005.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moonen CT, Vervoort J, Müller F. Biochemistry. 1984;23:4868–4872. doi: 10.1021/bi00316a008. [DOI] [PubMed] [Google Scholar]
- 16.Gruber TD, Borrok MJ, Westler WM, Forest KT, Kiessling LL. J. Mol. Biol. 2009;391:327–340. doi: 10.1016/j.jmb.2009.05.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yuan Y, Wen X, Sanders DA, Pinto BM. Biochemistry. 2005;44:14080–14089. doi: 10.1021/bi0513406. [DOI] [PubMed] [Google Scholar]
- 18.Lennon BW, Williams CH, Ludwig ML. Protein Sci. 1999;8:2366–2379. doi: 10.1110/ps.8.11.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan Y, Bleile DW, Wen X, Sanders DA, Itoh K, Liu HW, Pinto BM. J. Am. Chem. Soc. 2008;130:3157–3168. doi: 10.1021/ja7104152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chad JM, Sarathy KP, Gruber TD, Addala E, Kiessling LL, Sanders DA. Biochemistry. 2007;46:6723–6732. doi: 10.1021/bi7002795. [DOI] [PubMed] [Google Scholar]
- 21.Zacharias N, Dougherty DA. Trends Pharmacol. Sci. 2002;23:281–287. doi: 10.1016/s0165-6147(02)02027-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.