Abstract
The Drosophila central brain is composed of approximately 100 paired lineages, with most lineages comprising 100–150 neurons. Most lineages have a number of important characteristics in common. Typically, neurons of a lineage stay together as a coherent cluster and project their axons into a coherent bundle visible from late embryo to adult. Neurons born during the embryonic period form the primary axon tracts (PATs) that follow stereotyped pathways in the neuropile. Apoptotic cell death removes an average of 30–40% of primary neurons around the time of hatching. Secondary neurons generated during the larval period form secondary axon tracts (SATs) that typically fasciculate with their corresponding primary axon tract. SATs develop into the long fascicles that interconnect the different compartments of the adult brain. Structurally, we distinguish between three types of lineages: PD lineages, characterized by distinct, spatially separate proximal and distal arborizations; C lineages with arborizations distributed continuously along the entire length of their tract; D lineages that lack proximal arborizations. Arborizations of many lineages, in particular those of the PD type, are restricted to distinct neuropile compartments. We propose that compartments are ‘scaffolded” by individual lineages, or small groups thereof. Thereby, the relatively small number of primary neurons of each primary lineage set up the compartment map in the late embryo. Compartments grow during the larval period simply by an increase in arbor volume of primary neurons. Arbors of secondary neurons form within or adjacent to the larval compartments, resulting in smaller compartment subdivisions and additional, adult specific compartments.
Keywords: Drosophila, brain, lineage, pathfinding, connectivity
Introduction
The Drosophila CNS, comprised of the ventral nerve cord and brain, is formed in the early embryo through successive waves of neuroblast (NB) delamination followed by several cycles of cell division. One brain hemisphere comprises approximately 100 neuroblasts, which have been mapped topologically and with regard to their pattern of gene expression in previous papers (Younossi-Hartenstein et al., 1996; Urbach and Technau, 2003; Sprecher et al., 2007). During the embryonic period, each neuroblast produces a lineage of 10 to 16 cells, the primary neurons and glia. Similar to what had been found previously in developing brains of other insect species (Truman, 1990), we observed that neurons belonging to the same lineage remain clustered together; what is more, their axons form a coherent bundle, the primary axon tract (PAT; Younossi-Hartenstein et al., 2006). During late embryogenesis primary neurons form dendritic and axonal arborizations that, together with sheath-like processes formed by glial cells, establish the neuropile compartments of the larval brain. After a period of mitotic quiescence that lasts until midlarval development, neuroblasts resume their activity, undergoing between an estimated 40 to 75 additional rounds of mitosis that produce secondary neurons (Ito and Hotta, 1992; Dumstrei et al., 2003a; Bello et al., 2008). Similar to primary axons, axons of a given secondary lineage fasciculate with each other, thereby forming discrete bundles (secondary axon tracts, or SATs) whose invariant trajectory enabled us to map all brain lineages for the late larval brain (Dumstrei et al., 2003a, Pereanu and Hartenstein, 2006). Most secondary neurons do not differentiate in the larva. Thus, SATs remain unbranched bundles until the pupal period, when secondary lineages form proximal branches (dendrites) and terminal branches (axons) that, together with remodeled primary neurons become integrated, into the adult brain.
The Drosophila brain is characterized functionally and anatomically by distinct compartments. Compartments are rich in terminal neurite branching and synapses; they are the neuropile domains in which signal processing takes place. Examples of fly brain compartments are the calyx of the mushroom body, the antennal lobe, or central complex (Strausfeld, 1976). Neuropile compartments are surrounded by more or less dense layers of glial cells, as well as bundles of long fibers that interconnect compartments (Younossi-Hartenstein et al., 2003; Pereanu et al., 2009). It has become clear that at least in some cases, lineages and compartments are related in a straightforward manner. The mushroom body, to give but one example, is formed by four lineages. If the neuroblasts of these lineages are ablated, the compartments of the mushroom body (calyx, peduncle, lobes) are also missing (Ito et al., 1997; Malun, 1998). But how typical is the mushroom body and its lineages for the central brain in general?
To answer this question, and learn about the structure of lineages in relationship to compartments in general, we undertook an analysis of the development of many different brain lineages from the embryonic to adult stage. In previous works, primary lineages of the Drosophila thoracic and abdominal neuromeres were studied by applying DiI to individual neuroblasts in the early embryo (Bossing et al., 1996; Schmidt et al., 1997; Schmid et al., 1999). All lineages labeled in this manner were imaged in late (stage 16) embryos, a stage when the primary axon tracts have formed, but little terminal branching has occurred. Therefore, the manner in which individual lineages contribute to discrete domains within the neuropile of the embryonic ventral nerve cord was not further studied. More recently, a detailed map of the secondary lineages of thoracic neuromeres was published by Truman and colleagues (2004). Similar to the brain lineages (Pereanu and Hartenstein, 2006), lineages of the ventral nerve cord form coherent clusters that each emit an SAT. SATs followed reproducible pathways within the neuropile, which served as the main criterion for identification. Like their counterparts in the brain, secondary lineages of the nerve cord postpone terminal arborization until metamorphosis, and the characteristics of this process have not yet been elucidated.
In order to image individual lineages, Drosophila developmental genetics offers two alternative approaches to the injection of dyes. One is to generate positively labeled clones by the flip-out technique (Ito et al., 1997) or MARCM technique (Lee and Luo, 2001). The second technique employs Gal4-driver constructs (Brand and Perrimon, 1993) that are stably expressed in one or a few lineages. Clonal analysis targets random lineages and does not allow one to follow a given lineage throughout development. Specific Gal4-lines do not have this disadvantage, but so far have been described for only very few lineages, including the four mushroom body lineages (Ito et al., 1997), the atonal-positive BLD5 lineage (Hassan et al., 2000; Zheng et al., 2006), three lineages associated with the antennal lobe (Stocker et al., 1997; Lai et al., 2008), and several lineages forming the fan-shaped body (Ito and Awasaki, 2008). Active screens are under way to generate a library of markers that covers all lineages (Pfeiffer et al., 2008). In this paper we have employed the clonal approach and a few select markers to address a number of unexplored properties of neuronal lineages. What is the dynamics of birth and growth of lineages in the embryo? Are lineages affected by cell death? How do the tracts formed by primary neurons and secondary neurons of one and the same lineage relate to each other? Do secondary axons fasciculate with their primary counterparts, or do secondary axons target neuropile compartments independent of the corresponding primary axons? How do lineages and compartment boundaries relate? It is known for the lineages that form the mushroom body that dendritic and axonal arborizations are highly resticted: Dendritic arbors of all mushroom body neurons, formed by four lineages, are restricted to the small compartment called calyx; all axons are restricted to the peduncle and lobes. But are the four mushroom body lineages an exception, or the rule? Similarly, do all lineages show defined input and output region like the mushroom body lineages? Answers to these questions will be helpful for future studies following the lineage-based approach to brain connectivity.
Material and Methods
Markers
The brain neuropile was labeled with an antibody against Drosophila N-cadherin (DNcad; Iwai et al., 1997; Developmental Studies Hybridoma Bank) or against Drosophila Bruchpilot (Brp; Developmental Studies Hybridoma Bank Nc82). Both proteins are enriched in synapses and serve as markers for compartments. Glial cells were labeled by Nrv2-Gal4+UAS-GFP (Sun et al., 1999). Secondary neurons and axon tracts were labeled by antibodies against Drosophila Neurotactin (Nrt; de la Escalera et al., 1990; Hortsch et al., 1990; Developmental Studies Hybridoma Bank BP106) and DE-cadherin (Dumstrei et al., 2003a, b). The Nrt epitope appears on all neurons, but in a stage specific manner. Differentiated primary neurons do not express it any longer in the late larva, so that only the emerging secondary neurons are labeled. Embryonic primary axon tracts were labeled by UAS-Synaptobrevin-GFP (Estes et al., 2000; kindly provided by Dr. K. Ito) driven by elav-Gal4 (available from the Bloomington Stock Center). As driver lines for specific lineages we used engrailed-Gal4 (enGal4; Tabata et al., 1995) and period-Gal4 (perGal4; Kaneko and Hall, 2000).
Fly stocks and egg collections
As wild-type stock we used Oregon R. Flies were grown under standard conditions at room temperature (25°C). Egg collections were done on yeasted apple juice agar plates (Ashburner, 1989). Hatching larvae were transferred to Petri dishes filled with standard fly food at a density of three to five individuals per cc of food. Larvae were collected at desired time points and brains were dissected in PEMS (.1M Pipes, 2mM MgSO4, 1mM EGTA, pH7.0) buffered 4%Formaldehyde.
To label individual lineages derived from single brain neuroblasts with GFP we used the FLP/FRT system (Golic et al., 1997), and screened the brains using confocal microscopy. We used a fly stock containing the construct UAS >stop>CD8GFP (kindly provided by Dr. Kei Ito) recombined with a construct containing the heat-shock promoter driving flipase. Heat shocking embryos activates flipase which in-turn drives homologous recombination between the FRT sites. This removes the stop sequence upstream of GFP and allows expression of GFP driven by any appropriate GAL4 driver. We used a membrane targeted version of GFP so that cells bodies and neuronal projections could be clearly visualized. We chose the armadillo-GAL4 driver (Bloomington Stock Center) as it is active well before NB delamination. We found that a heat shock period of 20 min was an ideal time period to label 1 or 2 separate NBs within the brain. Eggs were collected for 45 min and then allowed to develop at 25°C. To label the entire lineage from one NB, heat shock was applied at 3 hours after egg collection. For screening, embryos were aligned in numbered rows similar to the method used for egg injection. We found that GFP expression could be detected at about stage 12 (staging according to Campos-Ortega and Hartenstein, 1997). We screened embryos for GFP expression and only used embryos with 1 or 2 well separated labeled cells in our analysis.
For the generation of selected GFP labeled clones among the engrailed-positive lineages, a stock containing a recombinant chromosome was constructed: enGal4 UAS mCD8::GFPLL, UAS-nlslacZ20b on chromosomal arm 2R and FRT 82B tubP-GAL80LL3 on chromosome 3. To generate MARCM clones (Lee and Luo, 2001), +; UAS-mCD8::GFPLL5, UAS-nlslacZ20b; FRT82B (Bello et al., 2003) males were crossed to females of the MARCM driver stock hs-FLP; en-GAL4 UAS mCD8::GFPLL, UAS-nlslacZ20b; FRT82B tubP-GAL80LL3, resulting in wild-type clones of GFP-expressing cells. For clones in the period-expressing lineages, a stock containing the following was constructed: perGAL4,UAS mCD8GFP on chromosome 2 and FRT19A,hsFLP,tubGAL80 on the X chromosome. To visualize MARCM clones in the period lineages, the MARCM driver stock FRT19A,hsFLP,tubGAL80/y ; per-GAL4,UAS mCD8GFP (males) were crossed to FRT19A homozygous females (gift from Chris Doe; Rolls and Doe, 2004). For MARCM experiments (Lee and Luo, 2001), embryos of the appropriate genotype were collected on standard cornmeal/yeast/agar medium supplemented with live yeast over a 4 hour time window and raised at 25°C for 21 to 25 hours before heat-shock treatment. Heat-shock induction of FLP was done at 37°C for 60 minutes.
Immunohistochemistry and histology
The Nc82, BP106, BP104 and anti-DNcad antibodies were diluted 1:20, 1:10, 1:20, and 1:25, respectively. Secondary antibodies were Cy3 conjugated anti-rat Ig (Jackson Laboratories) and FITC-conjugated anti-mouse Ig (Jackson Laboratories) used at a 1:100 dilution. Standard antibody labeling procedures were followed (e.g., Ashburner, 1989). Some preparations were counterstained with the nuclear marker Sytox green (Molecular Probes; added at a concentration of 1:10,000 to one of the final washing steps before mounting). For histology and electron microscopy, larval brains were dissected and fixed in 2% glutaraldehyde in PBS for 20 minutes, followed by a postfixation for 30 minutes in a mixture of 1% osmium tetroxide and 2% glutaraldehyde in 0.15 M cacodylate buffer (on ice). Specimens were washed several times in PBS and dehydrated in graded ethanol and acetone (all steps on ice). Preparations were left overnight in a 1:1 mixture of Epon and acetone and then for 5–10 hours in unpolymerized Epon. They were transferred to molds, oriented, and placed at 60°C for 24 hours to permit polymerization of the Epon. Blocks were sectioned (60nm). Sections were mounted on net grids (Ted Pella) and treated with uranyl acetate and lead citrate. Samples were sectioned by the Microscopic Techniques Laboratory and examined using the JEOL 100CX transmission electron microscope at the Electron Microscope Laboratory of the UCLA Brain Research Institute.
Cell counts
We followed two independent approaches to count cells in embryonic and larval brains. (1) The average cell diameter was calculated from measurements taken from confocal sections (five brains, several hundred neurons). Assuming the shape of a truncated octahedron (this type of polyhedron comes closest to a sphere and at the same time tessellates into a solid volume) the average volume of a neuron was determined. The overall volume of the brain cortex was measured from 3D segmentations (see below) and divided by the neuronal volume. Note that neurons and glial cells will be both included in this number; glia accounts for less than 5% of the overall cell number in the late embryonic/early larval brain (Hartenstein et al., 1998; Pereanu et al., 2005). (2) Nuclei were automatically segmented from each section of a stack with labeled nuclei using the watershed algorithm, as implemented in ImageJ’s Find Maxima function. Nuclei from cells that are small and/or near each other were frequently observed to be fused, having no distinguishable border between them. In order to avoid undercounting, each section of the stack was first convolved (using ImageJ’s built-in Convolve function) using a kernel designed for the particular size of nucleus in the dataset. The number of maxima was then counted for the stack and divided by the number of sections in which a single nucleus appears. The program was run on five representative L1 and late embryonic brains in which neurons were labeled with the nuclear marker Sytox. The results of both methods coincided closely: Calculating neurons based on volume yielded 2627 cells for embryo, and 1730 for early larva; the corresponding numbers resulting from automatic segmentation and were 2576 and 1828, respectively.
Generation of Three-Dimensional Models
Staged Drosophila larval and adult brains labeled with suitable markers were viewed as whole-mounts by confocal microscopy [Biorad MRC 1024ES microscope using Biorad Lasersharp version 3.2 software; lenses: 40× oil (numerical aperture 1.0; WD 0.17). Complete series of optical sections were taken at 2-µm intervals. Digitized images of confocal sections were imported into the Amira (www.amiravis.com) program. Since sections were taken from focal planes of one and the same preparation, there was no need for alignment of different sections. All models were generated using the Amira software package. Surface rendered digital atlas models were created by manually labeling each lineage and neuropile compartment within a series of confocal images.
Results
Growth of primary lineages in the Drosophila embryo
The embryonic brain is formed by an invariant set of approximately 100 neuroblasts which proliferate in a stem cell mode, each neuroblast producing a lineage comprising neurons and (in some some cases) glial cells (Younossi-Hartenstein et al., 1996; 2006; Urbach and Technau, 2003). Cells produced during the embryonic period are referred to as primary neurons, those added in the larva represent the secondary neurons. Primary neurons start differentiating between nine and twelve hours of embryogenesis (stage 13–15). At this stage, lineages can be recognized as cone shaped cell clusters distributed rather evenly over the periphery of the brain. Axons formed by neurons of the same lineage fasciculate and form a simple, unbranched bundle, called primary axon tract (PAT). Axonal and dendritic branches of the PATs form the emerging neuropile compartments. Using location and axonal trajectory as a criterion, primary lineages and the compartments they are topologically associated with were subdivided into ten discrete classes called dorso-posterior medial (DPM), dorso-posterior lateral (DPL), dorso-anterior medial (DAM), dorso-anterior lateral (DAL), centro-posterior medial (CM), centro-posterior (CP), mushroom body (MB), baso-posterior lateral (BL), baso-anterior (BA) and tritocerebral (TR). These classes of lineages and their characteristic trajectories are illustrated in Figure 1 (Younossi-Hartenstein et al., 2006; TR lineages not shown).
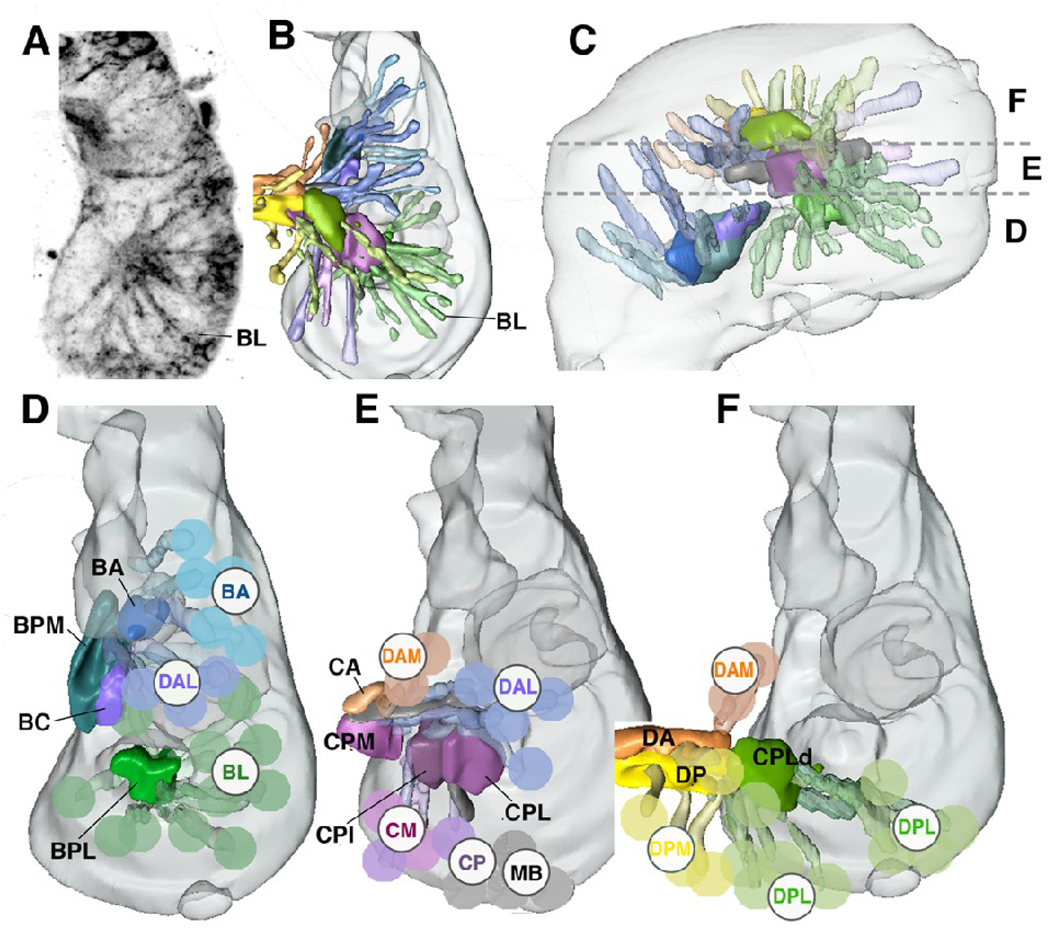
Fig.1.
Projection of groups of neural lineages of the Drosophila central brain.
A: Z-projection of horizontal confocal sections of a late stage 15 brain hemisphere, labeled with UAS-synaptobrevin-GFP driven by elav-Gal4 which visualizes primary lineages. Medial is to the left, anterior down. Z-projection was generated by combining ten contiguous focal planes taken at 1µm increment; it represents a “slice” of the brain containing the ventral lineages and compartments. Lineages can be recognized by characteristically positioned primary axon tracts; one tract, formed by one of the BL lineages, is shown as example. B, C: 3D digital models of late embryonic brain hemisphere; B presents dorsal view (anterior to the top), E shows lateral view (anterior to the left). Brain is outlined in gray. Primary axon tracts of groups of primary lineages are rendered in different colors. Emerging compartments are rendered in the same colors as the groups of lineages that most strongly contribute to (scaffold) the corresponding compartment. The same color key is applied throughout panels B–F. Horizontal hatched lines in C demarcate boundaries between the dorsal cerebrum (F), middle cerebrum (E), and ventral cerebrum (D). Groups of lineages located at these levels are shown separately in a dorsal view in the models presented in panels D–F. Here, aside from the primary axon tract, a lineage is schematically represented by a colored sphere and identified by acronyms (BA baso-anterior; BL baso-lateral; CM centro-medial; CP centro-posterior; DAM dorso-anterior medial; DAL dorso-anterior lateral; DPL dorso-posterior lateral; DPM dorso-posterior medial; MB mushroom body; for detail, see Younossi-Hartenstein et al., 2006). Abbreviations of compartments are identified by black capital letter without sphere: BA baso-anterior; BC baso-central; BPL baso-posterior lateral; BPM baso-posterior medial; CA centro-anterior; CPI centro-posterior intermediate; CPL centro-posterior lateral; CPLd dorsal domain of CPLd; CPM centro-posterior medial; DA dorso-anterior; DP dorso-posterior. Note that in this and the following figures, lineages of th tritocerebrum, which have been insufficiently mapped up to the present point, are not shown.
Bar: 20µm
In order to study the structure and growth dynamics of neuronal lineages in embryos, labeled clones of primary neurons were generated by activating GFP through heat pulse induced flip-out (Golic et al., 1997) at stage 9 (3–4h post egg laying), the beginning of neuroblast proliferation. Clone sizes and PATs were assayed at three stages of embryonic development, stage 13/14 (9–10h), stage 15/16early (12–13h), and stage 16late/17 (15–17h). Based on their position and axon trajectory we were able to assign most clones to the discrete classes defined previously (Younossi-Hartenstein et al., 2006). Figure 2 illustrates representative clones for the different groups of lineages and documents clone sizes and PAT lengths. Clone sizes were relatively uniform and ranged around eight to ten cells at stage 13/14, and ten to twelve cells at stage 16late/17 (Fig.2).
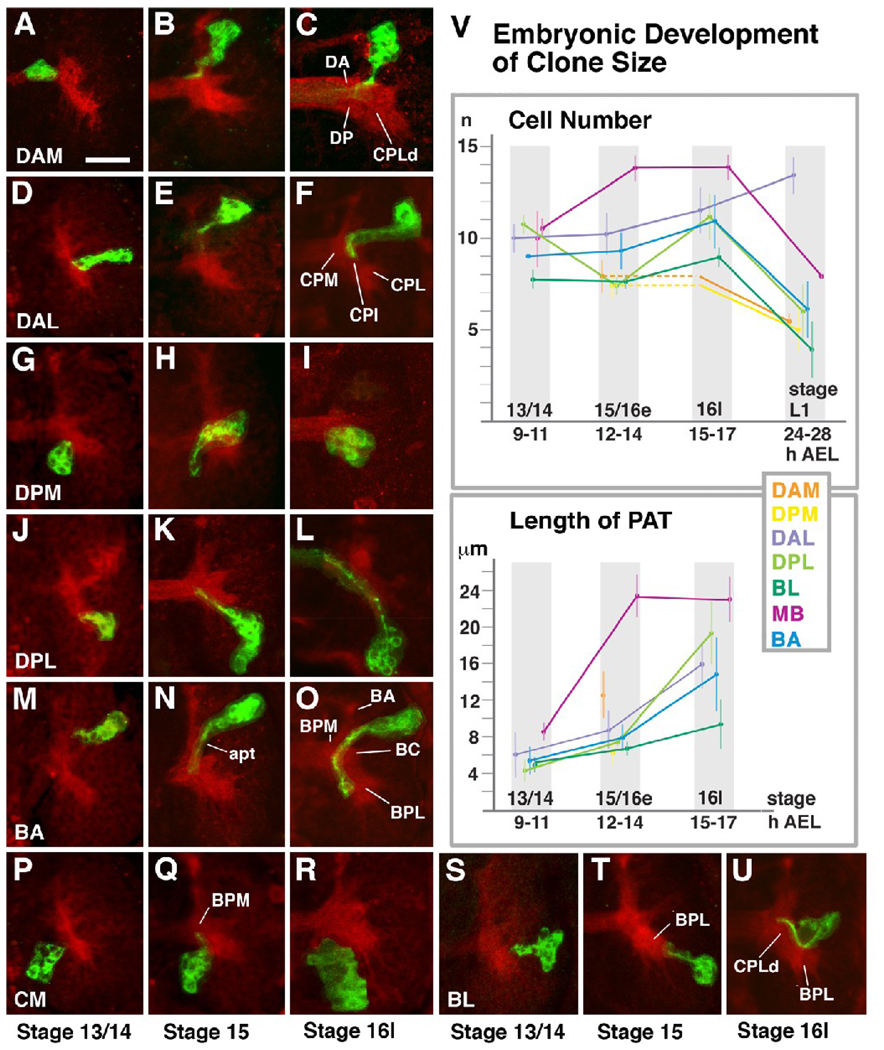
Fig.2.
Morphogenesis of brain lineages during the embryonic period. Panels A–U show Z-Projections of horizontal confocal sections of right brain hemisphere (anterior to the top, lateral to the right). Neuropile was labeled with anti-DNcad (red). Individual lineages (“primary clones”) were labeled by inducing mCD8-GFP (green) in embryonic neuroblasts at around 5h post fertilization. Each row (A–C, D–F, etc) shows clone representing a given group of lineages, indicated at the lower left corner (e.g., DAM, DAL, etc; see Fig.1). Left panel of a given row shows clone at stage 13/14 (9–11h); middle panel depicts clones fixed at stage 15/early 16 (12–14h); clones illustrated in right panel were fixed at 15–17h (stage late16/ early 17). Graphs in panel V quantify average number of cells per clone of a given group (upper graph) and average length of axon tract (PAT) at different stages of development. For the measurements, 150 clones were analyzed quantitatively (BA: 20; BL: 32; CM: 4; DAL: 26; DAM: 10; DPL: 34; DPM: 9; MB: 15). For a given stage, the number of data points ranged between three and 12. Somata could be individually counted because the GFP expression leaves the cell nucleus free of label, or labeled weakly. Error bars in the curves reflect one standard deviation. For some families of lineages we did not have sufficient representation at certain stages. For example, in case of DAM and DPM we could score clones only at stage 15/16e and L1. We used the T-test to compare clone sizes and PAT length. To label the difference between averages as significant, a P-value < 0.05 was assumed. Colors represent different groups of lineages; color key is given at top of lower graph. At all stages, lineages of the MB, DAL and DPL group are significantly larger than lineages of BL, DAM and DPM (p=0.05); differences in cell numbers among MB/DAL/DPL/BA, or DAM/DPM/BL/BA, are not significant. The change in clone size over time is significant for all lineage groups between late embryo (stage 16l) and larva. Other than that, only the increase in size of MB lineages between stage 13/14 and 15/16e is significant, as well as the dip in size of DPL lineages. MB neuroblasts divide even in the late embryo, which explains the first change; we have no explanation for the dip in the DPL curve. In regard to PAT length (lower graph), differences among lineage groups at stage 13/14 are not significant. The increase in PAT length between stage 15/16e and 16l is significant (p=0.05) for all groups. For more details see text.
For abbreviation of lineages and neuropile compartments see legend of Fig.1. Other abbreviations: apt antenno-protocerebral tract, AEL after egg laying
Bar: 20µm
Primary axon tracts start to form at stage 13/14 and, for the large majority of lineages, increase to an average length that lies in the range of 15–20µm in late embryos (Fig.2V). Up to stage 15/16early, PATs are more or less straight, unbranched fiber bundles that extend radially from the cell body cluster into the neuropile. In the late embryo (stage 16late/17; panels of third column in Fig.2), most PATs have formed short secondary branches and grown to their final length and characteristic shape. It is worth mentioning that the lineages of all classes behave rather uniformly in regard to axonal extension (Fig.2V). Thus, during the first three hours of neuronal differentiation, between stage 13 and 15, PATs grow slowly at a rate of 1–2µm per hour. This estimate was calculated by dividing the difference in average length of PATs between these stages by the number of hours separating them. Only mushroom body lineages show a faster axon growth. By stage 15, PATs of all lineages range between 6 and 12µm in average length. Axon extension speeds up slightly during the time interval between stage 15 and 17, when the large majority of PATs reach a length that falls within the range of 10–20µm.
Aside from neural clones, we also obtained glial clones at a relatively low frequency. They measure between two and eight cells and represented all three main classes of glia (surface glia, cortex glia, neuropile glia; data not shown).
Apoptotic cell death in primary lineages
Previous studies had reported apoptotic cell death in the nervous system of late embryos (Abrams et al., 1993; Rogulja-Ortmann et al., 2007), and our clonal data for early larval brains yielded significantly smaller numbers of neurons per lineage compared to embryos (see below). To address the question of cell death between the embryonic and larval period we first performed cell counts in brain hemispheres (Fig.3). The number of neurons and glia per brain hemisphere, counted for five specimens, measures approximately 2600 in the late embryo (stage 16) and 1600 in the L1 (for counting methods, see Materials and Methods). This 40% reduction is closely paralleled by a reduction in clone size (late embryo: average of 10.2, standard deviation 2.6; L1 larva: average of 6.4, standard deviation 3.3; Fig.2V). Likewise, one observes (at roughly constant neuronal cell body volume) a decrease in volume of the brain cortex (Fig.3A–E): In late embryos, the cortex measures approximately 180,000µm3; in the L1 larva it is 130,000µm3(both hemispheres combined) As the cortex volume decreases, the neuropile volume increase from approximately 18,000µm3 to over 40,000µm3. Overall, the brain size remains constant from late embryo to L1 larva.
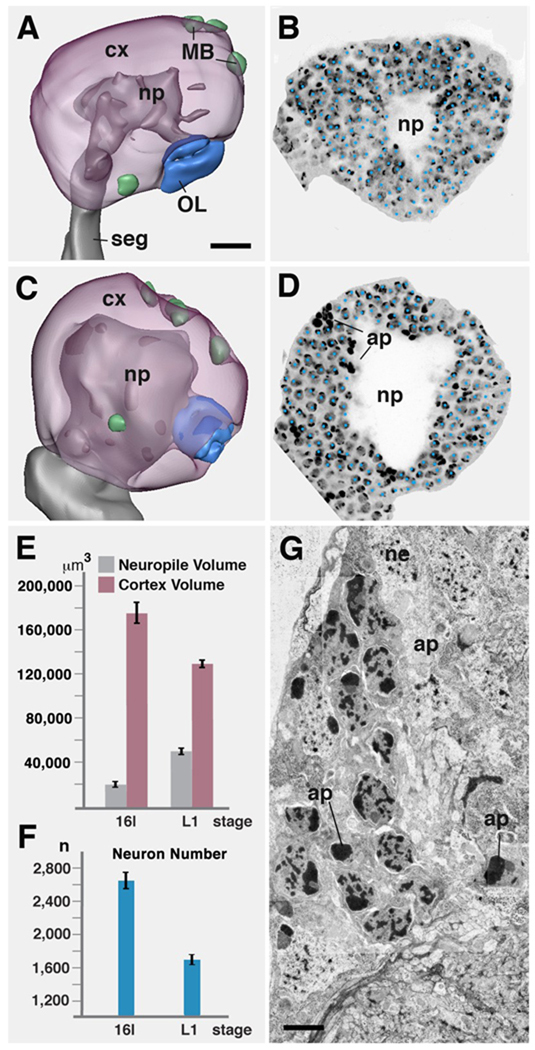
Fig.3.
Apoptotic cell death of primary neurons at the embryo-larva transition. A, B: stage 16 embryonic brain hemisphere. A shows 3D digital model (lateral view, anterior to the left) in which neuropile (np) is rendered grey, cortex (cx) transparent magenta. Active neuroblasts (green) are associated with the mushroom body (MB) and basal-anterior compartment. Optic lobe primordium (OL) is rendered blue. B shows a parasagittal confocal section in which nuclei of neurons and glial cells were labeled with Sytox (individually marked by blue dots). C, D: Representation of first instar larval brain hemisphere as digital 3D model (C) and confocal section (D; same scale as A and B). Note increase in diameter of neuropile, and concomitant decrease in thickness of cortex. Dense bodies in D (ap) are pycnotic nuclei of apoptotic neurons/glial cells. E: Graph comparing volume of neuropile and cortex between stage 16 embryo (left bars) and first instar larva (right bars; both hemispheres combined). F: Graph comparing number of cells in cortex of one brain hemisphere between stage 16 embryo (left) and first instar larva (right). G: Electron micrograph of section of first instar brain cortex, showing somata of neurons (ne) and apoptotic nuclei (ap).
Bars: 10µm (A); 2.5µm (G)
The decrease in neuron number is due to apoptotic cell death, which can be directly observed in confocal or TEM sections of L1 brains (Fig.3D, G). In both, pyknotic cells and cell fragments are scattered throughout the cortex. Previous studies had described cell death in the CNS of late embryos (Abrams et al., 1993; Rogulja-Ortmann et al., 2007), but not further quantified it or commented on the exact distribution of apoptotic cells. In our material, the apoptotic cells occur in clusters, often at the cortex-neuropile boundary, but also in the center of the cortex or, more infrequently, at the cortex surface. The clustered occurrence of apoptotic figures suggests that cell death is uneven for different lineages: some lineages maybe reduced by a major fraction or wiped out entirely, whereas others could be not affected at all.
Arborization of primary lineages in the larval brain
GFP-labeled clones provided information about the branching pattern of primary lineages in the larval brain. Thus, the significant increase in neuropile volume reported above is caused by an increase in neurite length and neurite arborization of primary neurons. Clones visualized in L1 brains resemble embryonic clones in that cell bodies are still clustered together and produce axons that fasciculate in primary axon tracts. However, the sparse branching seen in late embryos has increased dramatically (compare panels A/B, D/E, G/H, and J/K of Fig.4, or panels A/B and G/H of Fig.5). Among the different primary lineages represented by clones we can distinguish three major patterns of arborization (Fig.4T). The first pattern is characterized by distinct, spatially separate proximal and distal arborizations. We call lineages showing this arborization pattern “PD lineages”. Examples are the BA lineages that comprise projection neurons with proximal dendrites in the BA compartment (antennal lobe) and distal axonal arborizations in the CPLd compartment (the primordium of the lateral horn; Pereanu et al., 2009; Fig.4J–K). Also the four mushroom body lineages with proximal dendrites in the calyx and distal axons in the lobes are PD lineages (note, however, that in the mushroom body neurons synapses occur at all levels along the peduncle as well; Fahrbach, 2006). The second type of primary lineages observed in L1 have arborizations distributed more or less evenly along the entire length of the PAT (C lineages). Examples are the engrailed-positive DPLam lineage (Fig.5B), as well as the majority of DPL and BL lineages visualized as clones (Fig.4E, Q). Lineages of the third type lack proximal arborizations; their fiber tract extends for a considerable distance into the neuropile before branching into a more or less complex distal arbor (D lineages). Examples for D lineages are the per-positive BAmv lineage (Fig.5Z), or the DPM lineage in Fig.4N.
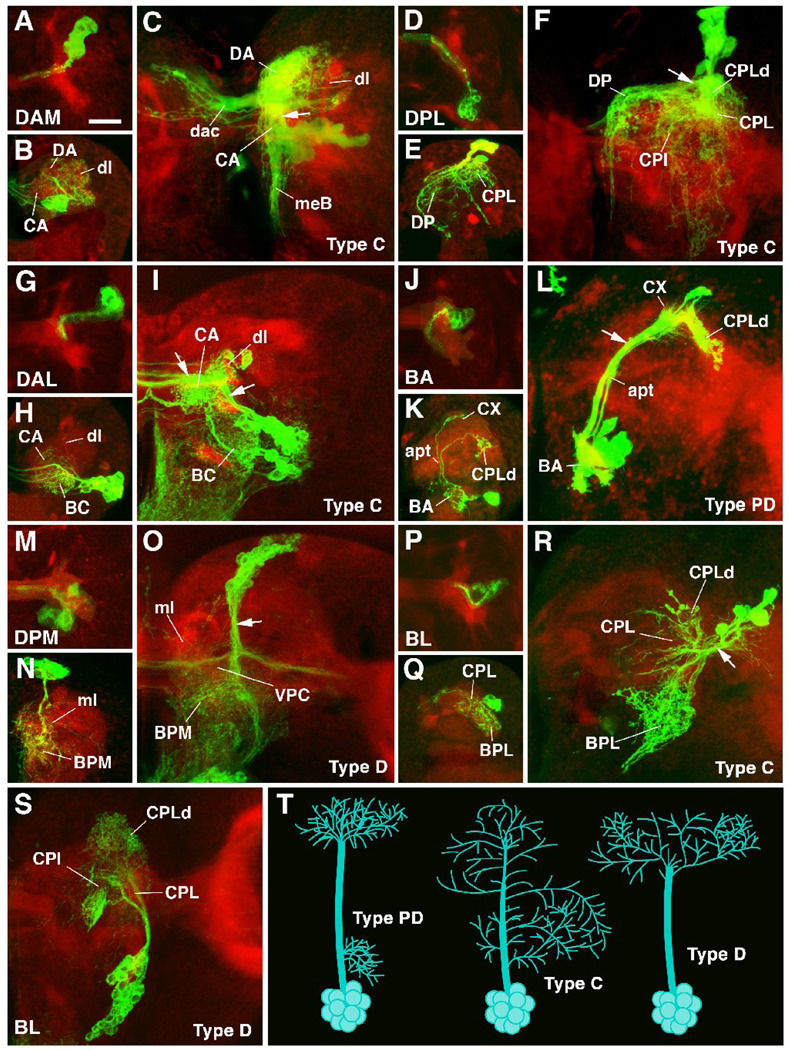
Fig.4.
Morphogenesis of lineages from embryo to larva. All panels are Z projections of confocal sections of brain hemisphere labeled with anti-DNcadherin (red; neuropile) and antiGFP to visualize individual lineages in which GFP was activated in early embryo by the FLP;FRT technique. Except for S and T, all panels are arranged in triplets (A–C, D–F, G–I, J–L, M–O, P–R) that correspond to lineage groups. In each triplet, the lineage group is indicated in first panel (e.g., panels A–C show lineage belonging to the DAM group). The first panel of each triplet (e.g., A) represents stage 16 embryonic brain (dorsal view, anterior up); the second panel (e.g., B) represents first instar larval brain (anterior view; dorsal up); the third panel (e.g., C) shows late third instar brain (anterior view, dorsal up). Panel T provides a schematic representation of different types of lineages encountered in brain (PD: separate proximal and distal arborization; C: continuous arborization; D: distal arborization). Note for all lineages shown the massive increase in arborization of primary neurons that occurs between late embryo and early larva (i.e., compare first and second panel of each triplet). Also, note presence of secondary neurons and secondary axon tracts in the lineages shown for third instar larva (indicated by white arrow in third panel of each triplet). A–C: DAM lineages. Cell bodies located in antero-medial-dorsal cortex. Typically C lineages, with diffuse crossed (dac dorsal anterior commissure) arborization in DA and CA compartments, and descending axons through median bundle (meB) to tritocerebrum and ventral nerve cord. D–F: DPL lineages. Cell bodies in dorso-lateral cortex. Typically C lineages with ventrally and medially directed branches in CPL, CPI, CPLd and DP compartments. Some commissural fibers (see panel D). G–I: DAL lineages. Cell bodies in antero-lateral cortex. Typically PD and C (as shown here). Arborization in BC, CPI and CPM. Often with commissural branches (H, I). J–L: BA lineages. Cell bodies in ventro-anterior cortex. Typically PD lineages (shown here) or D lineages. Examples shown are projection neuron lineages with proximal dendrites in the BA compartment (presumptive antennal lobe), axonal path along antenno-protocerebral tract (apt), and axonal arborizations in calyx (CX) and CPLd compartment (presumptive lateral horn). M–O: DPM lineages. Cell bodies in dorso-posterior medial cortex. Contains all types of lineages; D lineage shown here. Terminal arborizations in BPM and BPL compartments; crossing branches in VPC commissure. ml medial lobe of mushroom body. P–R: BL lineages. Cell bodies in dorso-lateral cortex. Mostly C lineages with branches connecting BPL compartment and CPLd. Note that for the third instar brain, four subgroups of BL lineages were recognized on the basis of SAT projection (BLA, BLD, BLP, BLV; Pereanu and Hartenstein, 2006). In the much smaller L1 brain, BL lineages as such are clearly distinguishable, but cannot yet assigned unambiguously to these subclasses. S: BLV lineage. Cell bodies in ventro-lateral cortex, branches in CPL and CPLd compartments.
Bar 20µm
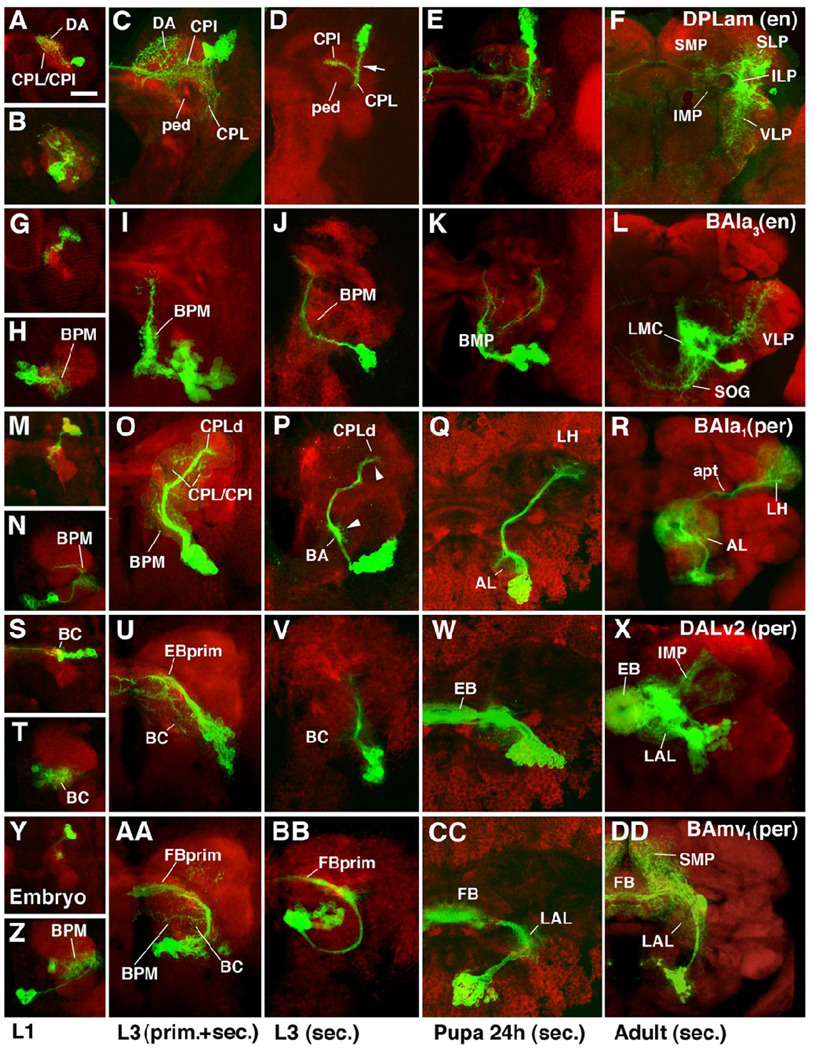
Fig.5.
Morphogenesis of selected lineages marked by specific Gal4 driver lines. Panels represent Z projections of confocal sections of brain hemisphere labeled with anti-DNcadherin (red; neuropile) and antiGFP to visualize individual lineages in which UAS-mCD8-GFP is driven by driver line engrailed-Gal4 (A–L) or period-Gal4 (M-DD). Panels are organized such that each row shows one specific lineage at four different developmental stages, indicated at bottom of figure. The first panel (e.g., A) shows embryo, the second (e.g., B) first instar larva; the third (e.g., C) late third instar in which both primary and secondary neurons are labeled; the fourth (e.g., D) late third instar with labeling of only secondary neurons; the fifth (e.g., E) pupa 24h after puparium formation; the sixth (e.g., F) adult. Except for panels depicting embryos, which present dorsal view, all panels show anterior view. A–F: Engrailed–positive DPLam lineage; type C. Primary neurons arborize widely in CPL, CPI, DP/DA, and CA compartments (A–C). Secondary axon tract (arrow in D) follows primary neurons into CPL compartment; here it branches into medial branch that crosses over peduncle (ped) into CPI, and a ventral branch remaining in CPL. Branches of secondary neurons develop along these axes (E, F), filling inferior medial and inferior lateral protocerebrum (IMP, ILP; descendants of CPI and CPL, respectively). Peripherally, branches continue into neighboring regions of superior lateral protocerebrum (SLP) and ventro-lateral protocerebrum (VLP). Interestingly, there seems to be no arborization of secondary neurons in superior medial protocerebrum (SMP), the descendant of the DA compartment that was filled with primary neurites of the DPLam lineage (compare panels C and F). G–L: Engrailed-positive BAla3 lineage. Both primary and secondary components of this lineage project long undivided axon tract posteriorly (D lineage; see Fig.4) and branch in BPM compartment. In addition, some secondary branches project latero-dorsally into VLP, and ventro-medially into subesophageal ganglion (SOG). M–R: Period-positive BAla1 lineage; type PD. Primary neurons project to BPM compartment and the region around the peduncle (CPI, CPL). Secondary neurons represent the class of multiglomerular antennal projection neurons. They form proximal arborizations (dendrites) in antennal lobe (AL in Q and R), and axonal arborizations in lateral horn (LH). Note how proximal and distal arbors are prefigured by tufts of filopodia along secondary axon tract in larva (arrowheads in P). S–X: Period-positive DALv2 lineage; type PD. Primary arborizations in BC compartment and the primordium of the ellipsoid body (EBprim), which in larva forms part of CPM compartment. Secondary neurons of this lineage form majority of ring neurons of ellipsoid body (EB). Other neurons of the same lineage form additional arborizations in lateral accessory lobe (LAL; descendant of BC compartment) and inferior medial protocerebrum (IMP). Y-DD: Period-positive BAmv1 lineage. D lineage with conspicuous crescent-shaped tract, projecting first posteriorly, then dorsolaterally, and finally dorso-medially towards the primordium of the fan-shaped body (FBprim), which forms part of the CPM compartment of the larval brain. Arborizations in BC, BPM and FBprim. Secondary axons follow the same trajectory and branch in LAL, fan-shaped body (FB), and superior medial protocerebrum (SMP).
Bar: 20µm
In PD lineages, arborizations are spatially confined to individual neuropile compartments, such as the antennal lobe and CPLd (Fig.4J–L; 5Q, R), or calyx and lobes (in case of the mushroom body; not shown). The overall neuropile volume occupied by C lineages and D lineages is typically larger than that of PD lineages and comprises several adjacent compartments. However, one can distinguish in all C lineages a “principal domain” of arborization to which the large majority of branches is confined, and a “peripheral domain” that receives a few, long neurites. Examples are shown in Fig.4: the principal domain of the DPL lineage (Fig.4D–F) is the CPLd and DP compartment; the BPL and BPM compartments receive a few peripheral long neurites. The principal domain of the DPMm lineage (type D) is the BPM compartment, with sparse branches to the DP. The principal compartment of most primary BL lineages (type C; Fig.4Q, R) is the BPL compartment, with peripheral branches to CPLd and DP.
In late third instar (L3) brains, clones induced in the embryo comprise two very different elements: the differentiated primary neurons, and the undifferentiated secondary neurons. The arborization formed by primary neurons has grown in overall size, but otherwise shows a pattern that is very similar to that seen in L1 brains. This is exemplified by the L1 and L3 clones shown side by side in Fig. 4E/F and 4K/L, or Fig.5B/C,H/I, T/U, and Z/AA. In some instances, the primary neuronal arborization grows in overall volume, but not density of terminal branches. In this case, arborizations at the later stage (L3) look rather sparse. An example is shown with the period-positive DALv2 lineage, where primary neurons in L1 fill the small BC compartment “solidly” with fine arborizations. By comparison, the grown BC compartment of L3, although still receiving DALv2 branches throughout its entire volume, looks relatively empty. In other cases, not only does the overall arborization of a lineage grow with the compartment(s) it is located in, but it keeps up with producing more fine branches, so that the density of innervation is maintained. An example is shown with the DPLc lineage in Fig.4E/F, and the engrailed-positive DPLam lineage in Fig.5B/C.
Secondary axon tracts fasciculate with primary axons
Drosophila brain neuroblasts produce two subsets of progeny, the primary neurons and secondary neurons. The microenvironment with which these classes of neurons interact is quite different, and we wanted to establish in how far the trajectory of PATs and the axons formed by secondary neurons coincide. In the late larval brain, secondary neurons form clusters of small, closely packed cells that are located adjacent to primary neurons in more superficial strata of the cortex. Axons of these neurons also fasciculate and form the secondary axon tracts (SATs; Dumstrei et al., 2003a, b; Pereanu and Hartenstein, 2006). In mixed primary/secondary lineages visualized in clones or by specific drivers, the two cell types can be told apart by the fact that primary neurons are larger in diameter, are located more deeply in the cortex, and are less tightly packed than secondary neurons (the latter probably due to the fact that primary neurons are individually “packaged” by cortex glia, whereas secondary neurons are surrounded by glia as a group; Pereanu et al., 2005; see also below).
Our results demonstrate that in all clones induced in embryos (comprising both primary and secondary neurons), as well as in the lineages visualized by specific Gal4 driver lines, primary and secondary axons fasciculate with each other, or are at least extend very close to each other (in Fig.4C, I, L, O the SAT is pointed out by white arrow). This implies that secondary axons may use primary axons as “guiding substrate”. It also shows that the primary and secondary neurons of a given lineage are related in terms of connectivity and function. The close relationship between PAT and SAT is shown even clearer in Fig.5 and Fig 6 where, due to the availability of lineage specific drivers, the shape of purely secondary lineages (visualized by inducing clones after hatching) is compared to that of the corresponding mixed primary/secondary lineages (compare Fig.5C/D, I/J, O/P, U/V, AA/BB). In all cases, SATs represent unbranched tracts that follow the same pathway as the corresponding primary axons. Figure 6 shows four different planes of the engrailed-positive DPLam lineage. Panels of the left column show only the secondary component of this lineage; the middle column represents secondary plus primary component. The top panels (Fig 6A, E) show sections of the superficial cortex where only secondary neurons are located. Close to the cortex-neuropile boundary (Fig 6B–F) are the large primary neurons which are visible only in panel Fig.6F. Note presence of secondary axon tract closely adjacent to primary neurons. G–H are sections of the superficial neuropile where primary and secondary axon tract are adjacent to each other. In the deep neuropile, primary axons branch; the secondary axon tract remains a compact bundle.
Fig.6.
Spatial relationship between primary and secondary neurons. A–D: Z-projections of confocal sections of part of third instar brain labeled with antibody Nc82 (white; labels neuropile). Secondary neurons (only) of the engrailed-positive DPLam lineage are visualized by MARCM where heat shock to induce recombination was given shortly after larval hatching. Panels show focal planes at different depth, indicated in schematic shown in panel I. A shows section near the cortex (cx) surface, containing the somata of secondary neurons. B represents deep cortex, C superficial neuropile, and D central neuropile. In B–D, secondary axon tract formed by secondary neurons of DPLam lineage is indicated by white arrow. Note branching of SAT in central neuropile (D). E–H, J–L: Z-projections of confocal sections of third instar brain hemisphere triple labeled with anti-DNcadherin (white in E–H; labels neuropile), anti-Neurotactin (secondary neurons; red in E–H, white in J–L), and anti-GFP to image the primary and secondary neurons of the DPLam lineage in which UAS mCD8-GFP is driven by engrailed-Gal4. These panels show brain slices at similar levels as adjoining panels A–D; levels indicated in schematic of panel I. Superficial panel (E) shows secondary DPLam neurons (sn), expressing both GFP and Neurotactin. In deep cortex (F, J) the primary neurons (pn), labeled with GFP but not neurotactin, are visible. In superficial neuropile (G, K), the primary axon tract (PAT) and secondary axon tract (SAT; white arrow) can be seen side by side. The PAT expresses only GFP. The SAT expresses both Neurotactin and GFP. In deep neuropile (H, L), labeling of SAT is faint, but still visible (see L). Primary axons have diverged to form primary axonal arborization (par).
Bar: 10µm
Secondary axons remain fasciculated and become the long fiber bundles of the adult brain
Secondary axon tracts form conspicuous fiber bundles in the larval brain. Are these bundles the forerunners of the axon fascicles that interconnect different compartments of the adult brain? Do arborizations formed by the neurons of one lineage during metamorphosis focus on specific brain compartments? At present, specific molecular markers (i.e., Gal4 driver lines) exist for only about 20% of the lineages of the central brain, which then can be followed into the adult stage, and more work is required even for these 20 lineages to document in detail the changes in terminal arborization that occur during metamorphosis. However, the analysis of the material we have assembled to date points out a number of generally valid principles of lineage maturation between larval and adult stage. We refer in the following to three lineages as examples, BAla1, BAmv1, and DALv2, which are shown in Figure 5 and Figure 7. Figure 5 (panel P-R for BAla1, panels V-X for DALv2, and panels BB-DD for BAmv1) shows Z-projections of the entire lineage; in Figure 7, separate sections of the labeled lineages along the ap axis are depicted.
the SAT that exists in the late larva corresponds already in length and trajectory to the SAT visible in the adult. Note, for example, BAla1 (multiglomerular projection neurons of the antennal lobe; see also Lai et al., 2008). BAla1 somata are located in the antero-ventral cortex, close to the BA compartment that will become the antennal lobe. The SAT initially extends along the common antenno-protocerebral tract. It then deviates laterally, passes underneath the peduncle, and terminates in a discrete small domain of the CPLd compartment from which the lateral horn develops (Pereanu et al., 2009). A fiber bundle with exactly these properties also appears in the adult brain. What is added are proximal arborizations in the antennal lobe, and distal arborizations in the lateral horn (Fig.5R).
The elaboration of terminal arborizations seems to occur rather synchronous in all lineages observed. Thus, in the larva, only the SAT is present; in many lineages, tufts of short filopodia foreshadow the occurrence of terminal branches (arrowhead in Fig.5P for BAla1, in Fig.5BB for BAmv1). At 24h after puparium formation, all lineages show short fibers that have grown out into the respective compartments (Fig.5E, K Q, W, CC); these fibers grow in length and terminal branch density to reach the adult configuration (Fig.5F, L, R, X, DD).
Somata belonging to one lineage stay close together in the adult cortex. The cortex volume occupied by one lineage is hardly any larger in the adult than in the larva. This implies that there is little tangential cell migration or intermingling of neurons in the Drosophila brain.
The axons of one lineage remain fasciculated in the adult brain. Since the SATs are typically surrounded by glia, and lack synapses, they appear as distinct signal-negative “holes” or impressions on confocal sections of preparations labeled with antibodies against synaptic proteins, like Bruch pilot (Nc82), Synaptotagmin, or DNcadherin. This is shown for the BAla1, BAmv1 and DALv2 lineage in Fig.7. Panels of the left column show confocal sections of a brain labeled only with Nc82 at different antero-posterior levels; columns E–H, I–J, and K–L show sections at corresponding levels of brains in which one lineage is labeled by GFP. Note that the SAT of a lineage along its entire trajectory creates a Nc82-negative impression that is invariant from one brain to the next. For example, the Nc82-negative impression occupied by fibers of the BAla 1 lineage as they enter the antennal lobe from ventro-laterally (Fig.7E) can be identified in the section shown in Fig.7A (green arrow labeled “E”); similarly, the trajectories of the BAla1 tract can be followed when it leaves the antenno protocerebral tract and turns laterally (compare Fig.7B/F, C/G, D/H. The same holds true for the SAT of the other two lineages shown: the SAT of DALv2 forms part of the lateral ellipsoid fascicle (LEF) shown in Fig.7B and J; BAmv1 axons initially form part of the longitudinal ventral fascicle (loV in Fig.7K; see green arrow with “K” pointing at corresponding Nc82-negative impression in Fig.7A); further posteriorly, the SAT of BAmv1 curves dorsally (Fig.7L and arrow with “L” in Fig.7B) and the laterally to join the LEF. Based on these findings we predict that, after taking into account the morphogenetic changes of metamorphosis (growth of neuropile compartments), we can identify in the adult brain the same pattern of SATs described for the larval brain (Pereanu and Hartenstein, 2006). These SATs represent the long fiber bundles interconnecting the different compartments of the adult brain.
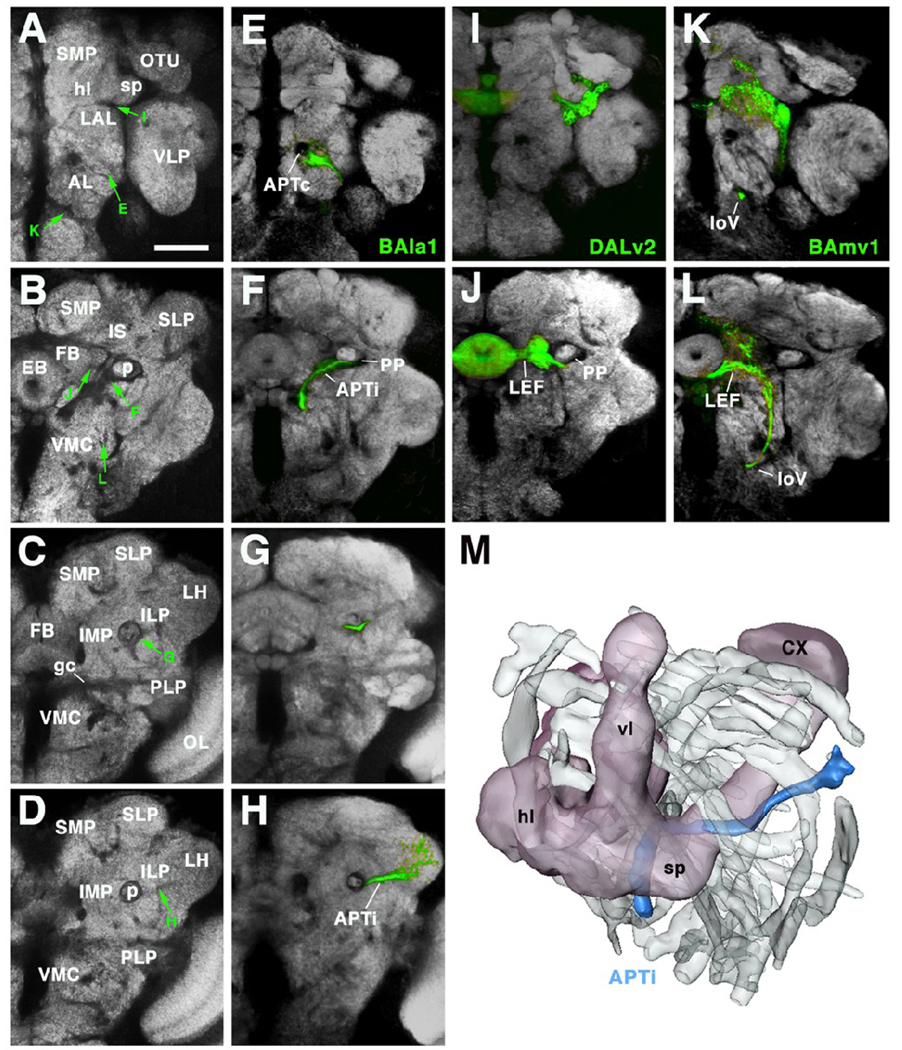
Fig.7.
Secondary axon tracts develop into long fiber bundles of adult brain. A–L show frontal confocal sections of adult brain hemisphere labeled with antibody Nc82 (neuropile; white). Rows correspond to the antero-posterior level of brain slice shown. Top row (A, E, I, K) corresponds to anterior level, showing optic tubercle (OTU) and horizontal lobes of mushroom body (hl). Slices shown in second row (B, F, J, L) are slightly more posteriorly, at level of ellipsoid body (EB). Third row taken at level of fan-shaped body (FB), bottom row at level of lateral horn (LH). In panels of left column (A–D) Nc82 was only marker used. In panels of second column (E–H), the secondary neurons of the BAla1 lineage are labeled by GFP (driven by period-Gal4). Third column (I, J) shows period-positive DALv2 lineage; fourth column (K, L) shows period-positive BAmv1 lineage. Note that the coherent secondary axon tracts of lineages now form long fiber bundles which are visible as Nc82-negative (i.e., synapse-free) “tunnels”, indicated by green arrows in panels of left column. Axons of BAla1 join the common antenno-protocerebral tract (APTc; E), a massive, Nc82-negative landmark. At the level of the ellipsoid body (F) BAla1 axons diverge laterally and then pass underneath the peduncle (p; in F and G), forming the medio-lateral APT (APTl). Further posteriorly, this tract continues straight towards the lateral horn (D, H). Axons of DALv2 (I, J) pass underneath the horizontal lobe of the mushroom body (I; see arrow in A) and then continue medially towards the ellipsoid body (J, B), forming the lateral ellipsoid fascicle (LEF). BAmv1 axons grow posteriorly as part of the longitudinal ventral fascicle (loV; panels K, L, C, D). Subsequently they turn upward laterally, then medially, to join the LEF at its posterior face (L). M: 3D digital model of left hemisphere of adult brain, shown in antero-dorso-lateral view. Outlines of all long fascicles that produce nc82-negative impressions of more than 1.5µm diameter are shown shaded grey. One bundle, the APTl (marked by the BAla1 lineage; see panels E–H) is shown in color (blue). The mushroom body is shown for orientation (CX calyx; hl horizontal lobes; vl vertical lobes; sp spur). Other abbreviations:
AL antennal lobe; gc great commissure; ILP inferior lateral protocerebrum; IMP inferior medial protocerebrum; LAL lateral accessory lobe; LH lateral horn; OL optic lobe; OTU optic tubercle; PLP postero-lateral protocerebrum; SLP superior lateral protocerebrum; SMP superior medial protocerebrum; VLP ventro-lateral protocerebrum; VMC ventro-medial cerebrum
Bar: 40µm
Lineages are not spatially segregated by glial sepata in the adult brain cortex
It was stated above that neuronal somata belonging to one lineage lie close to each other in the brain cortex. This could imply that dedicated glial septa form around the neurons of a given lineage. Such relationship between glia and lineages is mentioned in the literature (e.g., Staudacher, 1998). However, double labeling of adult brains with an antibody against neurons of a few lineages (anti-Engrailed; Fig.8A) and a marker for glia (Nrv2-Gal4>UAS-GFP; Fig.8B) demonstrates that no such specialized boundary exists. Thus, thin processes formed by cortex glia surround each individual neuronal cell body (Fig.8C). At the boundary between two lineages (arrow in Fig.8C, pointing at border between En-positive and En-negative cells, which belong to different lineages) no thickening of the glial septa can be detected. The same applies to the primary neurons found in the larva, which are also individually wrapped by fine glial processes (Fig.8D, E). Only clusters of young, undifferentiated secondary neurons, along with the neuroblast that has produced them, are contained within “communal” glial chambers which form a prominent feature of the superficial cortex of late larvae (Pereanu et al., 2005; Fig.8E). As secondary neurons mature cortex glial processes extend around them. Thus, already at the late larval stages, all cells of the deep cortex, including primary and old secondary neurons, are individually wrapped by glia.
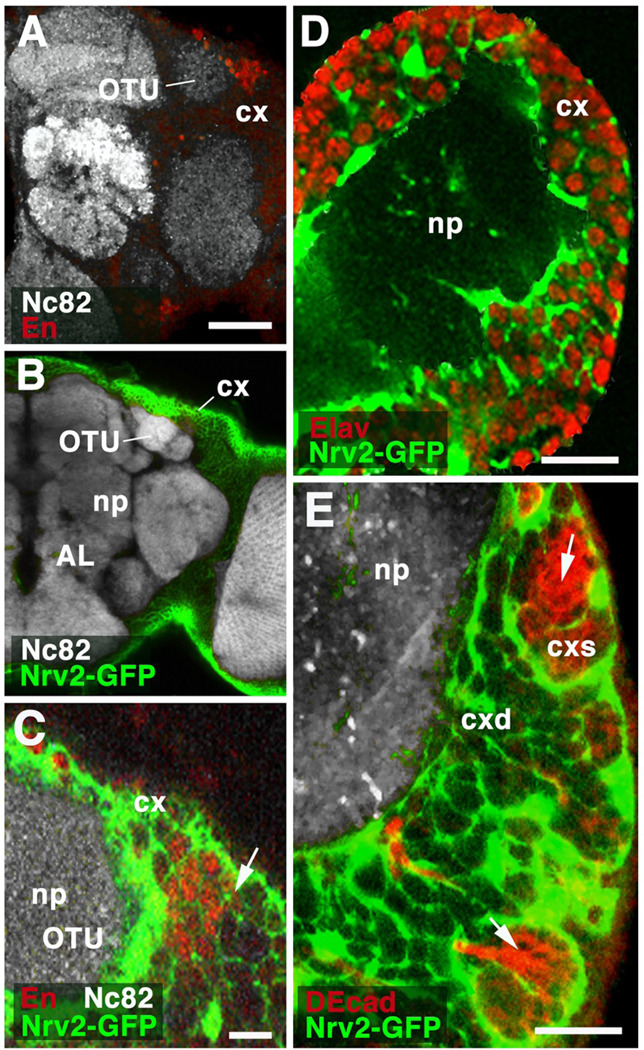
Fig.8.
Relationship of glia and lineages. A–C: Z-projections of frontal sections of adult brain hemisphere, labeled with antibody Nc82 (neuropile; np). In A, anti-Engrailed shows cell bodies of DPLam lineage, located in cortex (cx) flanking the optic tubercle (OTU). In B and C, cortex glia is labeled by GFP driven by Nrv2-Gal4. C presents a high magnification view of the dorso-lateral cortex containing the Engrailed-positive DPLam neurons (red). Cortex glia (green) forms sheaths around each individual neuron. No specialized glial layer demarcates the boundary (arrow) between labeled neurons of DPLam lineage and their unlabeled neighbors, which belong to different lineages. D: Z-projection of frontal section of first instar brain hemisphere. Neuronal nuclei are labeled by anti-Elav; cortex glia is labeled by Nrv2-Gal4>UAS-GFP. At this stage, all neurons in brain cortex (cx) are differentiated, primary neurons. As in adult (see C), each individual differentiated neuron is surrounded by cortex glial lamella. E: Z-projection of frontal confocal sections of late third instar brain hemisphere. As in B – D, cortex glia is labeled (green). Newly born ganglion mother cells and secondary neurons, located in superficial cortex (cxs), are labeled by anti-DEcadherin (red). Note that these undifferentiated secondary neurons form clusters which are enclosed in large glial chambers (arrow). As secondary neurons mature in deep cortex (cxd), they become individually enclosed by glial lamella. Other abbreviations: AL antennal lobe; cxd deep cortex
Bars: 40µm (A, B); 10µm (C, D, E)
Discussion
The peculiar mode of generating fixed lineages of neurons from a small number of stem cell-like neuroblasts, so far found only in insects and crustaceans, has been studied in great detail since many decades (Campos-Ortega and Hartenstein, 1985; Stollewerk and Simpson, 2005). More recent genetic studies on neuroblasts in Drosophila have contributed significantly to the identification of molecular factors involved in the asymmetric distribution of cellular components, or the orientation of the mitotic spindle, during cell division (Brody and Odenwald, 2002; Yu et al., 2006; Chia et al., 2008). Less attention was given to the morphological characteristics of the neurons that formed part of the cell lineages derived from individual neuroblasts. In this paper we have addressed several aspects of the structural development of the lineages forming the Drosophila brain. Using GFP labeled clones, as well lineage-specific GFP expression by Gal4 driver lines, we followed the growth of lineages from embryo to adult, focusing on cell number, neurite trajectory and neurite arborization pattern. Our main interest was to identify attributes that lineages had in common, or that neurons belonging to a given lineage had in common, rather than emphasizing the diversity of neuronal phenotypes.
Pattern of growth and projection of embryonic (primary) lineages
Brain neuroblasts segregate from the procephalic (head) neurectoderm during early embryogenesis. They form a layer of approximately 100 cells; within this layer, neuroblasts with a specific genetic identity (i.e., pattern of expression of a certain set of transcription factors) always occupy the same position (Younossi-Hartenstein et al., 1996; Urbach and Technau, 2003).
Neuroblasts undergo 5–8 divisions in the embryo (Hartenstein et al., 1987), producing that many ganglion mother cells; each ganglion mother cell divides once and forms two neurons or glial cells. It should be mentioned that one should expect exceptions to this general rule; for example, in the ventral nerve cord, atypical neuroblasts like the MP cells derived from the mesectoderm divide only once (Bossing and Technau, 1994). One neuroblast of the head, characterized by Rulifson and collaborators (Wang et al., 2007), appears to produce only 8 neurons that all express Dilp. However, as observed for the ventral cord, the large majority of brain neuroblasts divide at intervals of 50–60min after their segregation, and therefore should produce 10–16 labeled cells by the end of embryogenesis (Younossi-Hartenstein et al., 1996). The clone sizes we obtained for late embryos were indeed quite uniform, but were consistently smaller than the expected value, most of them ranging between 8 and 12 cells. A similar discrepancy has been observed in previous studies (Mandal et al., 2004; Ward and Skeath, 2000) using embryonic Flipout clones. For example, mesodermal clones that should comprise a total of 8–16 cells (based on four mesodermal mitoses; Bate, 1993) are only two to six cells in average (Mandal et al., 2004). The smaller than expected clone size is most likely due to the time interval required for building up a sufficient level of Flipase. For both mesodermal and brain clones, expected and observed clone size differ by about four cells, corresponding to two rounds of mitosis. Assuming a cell cycle length of about one hour, these data point at a two-hour lag phase required for Flp build-up. In other words, heat-shocking at around 3hrs (and one cannot go earlier than that, because it would interfere with gastrulation) will lead to flip-out around 5–6hrs, the time when most neuroblasts, or mesodermal cells, have already divided twice.
Aside from rather equal sizes, embryonic clones also behaved quite uniformly in regard to the pattern of axonal projection. Neurons of one lineage invariantly form a tight cluster and send axons in a single bundle that we called primary axon tract (PAT; Younossi Hartenstein et al., 2006). PATs were always directed radially away from the surface. Up until stage 15, PATs remained short and unbranched. They entered the minuscule embryonic neuropile at the location closest to their cluster of origin and stopped near the center of the brain; many lineages located dorsally (DPL, DPM) projected towards the commissure connecting the two brain hemispheres. Only during late embryonic stages (late 16, 17; 15 hours after fertilization) thin neurites with higher order branches became visible. The fact that we did not see longer axons (e.g., the pioneer axons establishing the major embryonic brain tracts; Therianos et al., 1995; Nassif et al., 1998) at an earlier stage is most likely due to the fact, discussed above, that our clones did not included the first born neurons. For the ventral nerve cord, most of the pioneer axons (e.g., acc, pcc, RP2) are formed by neurons that are the first born in their lineage. Accordingly, if lineages are visualized by dye injection (Bossing et al., 1996; Schmidt et al., 1997), they are in most cases comprised of two different types of neurons. One type has larger neurons with long axons that often project in quite different directions (like acc and pcc in the Nb1.1 lineage); these neurons are typically located deep, adjacent to the neuropile. The second type comprises smaller neurons, located more superficially in the cortex, and forming shorter and more uniform projections. The first type represents neurons born first; the second type later born neurons. Using Flip-out induced labeling of lineages, we only see the later born neurons.
Cell death and cell rearrangement in the brain cortex
We show that a significant number of primary neurons undergo apoptosis at the embryo-to-larva transition. Cell death in the developing arthropod CNS has been described frequently (Abrams et al., 1993; Harzsch et al., 1999; Ganeshina et al., 2000); our data, based on both single clone sizes as well as cell counts, put the ratio of cell death occurring at the embryo-to-larva transition at 40%. As a result of this massive amount of cell death, as well as local cell body rearrangements, the cortex thickness shrinks considerably from late embryo to early larva, whilst the volume of the neuropile increases due to neurite branching. The same phenomenon can be observed during the pupal phase: before onset of pupation, a stage when most neuroblasts have finished or are close to finish dividing and the number of neurons is maximal, the cortex measures 10–12 neuron diameters in thickness (Dumstrei et al., 2003b). In the adult brain, the cortex is very uneven in thickness. It is stretched out into a thin layer of 1–2 cell diameters thickness over most of the brain surface (VH, unpublished observation). At certain locations, in particular in crevices in between compartments that form protrusions at the posterior and anterior brain surface (e.g., calyx, optic tubercle, antennal lobe, ventro-lateral protocerebrum), the cortex is 5–8 cell diameters in thickness. During the same period, the neuropile volume increases dramatically. Precise neuron cell counts do not exist yet for the late larval and adult stages. Based on studies that focus on individual lineages (e.g., BLVa1–3, VH and SH, unpublished) it is clear that apoptotic cell death eliminates a major fraction of the secondary neurons; whether it amounts to 40%, as in case of primary neurons during the embryo-larval transition, remains to be seen.
The thinning of the brain cortex observed in the embryo-larval and the larval-adult transition is accompanied by rearrangement of neuronal cell bodies. These can be appreciated best when visualizing individual lineages. As might be expected, neuronal cell bodies of a given lineage form radially oriented, wedge- or column shaped clusters in the late embryo (primary lineages) or late larva (secondary lineages). In early larvae, primary neurons of a lineage are more spread out tangentially; the same happens to secondary neurons in the adult brain cortex. That being said, cell bodies of a lineage do stay together as one cluster. There does not appear to be a large scale intermingling of cell bodies of different lineages.
Another aspect of the architecture of the brain cortex that we addressed in this study is the delineation, or absence thereof, of lineages by glial cells. The notion appears in the literature (e.g., Staudacher, 1998) that specialized glial boundaries separate clusters of cells that belong to different lineages. This is definitely not the case in Drosophila, as shown here for the cells of the engrailed-positive DPLam lineage. Cortex glia forms a meshwork of processes, called trophospongium (Hoyle et al., 1986; Buchanan and Benzer, 1993; Pereanu et al., 2005), that wrap all neuronal cell bodies individually. The trophospongium at the boundary between cells of different lineages does not appear to be thicker, or specialized in any other way. Only at the larval stage when secondary neurons are being generated, clusters of newly born ganglion mother cells and neurons, located at the surface around the neuroblasts, are enclosed as a group (a sublineage) by cortex glia; as these cells are pushed away from the neuroblast by subsequent divisions, they become individually wrapped by cortex glia (Dumstrei et al., 2003b; Pereanu et al., 2005).
Terminal arborization of primary and secondary lineages: Relationship of lineages and compartments
Anatomical work on insect brains, employing methods such as Golgi silver impregnation, dye backfilling and injection, or antibody labeling, have visualized the arborization patterns of a large number of individual neurons that vary enormously in size and shape. They include giant neurons with arborizations reaching throughout most compartments, as well small neurons with arbors restricted to fractions of a single compartment (Strausfeld, 1976). To date, no systematic effort was undertaken to investigate how parameters like cell size or cell shape are represented numerically (are there 90% of giant neurons and 10% of dwarfs, or vice versa?) or related to development (are giants always born earlier than dwarfs? Do they come from different lineages?). Our paper does not provide a definitive answer to these questions, but presents a step along the way that will, hopefully soon, arrive at these answers.
Neurons of most of the brain lineages visualized by clones or specific Gal4 drivers share a common projection pattern and, most likely, terminal arborization pattern. We define three main types of lineages, based on their geometry: PD lineages with well defined proximal and distal arborizations (e.g., MB lineages, BAla1 antennal projection neurons, DALv2 ellipsoid body neurons), C lineages where proximal and distal projections blend into each other (e.g., DPLam), and D lineages which lack proximal arborizations (e.g.BAla3). At both larval and adult stages, the arborization of most lineages is restricted to a minor fraction of the neuropile volume (5–20%). Many PD lineages and some C lineages outline discrete compartments; aside from the well studied calyx and lobes of the mushroom body, defined by the four MB lineages (Ito et al., 1997), one can point at the BA lineages that generate the antennal projection neurons (Lai et al.., 2008), the primary DALv2 linage whose proximal arbors are restricted to the larval BC compartment (this paper), many of the primary BL lineages that outline the BPL compartment (this paper), or the BAla3 lineage that is mostly restricted to the BPM (Kumar et al., 2009). Proximal arborizations of the secondary DPMpm, DPMm, and CM4 lineages define the protocerebral bridge; distal arborizations produce the fan-shaped body and the noduli (Ito and Awasaki, 2008). Proximal arbors of the secondary DALv2 define the “bulbs” (formerly called “lateral triangle”), which represent the input domain of the ellipsoid body. Distal secondary DALv2 neurons generate the ellipsoid body. We propose that lineages such as MB, BAla/Bald, DPMm/DPMpm/CM4, or DALv2 act to “scaffold” these compartments. According to this hypothesis, each compartment, A’, has its own “scaffolding lineage”, A (or set of scaffolding lineages). A “scaffolding lineage” would then be defined in the following manner: (i) during development, the outgrowth of neurites from a lineage A actually creates the compartment A’. If A is deleted, A’ also does not form. This has been shown experimentally for the calyx, the compartment scaffolded by the four MB lineages (Ito et al., 1997). (ii) The arborization of lineage A forms a dense matrix of terminal axons on which synapses of A neurons themselves, as well as extrinsic neurons that enter compartment A’ from the outside are made. Again, the calyx provides an example in this case: electron microscopic investigations have shown that well over two thirds of the postsynaptic terminal neurites belong to neurons of the MB lineages (Yasuyama et al., 2002).
Our data suggest that a lineage-directed approach may facilitate the analysis of Drosophila brain development and structure considerably. This requires, first and foremost, to identify suitable Gal4 driver lines whose expression is restricted to one or a few lineages, and which then can be used to label, ablate, activate, or otherwise manipulate this lineage. Large scale screens are underway to attain this goal (Pfeiffer et al., 2008). Lineage-specific driver lines will also be crucial for the next step of the analysis, which looks at individual neurons within lineages. Published data suggest that many lineages can be subdivided “vertically” into two hemilineages, and “horizontally” into several sublineages (Ito and Awasaki, 2008). Neuroblasts produce series of ganglion mother cells (GMCs), each of which divides into an “a” and a “b” daughter cell. It has been shown for many lineages of the thoracic ganglia that all neurons of the a-hemilineage share properties that are different from neurons of the b- hemilineage (Jim Truman, pers. comm..). In some cases, a and b- hemilineages produce different SATs; or one complete hemilineage undergoes apoptosis, whereas the other one survives. Preliminary data suggest that hemilineages also exist for the brain (VH and SP, unpublished).
Sublineages are groups of neurons born during a defined time interval. For example, the primary neurons represent one significant sublineage for each (hemi)lineage; most likely, as discussed above, the primary neurons of most lineages can be further subdivided into smaller subgroups. It has been shown that the lineages scaffolding the fan-shaped body comprise multiple sublineages with different distal arborizations. The fan-shaped body is subdivided into six horizontal layers and eight vertical columns. All neurons of a given lineage (e.g., DPMpm) project to two distinct columns. Within a given lineage, neurons born at different times form sublineages that target different layers within the columns (Ito and Awasaki, 2008). We have preliminary data that DALv2 forms sublineages whose distal arbors define the discrete layers within the ellipsoid body (VH and SP, unpublished). Other lineages may form more overlapping projections; for example, each neuron of one of the MB lineages forms dendritic arbors that spread throughout most of the neuropile volume occupied by the lineage as a whole (Ito et al., 1997).
Axonal projection of lineages: Relationship of PATs, SATs and adult brain fascicles
The neurons of the primary lineages formed during embryogenesis form primary axon tracts (PATs) with invariant and characteristic trajectories. Note that, following a widely upheld convention, we call the unbranched processes that initially grow out from neurons “axons”, irrespective of the fact that eventually, they will form terminal side branches that carry postsynaptic sites, presynaptic sites, or both. For some lineages, PATs can be still recognized in the differentiated larval neuropile. That is particularly true for lineages that form long PATs, like the antenno-protocerebral tract (see Fig.3K, L), central anterior protocerebral tract (CAPT), or various commissural tracts. In other cases, the individual axons of one lineage may disperse to a certain extent and form a loose bundle (see for example the DPL lineage in Fig.3E, F). Once secondary axon tracts (SATs) form, they always extend in close proximity to the PATs of the corresponding lineage. This confirms that, even though several days pass between the time when it has finished producing its primary lineage and starts forming its secondary lineage, a neuroblast remains stationary, in close contact with the primary neurons. As a result, secondary neurons are born in contact with their older, primary siblings; the first primary axons they encounter when growing out their own axons are the ones belonging to these siblings.
In the neuropile, SATs of several lineages form thicker bundles, or fascicles. We will use the term fascicle because according to convention it denotes a bundle of nerve fibers with different directionality and endings. In our previous characterization of secondary lineages (Pereanu and Hartenstein, 2006) we have characterized the most prominent fascicles. We show here that the larval fascicles, formed by discrete sets of SATs, persist throughout metamorphosis and become the brain fascicles of the adult. In adult brain preparations labeled with markers for synapses, the fascicles (formed by long axons lacking synapses) stand out as signal-negative spaces.
Being able to follow fascicles from the larval period onward will help to unravel the connectivity of the adult brain. At present, unlike for vertebrate brains, our knowledge of the "macro-connectivity" (i.e., fiber bundles connecting different brain compartments) of the insect brain is very rudimentary. We only know about a few fiber bundles, such as the antenno-protocerebral tract connecting the antennal compartment with the calyx. Other tracts have been tentatively named on section-based maps of the adult brain (e.g., Strausfeld, 1976), but the beginning and ending of these tracts largely remains unclear, and the tract names suggested are not in wide use. The analysis of the secondary axon tracts will allow for a new and systematic effort to unravel macroconnectivity of the brain.
Neural lineages in Drosophila and other animals
One of the key characteristics of neural development in Drosophila (and probably insects in general) is that neurons of a lineage form a relatively coherent unit, where somata, axons and major parts of their arborizations stay together and define discrete modules of the brain neuropile. In the few cases where experimental studies were done (Ito et al., 1997; Stocker et al., 1997), removal of a particular lineage leads to the absence of the corresponding module; there is little or no regulation. How widespread are these insect-type neural lineages in the animal kingdom? Do they exist in vertebrates?
The second question can be answered in the negative. A number of fate mapping studies where the birth and migration of neurons was followed were done by the infection of progenitors with reporter-construct carrying viruses or mitotic recombination (Tan et al., 1998; Mathis and Nicolas, 2000; 2006). These experiments indicate that clones of cells derived from individual neural progenitors do not form structural modules where cell bodies all adhere to each other, and axons form coherent bundles. Vertebrate neural progenitors (“neural stem cells”) divide within the ventricular layer, or, in the forebrain, the subventricular zone of the embryonic neural tube. Subsequently, guided by radial glia (specialized neuroepithelial cells that later become astrocytes; Mission et al., 1991), neurons migrate preferentially radially. In a mouse study where labeled stem cells were integrated into the 10.5 day anterior neural tube (presumptive forebrain), the authors found that these cells undergo 9–11 rounds of mitosis every 10 hours, producing clones of approximately 600 pyramidal cells. (One might point out that this figure lies in the same ballpark as the number of neurons produced by one insect neuroblast. In Drosophila, most lineages consist of 150–200 neurons; some lineages have more than 500 neurons; Bello et al., 2008). The neurons of one clone were relatively close to each other, but left in between them large spaces filled with unlabeled neurons, indicating that cells of many neighboring clones intermingle. More importantly, the dendritic or axonal projections of a clone did not form coherent bundles. Aside from the radial clones of pyramidal cells, the authors found clones of tangentially migrating neurons. These clones correspond to the interneurons, which are born in a restricted domain of the ventral forebrain (eminence), from where they spread out tangentially to populate the different areas of the cortex (Zhu et al., 1999).
One can conclude that vertebrate neural lineages, at least up to a certain point in development, appear “open”: Cell bodies of numerous lineages intermingle within a given volume of the brain neuropile, and projections of members of a given lineage do not form distinct bundles or compartments. Outside arthropods and vertebrates, little is known about the relationship between neural architecture and neural development. In various protostome taxa, neuronal cell bodies in the cortex of the central nervous system are clustered around axon bundles (e.g., plathelminthes: Morris et al., 2004); whether these clusters of neurons correspond to lineages remains to be shown.
Outlook
The fact that the Drosophila nervous system is composed of structural modules that are in many cases defined by discrete lineages offers the exciting possibility of getting closer at the link between genes and behavior. In a developmental sense, lineages represent “units of gene expression”. The expression pattern of more than fifty transcription factors in specific embryonic neuroblasts has been described (Urbach and Technau, 2003). In the embryo, a given transcription factor becomes active in one, or a small number of, neuroblasts; a particular neuroblast thereby acquires a “genetic address”, consisting of specific sets of transcription factors. It is thought that this genetic address will essentially be involved in shaping the morphology and function of that lineage. Lineages also represent to some extent structural modules of the brain. It stands to reason that in many cases, a lineage with its highly restricted dendritic arborization and axonal pathway, will be involved in a single or a limited number of behaviors, or, probably more accurately, “behavioral subroutines”. If that is indeed correct, one can manipulate the structural module scaffolded by a given lineage, and thereby address its function, and what aspects of that function is controlled by a given gene expressed in the lineage. For example, completely removing that lineage by driving a cell death-inducing gene, using a lineage specific promoter, would eliminate the module. Behavioral assays may show, in most cases, the deterioration of certain behavioral subroutines. Using the same driver to modify the expression of genes specifically found in a particular lineage, one might be able to manipulate the genes one at a time, to find that one gene may be simply involved in increasing the cell number in that lineage, or the density of synapses in the scaffolded compartment, etc. By systematically following this approach for each one of the ~100 lineages of the brain the expectation is that we will arrive at a much better understanding of how genes control behavior.
Acknowledgements
We thanks Drs.K. Ito and C. Doe for reagents. This work was supported by NIH Grant R01 NS054814 to VH.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abrams JM, White K, Fessler LI, Steller H. Programmed cell death during Drosophila embryogenesis. Development. 1993;117(1):29–43. doi: 10.1242/dev.117.1.29. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Drosophila. A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 1989. pp. 214–217. [Google Scholar]
- Bate M. The Mesoderm and its Derivatives. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila. Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- Bello BC, Hirth F, Gould AP. A pulse of the Drosophila Hox protein Abdominal-A schedules the end of neural proliferation via neuroblast apoptosis. Neuron. 2003;37(2):209–219. doi: 10.1016/s0896-6273(02)01181-9. [DOI] [PubMed] [Google Scholar]
- Bello BC, Izergina N, Caussinus E, Reichert H. Amplification of neural stem cell proliferation by intermediate progenitor cells in Drosophila brain development. Neural Develop. 2008;19(3):5. doi: 10.1186/1749-8104-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossing T, Technau GM. The fate of the CNS midline progenitors in Drosophila as revealed by a new method for single cell labelling. Development. 1994;120(7):1895–1906. doi: 10.1242/dev.120.7.1895. [DOI] [PubMed] [Google Scholar]
- Bossing T, Udolph G, Doe CQ, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. I. Neuroblast lineages derived from the ventral half of the neuroectoderm. Dev Biol. 1996;179(1):41–64. doi: 10.1006/dbio.1996.0240. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118(2):401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brody T, Odenwald WF. Cellular diversity in the developing nervous system: a temporal view from Drosophila. Development. 2002;129(16):3763–3770. doi: 10.1242/dev.129.16.3763. [DOI] [PubMed] [Google Scholar]
- Buchanan RL, Benzer S. Defective glia in the Drosophila brain degeneration mutant drop-dead. Neuron. 1993;10(5):839–850. doi: 10.1016/0896-6273(93)90200-b. [DOI] [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. Development of the Nervous System. In: Kerkut GA, Gilbert LI, editors. Comprehensive Insect Physiology, Biochemistry, and Pharmacology. Vol.5. Oxford: Pergamon; 1985. pp. 49–85. [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The Embryonic Development of Drosophila melanogaster. 2nd Edition. Springer; 1997. [Google Scholar]
- Chia W, Somers WG, Wang H. Drosophila neuroblast asymmetric divisions: cell cycle regulators, asymmetric protein localization, and tumorigenesis. J.Cell Biol. 2008;180(2):267–272. doi: 10.1083/jcb.200708159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Escalera S, Bockamp EO, Moya F, Piovant M, Jiménez F. Characterization and gene cloning of neurotactin, a Drosophila transmembrane protein related to cholinesterases. EMBO J. 1990;9(11):3593–3601. doi: 10.1002/j.1460-2075.1990.tb07570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Nassif C, Hartenstein V. Early development of the Drosophila brain. V. Pattern of postembryonic neuronal lineages expressing Shg/DE cadherin. J. Comp. Neurol. 2003a;455(4):451–462. doi: 10.1002/cne.10484. [DOI] [PubMed] [Google Scholar]
- Dumstrei K, Wang F, Tepass U, Hartenstein V. Role of DE-cadherin in neuroblast proliferation, neural morphogenesis and axon tract formation in Drosophila larval brain development. J. Neurosci. 2003b;23(8):3325–3335. doi: 10.1523/JNEUROSCI.23-08-03325.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estes PS, Ho GL, Narayanan R, Ramaswami M. Synaptic localization and restricted diffusion of a Drosophila neuronal synaptobrevin-green fluorescent protein chimera in vivo. J. Neurogenet. 2000;13(4):233–255. doi: 10.3109/01677060009084496. [DOI] [PubMed] [Google Scholar]
- Fahrbach SE. Structure of the mushroom bodies of the insect brain. Annu. Rev. Entomol. 2006;51:209–232. doi: 10.1146/annurev.ento.51.110104.150954. [DOI] [PubMed] [Google Scholar]
- Ganeshina O, Schäfer S, Malun D. Proliferation and programmed cell death of neuronal precursors in the mushroom bodies of the honeybee. J. Comp. Neurol. 2000;417(3):349–365. [PubMed] [Google Scholar]
- Golic MM, Rong YS, Petersen RB, Lindquist SL, Golic KG. FLP-mediated DNA mobilization to specific target sites in Drosophila chromosomes. Nucleic Acids Res. 1997;25(18):3665–3671. doi: 10.1093/nar/25.18.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenstein V, Rudloff E, Campos-Ortega JA. The pattern of proliferation of the neuroblasts in the wildtype embryo of Drosophila melanogaster. Wilhelm Roux's Arch. Dev. Biol. 1987;196:473–485. doi: 10.1007/BF00399871. [DOI] [PubMed] [Google Scholar]
- Harzsch S, Miller J, Benton J, Beltz B. From embryo to adult: persistent neurogenesis and apoptotic cell death shape the lobster deutocerebrum. J. Neurosci. 1999;19(9):3472–3485. doi: 10.1523/JNEUROSCI.19-09-03472.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan BA, Bermingham NA, He Y, Sun Y, Jan YN, Zoghbi HY, Bellen HJ. atonal regulates neurite arborization but does not act as a proneural gene in the Drosophila brain. Neuron. 2000;25(3):549–561. doi: 10.1016/s0896-6273(00)81059-4. [DOI] [PubMed] [Google Scholar]
- Hortsch M, Patel NH, Bieber AJ, Traquina ZR, Goodman CS. Drosophila neurotactin, a surface glycoprotein with homology to serine esterases, is dynamically expressed during embryogenesis. Development. 1990;110(4):1327–1340. doi: 10.1242/dev.110.4.1327. [DOI] [PubMed] [Google Scholar]
- Hoyle G, Williams M, Phillips C. Functional morphology of insect neuronal cell-surface/glial contacts: the trophospongium. J. Comp. Neurol. 1986;246(1):113–128. doi: 10.1002/cne.902460108. [DOI] [PubMed] [Google Scholar]
- Ito K, Hotta Y. Proliferation pattern of postembryonic neuroblasts in the brain of Drosophila melanogaster. Dev. Biol. 1992;149(1):134–148. doi: 10.1016/0012-1606(92)90270-q. [DOI] [PubMed] [Google Scholar]
- Ito K, Awano W, Suzuki K, Hiromi Y, Yamamoto D. The Drosophila mushroom body is a quadruple structure of clonal units each of which contains a virtually identical set of neurones and glial cells. Development. 1997;124(4):761–771. doi: 10.1242/dev.124.4.761. [DOI] [PubMed] [Google Scholar]
- Ito K, Awasaki T. Clonal unit architecture of the adult fly brain. Adv. Exp. Med. Biol. 2008;628:137–158. doi: 10.1007/978-0-387-78261-4_9. [DOI] [PubMed] [Google Scholar]
- Iwai Y, Usui T, Hirano S, Steward R, Takeichi M, Uemura T. Axon patterning requires DN-cadherin, a novel neuronal adhesion receptor, in the Drosophila embryonic CNS. Neuron. 1997;19(1):77–89. doi: 10.1016/s0896-6273(00)80349-9. [DOI] [PubMed] [Google Scholar]
- Kaneko M, Hall JC. Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J. Comp. Neurol. 2000;422(1):66–94. doi: 10.1002/(sici)1096-9861(20000619)422:1<66::aid-cne5>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Kumar A, Fung S, Lichtneckert R, Reichert H, Hartenstein V. The arborization pattern of engrailed-positive neural lineages reveal neuromere boundaries in the Drosophila brain neuropile. J. Comp. Neurol. 2009 doi: 10.1002/cne.22112. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai SL, Awasaki T, Ito K, Lee T. Clonal analysis of Drosophila antennal lobe neurons: diverse neuronal architectures in the lateral neuroblast lineage. Development. 2008;135(17):2883–2893. doi: 10.1242/dev.024380. [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci. 2001;24(5):251–254. doi: 10.1016/s0166-2236(00)01791-4. [DOI] [PubMed] [Google Scholar]
- Malun D. Early development of mushroom bodies in the brain of the honeybee Apis mellifera as revealed by BrdU incorporation and ablation experiments. Learn Mem. 1998;5(1–2):90–101. [PMC free article] [PubMed] [Google Scholar]
- Mandal L, Banerjee U, Hartenstein V. Evidence for a fruit fly hemangioblast and similarities between lymph-gland hematopoiesis in fruit fly and mammal aortagonadal-mesonephros mesoderm. Nat. Gen. 2004;36(9):1019–1023. doi: 10.1038/ng1404. [DOI] [PubMed] [Google Scholar]
- Mathis L, Nicolas JF. Different clonal dispersion in the rostral and caudal mouse central nervous system. Development. 2000;127(6):1277–1290. doi: 10.1242/dev.127.6.1277. [DOI] [PubMed] [Google Scholar]
- Mathis L, Nicolas JF. Clonal origin of the mammalian forebrain from widespread oriented mixing of early regionalized neuroepithelium precursors. Dev Biol. 2006;293(1):53–63. doi: 10.1016/j.ydbio.2005.12.055. [DOI] [PubMed] [Google Scholar]
- Mission JP, Takahashi T, Caviness VS., Jr Ontogeny of radial and other astroglial cells in murine cerebral cortex. Glia. 1991;4(2):138–148. doi: 10.1002/glia.440040205. [DOI] [PubMed] [Google Scholar]
- Morris J, Nallur R, Ladurner P, Egger B, Rieger R, Hartenstein V. The embryonic development of the flatworm Macrostomum sp. Dev. Gen. Evol. 2004;214(5):220–239. doi: 10.1007/s00427-004-0406-4. [DOI] [PubMed] [Google Scholar]
- Nassif C, Noveen A, Hartenstein V. Embryonic development of the Drosophila brain I. The pattern of pioneer tracts. J. Comp. Neur. 1998;402(1):10–31. [PubMed] [Google Scholar]
- Pereanu W, Shy D, Hartenstein V. Morphogenesis and proliferation of the larval brain glia in Drosophila. Dev. Biol. 2005;283(1):191–203. doi: 10.1016/j.ydbio.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Pereanu W, Hartenstein V. Neural lineages of the Drosophila brain: A 3D digital atlas of the pattern of lineage location and projection at the late larval stage. J. Neurosci. 2006;26(20):5534–5553. doi: 10.1523/JNEUROSCI.4708-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereanu W, Jennett A, Younossi-Hartenstein A, Kumar A, Reichert H, Hartenstein V. A development-based compartmentalization of the Drosophila central brain. 2009 doi: 10.1002/cne.22376. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, Murphy C, Scully A, Carlson JW, Wan KH, Laverty TR, Mungall C, Svirskas R, Kadonaga JT, Doe CQ, Eisen MB, Celniker SE, Rubin GM. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl. Acad. Sci. U S A. 2008;105(28):9715–9720. doi: 10.1073/pnas.0803697105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogulja-Ortmann A, Lüer K, Seibert J, Rickert C, Technau GM. Programmed cell death in the embryonic central nervous system of Drosophila melanogaster. Development. 2007;134(1):105–116. doi: 10.1242/dev.02707. [DOI] [PubMed] [Google Scholar]
- Rolls MM, Doe CQ. Baz, Par-6 and aPKC are not required for axon or dendrite specification in Drosophila. Nat Neurosci. 2004;7(12):1293–1295. doi: 10.1038/nn1346. [DOI] [PubMed] [Google Scholar]
- Schmid A, Chiba A, Doe CQ. Clonal analysis of Drosophila embryonic neuroblasts: neural cell types, axon projections and muscle targets. Development. 1999;126(21):4653–4689. doi: 10.1242/dev.126.21.4653. [DOI] [PubMed] [Google Scholar]
- Schmidt H, Rickert C, Bossing T, Vef O, Urban J, Technau GM. The embryonic central nervous system lineages of Drosophila melanogaster. II. Neuroblast lineages derived from the dorsal part of the neuroectoderm. Dev Biol. 1997;189(2):186–204. doi: 10.1006/dbio.1997.8660. [DOI] [PubMed] [Google Scholar]
- Sprecher S, Reichert H, Hartenstein V. Gene expression patterns in primary neuronal clusters of the Drosophila embryonic brain. Gene Expr. Patterns. 2007;7(5):584–595. doi: 10.1016/j.modgep.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudacher E. Distribution and morphology of descending brain neurons in the cricket Gryllus bimaculatus. Cell Tissue Res. 1998;294(1):187–202. doi: 10.1007/s004410051169. [DOI] [PubMed] [Google Scholar]
- Stocker RF, Heimbeck G, Gendre N, de Belle JS. Neuroblast ablation in Drosophila P[GAL4] lines reveals origins of olfactory interneurons. J. Neurobiol. 1997;32(5):443–456. doi: 10.1002/(sici)1097-4695(199705)32:5<443::aid-neu1>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Stollewerk A, Simpson P. Evolution of early development of the nervous system: a comparison between arthropods. Bioessays. 2005;27(9):874–883. doi: 10.1002/bies.20276. [DOI] [PubMed] [Google Scholar]
- Strausfeld N. Atlas of an Insect Brain. Berlin: Springer; 1976. [Google Scholar]
- Sun BH, Xu PZ, Salvaterra PM. Dynamic visualization of nervous system in live Drosophila. Proc. Natl. Acad. Sci. USA. 1999;96(18):10438–10443. doi: 10.1073/pnas.96.18.10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata T, Schwartz C, Gustavson E, Ali Z, Kornberg TB. Creating a Drosophila wing de novo, the role of engrailed, and the compartment border hypothesis. Development. 1995;121(10):3359–3369. doi: 10.1242/dev.121.10.3359. [DOI] [PubMed] [Google Scholar]
- Tan SS, Kalloniatis M, Sturm K, Tam PP, Reese BE, Faulkner-Jones B. Separate progenitors for radial and tangential cell dispersion during development of the cerebral neocortex. Neuron. 1998;21(2):295–304. doi: 10.1016/s0896-6273(00)80539-5. [DOI] [PubMed] [Google Scholar]
- Therianos S, Leuzinger S, Hirth F, Goodman CS, Reichert H. Embryonic development of the Drosophila brain: formation of commissural and descending pathways. Development. 1995;121(11):3849–3860. doi: 10.1242/dev.121.11.3849. [DOI] [PubMed] [Google Scholar]
- Truman JW. Metamorphosis of the central nervous system of Drosophila. J. Neurobiol. 1990;21(7):1072–1084. doi: 10.1002/neu.480210711. [DOI] [PubMed] [Google Scholar]
- Truman JW, Schuppe H, Shepherd D, Williams DW. Developmental architecture of adult-specific lineages in the ventral CNS of Drosophila. Development. 2004;131(20):5167–5184. doi: 10.1242/dev.01371. [DOI] [PubMed] [Google Scholar]
- Urbach R, Technau GM. Molecular markers for identified neuroblasts in the developing brain of Drosophila. Development. 2003;130(16):3621–3637. doi: 10.1242/dev.00533. [DOI] [PubMed] [Google Scholar]
- Wang S, Tulina N, Carlin DL, Rulifson EJ. The origin of islet-like cells in Drosophila identifies parallels to the vertebrate endocrine axis. Proc. Natl. Acad. Sci. U S A. 2007;104(50):19873–19878. doi: 10.1073/pnas.0707465104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward EJ, Skeath JB. Characterization of a novel subset of cardiac cells and their progenitors in the Drosophila embryo. Development. 2000;127(22):4959–4969. doi: 10.1242/dev.127.22.4959. [DOI] [PubMed] [Google Scholar]
- Yasuyama K, Meinertzhagen IA, Schürmann FW. Synaptic organization of the mushroom body calyx in Drosophila melanogaster. J. Comp. Neurol. 2002;445(3):211–226. doi: 10.1002/cne.10155. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Nassif C, Hartenstein V. Early neurogenesis of the Drosophila brain. J. Comp. Neur. 1996;370(3):313–329. doi: 10.1002/(SICI)1096-9861(19960701)370:3<313::AID-CNE3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Salvaterra P, Hartenstein V. Early development of the Drosophila brain IV. Larval neuropile compartments defined by glial septa. J Comp Neurol. 2003;455(4):435–450. doi: 10.1002/cne.10483. [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A, Shy D, Hartenstein V. The embryonic formation of the Drosophila brain neuropile. J. Comp. Neur. 2006;497(6):981–998. doi: 10.1002/cne.20884. [DOI] [PubMed] [Google Scholar]
- Yu F, Kuo CT, Jan YN. Drosophila neuroblast asymmetric cell division: recent advances and implications for stem cell biology. Neuron. 2006;51(1):13–20. doi: 10.1016/j.neuron.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Zheng X, Zugates CT, Lu Z, Shi L, Bai JM, Lee T. Baboon/dSmad2 TGF-beta signaling is required during late larval stage for development of adult-specific neurons. EMBO J. 2006;25(3):615–627. doi: 10.1038/sj.emboj.7600962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Li H, Zhou L, Wu JY, Rao Y. Cellular and molecular guidance of GABAergic neuronal migration from an extracortical origin to the neocortex. Neuron. 1999;23(3):473–485. doi: 10.1016/s0896-6273(00)80801-6. [DOI] [PubMed] [Google Scholar]