Abstract
Immunoglobulin D (IgD) is an enigmatic antibody isotype that mature B cells co-express with IgM through alternative RNA splicing. We found active T cell-dependent and T cell-independent IgM-to-IgD class switching in human upper respiratory mucosa B cells. This process required activation-induced cytidine deaminase and generated local and circulating IgD-producing plasmablasts reactive to respiratory bacteria. Circulating IgD bound to basophils through a calcium-mobilizing receptor that induced antimicrobial, opsonizing, inflammatory and B cell-stimulating factors including cathelicidin, interleukin-1, interleukin-4 and B cell-activating factor BAFF upon IgD cross-linking. By showing dysregulation of IgD class-switched B cells and IgD-armed basophils in autoinflammatory syndromes with periodic fever, our data indicate that IgD orchestrates an ancestral surveillance system at the interface between immunity and inflammation.
INTRODUCTION
Immunoglobulin D (IgD) has remained an enigmatic antibody class since its discovery more than forty years ago1. Because of its spotty presence in mammals and absence in birds, IgD was initially thought to be a recently evolved Ig isotype2. By showing that xenopous is orthologous to IgW, an antibody class found in cartilaginous fish and lungfish, previous studies demonstrate that IgD was present in the ancestor of all jawed vertebrates and arose together with IgM at the time of the emergence of the adaptive immune system, approximately 500 million years ago3. While IgM remains stable over evolutionary time, IgD shows greater structural plasticity and can be predominantly expressed as a transmembrane or secretory molecule in a species-specific manner4,5. One possible interpretation is that IgD has been preserved as a structurally flexible locus to complement the functions of IgM.
IgM and IgD are the first antibody isotypes expressed during B cell ontogeny. Bone marrow B cell precursors acquire surface IgM after assembling heavy (H) and light (L) chain variable region exons from prototypic variable (V), diversity (D) and joining (J) gene segments through an antigen-independent process mediated by recombination activating gene (RAG)-1 and RAG-2 proteins6. After leaving the bone marrow to colonize secondary lymphoid organs, B cells acquire surface IgD of the same specificity as surface IgM through alternative splicing of a pre-messenger RNA comprising V(D)J and both heavy chain constant μ (Cμ) and Cδ exons7. The significance of dual IgM and IgD expression remains unclear, because either isotype largely compensates for the loss of the other8-10.
After encountering antigen in secondary lymphoid organs, mature B cells transcriptionally down-regulate surface IgD11 and thereafter undergo somatic hypermutation (SHM) and class switch DNA recombination (CSR), two Ig gene-diversifying processes that require the DNA-editing enzyme activation-induced cytidine deaminase (AID)12. SHM introduces point mutations into VHDJH and VLJL exons, thereby providing the structural correlate for selection of high-affinity Ig variants by antigen13, whereas CSR substitutes the Cμ gene with Cγ, Cα or Cε, thereby generating secondary IgG, IgA and IgE isotypes with the same antigen binding specificity as IgM but additional effector functions14. Of note, CSR occurs through either a T cell-dependent (TD) follicular pathway involving engagement of the CD40 receptor on B cells by CD40 ligand (CD40L) on antigen-activated CD4+ T cells or through a T cell-independent (TI) extrafollicular pathway involving engagement of TACI and BAFF-R receptors on B cells by BAFF and APRIL, two CD40L-related tumor necrosis factor (TNF) family members released by antigen-activated dendritic cells, macrophages and mucosal epithelial cells15-21. Ultimately, antigen-experienced B cells generate antibody-secreting plasma cells and memory B cells22. These latter form new plasma cells upon exposure to previously encountered antigens. In general, plasma cell-derived IgG, IgA and IgE antibodies facilitate the elimination of invading pathogens by activating powerful Fc receptors that enhance the phagocytic, cytotoxic and pro-inflammatory functions of various innate immune cells, including granulocytes23.
Instead of switching from IgM to IgG, IgA or IgE, some B cells switch to IgD24, suggesting that IgD confers some functional advantage over IgM. The resulting IgD+IgM− plasma cells release highly mutated mono- and polyreactive IgD antibodies mostly containing λ light chains in the blood as well as respiratory, salivary, lacrimal and mammary secretions1,4,25-28. Secreted IgD might enhance immune protection by regulating B cell homeostasis and activation. Indeed, IgD-deficient mice have fewer B cells, delayed affinity maturation, and weaker production of IgG1 and IgE, two isotypes highly dependent on the cytokine interleukin-4 (IL-4; http://www.signaling-gateway.org/molecule/query?afcsid=A001262)8,9. Conversely, mice injected with a goat polyclonal anti-human IgD with potential agonistic activity produce more IgG1 and IgE and show robust IL-4 production by T cells and basophils29-31. These latter are a small granulocytic subset that triggers T and B cell responses by releasing IL-4 upon recognizing antigen via pre-bound IgE and IgG32-35. Whether IgD also facilitates antigen recognition by basophils remains unknown.
We found that human upper respiratory mucosa B cells generated local and circulating IgD+IgM− plasmablasts by undergoing Cμ-to-Cδ CSR through both TD follicular and TI extrafollicular pathways involving AID. Circulating IgD interacted with basophils through a calcium-fluxing receptor that induced antimicrobial, opsonizing, inflammatory and immunostimulating factors such as cathelicidin, pentraxin-3 (PTX3), IL-1, IL-4 and B cell-activating factor of the TNF family member (BAFF; http://www.signaling-gateway.org/molecule/query?afcsid=A000383). By showing dysregulation of IgD class-switched B cells and IgD-armed basophils in autoinflammatory syndromes, our data indicate that IgD orchestrates an ancestral surveillance system at the interface between immunity and inflammation. This system may not only monitor invasion by respiratory bacteria, but also regulate antibody responses.
RESULTS
IgD CSR occurs in the upper respiratory mucosa
Human B cells release IgD in the blood as well as respiratory, salivary, lacrimal and mammary secretions1,4,25,27. We utilized immunohistochemistry, light microscopy and flow cytometry to elucidate the geography and phenotype of IgD+IgM− B cells producing IgD. Unlike intestinal, hepatic, lymph nodal, splenic and hematopoietic tissues, tonsillar and nasal tissues harbored abundant IgD+IgM− B cells that had topographic and morphologic features distinct from those of conventional IgD+IgM+ B cells (Fig. 1a,b). These latter occupied the follicular mantle but not the germinal center of secondary lymphoid follicles, had a small size, round shape, scant cytoplasm and large nuclei, and expressed either κ or λ light chains on their surface in addition to the pan-B cell molecules CD19, CD20, CD21, CD22, CD24, CD39, CD40, major histocompatibility complex (MHC) class I and class II molecules, BAFF-receptor (BAFF-R), transmembrane activator and CAML interactor (TACI), and the transcription factor Pax5 (Fig. 1c and Supplementary Fig. 1 online).
Figure 1. Upper respiratory mucosa B cells generate IgD+IgM− plasmablasts by undergoing Cμ-to-Cδ CSR in situ.
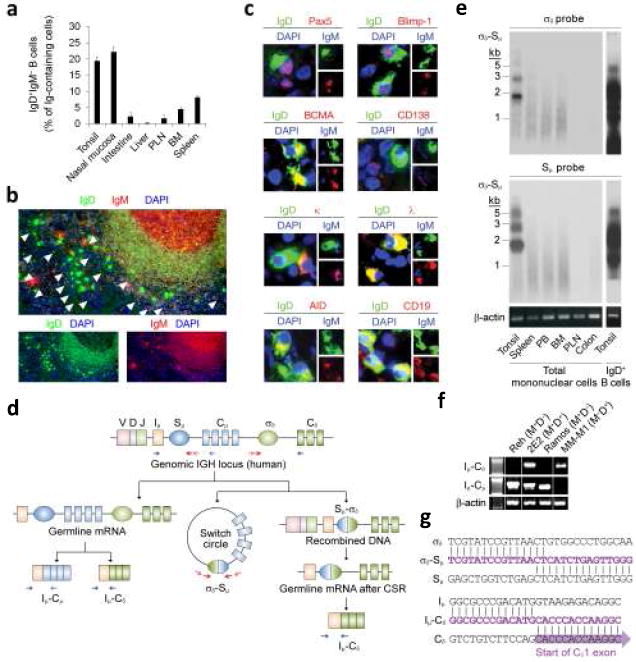
(a) Proportion of IgD+IgM− plasmablasts in various tissues calculated by immunofluorescence as described in Methods. PLN, peripheral lymph nodes; BM, bone marrow. (b) Tonsil tissue stained for IgD (green) and IgM (red). DAPI (blue) counterstains nuclei. Arrowheads point to IgD+IgM− plasmablasts. Original magnification ×10. (c) Representative IgD+IgM− B cells from tonsils stained for IgD (green), Pax-5, Blimp-1, BCMA, CD138, κ, λ, AID or CD19 (red), and IgM or DAPI (blue). (d) Diagram of CSR from Sμ to σδ. Germline Iμ-Cμ and Iμ-Cδ transcripts, σδ-Sμ switch circles, and post-switched Iμ-Cδ transcripts are shown. Arrows indicate primers. (e) Southern blots of σδ-Sμ switch circles PCR amplified from mononuclear cells of various tissues and hybridized with σδ or Sμ probes. Rightmost lane shows a typical multi-banded σδ-Sμ smear in enriched tonsillar IgD+ B cells. Kb, kilobases. (f) Germline Iμ-Cμ transcripts and germline or post-switch Iμ-Cδ transcripts in Reh IgD−IgM+ pre-B-like cells, 2E2 pre-germinal center IgD+IgM+ B cells, Ramos germinal center-like IgD−IgM+ B cells, and MM-M1 IgD+IgM− plasma cells. Genomic β-actin is a loading control. (g) Representative σδ-Sμ and Iμ-Cδ DNA sequences from tonsillar IgD+IgM− and IgD+IgM+ B cells, respectively. Panel a summarizes 1 of 3 experiments (bars indicate s.e.m.), whereas panels b, c, d, f, g and h show 1 of 5 experiments yielding similar results.
In contrast, IgD+IgM− B cells populated both germinal center and extrafollicular areas, had an ovaloid-elongated shape, medium-large size, abundant cytoplasm containing Cδ IgH chains preferentially coupled with λ light chains, and small and often picnotic eccentric nuclei. Consistent with prior data24,26,27, these IgD+IgM− plasmablasts accounted for up to 20-25% of antibody-forming plasma cells and for about 1.5-5% of total CD19+ B cells in tonsils. In spite of their plasma cell morphology, only a subset of IgD+IgM− B cells expressed B lymphocyte-induced maturation protein-1 (Blimp-1) and B cell maturation antigen (BCMA), and virtually none of them expressed CD138 (or syndecan-1), three hallmarks of mature plasma cells36. Furthermore, IgD+IgM− plasmablasts retained surface immunoglobulin as well as retained CD19, CD20, CD21, CD22, CD24, CD39, CD40, MHC class I and class II, BAFF-R, TACI and Pax5, which are usually down-regulated by terminally differentiated plasma cells36.
In addition to showing a bias for surface λ light chains, some or all IgD+IgM− plasmablasts expressed the activation molecule CD5 (ref. 37) the germinal center molecules CD10, CD38, CD77 and Ki-67 (ref. 24), the effector-memory molecule CD27 (ref. 38), as well as the DNA-editing enzyme AID, a hallmark of ongoing class switching18,19. Similar IgD+IgM− blasts were found in the peripheral blood (Supplementary Fig. 1 online), where they accounted for 0.5-1% of circulating CD19+ B cells. Our data indicate that the upper respiratory mucosa generates local and circulating IgD+IgM− plasmablasts specialized in IgD production. These cells express a complex phenotype that likely reflects a derivation from multiple follicular and extrafollicular precursors, including germinal center B cells.
Upper respiratory mucosa B cells undergo IgD CSR in situ
B cells express CH genes downstream of Cμ through a CSR reaction that relies on switch (S) regions14. Positioned upstream of each CH gene and an intronic (I) exon, S regions undergo germline transcription upon exposure of B cells to appropriate stimuli14. In addition to yielding IH-CH transcripts, germline transcription renders S regions substrate for DNA modifications by the CSR machinery, including AID14. Processing of these modifications into double-stranded DNA breaks is followed by fusion of these breaks through the non-homologous end joining pathway and looping-out deletion of the intervening DNA14. Ultimately, CSR generates post-switch Iμ-CH transcripts as well as an extrachromosomal S-S switch circle (Fig. 1d). Together with AID, switch circles are generally accepted as a marker of ongoing class switching, because they have a short half-life due to their rapid degradation by cellular nucleases12,14,18,19. In addition, switch circles do not replicate with the class switched B cell and therefore become rapidly diluted after CSR takes place.
Similar to pigs, sheep and cows39, humans have a Cδ gene preceded by a S-like σδ region that mediates CSR from Cμ to Cδ24,40. Using a nested polymerase chain reaction (PCR)-based strategy, we identified extrachromosomal σδ-Sμ switch circles in B cells from tonsils, but not peripheral blood, spleen, bone marrow, lymph nodes and intestine (Fig. 1e). CSR from Cμ to Cδ was associated with active Cδ gene transcription, because both the pre-germinal center IgD+IgM+ B cell line 2E2 and tonsillar follicular mantle IgD+IgM+ B cells contained chimeric germline Iμ-Cδ transcripts in addition to conventional germline Iμ-Cμ transcripts (Fig. 1f, g).
Although expressing Iμ-Cμ transcripts, neither the pre-B IgD−IgM+ cell line Reh nor the germinal center IgD−IgM+ B cell line Ramos contained Iμ-Cδ transcripts, suggesting that germline Cδ gene transcription is a developmentally regulated process beginning at a pre-germinal center stage of mature B cell differentiation. Finally, post-germinal center IgD+IgM− plasma cells lacked germline Iμ-Cμ transcripts, but contained post-switch Iμ-Cδ transcripts as a result of Sμ-to-σδ CSR-induced juxtaposition of the Iμ exon to Cδ exons. The identity of σδ-Sμ and Iμ-Cδ amplicons obtained from tonsillar IgD+ B cells was confirmed by DNA sequencing (Fig. 1g). Together with previous data showing AID expression by a fraction of tonsillar IgD+IgM− plasmablasts, these findings demonstrate that a subset of B cells actively undergo CSR from Cμ to Cδ in the microenvironment of the upper respiratory mucosa.
IgD CSR requires AID and occurs via multiple pathways
CSR usually involves engagement of CD40 on follicular B cells by CD40 ligand on T cells or engagement of TACI on extrafollicular B cells by BAFF or a proliferation-inducing ligand (APRIL) from innate immune cells20. Co-signals from cytokines are also required. IgD+IgM+ B cells induced σδ-Sμ switch circles, lost surface IgM, and secreted IgD upon exposure to CD40L, BAFF or APRIL plus either IL-15 and IL-21 (http://www.signaling-gateway.org/molecule/query?afcsid=A001258) or IL-2 and IL-21 (Fig. 2a-c and Supplementary Fig. 2 online), three cytokines produced by dendritic cells and T cells16,41,42. These effects were not due to expansion of IgD+IgM− plasmablasts present at the onset of the culture, because fresh IgD+IgM+ B cells lacked σδ-Sμ switch circles (not shown). In addition, CD40L, BAFF or APRIL alone or combined with either IL-2, IL-4, IL-10, IL-13, IL-15 or IL-21 elicited neither loss of surface IgM nor secretion of IgD in spite of being powerful inducers of B cell survival and proliferation (Supplementary Fig. 2 online and not shown). Of note, the specificity of in vitro induced σδ-Sμ switch circles was confirmed by sequencing (not shown).
Figure 2. Cμ-to-Cδ CSR occurs through both TD and TI pathways, requires AID, and leads to the production of IgD antibodies that bind to respiratory bacteria.
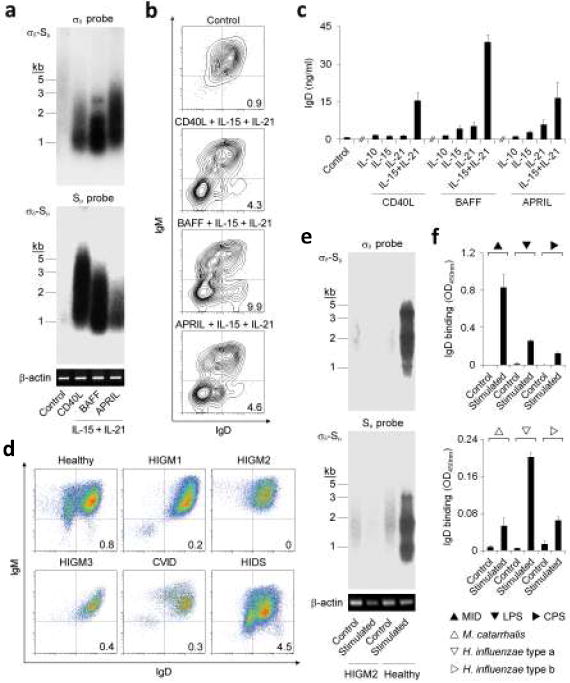
(a) σδ-Sμ switch circles from circulating IgD+IgM+ B cells stimulated with or without CD40L, BAFF or APRIL plus IL-15 and IL-21 for 4 d. Genomic β-actin is a loading control. Kb, kilobases. (b, c) Flow cytometric analysis of surface IgD and IgM and ELISA of secreted IgD from circulating IgD+IgM+ B cells stimulated as in a for 7 d. Control indicates medium alone. (d) Percentage of circulating IgD+IgM− B cells in a healthy donor and patients with TNFSF5 (HIGM1), AICDA (HIGM2), TNFRSF5 (HIGM3), TNFRSF13b (CVID) or MVK (HIDS) gene defects. (e) Induction of σδ-Sμ switch circles in circulating mononuclear cells from a HIGM2 patient and a control healthy donor incubated with medium alone (control) or BAFF, IL-15 and IL-21 (stimulated) for 4 d. An anti-IgM antibody was added to optimize the activation and expansion of IgD+IgM+ B cells. (f) Binding of secreted IgD to MID, CPS, LPS, M. catarrhalis, and H. influenzae type a or type b as determined by ELISA. IgD secretion was obtained by incubating circulating IgD+IgM+ B cells with or without BAFF, IL-15 and IL-21 for 7 d. Panels a, b, d and e show 1 of 5 experiments yielding similar results, whereas panels c and f summarize 3 experiments (bars indicate s.e.m.; *, p < 0.05).
The IgD-inducing function of CD40L, BAFF and APRIL was further studied in patients with primary immunodeficiencies. TNFSF5 and TNFRSF5 gene defects, which impair CD40L and CD40 signaling in patients with hyper-IgM type-1 (HIGM1) and HIGM3 syndromes43 respectively, led to a reduction of both circulating and follicular IgD+IgM− plasmablasts, but partially spared extrafollicular IgD+IgM− plasmablasts (Fig. 2d and Supplementary Fig. 3 online), suggesting that CD40L-CD40 interaction is indispensable for the generation of systemic but not mucosal IgD-producing effector-memory B cells.
TNFRSF13b gene defects, which impair TACI signaling in a subset of patients with common variable immunodeficiency (CVID)44,45, were also associated with a reduction of circulating IgD+IgM− plasmablasts. AICDA gene defects, which impair AID function in patients with HIGM2 syndrome43, caused a reduction of both circulating and follicular IgD+IgM− plasmablasts. This reduction was linked to a lack of functional AID protein, because B cells from HIGM2 patients neither induced σδ-Sμ switch circles nor underwent loss of surface IgD upon exposure to appropriate stimuli (Fig. 2e and Supplementary Fig. 3 online). Of note, HIGM2 as well as HIGM3 syndromes were associated with an increased generation of double-producing IgD+IgM+ plasmablasts at both follicular and extrafollicular mucosal sites, possibly reflecting the activation of a CSR-independent pathway for compensatory IgD synthesis. Thus, IgD CSR requires AID and occurs via both T cell-dependent (TD) and T cell-independent (TI) pathways in vivo.
IgD recognizes respiratory bacteria
Given their origin from the upper respiratory mucosa, IgD+IgM− plasmablasts may release IgD antibodies that recognize airborne bacteria. Consistent with this possibility, IgD secreted by stimulated IgD+IgM− plasmablasts bound to the Gram-negative bacteria Moraxella catarrhalis and Haemophilus influenzae type a and type b (Fig. 2f). As shown by others46, IgD also bound the virulence factor M. catarrhalis IgD-binding protein (MID, also known as Hag), which facilitates the adhesion of M. catarrhalis and H. influenzae to mucosal epithelial cells. Furthermore, secreted IgD reacted against lipopolysaccharide (LPS), a key parietal component of Gram-negative bacteria, and capsular polysaccharide (CPS), an essential protective factor for encapsulated bacteria such as H. influenzae. Thus, IgD from IgD+IgM− B cells recognizes respiratory bacteria and their products.
IgD binds to circulating basophils
Humans have 15-300 μg/ml of circulating IgD1,27, which likely originates from IgD+IgM− plasmablasts entering the bloodstream from inductive sites in the upper respiratory mucosa. To elucidate the function of circulating IgD, we examined its in vivo interaction with Fc receptor-blocked peripheral blood leukocytes by flow cytometry. Using a goat polyclonal antibody highly specific for human IgD and with no Fc portion and no IgG cross-reactivity, we found that basophils had abundant surface IgD, whereas T cells, NK cells, monocytes, myeloid dendritic cells, neutrophils, eosinophils and plasmacytoid dendritic cells exhibited no or little surface IgD (Fig. 3a and Supplementary Fig. 4 online). Basophils were consistently positive for surface IgD binding also when stained with a monoclonal anti-IgD (Supplementary Fig. 4 online).
Figure 3. IgD binds to basophils and mast cells in vivo.
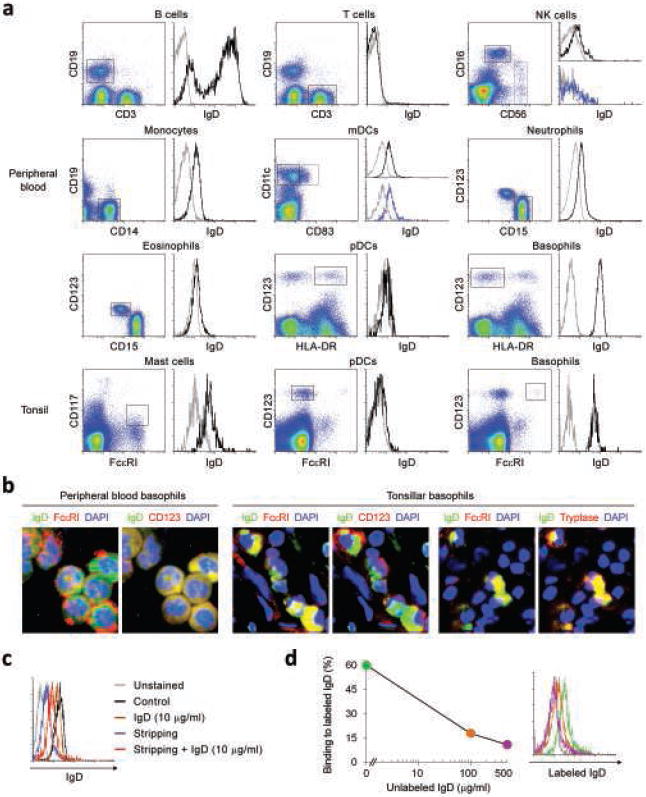
(a) Flow cytometric analysis of IgD on circulating or tonsillar CD19+CD3− B cells, CD19−CD3+ T cells, CD16highCD56low or CD16lowCD56high NK cells, CD19−CD14+ monocytes, CD11c+CD83− or CD11c+CD83+ myeloid dendritic cells (mDCs), CD15+CD123− neutrophils, CD15+CD123low eosinophils, CD123+HLA-DR+ or CD123+FcεRIlow plasmacytoid dendritic cells (pDCs), CD123+HLA-DR− or CD123+FcεRI+ basophils, and CD117+FcεRI+ mast cells. Gray and black histograms depict control unstained cells and IgD, respectively. A F(ab’)2 pAb was used to detect IgD. Dead cells were excluded using 7-AAD staining. (b) Immunofluoresence analysis of circulating and tonsillar basophils stained for IgD (green) and FcεRI, CD123 or tryptase (red). DAPI (blue) counterstains lobated nuclei of basophils. A F(ab’)2 pAb was used to detect IgD. (c) Flow cytometry of IgD levels on circulating basophils before (black histogram) and after exposure to exogenous monoclonal IgD (10 μg/ml, brown histogram), treatment with acidic buffer (blue histogram), and treatment with acidic buffer followed by addition of exogenous IgD (red histogram). A F(ab’)2 pAb was used to detect IgD. Unstained basophils were used as control (gray histogram) and dead cells were excluded using 7-AAD staining. (d) Binding of labeled monoclonal IgD to circulating basophils stripped of endogenous IgD and incubated with 0 μg/ml (green histogram), 100 μg/ml (orange histogram) or 500 μg/ml (purple histogram) of unlabelled monoclonal IgD. A F(ab’)2 pAb was used to detect IgD. Unstained basophils were used as control (gray histogram) and dead cells were excluded using 7-AAD staining. Panels a-d show 1 of 5 experiments yielding similar results.
Compared to polyclonal anti-IgD, monoclonal anti-IgD detected less IgD on circulating basophils as well as basophilic KU812 cells incubated with soluble IgD. In addition, monoclonal anti-IgD tendentially detected less IgD on circulating neutrophils, although there was inter-individual variability. These results likely reflected the wide range of serum IgD concentrations present in different individuals as well as the fact that monoclonal anti-IgD reagents allow a less complete detection of IgD due to their ability to detect only individual monomorphic Cδ epitopes and not multiple Cδ epitopes and conformations as the anti-IgD pAb does47. In this regard, transmembrane and secreted IgD preferentially associate with κ and λ light chains, respectively4. This phenomenon, known as “IgD paradox”, likely generates conformational differences in Cδ that further limit the recognition of secreted IgD by anti-IgD mAbs. Of note, one of the monoclonal anti-IgD reagents used in this study failed to effectively stain both soluble IgD bound on KU812 cells and transmembrane IgD on B cells (not shown), suggesting poor fluorochrome conjugation and/or intrinsically weak reactivity against Cδ.
IgD was detected not only on the surface, but also in the cytoplasm of circulating as well as mucosal basophils (Fig. 3b), suggesting that basophils internalize IgD-bound immune complexes. Mast cells, a mucosal immunocyte functionally related to basophils48,49, were also positive for IgD (Supplementary Fig. 5 online). Addition of exogenous monoclonal IgD did not increase the surface IgD binding on basophils, indicating saturation of binding sites by endogenous IgD (Fig. 3c). However, monoclonal IgD could bind to basophils after stripping of endogenous IgD. Finally, addition of increasing amounts of unlabeled monoclonal IgD progressively diminished the binding of labeled monoclonal IgD to basophils stripped of endogenous IgD (Fig. 3d).
The specific tropism of IgD for basophils and mast cells was confirmed in vitro by showing binding of physiological concentrations (from 0.5 to 50 μg/ml) of polyclonal or monoclonal IgD molecules to the pre-basophilic cell line KU812 and to the mastocytoid cell lines HMC-1 and LAD-2 (Fig. 4a and Supplementary Fig. 6 online). This binding was highly specific, because it was detected with both polyclonal and monoclonal anti-IgD reagents with no cross-reactivity for IgG. Except for the monocytic cell line U937, none of the T, NK and myeloid cell lines tested bound IgD. KU812 and LAD-2 cells up-regulated IgD binding upon exposure to IL-3 and/or IL-4 (Fig. 4b and Supplementary Fig. 7 online), two cytokines involved in basophil and mast cell differentiation48,49. In addition to augmenting their IgD-binding activity, IL-3 stimulated KU812 cells to increase surface expression of CD124 (IL-4 receptor) and FcεRI (IgE receptor) and decrease expression of CD117 (c-kit receptor), whereas IL-4-stimulated LAD-2 cells increased CD123 (IL-3 receptor) expression. Binding of labeled IgD to KU812 or HMC-1 cells was saturable and could be competed by unlabeled IgD, but not IgG or IgE, whereas IgA had a minimal inhibitory effect (Fig. 4c, d).
Figure 4. IgD binds to basophilic and mast cell lines in vitro.
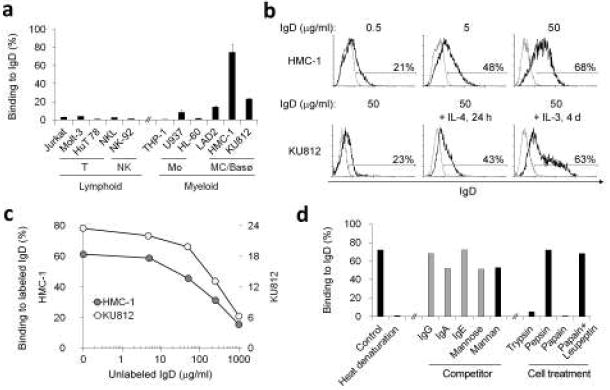
(a) Binding of monoclonal IgD (50 μg/ml) to various lymphoid and myeloid cell lines. A F(ab’)2 pAb was used to detect IgD. Mo, MC and Basø indicate monocytic, mast cell and basophilic cell lines, respectively. (b) Binding of increasing amounts (0.5, 5 and 50 μg/ml) of monoclonal IgD to the mast cell line HMC-1 and binding of monoclonal IgD (50 μg/ml) to the basophilic cell line KU812 before and after treatment with IL-4 for 1 d and with IL-3 for 4 d. Control indicates medium alone. A F(ab’)2 pAb was used to detect IgD. Gray histogram depicts background fluorescence of cells that were not incubated with monoclonal IgD. (c) Binding of fluorochrome-conjugated monoclonal IgD to HMC-1 or KU812 cells in the presence of increasing amounts (0, 5, 50, 250 or 1000 μg/ml) of unlabeled monoclonal IgD. (d) Binding of untreated or denatured monoclonal IgD (50 μg/ml) to HMC-1 cells pre-incubated or not with IgG, IgA, IgE, mannose, mannan, trypsin, pepsin, papain, or papain plus leupeptin. A F(ab’)2 pAb was used to detect IgD. In all the experiments dead cells were excluded using 7-AAD staining. Panel a summarizes 3 experiments (bars indicate s.e.m.), whereas panels b-d show 1 of 3 experiments yielding similar results.
Since both IgA and IgD are highly mannosylated50, IgD could utilize mannose to enhance its binding to basophils and mast cells. Accordingly, mannose and mannan slightly inhibited IgD binding to target cells. Finally, IgD binding was abolished by denaturing IgD or by pre-treating target cells with trypsin and papain, but not pepsin. Thus, IgD binds to basophils and mast cells through a protease-sensitive, IL-3 or IL-4-inducible receptor distinct from IgG, IgA and IgE receptors.
IgD binding to basophils is evolutionarily conserved
IgD is an ancestral antibody class expressed by early vertebrates, including teleost fish3-5. Knowing that channel catfish (Ictalurus punctatus) express both transmembrane and secreted IgD molecules5, it was relevant that the peripheral blood of some of these animals contained plasmablast-like IgD+IgM− B cells expressing transcripts for both transmembrane and secreted Cδ IgH chains, but not for transmembrane and secreted Cμ IgH chains (Supplementary Fig. 8 online). Flow cytometric studies with specific monoclonal antibodies showed catfish also exhibit circulating granular leukocytes armed with surface IgD but not IgM. These IgD+IgM− granulocytes contained neither transcripts for the T cell antigen receptor nor transcripts for transmembrane and secreted Cδ and Cμ IgH chains and did not react with a specific monoclonal antibody to neutrophils (not shown), suggesting their affiliation to a distinct granulocytic subset. These data suggest that binding of B cell-derived IgD to granulocytes is part of an evolutionarily conserved and potentially important immune pathway.
IgD-activated basophils produce B cell-activating factors
To elucidate its functions, we cross-linked IgD on basophils with a specific monoclonal anti-IgD in the presence or absence of the basophil maturation factor IL-3 (refs.48,49). Controls were performed with an isotype-matched irrelevant antibody or an antibody to IgE. IgE binds basophils through a high-affinity FcεRI receptor that triggers degranulation and histamine release upon cross-linking48,49. Unlike IgE cross-linking, IgD cross-linking elicited neither surface up-regulation of the granular molecule CD63 nor histamine release regardless of the presence of IL-3 (Fig. 5a,b and Supplementary Fig. 9 online). Although less efficient than IgE cross-linking in inducing membrane-bound CD40L and BAFF, IgD cross-linking was more efficient than or as efficient as IgE cross-linking in inducing soluble IL-4, IL-13 and BAFF, membrane-bound APRIL, and soluble IL-8 and CXCL10 (Fig. 5c and Supplementary Fig. 9 online), two chemokines active on monocytes, neutrophils and T cells. These effects were potentiated by IL-3.
Figure 5. Basophils release immunostimulating and pro-inflammatory factors upon IgD cross-linking.
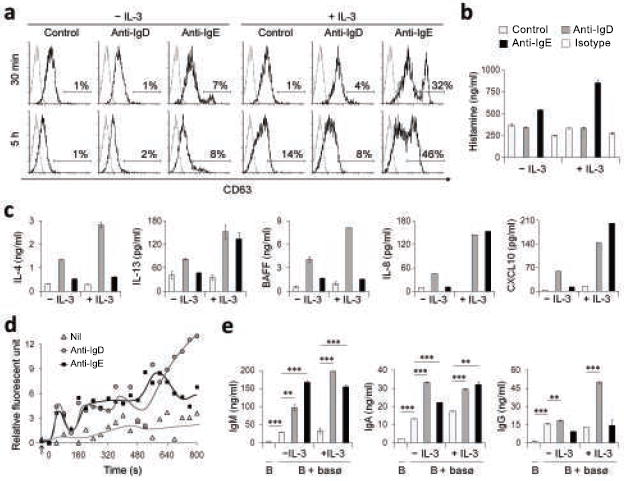
(a) Flow cytometry of CD63 on basophils exposed to microbeads alone (control), microbead-bound monoclonal anti-IgD, or microbead-bound monoclonal anti-IgE for 30 min or 5 h in the presence or absence of IL-3. (b) ELISA of histamine from basophils exposed to microbeads alone (open bar), microbead-bound monoclonal anti-IgD (gray bar), microbead-bound monoclonal anti-IgE (black bar), or microbead-bound isotype-matched control monoclonal antibody (striped bar) for 30 min in the presence or absence of IL-3. (c) ELISA of IL-4, IL-13 or BAFF from basophils exposed to microbeads alone (open bar), microbead-bound monoclonal anti-IgD (gray bar), or microbead-bound monoclonal anti-IgE (black bar) for 16 h in the presence or absence of IL-3. IL-8 and CXCL10 were measured after 48 h. (d) Intracellular Ca2+ levels of basophils treated as in c. The arrow indicates addition of the cross-linking reagent and 0 sec indicates the start of the kinetic measurement. (e) IgM, IgA and IgG production by peripheral blood IgD+IgM+ B cells exposed for 7 d to basophils treated as in c. Panels a and d show 1 of 3 experiments yielding similar results, whereas panels b, c and e summarize 3 experiments (bars indicate s.e.m.; *, p < 0.01; **, p < 0.001).
Activated basophils usually release mediators by undergoing calcium-dependent degranulation upon IgE cross-linking by antigen48,49. Although unable to induce degranulation, IgD cross-linking triggered intracellular calcium fluxes more sustained than those induced by IgE (Fig. 5d). The basophil-stimulating activity of IgD was further confirmed by experiments showing that, in the presence of IgD cross-linking, basophils pre-treated or not with IL-3 acquired the capability of inducing IgM secretion as well as IgG and IgA class switching in B cells (Fig. 5e). Of note, the B cell-licensing function of IgD-cross-linked basophils was largely dependent on BAFF and APRIL, because it was abolished by a soluble TACI-Ig decoy receptor (not shown). As shown by others32, IgE cross-linked basophils elicited B cell activation mostly via CD40L (not shown). Thus, IgD stimulates basophils through a calcium-fluxing receptor that induces multiple cytokines, including the B cell-activating factors IL-4, IL-13, BAFF and APRIL.
IgD-stimulated basophils produce antimicrobial factors
In addition to adaptive B cell responses, IgD-activated basophils may trigger innate antimicrobial responses48,49. Indeed, IgD-cross-linked basophils induced antimicrobial, opsonizing and alarm-signaling factors51-53, including DEFB103A, CAMP, SPAG11A/F, SPAG11D/G, PTX3 and CRP transcripts for β-defensin 3, cathelicidin, sperm associated antigen 11 (SPAG11) A/F isoforms, SPAG11 D/G isoforms, PTX3, and C-reactive protein (CRP), respectively (Fig. 6a). In addition, IgD-stimulated basophils released the cathelicidin-derived peptide LL-37 (Fig. 6b). Consistent with these results, supernatants from IgD-cross-linked basophils inhibited the replication of M. catarrhalis and H. influenzae (Fig. 6c). In contrast, IgE cross-linked basophils had little or no antimicrobial activity. Thus, IgD cross-linking activates powerful antimicrobial, opsonizing and alarm-signaling programs in basophils.
Figure 6. Basophils release antimicrobial factors upon IgD cross-linking.
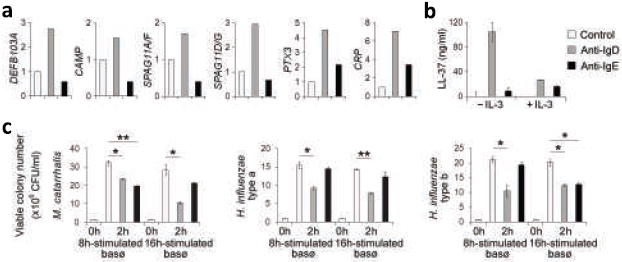
(a) QRT-PCR of DEFB103A, CAMP, SPAG11A/F, and SPAG11D/G transcripts from basophils exposed to microbeads alone (control, open bar), microbead-bound monoclonal anti-IgD (gray bar), or microbead-bound monoclonal anti-IgE (black bar) for 6 h. PTX3 and CRP transcripts were measured after 16 h. mRNAs were normalized to ACTB mRNA. (b) ELISA of LL-37 from basophils stimulated as in a in the presence or absence of IL-3 for 8 h. (c) Growth of Moraxella catarrhalis and Haemophilus influenzae type-a and type-b upon 2-h exposure to culture supernatants from basophils stimulated as in a for 8 h or 16 h. CFU, colony forming unit. Panels a-c summarize 3 experiments (bars indicate s.e.m.; *, p < 0.03; **, p < 0.02; ***, p < 0.01).
Basophils are dysregulated in hyper-IgD patients
To elucidate the relationship between IgD and basophils in vivo, we analyzed IgD class-switched B cells and IgD-armed basophils from patients with hyper-IgD syndrome (HIDS), TNF receptor-associated periodic fever syndrome (TRAPS), Muckle-Wells syndrome (MWS), and periodic fever-aphthous stomatitis-pharyngitis-cervical adenitis (PFAPA) syndrome. Although characterized by diverse genetic defects, these autoinflammatory disorders share a perturbation of the mechanisms that initiate and control the inflammatory reaction. Such a perturbation causes periodic attacks of fever and tissue damage as well as abnormal IL-1 and TNF release and elevated IgD production4,54.
Compared to healthy subjects, HIDS, TRAPS, MWS, and PFAPA syndrome patients had more circulating and mucosal IgD+IgM− plasmablasts (Fig. 7a,b and Supplementary Fig. 10 online), suggesting that switching from IgM to IgD is augmented in autoinflammatory syndromes. These disorders were also associated with fewer circulating but more mucosal IgD-armed basophils and probably mast cells (Fig. 7c,d and Supplementary Fig. 10 online), suggesting that elevated IgD production enhances mucosal homing and proliferation of basophils and mast cells. Consistent with this possibility, mucosal basophils exhibited signs of hyperactivation, including strong BAFF, APRIL and LL-37 expression.
Figure 7. Increased IgD class-switched plasmablasts and IgD-armed basophils in inflamed tissues from patients with periodic fever syndromes.
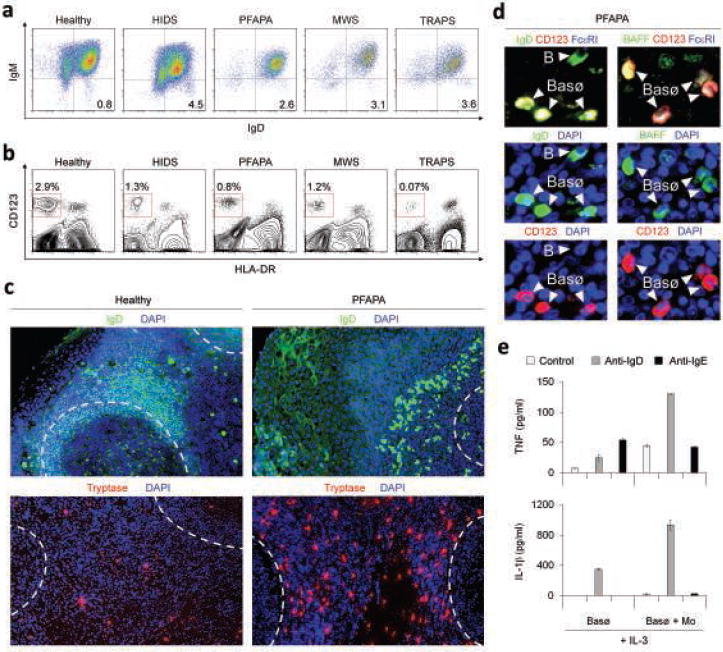
(a, b) Flow cytometric analysis of circulating IgD+IgM− B cells and CD123+HLA-DR− basophils in a healthy subject and hyper-IgD patients with MVK (HIDS), unknown (PFAPA), NALP3 (MWS) and TNFRSF1A (TRAPS) gene defects. Numbers indicate percentage of IgD+IgM− B cells in CD19+ B cells and of CD123+HLA-DR− basophils in mononuclear cells. (c) Immunofluorescence analysis of a tonsil tissue specimen from healthy individual or a PFAPA syndrome patient stained for IgD (green), tryptase (red) and DAPI (blue). Dashed lines demarcate lymphoid follicles. (d) BAFF (green) expression by basophils co-stained for IgD (green), CD123 (red) and FcεRI (blue). Arrowheads show both IgD+CD123− plasmablasts (B) and IgD+CD123+ basophils (Basø). (e) ELISA of TNF and IL-1β from basophils cultured with IL-3 and microbeads alone (control, open bar), microbead-bound monoclonal anti-IgD (gray bar) or microbead-bound monoclonal anti-IgE (black bar) for 48 h in the presence or absence of monocytes. Panels a-d show 1 of 3 experiments yielding similar results, whereas panel e summarizes 3 experiments (bars indicate s.e.m.).
Knowing that IL-1β and TNF are IgD-inducible pro-inflammatory cytokines involved in autoinflammatory syndromes54,55, we wondered whether IgD triggering increases IL-1 and TNF production by basophils. Upon IgD cross-linking, IL-3-treated basophils from healthy subjects released both IL-1β and TNF and this release further increased in the presence of monocytes (Fig. 7e), a cell type heavily involved in inflammation. In contrast, IgE cross-linking had little or no IL-1- and TNF-inducing activity. Overall, our data indicate that IgD and basophils orchestrate an ancestral surveillance system at the interface between immunity and inflammation (Supplementary Fig. 11 online). A dysregulation of this system may contribute to the pathogenesis of autoinflammatory syndromes with periodic fever.
DISCUSSION
In this study we have reported that human B cells from the upper respiratory mucosa actively class switch from Sμ to σδ, thereby generating local and circulating IgD antibodies highly reactive to respiratory bacteria. Circulating IgD bound to basophils through a calcium-mobilizing receptor that activated antimicrobial, opsonizing, pro-inflammatory and B cell-stimulating programs upon cross-linking. Both IgD class-switched B cells and IgD-armed basophils were dysregulated in patients with autoinflammatory syndromes and periodic fever, indicating that IgD orchestrates an ancestral surveillance system at the interface between immunity and inflammation.
Human B cells release IgD antibodies in the blood as well as respiratory, salivary, lacrimal and mammary secretions1,4,25-28. The regulation and function of these antibodies remain unknown. We found a large fraction of IgD+IgM− plasmablasts in the upper respiratory mucosa. These plasmablasts originated in situ from an active process of Sμ-to-σδ CSR that involved germline Iμ-Cδ transcription, required AID expression, and occurred through either a TD follicular pathway involving engagement of CD40 on B cells by CD40L on T cells or a TI extrafollicular pathway involving engagement of TACI on B cells by BAFF or APRIL from innate immune cells, possibly including epithelial and dendritic cells18-20.
The composite ontogeny of mucosal IgD+IgM− plasmablasts was consistent with the fact that they expressed phenotypic traits usually associated with extrafollicular activated B cells or follicular germinal center B cells. Like these B cell types, mucosal IgD+IgM− plasmablasts produce both polyreactive and monoreactive IgD antibodies encoded by unmutated and mutated V(D)J genes, respectively28, possibly reflecting the need of the upper respiratory mucosa to mount maximally diversified IgD responses for frontline defense purposes. Such IgD responses likely entail the stimulation of specific λ+ B cell precursors by a unique cocktail of mucosal signals comprising IL-21 and IL-2 or IL-21 and IL-15, three cytokines released by dendritic cells and T cells16,18,41,42. Together with CD40L, BAFF and APRIL, these cytokines might account for the massive V(D)J gene diversification and oligoclonal expansion of IgD+IgM− B cells previously observed in tonsils24,26. Why these cells are strongly biased for λ light chain expression remains unclear. One possibility is that the formation of fully functional soluble IgD antibodies requires pairing of Cδ IgH with λ light chains.
IgD+IgM− plasmablasts in the peripheral blood are likely in transit from inductive sites in the upper respiratory mucosa to distant mammary, salivary, lacrimal, respiratory, tubal and auditive sites27,38,56. At these effector sites, soluble IgD antibodies might enhance immune protection against local pathogens and commensal bacteria. Indeed, IgD released by stimulated IgD+IgM− B cells showed strong binding activity against respiratory bacteria and their products, including M. catarrhalis, H. influenzae, LPS, CPS and MID. Although unable to utilize the polymeric Ig receptor27, which translocates polymeric IgA and IgM from the basolateral to the apical surface of epithelial cells, monomeric IgD could reach mucosal surfaces via an alternative epithelial transporter, perhaps similar to that mediating transcytosis of monomeric IgG27. Once on the mucosal surface, IgD could take advantage of its poor complement-inducing activity to mediate immune exclusion in a non-inflammatory fashion4. Alternatively, IgD may confine its defensive functions to the subepithelial area of the upper respiratory tract.
In addition to mediating mucosal immunity, IgD may enhance systemic immunity by ‘educating’ circulating innate effector cells as to the antigenic composition of the upper respiratory tract. Indeed, we detected abundant IgD on basophils, but not on other leukocytes, except B cells. In the presence of IgD cross-linking, basophils acquired the ability to inhibit the growth of respiratory bacteria, presumably through the induction of β-defensin 3, SPAG11 isoforms, PTX3, CRP and cathelicidins. These antimicrobial and opsonizing factors also have immunostimulating and alarm-inducing activity51-53. Thus, IgD-armed basophils may function as circulating sentinels capable of triggering rapid innate and adaptive immune responses upon sensing pathogens from the upper respiratory tract.
The primordial nature of this immune surveillance system was confirmed by the presence of both IgD-producing (IgM-negative) plasmablasts and IgD-armed granulocytes in channel catfish. That IgD must mediate some important and evolutionarily conserved functions complementary to those of IgM was also suggested by the presence of double-producing IgD+IgM+ plasmablasts in patients with HIGM syndromes. This compensatory pathway for IgD production could explain why serum IgD is conserved or even elevated in HIGM and other primary immunodeficiencies resulting from an overall or selective impairment of CSR, including CVID and selective IgA deficiency4,25,27.
Consistent with the B cell-activating, antibody-inducing and basophil-stimulating properties of IgD and anti-IgD in vivo29,30,57, we found that basophils up-regulated the production of IL-4, IL-13, BAFF and APRIL upon IgD cross-linking in vitro. This effect correlated with the acquisition of B cell-licensing functions, including the capability to elicit IgM secretion as well as IgG and IgA class switching. In addition to promoting TI humoral immunity, IL-4 and IL-13 from IgD-activated basophils may initiate TD humoral immunity by triggering the formation of TH2 (CD4+ T helper type-2) cells with B cell-helper activity34,35. Thus, it is likely that basophils respond to IgD-reactive pathogens by eliciting not only innate, but also adaptive immune responses. Such responses would take place in both systemic and mucosal districts. Indeed, while earlier work shows migration of basophils to draining lymph nodes34, we detected basophils in the upper respiratory mucosa, particularly in the presence of inflammation.
Our finding that IgD-stimulated basophils release an obligatory B cell survival factor such as BAFF would provide a mechanistic explanation to prior studies showing that IgD deficiency leads to a contraction of the peripheral B cell compartment in mice8,58. As suggested by the poly- and autoreactivity of a large fraction of IgD antibodies28, basophils could undergo tonic or intermittent release of BAFF upon sensing circulating foreign or autologous antigens through pre-bound IgD. The ensuing receptor-dependent internalization of antigen-IgD complexes might account for the cytoplasmic accumulation of IgD observed in ex vivo isolated basophils.
In addition to basophils, IgD bound to mast cells through an IL-4-inducible calcium-fluxing receptor distinct from IgG, IgA and IgE receptors. Indeed, basophil-like and mast cell-like lines pre-treated with IgG, IgA and IgE retained IgD binding activity. A minimal but reproducible inhibition was induced by IgA, the only antibody isotype together with IgD exhibiting CH region-associated O-linked carbohydrates50, which may therefore contribute to the binding of IgD to its receptor sites. The basophilic cell line KU812 increased IgD binding in response to IL-3, a mast cell-derived cytokine that enhances basophil activation and recruitment to lymphoid and mucosal effector sites48,49. Since IL-3 also enhances basophil activation by IgD, basophils and mast cells may form an IgD-mediated IL-3-dependent axis for the amplification of both systemic and mucosal immune responses.
The interaction of IgD with basophils and mast cells was sensitive to the proteolytic activity of trypsin and papain, but not to that of pepsin. Previous studies show that papain initiates IgE responses by inducing IL-4 release from basophils through an enzymatic mechanism34. One possibility is that IgD and papain interact with similar receptors on basophils. Importantly, IgD clearly stimulates basophils through a pathway distinct from that induced by IgE. Indeed, in spite of eliciting IgE-like intracellular calcium fluxes, IgD cross-linking did not induce degranulation and histamine release. Furthermore, IgD was less effective than IgE cross-linking in up-regulating surface CD40L, a powerful class switch-inducing factor32. Conversely, IgD was generally more effective than IgE cross-linking in inducing antimicrobial, immunostimulating and pro-inflammatory factors such as IL-1β, TNF, IL-8 and CXCL10.
The pro-inflammatory function of IgD was further suggested by the analysis of autoinflammatory syndromes, a group of disorders characterized by periodic attacks of fever and inflammation as well as exaggerated IL-1β and IgD production4,54. We found more class-switched IgD+IgM− plasmablasts and more mucosal IgD-armed basophils in patients with HIDS, TRAPS, MWS or PFAPA syndrome. In these subjects, mucosal basophils could enhance inflammation by releasing IL-1β and LL-37 in response to microbial antigens with IgD-binding activity. Consistent with this possibility, periodic fever is often triggered by infections or immunization, particularly in HIDS patients54. Similar IgD-binding antigens could also increase basophil expression of B cell-activating and class switch-inducing factors such as CD40L, BAFF and APRIL. This effect could account for the elevated serum IgG and IgA titers often observed in individuals with HIDS, TRAPS, MWS and PFAPA syndrome4,54. Finally, the fact that IgD predominantly exerts its activity as a basophil-bound antibody could explain why periodic attacks of fever and inflammation do not predictably correlate with serum IgD in patients with HIDS54.
In summary, human B cells may produce IgD to instruct basophils as to the antigenic composition of the upper respiratory tract. This evolutionarily conserved immune surveillance system would not only monitor systemic invasion by airborne pathogens, but also regulate B cell homeostasis, antibody production and inflammation.
METHODS
Fresh cells
Peripheral blood mononuclear cells from buffy coats of healthy subjects were purchased at the New York Blood Center. Additional mononuclear cells were isolated from the peripheral blood of patients with primary immunodeficiencies, mastocytosis or autoinflammatory syndromes. Tonsillar and pulmonary mononuclear cells were obtained from tissue specimens of patients undergoing resection of hypertrophic tonsils or lung neoplastic lesions at New York Presbyterian Hospital-Weill Cornell Medical Center. The Institutional Review Board of Weill Medical College of Cornell University approved the use of blood, tonsil and lung specimens for this study, and patients provided informed consent. IgD+IgM+ B cells and CD14+ monocytes were magnetically sorted from peripheral blood and tonsillar mononuclear cells as reported16. In the experiments involving induction of σδ-Sμ switch circles and loss of surface IgM, peripheral blood IgD+IgM+ B cells were depleted of antigen-experienced plasmablasts using CD27-targeting microbeads (Miltenyi Biotec). For morphological studies, IgD+IgM+ and IgD+IgM− B cells were FACSorted from tonsillar mononuclear cells. Granulocytes were separated from peripheral blood mononuclear cells using Histopaque-1119 and Histopaque-1077 double gradients (Sigma). Untouched basophils were purified by negative selection from peripheral blood mononuclear cells using a Basophil Isolation Kit (Miltenyi Biotec). Mast cells were enriched from lung mononuclear cells using a biotin-conjugated AER-37 monoclonal antibody (mAb) to FcεRI (eBiosciences) and anti-biotin microbeads Miltenyi Biotec). Catfish peripheral blood lymphocytes were obtained and stained as reported5,59. Catfish IgM and IgD were detected with 9E1 and 7D11 mAbs, respectively.
Cell lines
The T cell lines Jurkat and Molt-3, the B cell lines Reh, 2E2 and Ramos, the myeloid cell lines THP-1, U937 and HL60, and the pre-basophilic cell line KU812 were cultured in complete RPMI 1640 medium (Gibco). The T cell line HuT78 was cultured in complete Iscove’s DMEM (Mediatech). The NK cell line NKL was cultured in complete RPMI 1640 medium further supplemented with 1% non-essential amino acids (Mediatech) and 200 U/ml IL-2. The NK cell line NK-92 was cultured in MEM-α medium with 2 mM glutamine, 1.5 mg/ml NaHCO3, 0.1 mM β-mercaptoethanol, 0.02 mM folate, 0.2 mM inositol, 12.5% FBS and 12.5% horse serum. The mastocytoid cell line LAD2 (from A. Kirshenbaum and D. Metcalfe, National Institutes of Health) was cultured in serum-free Stem Pro-34 medium (Invitrogen) containing 100 ng/ml of stem cell factor (Peprotech). The mastocytoid cell line HMC-1 was cultured in Iscove’s DMEM (Mediatech) with 10% FBS, 2 mM glutamine, 0.02% α-thioglycerol and antibiotics.
Tissues and immunohistochemistry
Frozen and paraffin-embedded tissue sections from tonsils, nasal cavities, intestine, liver, lymph nodes, bone marrow and spleen from healthy subjects or patients with primary immunodeficiencies or autoinflammatory syndromes were stored at −80 °C and 25 °C, respectively. Sections were blocked with a saturating concentration of purified human IgG and stained for immunohistochemical analysis with various combinations of antibodies (Supplementary Table 1 online) as described previously18. Primary antibodies with irrelevant binding activity and appropriate secondary reagents were utilized to test the specificity of tissue stainings as previously described33. The proportion of IgD+IgM− plasmablasts in different organs was determined by sequentially staining tissue sections with a polyvalent goat polyclonal antibody (pAb) to human Igs (Cappel), an Alexa Fluor 647-conjugated secondary pAb to goat Igs, a fluorescein-conjugated goat F(ab’)2 pAb to human IgD (Southern Biotech), and a biotin-conjugated goat F(ab’)2 pAb to human IgM. Nuclei were visualized with DAPI, 4’,6-diamidine-2’-phenylindole dihydrochloride (Boehringer Mannheim). The following formula was used: number of cytoplasmic IgD+IgM− B cells / number of cytoplasmic Ig+ B cells × 100. Sections adjacent to those analyzed for IgM, IgD and total Ig were stained with 7G3 mAb to CD123. IgD+ cells with no clear plasmacytoid morphology, but with granular cytoplasm, lobated nucleus and/or CD123 expression were scored as basophils and therefore excluded from the analysis. Follicular mantle IgD+IgM− B cells were also excluded from the analysis. These B cells have bright IgD and dim IgM expression due to the higher stability and less rapid turnover of V(D)J-Cδ transcripts compared to V(D)J-Cμ transcripts4. Pseudocolor images were composed using Metamorph 7.5 (Molecular Device) and edited using Photoshop CS2 (Adobe).
Patients
HIGM patients had various TNFSF5, AICDA or TNFRSF5 gene mutations impairing the function of CD40L, AID and CD40, respectively43. CVID patients had a heterozygous C104R mutation in the TNFRSF13b gene. This mutation prevents the binding of BAFF and APRIL to TACI44,45. The MWS patient had a heterozygous C1043T mutation in the NALP3 gene. This mutation enhances the activity of cryopyrin, a key component of the inflammasome54. TRAPS patients had heterozygous G173A, G362A, or T123G mutations in the TNFRSF1A gene. These mutations prevent cleavage of the extracellular domain of TNF receptor type-1, thereby impairing feedback inhibition of the pro-inflammatory cytokine TNF by its soluble decoy receptor54. HIDS patients had a homozygous G1129A mutation or compound G632A and G1129A mutations of the MVK gene, which encodes mevalonate kinase. The mechanism by which these mutations cause inflammation remains unknown. PFAPA patients had no molecular diagnosis, except one, who was reported to have a heterozygous mutation in the MEFV gene encoding pyrin (also known as marenostrin), a negative regulator of the inflammasome54.
Bacteria
M. catarrhalis BAA-1425, H. influenzae type-a 9006, and H. influenzae type-b 9795 (American Type Cell Culture) were cultured according to the manufacturer’s instructions.
Cultures and reagents
Cultures were performed in complete RPMI medium supplemented with 10% (volume/volume) bovine serum. Purified B cells were incubated with CD40L (Amgen), 500 ng/ml; BAFF (Alexis), 500 ng/ml; APRIL MegaLigand (Alexis), 500 ng/ml; IL-2 (Peprotech), 100 ng/ml; IL-10 (Peprotech), 50 ng/ml; IL-15 (Sigma), 50 ng/ml; and IL-21 (R&D Systems), 100 ng/ml. Cultures involving HIGM2 specimens were carried out with peripheral blood mononuclear cells instead of purified B cells due to the scarce amount of blood available. In these cultures, B cell activation was optimized by using 5 μg/ml goat F(ab’)2 pAb to human IgM (Southern Biotech). Purified basophils with Fc receptors already blocked during the purification step were incubated with or without 30 ng/ml of IL-3 (Peprotech) for 4 h. These basophils were stimulated with 5 μl of anti-mouse IgG-coated microbeads (Miltenyi Biotec) mixed with one of the following antibodies: a control IgG2a mAb with irrelevant binding activity (Santa Cruz Biotechnology), 25 μg/ml; a mouse IgG2a IA6-2 mAb to IgD (BD Biosciences), 25 μg/ml; or a mouse IgG2a G7-18 mAb to IgE (BD Biosciences), 2 μg/ml. Basophils stimulated with equal amounts of microbeads only were included as an additional control. Basophil-B cell co-cultures were set up by incubating IgD+IgM+ B cells with basophils activated as described above for 1 h (1:1 ratio). In these co-cultures, basophils were extensively washed to eliminate excess mAb to IgD or IgE before B cell addition.
Flow cytometry
Cells were incubated with an Fc blocking reagent (Miltenyi Biotec) or saturating concentrations of purified human IgG and stained at 4 °C with appropriate combinations of pAbs and mAbs to various surface antigens (Supplementary Table 1 online). 7-AAD was routinely used to exclude dead cells from the analysis. All gates and quadrants were drawn to give ≤ 1% total positive cells in the sample stained with control antibodies. Events were acquired on a FACS Calibur or BD LSR II (BD Biosciences) and analyzed by FlowJo (Tree Star). Tonsillar IgD+IgM− plasmablasts were identified within an electronic gate containing CD19+ mononuclear cells.
Enzyme-linked immunsorbent assay (ELISA) and cytometric bead array
IgD (Bethyl Laboratories), histamine (Neogen Corporation), IL-1β, IL-4, IL-13, APRIL and TNF (Bender Medsystems) were measured by ELISAs as instructed by the manufacturer. IL-8 and CXCL10 were measured using Human TH1–TH2 Cytometric Bead Array (BD Biosciences). IgM, IgG, IgA and BAFF were detected by standard ELISAs as described16, whereas IgE was measured by ELISA using a mAb to human IgE (G7-18, BD Biosciences) as capture antibody and a biotinylated mAb to human IgE (G7-26, BD Biosciences) as detection antibody. LL-37 was measured by ELISA using a mouse mAb to LL-37 (3D11, Hycult Biotechnology) as capture antibody and a rabbit pAb to LL-37 (PANATecs GmbH) as detection antibody, followed by a pAb to rabbit IgG conjugated to horseradish peroxidase (Santa Cruz Biotechnology). Human LL-37 peptide (PANATec GmbH) was used to generate a standard curve. Supernatants from basophil-containing cultures were centrifuged at 18,000 g for 20 min twice and adsorbed with mouse IgG before using them for ELISAs.
Genomic PCR, Southern blotting, cloning and sequencing
Active CSR from IgM to IgD was determined by PCR amplifying extrachromosomal σδ-Sμ reciprocal DNA recombination products instead of ligated chromosomal Sμ-σδ DNA products, which could be inflated by the proliferation of the class-switched B cells. Genomic DNA was extracted using the QIAamp DNA Mini Kit. σδ-Sμ switch circles were amplified from genomic DNA using elongase (Invitrogen) and a σδ sense primer 5’-TCATCATTGCCCAGATGCTAGGGCT-3’ coupled with an Sμ antisense primer 5’-TGAGTGCCCTCACTACTTGCGTCCCG-3’ under the following conditions: initial denaturation for 30 s at 94 °C, followed by 35 cycles of denaturation for 30 s at 94 °C, annealing for 30 s at 62 °C, and extension for 7 min at 68 °C, and a final extension for 15 min at 68 °C. Products were resolved in a 1% agarose gel, transferred overnight onto nylon membranes, and hybridized with two 32P-labeled probes specific to the 3’ portion of Sμ and the 5’ portion of σδ respectively. To further verify the specificity of PCR-amplified products, a second PCR was performed under the same conditions using the σδ sense primer 5’-TCATCATTGC CCAGATGCTAGGGCT-3’ coupled with a nested Sμ antisense primer 5’-CAGACTGTCATGGCTATCA GGGGTGGCGGGG-3’. PCR products longer than 1.5 kb were purified, ligated into pCR2.1 TOPO vectors, and transformed into Escherichia coli using a TOPO TA cloning kit (Invitrogen). Plasmids in selected colonies were purified and sequenced. Sequences were aligned with human genomic DNA sequences to identify regions spanning both σδ and Sμ.
RT-PCR
Active germline Cδ gene transcription and post-switch juxtaposition of chromosomal Iμ and Cδ exons were evaluated through the analysis of chimeric Iμ-Cδ transcripts in pre-switched and post-switched B cells, respectively. Total RNA was extracted using the QIAamp DNA Mini Kit and RNeasy Mini Kit (Qiagen) and cDNA was synthesized as previously described16. Iμ-Cδ transcripts were amplified from cDNA using Taq polymerase with an Iμ sense primer 5’-GTGATTAAGGAGAAACACTTTGAT-3’ and a Cδ antisense primer 5’-CTGGCCAGCGGAAGATCTCCTTCTT-3’ under the following conditions: initial denaturation for 5 min at 94 °C, followed by 25 cycles of denaturation for 1 min at 94 °C, annealing for 1 min at 60 °C and extension for 1 min at 72 °C, and a final extension for 7 min at 72 °C. Bands were detected as described above, using 32P-labeled probes specific for Iμ and Cδ exons. RT-PCR products were cloned and sequenced to identify regions spanning both Iμ and Cδ.
QRT-PCR
Total RNA was extracted from basophils using TRIzol (Invitrogen). cDNA synthesis and QRT-PCR were performed using various primer pairs (Supplementary Table 2 online) as described18.
Calcium flux assay
Basophils (2.5 × 106) were resuspended in 1 ml of phosphate buffer (pH 7.4) solution containing 0.5% bovine serum albumin and 2 mM ethylenediaminetetraacetic acid (EDTA). Cells were loaded with 5 μM Fura-3/AM (Sigma) for 30 min at 37 °C. Excess Fura-3/AM was removed by washing cells twice in phosphate buffer solution. Cells were then stimulated as reported in the text. Intracellular Ca2+ dynamics was monitored following the addition of stimulants in a plate reader with a 336 nm laser and a 495 nm filter. Readings were recorded every 40 seconds.
IgD binding assay
Polyclonal or monoclonal IgD antibodies purified from the plasma of healthy individuals or multiple myeloma patients (Athens Research & Technology) were incubated at the indicated concentrations with various cell types (1 × 105) for 20 min at 4 °C. Binding assays were performed with a concentration of IgD within or below the physiological range of serum IgD concentrations (15-300 μg/ml). Cells were then washed, stained with fluorescein-conjugated F(ab’)2 pAb to IgD (Southern Biotech), washed and analyzed by flow cytometry. This F(ab’)2 antibody had no cross-reactivity to other immunoglobulin classes (Supplementary Figure 6 online) and could not bind to Fc receptors due to the lack of its Fc portion. Cells stained with this pAb to IgD in the absence of purified IgD were used as background control (Supplementary Figure 6 online). Alternatively, cells were stained with fluorescein-conjugated mouse IA6-2 or IgD26 mAbs to IgD.
Competitive IgD binding assays
Monoclonal IgD purified from the plasma of a multiple myeloma patient was labeled with Alexa Fluor 488 using a Microscale Protein Labeling kit (Molecular Probes) and dialyzed in phosphate buffer (pH 7.4) solution overnight to remove excess fluorochrome. Freshly isolated peripheral blood basophils were stripped of bound endogenous IgD by incubation in an acidic sodium citrate buffer (40 mM sodium citrate, 140 mM NaCl, pH 3.0) for 3 min. Then, cells were quickly spun down to remove the acidic buffer and PBS was added to the cells to neutralize the pH. Basophils stripped of endogenous bound IgD were incubated with 0, 100 or 500 μg/ml unlabelled monoclonal IgD, followed by 40 μg/ml Alexa Fluor 488-labelled monoclonal IgD. After incubation, cells were washed and analyzed by flow cytometry. 7-AAD was used to exclude dead cells.
Statistical analysis
Values were expressed as mean ± standard error of the mean (s.e.m.). Statistical significance was assessed by a one-tailed unpaired Student’s t-test.
Supplementary Material
Acknowledgments
We thank A. Kirshenbaum and D. Metcalfe (National Institutes of Health) for the mast cell line LAD2, R. Silver, R. Schreiner and F. Diaz (Weill Medical College of Cornell University) for primary lung mast cells and discussion on transcytosis assays. Supported by the National Institutes of Health (RO1 AI057653, RO1 AI057653 supplement, and RO1 AI074378 to A.C.; and funds from T32 AI07621 to W.X.), Cancer Research Institute (fellowship to P.S.), The Irma T. Hirschl Charitable Trust (Scholar Award to A.C.), and Fondazione C. Golgi and Centro Immunodeficienze Mario Di Martino (funds to A.P.).
Footnotes
AUTHOR CONTRIBUTIONS K.C. designed and performed research, and wrote the paper; W.X. performed research and discussed data; B. He performed research and discussed data; P.A.S. discussed data; A. Chiu provided samples and discussed data; B. Huang provided reagents and performed research; M.C., J.L., C.C.R. and J.B. provided blood and tissue samples and discussed data; M.W., E.B., E.S.E., N.W.M., and P.R. performed research and discussed data; K.R. provided reagents; and A.Cerutti designed research, discussed data and wrote the paper.
COMPETING INTERESTS STATEMENT The authors declare no competing financial interests.
References
- 1.Rowe DS, Fahey JL. A new class of human immunoglobulins. J Exp Med. 1965;121:171–199. doi: 10.1084/jem.121.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Butler JE, Sun J, Navarro P. The swine Ig heavy chain locus has a single JH and no identifiable IgD. Int Immunol. 1996;8:1897–1904. doi: 10.1093/intimm/8.12.1897. [DOI] [PubMed] [Google Scholar]
- 3.Ohta Y, Flajnik M. IgD, like IgM, is a primordial immunoglobulin class perpetuated in most jawed vertebrates. Proc Natl Acad Sci U S A. 2006;103:10723–10728. doi: 10.1073/pnas.0601407103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Preud’homme JL, et al. Structural and functional properties of membrane and secreted IgD. Mol Immunol. 2000;37:871–887. doi: 10.1016/s0161-5890(01)00006-2. [DOI] [PubMed] [Google Scholar]
- 5.Bengten E, et al. The IgH locus of the channel catfish, Ictalurus punctatus, contains multiple constant region gene sequences: different genes encode heavy chains of membrane and secreted IgD. J Immunol. 2002;169:2488–2497. doi: 10.4049/jimmunol.169.5.2488. [DOI] [PubMed] [Google Scholar]
- 6.Schlissel MS. Regulating antigen-receptor gene assembly. Nat Rev Immunol. 2003;3:890–899. doi: 10.1038/nri1225. [DOI] [PubMed] [Google Scholar]
- 7.Maki R, et al. The role of DNA rearrangement and alternative RNA processing in the expression of immunoglobulin delta genes. Cell. 1981;24:353–365. doi: 10.1016/0092-8674(81)90325-1. [DOI] [PubMed] [Google Scholar]
- 8.Nitschke L, Kosco MH, Kohler G, Lamers MC. Immunoglobulin D-deficient mice can mount normal immune responses to thymus-independent and -dependent antigens. Proc Natl Acad Sci U S A. 1993;90:1887–1891. doi: 10.1073/pnas.90.5.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roes J, Rajewsky K. Immunoglobulin D (IgD)-deficient mice reveal an auxiliary receptor function for IgD in antigen-mediated recruitment of B cells. J Exp Med. 1993;177:45–55. doi: 10.1084/jem.177.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lutz C, et al. IgD can largely substitute for loss of IgM function in B cells. Nature. 1998;393:797–801. doi: 10.1038/31716. [DOI] [PubMed] [Google Scholar]
- 11.Monroe JG, Havran WL, Cambier JC. B lymphocyte activation: entry into cell cycle is accompanied by decreased expression of IgD but not IgM. Eur J Immunol. 1983;13:208–213. doi: 10.1002/eji.1830130306. [DOI] [PubMed] [Google Scholar]
- 12.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 13.Odegard VH, Schatz DG. Targeting of somatic hypermutation. Nat Rev Immunol. 2006;6:573–583. doi: 10.1038/nri1896. [DOI] [PubMed] [Google Scholar]
- 14.Chaudhuri J, Alt FW. Class-switch recombination: interplay of transcription, DNA deamination and DNA repair. Nat Rev Immunol. 2004;4:541–552. doi: 10.1038/nri1395. [DOI] [PubMed] [Google Scholar]
- 15.Aruffo A, et al. The CD40 ligand, gp39, is defective in activated T cells from patients with X-linked hyper-IgM syndrome. Cell. 1993;72:291–300. doi: 10.1016/0092-8674(93)90668-g. [DOI] [PubMed] [Google Scholar]
- 16.Litinskiy MB, et al. DCs induce CD40-independent immunoglobulin class switching through BLyS and APRIL. Nat Immunol. 2002;3:822–829. doi: 10.1038/ni829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castigli E, et al. TACI and BAFF-R mediate isotype switching in B cells. J Exp Med. 2005;201:35–39. doi: 10.1084/jem.20032000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu W, et al. Epithelial cells trigger frontline immunoglobulin class switching through a pathway regulated by the inhibitor SLPI. Nat Immunol. 2007;8:294–303. doi: 10.1038/ni1434. [DOI] [PubMed] [Google Scholar]
- 19.He B, et al. Intestinal bacteria trigger T cell-independent immunoglobulin A2 class switching by inducing epithelial-cell secretion of the cytokine APRIL. Immunity. 2007;26:812–826. doi: 10.1016/j.immuni.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 20.Cerutti A. The regulation of IgA class switching. Nat Rev Immunol. 2008;8:421–434. doi: 10.1038/nri2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Castigli E, et al. TACI is mutant in common variable immunodeficiency and IgA deficiency. Nat Genet. 2005;37:829–834. doi: 10.1038/ng1601. [DOI] [PubMed] [Google Scholar]
- 22.McHeyzer-Williams MG, Ahmed R. B cell memory and the long-lived plasma cell. Curr Opin Immunol. 1999;11:172–179. doi: 10.1016/s0952-7915(99)80029-6. [DOI] [PubMed] [Google Scholar]
- 23.Stavnezer J. Antibody class switching. Adv Immunol. 1996;61:79–146. doi: 10.1016/s0065-2776(08)60866-4. [DOI] [PubMed] [Google Scholar]
- 24.Arpin C, et al. The normal counterpart of IgD myeloma cells in germinal center displays extensively mutated IgVH gene, Cμ-Cδ switch, and λ light chain expression. J Exp Med. 1998;187:1169–1178. doi: 10.1084/jem.187.8.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plebani A, et al. IgM and IgD concentrations in the serum and secretions of children with selective IgA deficiency. Clin Exp Immunol. 1983;53:689–696. [PMC free article] [PubMed] [Google Scholar]
- 26.Liu YJ, et al. Normal human IgD+IgM− germinal center B cells can express up to 80 mutations in the variable region of their IgD transcripts. Immunity. 1996;4:603–613. doi: 10.1016/s1074-7613(00)80486-0. [DOI] [PubMed] [Google Scholar]
- 27.Brandtzaeg P, et al. The B-cell system of human mucosae and exocrine glands. Immunol Rev. 1999;171:45–87. doi: 10.1111/j.1600-065X.1999.tb01342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koelsch K, et al. Mature B cells class switched to IgD are autoreactive in healthy individuals. J Clin Invest. 2007;117:1558–1565. doi: 10.1172/JCI27628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conrad DH, Ben-Sasson SZ, Le Gros G, Finkelman FD, Paul WE. Infection with Nippostrongylus brasiliensis or injection of anti-IgD antibodies markedly enhances Fc-receptor-mediated interleukin 4 production by non-B, non-T cells. J Exp Med. 1990;171:1497–1508. doi: 10.1084/jem.171.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seder RA, et al. Mouse splenic and bone marrow cell populations that express high-affinity Fc epsilon receptors and produce interleukin 4 are highly enriched in basophils. Proc Natl Acad Sci U S A. 1991;88:2835–2839. doi: 10.1073/pnas.88.7.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshimoto T, Bendelac A, Watson C, Hu-Li J, Paul WE. Role of NK1.1+ T cells in a TH2 response and in immunoglobulin E production. Science. 1995;270:1845–1847. doi: 10.1126/science.270.5243.1845. [DOI] [PubMed] [Google Scholar]
- 32.Gauchat JF, et al. Induction of human IgE synthesis in B cells by mast cells and basophils. Nature. 1993;365:340–343. doi: 10.1038/365340a0. [DOI] [PubMed] [Google Scholar]
- 33.Min B, et al. Basophils produce IL-4 and accumulate in tissues after infection with a Th2-inducing parasite. J Exp Med. 2004;200:507–517. doi: 10.1084/jem.20040590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sokol CL, Barton GM, Farr AG, Medzhitov R. A mechanism for the initiation of allergen-induced T helper type 2 responses. Nat Immunol. 2008;9:310–318. doi: 10.1038/ni1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Denzel A, et al. Basophils enhance immunological memory responses. Nat Immunol. 2008;9:733–742. doi: 10.1038/ni.1621. [DOI] [PubMed] [Google Scholar]
- 36.Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133–169. doi: 10.1146/annurev.immunol.26.021607.090241. [DOI] [PubMed] [Google Scholar]
- 37.Bekeredjian-Ding I, et al. TLR9-activating DNA up-regulates ZAP70 via sustained PKB induction in IgM+ B cells. J Immunol. 2008;181:8267–8277. doi: 10.4049/jimmunol.181.12.8267. [DOI] [PubMed] [Google Scholar]
- 38.Klein U, Rajewsky K, Kuppers R. Human immunoglobulin (Ig)M+IgD+ peripheral blood B cells expressing the CD27 cell surface antigen carry somatically mutated variable region genes: CD27 as a general marker for somatically mutated (memory) B cells. J Exp Med. 1998;188:1679–1689. doi: 10.1084/jem.188.9.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, et al. Artiodactyl IgD: the missing link. J Immunol. 2002;169:4408–4416. doi: 10.4049/jimmunol.169.8.4408. [DOI] [PubMed] [Google Scholar]
- 40.Kluin PM, et al. IgD class switching: identification of a novel recombination site in neoplastic and normal B cells. Eur J Immunol. 1995;25:3504–3508. doi: 10.1002/eji.1830251244. [DOI] [PubMed] [Google Scholar]
- 41.Granucci F, et al. Inducible IL-2 production by dendritic cells revealed by global gene expression analysis. Nat Immunol. 2001;2:882–888. doi: 10.1038/ni0901-882. [DOI] [PubMed] [Google Scholar]
- 42.Leonard WJ, Spolski R. Interleukin-21: a modulator of lymphoid proliferation, apoptosis and differentiation. Nat Rev Immunol. 2005;5:688–698. doi: 10.1038/nri1688. [DOI] [PubMed] [Google Scholar]
- 43.Cunningham-Rundles C, Ponda PP. Molecular defects in T- and B-cell primary immunodeficiency diseases. Nat Rev Immunol. 2005;5:880–892. doi: 10.1038/nri1713. [DOI] [PubMed] [Google Scholar]
- 44.Garibyan L, et al. Dominant-negative effect of the heterozygous C104R TACI mutation in common variable immunodeficiency (CVID) J Clin Invest. 2007;117:1550–1557. doi: 10.1172/JCI31023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang L, et al. Transmembrane activator and calcium-modulating cyclophilin ligand interactor mutations in common variable immunodeficiency: clinical and immunologic outcomes in heterozygotes. J Allergy Clin Immunol. 2007;120:1178–1185. doi: 10.1016/j.jaci.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Riesbeck K, Nordstrom T. Structure and immunological action of the human pathogen Moraxella catarrhalis IgD-binding protein. Crit Rev Immunol. 2006;26:353–376. doi: 10.1615/critrevimmunol.v26.i4.40. [DOI] [PubMed] [Google Scholar]
- 47.Vladutiu AO. Immunoglobulin D: properties, measurement, and clinical relevance. Clin Diagn Lab Immunol. 2000;7:131–140. doi: 10.1128/cdli.7.2.131-140.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dawicki W, Marshall JS. New and emerging roles for mast cells in host defence. Curr Opin Immunol. 2007;19:31–38. doi: 10.1016/j.coi.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 49.Karasuyama H, Mukai K, Tsujimura Y, Obata K. Newly discovered roles for basophils: a neglected minority gains new respect. Nat Rev Immunol. 2008 doi: 10.1038/nri2458. [DOI] [PubMed] [Google Scholar]
- 50.Gala FA, Morrison SL. The role of constant region carbohydrate in the assembly and secretion of human IgD and IgA1. J Biol Chem. 2002;277:29005–29011. doi: 10.1074/jbc.M203258200. [DOI] [PubMed] [Google Scholar]
- 51.Yamaguchi Y, et al. Identification of multiple novel epididymis-specific beta-defensin isoforms in humans and mice. J Immunol. 2002;169:2516–2523. doi: 10.4049/jimmunol.169.5.2516. [DOI] [PubMed] [Google Scholar]
- 52.Oppenheim JJ, Yang D. Alarmins: chemotactic activators of immune responses. Curr Opin Immunol. 2005;17:359–365. doi: 10.1016/j.coi.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 53.Garlanda C, Bottazzi B, Bastone A, Mantovani A. Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu Rev Immunol. 2005;23:337–366. doi: 10.1146/annurev.immunol.23.021704.115756. [DOI] [PubMed] [Google Scholar]
- 54.Ryan JG, Kastner DL. Fevers, genes, and innate immunity. Curr Top Microbiol Immunol. 2008;321:169–184. doi: 10.1007/978-3-540-75203-5_8. [DOI] [PubMed] [Google Scholar]
- 55.Drenth JP, Goertz J, Daha MR, van der Meer JW. Immunoglobulin D enhances the release of tumor necrosis factor-alpha, and interleukin-1 beta as well as interleukin-1 receptor antagonist from human mononuclear cells. Immunology. 1996;88:355–362. doi: 10.1046/j.1365-2567.1996.d01-672.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johansen FE, et al. Regional induction of adhesion molecules and chemokine receptors explains disparate homing of human B cells to systemic and mucosal effector sites: dispersion from tonsils. Blood. 2005;106:593–600. doi: 10.1182/blood-2004-12-4630. [DOI] [PubMed] [Google Scholar]
- 57.Xue B, et al. Physiology of IgD. IV. Enhancement of antibody production in mice bearing IgD-secreting plasmacytomas. J Exp Med. 1984;159:103–113. doi: 10.1084/jem.159.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schiemann B, et al. An essential role for BAFF in the normal development of B cells through a BCMA-independent pathway. Science. 2001;293:2111–2114. doi: 10.1126/science.1061964. [DOI] [PubMed] [Google Scholar]
- 59.Miller NW, et al. Development and characterization of channel catfish long term B cell lines. J Immunol. 1994;152:2180–2189. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


