Meiosis takes the genetic variation from the parents and scrambles it before passing it on to the progeny. This partitioning is accomplished by two phenomena. First, because the genome is fractionated into chromosomes, the random segregation of nonhomologues transmits different parental contributions to the meiotic end products. Second, crossing over between homologous pairs of chromosomes creates a mosaic of maternal and paternal contributions that are separated at anaphase I of meiosis. Recombination also ties together the homologues (Fig. 1) so that they can separate at this division with fidelity to avoid extra or missing chromosomes that would otherwise occur, a phenomenon leading to quite detrimental effects or even lethality in the subsequent generation. Thus, an understanding of meiotic pairing and recombination is central to many biological questions.
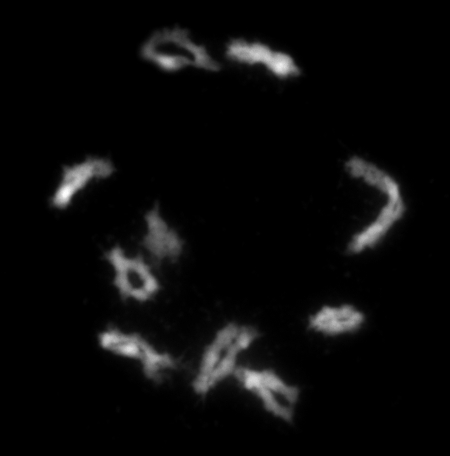
Fig. 1.
Pairing and recombination are required for the proper segregation of homologues. The 10 pairs of chromosomes in maize are depicted in late prophase of meiosis after successful pairing and recombination. Each pair has completed an homology search, and cross-overs hold together the bivalents in preparation for congression on the metaphase plate and separation in anaphase I of meiosis. Such pairing and recombination ensure the distribution of the homologues and prevents the production of meiotic end products with extra or missing chromosomes.
In this issue of PNAS, Ronceret et al. (1) define the product of the POOR HOMOLOGOUS SYNAPSIS (PHS1) gene that is involved in coordinating chromosome pairing and recombination in plants and provide clues to its function in both maize and Arabidopsis. The phs1 gene was first defined in maize from a mutation that exhibited pairing among nonhomologous chromosomes and generally poor synapsis (2). The genome of maize is largely composed of several families of retrotransposons resulting in a low gene density. Between different lines of maize, the intergenic space is often quite different (3). Thus, chromosome pairing and recombination are hypothesized to occur mainly between genes on the homologous pairs because the other sequences are usually distinct (2). Nonhomologous pairing can occur under many circumstances in maize but the orderly pairing and recombination is needed for the proper segregation of homologues under normal conditions. Because phs1 is defective in pairing and recombination functions, its study would provide insight into how these aspects of meiosis are coordinated in the early stages as the chromosomes pass through the successive initial stages of leptonema, zygonema, pachynema, diplonema, and diakinesis in preparation for the separation of homologues at metaphase and anaphase I (4).
Chromosome pairing and recombination appear to be intimately connected. Meiotic recombination is initiated by SPO11 to generate double-strand breaks (5, 6), which occur in the early leptotene stage in plants. The breaks are recognized by the MRN complex, which creates ssDNA overhangs (7, 8). The MRN complex is composed of MRE11, RAD50, and NBS1. The MRE11 protein binds DNA and with associated activities aids in the formation of the single-stranded overhangs (9). RAD50 is thought to play a structural role in this complex (10, 11). The NBS1 protein regulates MRE11 and attracts other proteins to the double-strand break. RAD51 and DMC1 associate with the single-stranded overhangs and facilitate the invasion of the complementary sequences on the homologous chromosome (12, 13). The breaks are repaired to produce cross-overs between homologous pairs or gene conversions. Knockouts of RAD50 and MRE11 in Arabidopsis lead to defects in homologue pairing, suggesting a link between pairing and recombination functions (7–11).
To investigate the action of the phs1 gene, its Arabidopsis orthologue was identified, RNAi constructs were generated for study, and a transposon insertion was obtained. Pairing defects were observed to some degree in pachynema but by diakinesis there was a predominance of single unpaired chromosomes (in contrast see Fig. 1 for normal). Interestingly, telomere clustering, which is customary in prezygonema (14) and involved with chromosome pairing, was normal in the mutants. Centromere clustering was also found in early zygonema that occurs before pairing of the chromosome arms and was not affected by the mutations. In the chromosomal regions that did exhibit pairing, the synaptonemal complex formation was studied and found to be similar to normal.
Two major conserved regions of the Arabidopsis protein were mutated and studied for their effects on pairing. The “conserved region 1” was found to be critical. Mutations in other portions of the protein were without mutant effects.
The localization of the recombination machinery proteins was then studied in the phs1 mutants in maize. The mutant did not show any change in the number of double-strand breaks but the number of RAD51 foci was clearly diminished. The critical finding was that the levels of RAD50 foci within the nuclei of mutant maize or Arabidopsis were dramatically reduced compared with normal. Abundant RAD50 protein was observed in the cytoplasm of the mutants. The normal time course and localization of PHS1 protein was shown to appear in the cytoplasm in leptonema and zygonema with progressive entry to the nucleus where discrete foci could be observed. After pachynema, the protein is not detectable.
To investigate the interrelationship of PHS1 with recombination, the distribution of PHS1 was studied in the Arabidopsis spo11 mutant and found to be normal. Given that SPO11 initiates double-strand breaks, these observations suggest that the entry of PHS1 into the nucleus is not triggered by the presence of the breaks. Thus, the conclusion from this study is that, because the RAD50 and MRE11 proteins do not contain nuclear localization signals, the product of PHS regulates the entry of RAD50 into the nucleus at the appropriate meiotic stages. The consequence of failure of this transport in the mutants is that the majority of double-strand breaks are not correctly resolved.
The phs1 gene does not appear to have orthologues outside of the plant kingdom. This realization led Ronceret et al. (1) to suggest that the gene product plays an important role in regulating the timing of entry of critical meiotic gene products into the nucleus. In plants, these meiotic gene products are expressed across a broad time frame in contrast to meiotic factors with a tight temporal transcriptional regulation in yeast. Thus, an important function of the phs1 gene is revealed in their study as a regulator of the timing of nuclear entry of critical meiotic proteins for the resolution of pairing and recombination.
Acknowledgments.
Work in our laboratory on related topics is supported by National Science Foundation Grants DBI 0423898 and DBI 0922703.
Footnotes
The authors declare no conflict of interest.
See companion article on page 20121.
References
- 1.Ronceret A, Doutriaux M-P, Golubovskaya IN, Pawlowski WP. PHS1 regulates meiotic recombination and homologous chromosome pairing by controlling the transport of RAD50 to the nucleus. Proc Natl Acad Sci USA. 2009;106:20121–20126. doi: 10.1073/pnas.0906273106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pawlowski WP, et al. Coordination of meiotic recombination, pairing, and synapsis by PHS1. Science. 2004;303:89–92. doi: 10.1126/science.1091110. [DOI] [PubMed] [Google Scholar]
- 3.Fu H, Dooner HK. Intraspecific violation of genetic colinearity and it implications in maize. Proc Natl Acad Sci USA. 2002;99:9573–9578. doi: 10.1073/pnas.132259199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pawlowski WP, Cande WZ. Coordinating the events of the meiotic prophase. Trends Cell Biol. 2005;15:674–681. doi: 10.1016/j.tcb.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 5.Grelon M, Vezon D, Gendrot G, Pelletier G. AtSPO11–1 is necessary for efficient meiotic recombination in plants. EMBO J. 2001;20:589–600. doi: 10.1093/emboj/20.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stacey NJ, et al. Arabidopsis SPO11–2 functions with SPO11–1 in meiotic recombination. Plant J. 2006;48:206–216. doi: 10.1111/j.1365-313X.2006.02867.x. [DOI] [PubMed] [Google Scholar]
- 7.Puizina J, Siroky J, Mokros P, Schweizer D, Riha K. Mre11 deficiency in Arabidopsis is associated with chromosomal instability in somatic cells and Spo11-dependent genome fragmentation during meiosis. Plant Cell. 2004;16:1968–1978. doi: 10.1105/tpc.104.022749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borde V. The multiple roles of the Mre11 complex for meiotic recombination. Chromosome Res. 2007;15:551–563. doi: 10.1007/s10577-007-1147-9. [DOI] [PubMed] [Google Scholar]
- 9.Daoudal-Cotterell S, Gallego ME, White CI. The plant Rad50–Mre11 protein complex. FEBS Lett. 2002;516:164–166. doi: 10.1016/s0014-5793(02)02536-x. [DOI] [PubMed] [Google Scholar]
- 10.Hopfner KP, et al. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature. 2002;418:562–566. doi: 10.1038/nature00922. [DOI] [PubMed] [Google Scholar]
- 11.Bleuyard JY, Gallego ME, White CI. Meiotic defects in the Arabidopsis rad50 mutant point to conservation of the MRX complex function in early stages of meiotic recombination. Chromosoma. 2004;113:197–203. doi: 10.1007/s00412-004-0309-1. [DOI] [PubMed] [Google Scholar]
- 12.Pawlowski WP, Golubovskaya IN, Cande WZ. Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests involvement of RAD51 in the meiotic homology recognition. Plant Cell. 2003;8:1807–1816. doi: 10.1105/tpc.012898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li J, et al. Functional analysis of maize RAD51 in meiosis and doublestrand break repair. Genetics. 2007;176:1469–1482. doi: 10.1534/genetics.106.062604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armstrong SJ, Franklin FC, Jones GH. Nucleolus-associated telomere clustering and pairing precede meiotic chromosome synapsis in Arabidopsis thaliana. J Cell Sci. 2001;114:4207–4217. doi: 10.1242/jcs.114.23.4207. [DOI] [PubMed] [Google Scholar]