Abstract
Defensins (e.g., human neutrophil peptides, or HNPs) contribute to innate immunity through diverse actions, including microbial killing; high concentrations are present in the lung in response to inflammation. Arginines are critical for HNP activity, which is decreased by their replacement with ornithine. ADP-ribosyltransferases (ARTs) catalyze transfer of ADP-ribose from NAD to an acceptor arginine in a protein substrate, whereas ADP-ribosylarginine hydrolases release ADP-ribose. ART1 on the surface of airway epithelial cells ADP-ribosylated HNP-1 specifically on arginines 14 and 24, with ADP-ribosylation altering biological activity. Di- and mono-ADP-ribosylated HNP-1 were isolated from bronchoalveolar lavage fluid (BALF) of patients with asthma and idiopathic pulmonary fibrosis (IPF), suggesting a role for ADP-ribosylation in disease. In the present study, we observed that ART1-catalyzed ADP-ribosylation of HNP-1 in vitro generated a product with ADP-ribose on arginine 24, and ornithine replacing arginine at position 14. We hypothesized that ADP-ribosylarginine is susceptible to a nonenzymatic hydrolytic reaction yielding ornithine. On incubation of di- or mono-ADP-ribosyl-HNP-1 at 37 °C, ADP-ribosylarginine was partially replaced by ornithine, whereas ornithine was not detected by amino acid analysis and mass spectrometry of unmodified HNP-1 incubated under the same conditions. Further, ornithine was produced from the model compound, ADP-ribosylarginine. BALF from an IPF patient contained ADP-ribosyl-HNP-ornithine as well as mono- and di-ADP-ribosylated HNP-1, consistent with in vivo conversion of arginine to ornithine. Targeted ADP-ribosylation of specific arginines by transferases, resulting in their replacement with ornithine, is an alternative pathway for regulation of protein function through posttranslational modification.
Keywords: posttranslational modification, NAD, bacterial toxins
Neutrophils, a critical component of the innate immune system, are recruited to airways in response to inflammation or infection (1). Neutrophil defensins [human neutrophil peptides (HNPs) 1–3], stored in azurophilic granules, are small cationic peptides whose main function is to defend the lung against pathogenic microorganisms (2). High levels of defensins have been found in patients with inflammatory lung diseases, such as idiopathic pulmonary fibrosis (IPF) (3) and cystic fibrosis (4). In addition to antimicrobial activities and other diverse functions (5), defensins interact with airway epithelial cells, increasing proliferation and stimulating wound repair (6). HNP1–3 are arginine rich and differ in sequence by one amino acid. The salt bridge formed by Arg5–Glu13 and three disulfide bridges are conserved, but not required, for antibacterial activity in vitro (7, 8). The arginines in HNP-1 are critical for maintaining activity (9). In addition, the low number of arginines in HD6 (human defensin 6 expressed in Paneth cells) may be responsible for its lack of antibacterial activity (10).
Mono-ADP-ribosylation is a posttranslational modification of proteins in which the ADP-ribose moiety of NAD is transferred to a specific amino acid. Several well-characterized mono-ADP-ribosyltransferases were identified in viruses, bacteria, and eukaryotes. The modification can be reversed by ADP-ribosyl- acceptor hydrolases, which cleave the ADP-ribose-acceptor bond (11, 12). Arginine-specific mono-ADP-ribosyltransferase-1 (ART1) is present on the apical surface of epithelial cells in human airways and is linked to the cell surface by a glycosylphosphatidylinositol (GPI) anchor (13, 14). ART1 modifies the arginines of several substrates, including HNP-1, thereby altering their activity (15, 16). ADP-ribosylation of HNP-1 decreased the antimicrobial and cytotoxic activities without affecting T-cell chemotaxis and IL-8 release from A549 lung carcinoma cells (17). In vitro, ART1 ADP-ribosylates HNP-1 on arginine 14 with a secondary site on arginine 24. Mono- and di-ADP-ribosylated HNP were isolated from the bronchoalveolar lavage fluid of IPF and asthma patients, consistent with a role for the modified HNP-1 in disease (18). In addition to the two modified forms, a third product separated by HPLC from the reaction of HNP-1, NAD, and ART1 was identified by mass spectrometry (MS) analysis as ADP-ribosylated HNP containing ornithine, a noncoded amino acid. Sequence analysis revealed ornithine as residue 14, with arginine 24 as the site of ADP-ribosylation.
We hypothesized that ADP-ribosylation of HNP-1 by ART1 was responsible for conversion of arginines to ornithine, suggesting a unique function for ADP-ribosylation. Because both modified forms of HNP-1 were found in bronchoalveolar lavage fluid, we investigated the possibility that ADP-ribosylation would change the primary sequence of HNP-1 in vivo, altering its activity. To determine the mechanism of ornithine production, we analyzed the HPLC-purified mono- or di-ADP-ribosylated-HNP-1 after incubation at pH 7 and 9. To verify that the amino acid sequence of HNP-1 was not critical for conversion of arginines to ornithine and to assess stability of the arginine-ADP-ribose bond, we examined the effect of incubating purified ADP-ribosylarginine on ornithine formation under the same conditions.
Results and Discussion
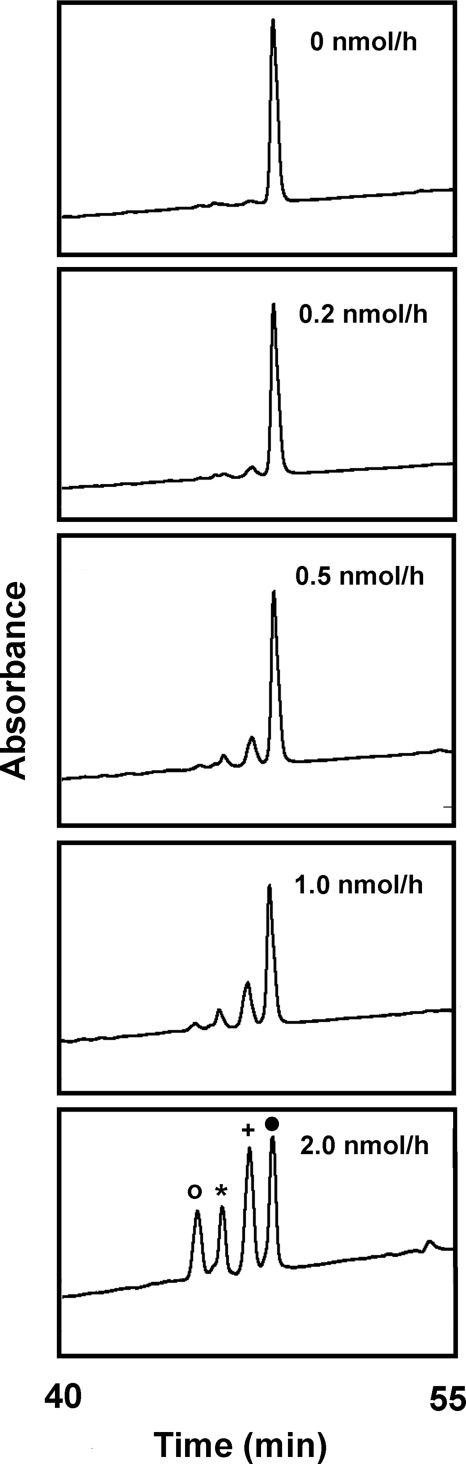
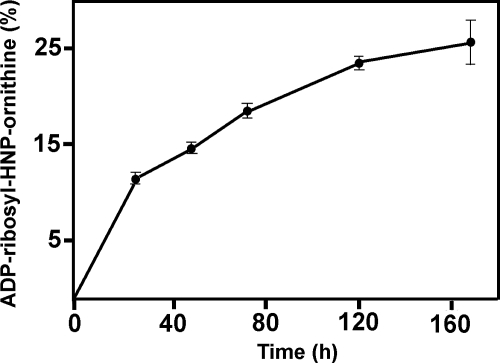
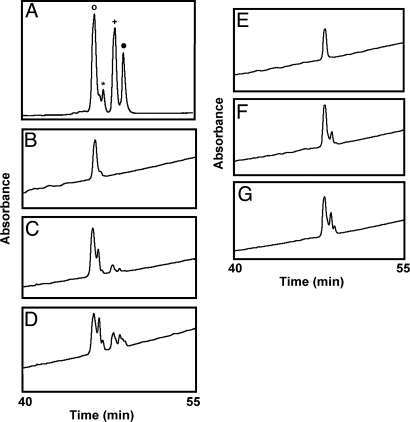
After HPLC separation of products of the ART1, HNP-1, and NAD reaction, MS analysis identified HNP-1, mono-ADP-ribosylated on arginine 14, and di-ADP-ribosylated-HNP-1, with a second modification on arginine 24. Incubation of ART1 (2 nmol/h activity) and HNP-1 for 24 h at 30 °C decreased the amount of di-ADP-ribosylated-HNP-1 and increased a fourth HPLC peak (Fig. 1). The purified 3,940-Da product, identified as ADP-ribosylated-HNP-ornithine by MS analysis, with ornithine mapped to position 14, was subjected to acid hydrolysis. Amino acid analyses confirmed the presence of ornithine in the modified but not in the substrate HNP-1 (Fig. 2). Because arginine-14 is the site of ADP-ribosylation, it appeared that ADP-ribosyl-arginine was the precursor of ornithine in HNP. Amounts of modified HNP-ornithine recovered in the reaction mix increased with incubation time reaching about 26% after 168 h, consistent with the nonenzymatic conversion of arginine to ornithine (Fig. 3). To verify that ART1 (or an enzymatic contaminant) was not required for ornithine formation, HPLC-purified mono- and di-modified HNP were incubated for 24 h at 37 °C at pH 7 or 9, and the reaction products analyzed by MS (Fig. 4 and Table 1). Di-ADP-ribosylated-HNP converted to mono-ADP-ribosylated HNP-ornithine of 3940 molecular mass at pH 7 and the amount was greater at pH 9. About 50% of di-ADP-ribosylated HNP-1 was converted to mono-ADP-ribosylated-HNP-1-ornithine at pH 9; when the ADP-ribose and ornithine were mapped to positions on the HNP-1, we observed ADP-ribosylated arginine 14 and ornithine at position 24 and ADP-ribosylarginine 24 and ornithine 14. Of note, with the in vitro-modified HNP-1, we did not find any di-ADP-ribosylated-HNP-1-ornithine. Purified mono-ADP-ribosylated HNP-1 was converted to HNP-1-ornithine, 3,399.5 Da under the same conditions at pH 7, but the amount was greater at pH 9. Since the two ADP-ribosylated HNP-1 products were isolated from the broncoalveolar lavage fluid (BALF) of patients with IPF (18), we expected that ADP-ribosylated-HNP-ornithine would be present in vivo. To look for formation of HNP-1-ornithine in vivo, we examined BALF samples from 7 patients with idiopathic pulmonary fibrosis as described (18). Briefly, 8 mL of BALF were applied to LC-18 Supelclean SPE tubes (Supelco) equilibrated in 10% isopropanol/0.1% TFA, washed and eluted with 50% isopropanol/0.1% TFA. The eluted proteins were vacuum concentrated before separation by RF-HPLC. In four of the seven IPF samples, a broad peak eluted at the retention time of HNP-1. MS analysis confirmed one of the samples contained HNP-1 and consisted of 38.8% HNP-1, 32.1% di-ADP-ribosylated HNP-1, 20.8% ADP-ribosyl-HNP-1, and 8.3% ADP-ribosyl-HNP-1-orninthine. These data are consistent with the in vivo alteration of HNP-1 primary sequence. Of note, as with in vitro ADP-ribosyl-HNP-1, in the in vivo-modified material, we did not see di-ADP-ribosylated HNP-1-ornithine, consistent with the fact that ornithine is not ADP-ribosylated by NAD:arginine ADP-ribosyltransferases. Experimentally, arginine and agmatine served as substrates for ART1, ornithine did not. We did not find HNP-1-ornithine in BALF from patients with asthma (n = 4); of the four patients, one had both di- and mono-ADP-ribosylated HNP-1. Thus, ADP-ribosylation of HNP-1 and HNP-1-ornithine were not seen in all patients with IPF or asthma.
Fig. 1.
RP-HPLC separation of reaction products from the incubation of HNP-1 with ART1. HNP-1 (3 nmol) was incubated overnight at 30 °C with 5 mM NAD in 150 μL of 50 mM potassium phosphate (pH 7.5) and 0, 0.2, 0.5, 1.0, and 2.0 nmol/h of ART1 activity as indicated. Reactions were terminated by addition of guanidine HCl (final concentration 6 M) before HPLC and analysis at absorbance 210 nm. Peaks were identified by MS as di-ADP-ribosylated-HNP-1 (○), ADP-ribosyl-HNP-ornithine (*), mono-ADP-ribosylated HNP-1 (+), and HNP-1 (●). Data are representative of 3 experiments.
Fig. 2.
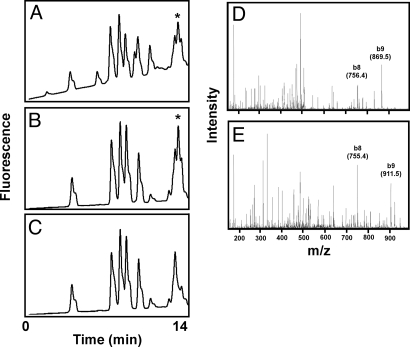
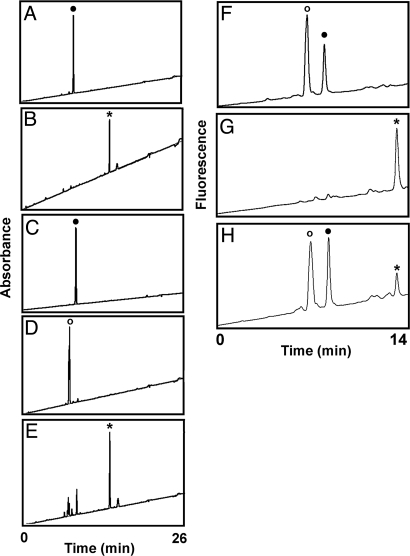
Amino acid analysis of purified ADP-ribosyl-HNP-ornithine. ADP-ribosyl-HNP-ornithine was purified by HPLC separation of the products from HNP-1 (10 nmol) and 5 mM NAD incubated with ART1 (12.8 nmol/h) as in Fig. 1, and its identity confirmed by MS analysis (Fig. 2D). Preparation for amino acid analysis was as described in Methods before OPA derivatization (Agilent Technologies) and separation by HPLC with monitoring by fluorescence (excitation 340 nm and emission 450 nm). (A) Purified ADP-ribosyl-HNP-1-ornithine (1 nmol), expected position of derivatized ornithine (indicated by ★). (B) HNP-1 (3 nmol) with ornithine (★) (23 pmol; Sigma) added after hydrolysis. (C) HNP-1 (3 nmol). (D and E) MS/MS spectrum of the tryptic peptide containing residues 6–14 from a digest of ADP-ribosyl-HNP-1. The b8 ion is generated from residues 6–13 and is therefore the same in the arg and orn peptides. The b9 ion is generated from residues 6–14 and contains either orn (D) or arg (E). The data are representative of 2 experiments.
Fig. 3.
Time course of ADP-ribosyl-HNP-ornithine production. HNP-1 (6 nmol) was incubated for the indicated times at 30 °C with 5 mM NAD and ART1 (5.8 nmol/h) in 150 μL of 50 mM potassium phosphate (pH 7.5). Reactions were terminated at the indicated time by addition of guanidine HCl (final concentration 6 M) before analysis by HPLC with monitoring of absorbance at 280 nm. ADP-ribosyl-HNP-ornithine was quantified as picomoles calculated from the area (mAu) under the peak identified as ADP-ribosyl-HNP-ornithine at 280 nm. The reported percent was based on the total number of picomoles of reaction products in the separation. Data are means ± SD from 4 experiments.
Fig. 4.
HPLC separation of purified modified HNP-1 after incubation at pH 7 or 9. Di-modified HNP-1 (6 nmol) and mono-modified HNP-1 (0.5 nmol) were isolated from the overnight incubation (30 °C) of HNP-1 (10 nmol) and 5 mM NAD with ART1 (12.8 nmol/h) in 50 mM potassium phosphate (pH 7.5). Identity and purity were confirmed by MS analysis. Purified di-modified HNP-1 or mono-modified HNP-1 was incubated as indicated at 37 °C for 24 h, except where noted, before separation by HPLC (see Methods), and monitored at 210 nm. (A) Products from the reaction of HNP-1 and NAD with ART1 (pH 7.5) overnight at 30 °C as described in Methods: di-ADP-ribosylated-HNP-1 (○), ADP-ribosyl-HNP-ornithine (*), mono-ADP-ribosylated-HNP-1 (+), and HNP-1 (●). (B) Di-modified HNP-1 at 0 time (pH 9). (C) Di-modified HNP-1 incubated at pH 7. (D) Di-modified HNP-1 incubated at pH 9. (E) Mono-modified HNP-1 at 0 time (pH 9). (F) Mono-modified HNP-1 incubated at pH 7. (G) Mono-modified HNP-1 incubated at pH 9. Data are representative of 2 experiments at pH 7 and 3 experiments at pH 9.
Table 1.
MS analysis of mono- and di-ADPribosyl-HNP-1 incubated at pH 7 and 9
| pH | Time, h | HNP | ADPR-HNP | HNP-orn | Di-orn-HNP | ADPR-HNP-orn | Di-ADPR-HNP | Total, % | |
|---|---|---|---|---|---|---|---|---|---|
| ADPR-HNP | 7 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 100 |
| ADPR-HNP | 7 | 24 | 0 | 82 ± 13 | 18 ± 13 | 0 | 0 | 0 | 100 |
| ADPR-HNP | 9 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 100 |
| ADPR-HNP | 9 | 24 | 0 | 61 ± 9 | 39 ± 9 | 0 | 0 | 0 | 100 |
| Di-ADPR-HNP | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 100 |
| Di-ADPR-HNP | 7 | 24 | 0 | 0 | 0 | 1 ± 1 | 32 ± 17 | 67 ± 18 | 100 |
| Di-ADPR-HNP | 9 | 0 | 0 | 0 | 0 | 0 | 13 | 87 | 100 |
| Di-ADPR-HNP | 9 | 24 | 0 | 0 | 0 | 9 ± 3 | 55 ± 3 | 36 ± 0 | 100 |
MS analysis of purified mono- and di-ADPribosyl-HNP-1 incubated at pH 7 and 9. HNP-1 (3,442 Da), ADP-ribosyl-HNP-1 (3,983 Da), HNP-ornithine (3,400 Da), Di-HNP-ornithine (3,358 Da), ADP-ribosyl-HNP-ornithine (3,941 Da), and di-ADP-ribosyl-HNP (4,525 Da). Data are the mean ± SD of 2 experiments on different preparations.
Effects of sequence and structure on the biological activity of the α-defensins have been studied by several investigators (19). After ADP-ribosylation by ART1, HNP-1 had reduced antimicrobial and cytotoxic activities but maintained its ability to recruit T lymphocytes and release IL-8 from A549 cells (17). To understand the cationicity of defensins, 3 of 4 arginines in HNP-1 (not arginine 5) were replaced with lysine or ornithine (9). Bactericidal activity was decreased as arginines were replaced by lysines or ornithines. These data suggested that in addition to ADP-ribosylation, conversion of arginine to ornithine can alter HNP-1 activity in vivo.
Arginase, found predominantly in liver, catalyses the hydrolysis of arginine to ornithine in the urea cycle. We considered the possibility that, although the enzyme is reported to require free arginine (20), HNP-1 could be a substrate for bovine liver arginase. HNP-1 (1.4 nmol), mono-ADP-ribosylated-HNP-1 (1 nmol), ADP-ribosylarginine (18 μM), and arginine (5 mM) were incubated with manganese-activated arginase (0.7 units) at pH 9.5 (in 200 μL, 37 °C, 10 min), conditions used for arginase hydrolysis of arginine to ornithine (21). Identification of the products by amino acid analysis and MS revealed ornithine only in the reaction that contained free arginine.
Ornithine was found in acid hydrolysates of human skin collagen and lens crystallins (22); the amounts increased with the age of the patients from whom the proteins were isolated. It was proposed that ornithine formation resulted from glycation of arginine by sugar moieties (Advanced Glycation End-products, AGEs), followed by time-dependent breakdown of the adduct to yield ornithine. In addition, because furornithine and Nδ-carboxymethyl-ornithine were also detected in the acid hydrolysates, ornithine formed in this manner appeared to be further glycated. Reducing sugars, such as ribose and ADP-ribose, produce protein glycation by reacting with a free amino group of lysine or arginine (23, 24). After the in vitro reaction of ribose with collagen, the hydrolysate contained α-NFC-1[Nδ-(4-oxo-5-dihydroimidazol-2-yl)-L-ornithine], a product of a modified arginine (25). Nonenzymatic modification of several proteins in vitro by sugars has been reported (26, 27), but ornithine in a protein primary sequence has been reported only in collagen as a result of age-related glycation. No ornithine was identified after incubation with free ADP-ribose by MS (18), and after incubation of HNP-1, which has no lysines in the native sequence, or arginine with free ADP-ribose by amino acid analysis (Fig. 5). In contrast to ADP-ribose glycation reactions with model conjugates, enzymatically, ADP-ribosylated arginine was stable at pH 9 for 30 min at 37 °C. ADP-ribose is released from the modified protein substrate chemically by incubation with hydroxylamine or by enzymatic cleavage by ADP-ribosylarginine-hydrolases (28–30).
Fig. 5.
Amino acid analysis following incubation of ADP-ribosylarginine at pH 7 and 9. ADP-ribosyl[14C]arginine was prepared and purified as described in Methods. ADP-ribosyl[14C]arginine, arginine, ADP-ribose plus arginine or ornithine (15 pmol) in 20 mM potassium phosphate (pH 7.5) adjusted to pH 9 by NaOH (measured by microelectrode) in 100 μL were incubated at 37 °C for 24 h except where noted. The samples were vacuum dried, dissolved in 25 μL 0.05% TFA (without acid hydrolysis) before amino acid analysis. The HPLC separation (see Methods) was monitored by UV detection at 338 nm. (A) ADP-ribose and arginine (●). (B) Ornithine (*) (Sigma). (C) ADP-ribosyl[14C]arginine incubated 24 h in 6N HCl at 37 °C. (D) ADP-ribosyl[14C]arginine (○) 0 time. (E) ADP-ribosyl[14C]arginine. Data are representative of 3 experiments. ADP-ribosyl[14C]arginine (1.8 nmol) in 20 mM potassium phosphate (pH 7) incubated for the indicated times at 37 °C before amino acid analysis monitored by fluorescence (340 nm excitation/450 emission). (F) ADP-ribosyl[14C]arginine (○) at 0 time. (G) Ornithine (*) (25 pmol). (H) ADP-ribosyl[14C]arginine after 24 h. Data are representative of 4 experiments.
Consistent with a nonenzymatic conversion of modified arginine to ornithine in HNP, ADP-ribosylarginine generated ornithine when incubated under the same conditions. Ornithine was observed by amino acid analysis without prior acid hydrolysis after incubation at pH 7 and 9 (37 °C) for 24 h. Arginine was cleaved from ADP-ribosylarginine incubated in 6N HCl at 37 °C for 24 h, but ornithine was not detected by amino acid analysis (Fig. 5). These data suggest that the amino acid sequence of HNP-1 is not required for conversion of modified arginines to ornithine. In contrast to the enzymatic cleavage of ADP-ribosylarginine by ADP-ribosyl-arginine hydrolase-1 (ARH1) at carbon 1′′ of ADP-ribose, which releases ADP-ribose from arginine, the nonenzymatic hydrolysis of ADP-ribosylarginine at the guanidino carbon of arginine produces ornithine.
In addition to altering the molecular charge, pKa, secondary structure, and biological activity, the presence of ornithine at the HNP-1 arginine site and the absence of ADP-ribose would prevent the modified protein from interacting with ADP-ribosyl-acceptor hydrolases or serving as a target for subsequent ADP-ribosylation. ADP-ribosyltransferases, such as cholera toxin, modify guanidine-containing compounds (e.g., arginine or arginine on peptides), not amino acids containing an amino group [e.g., lysine (31, 32) and ornithine (32)]. We found the same to be true of ART1. If ADP-ribosylated arginine is converted nonenzymatically to ornithine, then the level of ADP-ribosylated proteins may deteriorate over time. Moreover, the effect of ADP-ribosylation on signal transduction would be altered. We had reported that HNP-1 is specifically mono-ADP-ribosylated by ART1 on arginine 14, or di-ADP-ribosylated on arginines 14 and 24, suggesting that specific sites in HNP-1 were selected by ADP-ribosyltransferases for conversion to ornithine. We reported that the posttranslational modification of HNP-1 by ADP-ribose regulated its function. We have shown that this modification may be unstable, resulting in an HNP-ornithine peptide with altered function which may be relevant to innate immunity in the airway.
Methods
Human Subjects Protection.
The clinical protocol (99-H-0068) was approved by the National Heart, Lung, and Blood Institute Institutional Review Board. Written informed consent was obtained from all participants.
Preparation of mART1.
Rat mammary adenocarcinoma (NMU) cells transfected with plasmids containing mART1 were grown in Eagle's MEM with 10% FBS (Invitrogen) and Geneticin (G-418) 0.5 mg/mL. Cells were purchased from American Type Culture Collection. Protein released from the cells by PI-PLC, collected in the medium for ADP-ribosyltransferase activity (nmol/h), were assayed by quantifying the transfer of ADP-ribose to agmatine in standard assays as described (18).
Preparation of ADP-Ribosyl[14C]arginine.
CTA (60 μg), 30 mM DTT, 10 mM NAD, and 10 mM arginine (0.5 μCi14C/assay) with 30 μg ovalbumin in 20 mM potassium phosphate (pH 7.5; volume 300 μL) were incubated overnight at 30 °C. Reaction products were separated on a strong anion exchange (SAX) column (DuPont) by gradient elution (18). Radioactive peaks were collected, vacuum concentrated, and applied to a Discovery BioWide Pore C18 RF-HPLC column (Supelco) equilibrated for 15 min with HPLC water, 0.05% TFA (flow = 0.8 mL/min), followed by a 5-min linear gradient of 0% to 100% acetonitrile. Peaks, monitored by absorbance at 254 nm, and radioactivity identified as ADP-ribosyl[14C]arginine, were confirmed by MS analysis. Samples (25 μL) of ADP-ribosyl[14C]arginine were vacuum dried, dissolved in 200 μL 6N HCl (Fluka), and hydrolyzed under nitrogen (155 °C, 45 min) to release arginine. The hydrolysate was vacuum dried, dissolved in 25 μL 0.05% TFA, and subjected to OPA derivatization (Agilent Technologies) before HPLC separation (see text following). The arginine peak was quantified by absorbance at 338 nm and fluorescence (340 excitation/450 emission), then compared with a standard curve.
Amino Acid Analysis.
The indicated amount HNP-1 (Bachem) was vacuum dried, dissolved in 200 μL of 6N HCl plus 5 μL of 40 mM DTT before hydrolysis under nitrogen at 155 °C for 45 min. The hydrolysate was vacuum dried and solubilized in 25 μL water, 0.05% TFA before OPA (Agilent Technologies) precolumn automated derivatization. The conditions for the derivatization reaction and the HPLC separation (with the modification following) are described in Agilent Technologies Technical Note (publication no. 5980–1193EN). The HPLC column, Eclipse-AAA (4.6 × 150 mm, 5 μm particle size) was equilibrated with mobile phase A, 40 mM sodium dibasic phosphate buffer (pH 7.8) and amino acids were eluted with a linear gradient of 0% to 40% of phase B, acetonitrile/MeOH/water (45:45:10) for 1.9–15 min; 15–18.1 min gradient to 57% B, 18.1–18.6 min gradient to 100% B.
RF-HPLC Separation of HNP and ART1 Reaction Products.
Products of the reaction of HNP-1 (Bachem) and NAD catalyzed by ART1 were separated by reverse-phase HPLC on a Discovery BioWide Pore C18 column (Supelco) as described (18).
MS and Sequence Analysis.
HNP-1 was reduced, cleaved by trypsin, and analyzed by reverse-phase chromatography/mass spectrometry as described (17, 18), except that the reverse-phase column was a Zorbax 300SB-C18, 2.1 × 50 mm 3.5 μm, and the mass spectrometer was an Agilent model G1969 (Agilent Technologies) with a time-of-flight detector. Mass spectra were deconvoluted with the Agilent software, MassHunter version 2, and the fraction of each species was calculated from the areas of the deconvoluted peaks.
Preparation of Ornithine from ADP-Ribosyl[14C]arginine at pH 9 at 30 °C and 37 °C.
ADP-ribosyl[14C]arginine was prepared as described, followed by incubation at pH 9. Three samples were analyzed by amino acid analysis at 30 °C and 37 °C. The production of ornithine at 30 °C was 4.4 ± 0.03 pmol/μL, and at 37 °C was 5.1 ± 0.2 pmol/μL. Data are mean ± SEM of values from 3 samples.
Acknowledgments.
We thank Dr. Martha Vaughan and Dr. Gustavo Pacheco-Rodriguez (National Heart, Lung, and Blood Institute) for helpful discussions and critical review of the manuscript. These studies were supported by the Intramural Research Program of the National Institutes of Health, National Heart, Lung, and Blood Institute.
Footnotes
Author contributions: L.A.S., R.L.L., and J.M. designed research; L.A.S. and R.L.L. performed research; B.R.G. contributed new reagents/analytic tools; L.A.S., J.M., and R.L.L. analyzed data; and L.A.S. and J.M. wrote the paper.
The authors declare no conflict of interest.
References
- 1.Aarbiou J, Rabe KF, Hiemstra PS. Role of defensins in inflammatory lung disease. Ann Med. 2002;34:96–101. doi: 10.1080/07853890252953482. [DOI] [PubMed] [Google Scholar]
- 2.Bals R, Hiemstra PS. Innate immunity in the lung: How epithelial cells fight against respiratory pathogens. Eur Respir J. 2004;23:327–333. doi: 10.1183/09031936.03.00098803. [DOI] [PubMed] [Google Scholar]
- 3.Mukae H, et al. Raised plasma concentrations of alpha-defensins in patients with idiopathic pulmonary fibrosis. Thorax. 2002;57:623–628. doi: 10.1136/thorax.57.7.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Soong LB, Ganz T, Ellison A, Caughey GH. Purification and characterization of defensins from cystic fibrosis sputum. Inflamm Res. 1997;46:98–102. doi: 10.1007/s000110050114. [DOI] [PubMed] [Google Scholar]
- 5.Rehaume LM, Hancock RE. Neutrophil-derived defensins as modulators of innate immune function. Crit Rev Immunol. 2008;28:185–200. doi: 10.1615/critrevimmunol.v28.i3.10. [DOI] [PubMed] [Google Scholar]
- 6.van Wetering S, Tjabringa GS, Hiemstra PS. Interactions between neutrophil-derived antimicrobial peptides and airway epithelial cells. J Leukoc Biol. 2005;77:444–450. doi: 10.1189/jlb.0604367. [DOI] [PubMed] [Google Scholar]
- 7.Wu Z, Li X, de Leeuw E, Ericksen B, Lu W. Why is the Arg5-Glu13 salt bridge conserved in mammalian alpha-defensins? J Biol Chem. 2005;280:43039–43047. doi: 10.1074/jbc.M510562200. [DOI] [PubMed] [Google Scholar]
- 8.Lundy FT, et al. Antimicrobial activity of truncated alpha-defensin (human neutrophil peptide (HNP)-1) analogues without disulphide bridges. Mol Immunol. 2008;45:190–193. doi: 10.1016/j.molimm.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 9.Zou G, et al. Toward understanding the cationicity of defensins. Arg and Lys versus their noncoded analogs. J Biol Chem. 2007;282:19653–19665. doi: 10.1074/jbc.M611003200. [DOI] [PubMed] [Google Scholar]
- 10.Szyk A, et al. Crystal structures of human alpha-defensins HNP4, HD5, and HD6. Protein Sci. 2006;15:2749–2760. doi: 10.1110/ps.062336606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corda D, Di Girolamo M. Functional aspects of protein mono-ADP-ribosylation. EMBO J. 2003;22:1953–1958. doi: 10.1093/emboj/cdg209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koch-Nolte F, Kernstock S, Mueller-Dieckmann C, Weiss MS, Haag F. Mammalian ADP-ribosyltransferases and ADP-ribosylhydrolases. Front Biosci. 2008;13:6716–6729. doi: 10.2741/3184. [DOI] [PubMed] [Google Scholar]
- 13.Balducci E, et al. Selective expression of RT6 superfamily in human bronchial epithelial cells. Am J Respir cell Mol Biol. 1999;21:337–346. doi: 10.1165/ajrcmb.21.3.3638. [DOI] [PubMed] [Google Scholar]
- 14.Okazaki IJ, Moss J. Glycosylphosphatidylinositol-anchored and secretory isoforms of mono-ADP-ribosyltransferases. J Biol Chem. 1998;273:23617–23620. doi: 10.1074/jbc.273.37.23617. [DOI] [PubMed] [Google Scholar]
- 15.Corda D, Di Girolamo M. Mono-ADP-ribosylation: A tool for modulating immune response and cell signaling. Sci STKE. 2002;2002:PE53. doi: 10.1126/stke.2002.163.pe53. [DOI] [PubMed] [Google Scholar]
- 16.Di Girolamo M, Dani N, Stilla A, Corda D. Physiological relevance of the endogenous mono(ADP-ribosyl)ation of cellular proteins. FEBS J. 2005;272:4565–4575. doi: 10.1111/j.1742-4658.2005.04876.x. [DOI] [PubMed] [Google Scholar]
- 17.Paone G, et al. ADP ribosylation of human neutrophil peptide-1 regulates its biological properties. Proc Natl Acad Sci USA. 2002;99:8231–8235. doi: 10.1073/pnas.122238899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paone G, et al. ADP-ribosyltransferase-specific modification of human neutrophil peptide-1. J Biol Chem. 2006;281:17054–17060. doi: 10.1074/jbc.M603042200. [DOI] [PubMed] [Google Scholar]
- 19.Pazgier M, Li X, Lu W, Lubkowski J. Human defensins: Synthesis and structural properties. Curr Pharm Des. 2007;13:3096–3118. doi: 10.2174/138161207782110381. [DOI] [PubMed] [Google Scholar]
- 20.Jenkinson CP, Grody WW, Cederbaum SD. Comparative properties of arginases. Comp Biochem Physiol B Biochem Mol Biol. 1996;114:107–132. doi: 10.1016/0305-0491(95)02138-8. [DOI] [PubMed] [Google Scholar]
- 21.Bachetti T, et al. Arginase pathway in human endothelial cells in pathophysiological conditions. J Mol Cell Cardiol. 2004;37:515–523. doi: 10.1016/j.yjmcc.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Sell DR, Monnier VM. Conversion of arginine into ornithine by advanced glycation in senescent human collagen and lens crystallins. J Biol Chem. 2004;279:54173–54184. doi: 10.1074/jbc.M408946200. [DOI] [PubMed] [Google Scholar]
- 23.Jacobson EL, Cervantes-Laurean D, Jacobson MK. Glycation of proteins by ADP-ribose. Mol Cell Biochem. 1994;138:207–212. doi: 10.1007/BF00928463. [DOI] [PubMed] [Google Scholar]
- 24.Cervantes-Laurean D, Jacobson EL, Jacobson MK. Glycation and glycoxidation of histones by ADP-ribose. J Biol Chem. 1996;271:10461–10469. doi: 10.1074/jbc.271.18.10461. [DOI] [PubMed] [Google Scholar]
- 25.Paul RG, Avery NC, Slatter DA, Sims TJ, Bailey AJ. Isolation and characterization of advanced glycation end products derived from the in vitro reaction of ribose and collagen. Biochem J. 1998;330(Pt 3):1241–1248. doi: 10.1042/bj3301241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shapiro R, McManus MJ, Zalut C, Bunn HF. Sites of nonenzymatic glycosylation of human hemoglobin A. J Biol Chem. 1980;255:3120–3127. [PubMed] [Google Scholar]
- 27.Sun Y, Hayakawa S, Izumori K. Modification of ovalbumin with a rare ketohexose through the Maillard reaction: Effect on protein structure and gel properties. J Agric Food Chem. 2004;52:1293–1299. doi: 10.1021/jf030428s. [DOI] [PubMed] [Google Scholar]
- 28.Hsia JA, et al. Amino acid-specific ADP-ribosylation. Sensitivity to hydroxylamine of [cysteine(ADP-ribose)]protein and [arginine(ADP-ribose)]protein linkages. J Biol Chem. 1985;260:16187–16191. [PubMed] [Google Scholar]
- 29.Cervantes-Laurean D, Minter DE, Jacobson EL, Jacobson MK. Protein glycation by ADP-ribose: Studies of model conjugates. Biochemistry. 1993;32:1528–1534. doi: 10.1021/bi00057a017. [DOI] [PubMed] [Google Scholar]
- 30.Moss J, et al. Molecular and immunological characterization of ADP-ribosylarginine hydrolases. J Biol Chem. 1992;267:10481–10488. [PubMed] [Google Scholar]
- 31.Moss J, Vaughan M. Isolation of an avian erythrocyte protein possessing ADP-ribosyltransferase activity and capable of activating adenylate cyclase. Proc Natl Acad Sci USA. 1978;75:3621–3624. doi: 10.1073/pnas.75.8.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moss J, Vaughan M. Mechanism of action of choleragen. Evidence for ADP-ribosyltransferase activity with arginine as an acceptor. J Biol Chem. 1977;252:2455–2457. [PubMed] [Google Scholar]