Abstract
Targeted mRNA trafficking and local translation may play a significant role in controlling protein localization. Here we examined for the first time the localization of all (≈50) mRNAs encoding peroxisomal proteins (mPPs) involved in peroxisome biogenesis and function. By using the bacteriophage MS2-CP RNA-binding protein (RBP) fused to multiple copies of GFP, we demonstrated that >40 endogenously expressed mPPs tagged with the MS2 aptamer form fluorescent RNA granules in vivo. The use of different RFP-tagged organellar markers revealed 3 basic patterns of mPP granule localization: to peroxisomes, to the endoplasmic reticulum (ER), and nonperoxisomal. Twelve mPPs (i.e., PEX1, PEX5, PEX8, PEX11–15, DCI1, NPY1, PCS60, and POX1) had a high percentage (52%–80%) of mRNA colocalization with peroxisomes. Thirteen mPPs (i.e., AAT2, PEX6, MDH3, PEX28, etc.) showed a low percentage (30%–42%) of colocalization, and 1 mPP (PEX3) preferentially localized to the ER. The mPPs of the nonperoxisomal pattern (i.e., GPD1, PCD1, PEX7) showed ≪30% colocalization. mPP association with the peroxisome or ER was verified using cell fractionation and RT-PCR analysis. A model mPP, PEX14 mRNA, was found to be in close association with peroxisomes throughout the cell cycle, with its localization depending in part on the 3′-UTR, initiation of translation, and the Puf5 RBP. The different patterns of mPP localization observed suggest that multiple mechanisms involved in mRNA localization and translation may play roles in the importation of protein into peroxisomes.
Keywords: mRNA localization, peroxisomes
Eukaryotic cells are organized into separate compartments and structures, each with a distinctive set of proteins. In addition to targeting sequences (i.e., signal peptides, mitochondrial and peroxisomal targeting sequences) being embedded in proteins, directed mRNA localization and local translation may control intracellular protein targeting (1–3). mRNA localization is an efficient way to achieve protein localization, because a single mRNA molecule can serve as a template for multiple rounds of translation. As localized translation allows cells to quickly respond to changes in environmental conditions, it can be advantageous to localize mRNA, rather than protein, at the site of protein function (1–3).
One well-studied example of mRNA localization is that of ASH1 mRNA, which localizes to the bud tip in yeast and regulates mating-type switching (cell fate determination) (1, 3, 4). The mechanism by which ASH1 mRNA localizes involves cis sequences in the open-reading frame (ORF) and 3′-UTR, and several trans-acting factors, including She1–5 (1, 3, 4). The latter include the She2 RNA-binding protein (RBP) that binds ASH1 mRNA and She1/Myo4, a type V myosin that transports ribonucleoprotein (RNP) particles (5, 6). In addition, mRNAs encoding polarity and secretion factors (e.g., Sec4, Sro7, Cdc42) also target to the bud tip to facilitate cell growth (7). These mRNAs use the She machinery as well and, along with ASH1 mRNA, anchor to the endoplasmic reticulum (ER) and are transported to the incipient bud (7, 8). mRNA anchoring to the ER allows for the cotransport of both message and translation/translocation machinery, and is conserved through evolution (8). Another example of mRNA trafficking is to mitochondria. ATP2 mRNA targets to yeast mitochondria; impaired trafficking leads to respiratory deficiencies due to inefficient protein importation (9). Microarray analyses have demonstrated that ≈500 nuclear-encoded mRNAs localize to mitochondrion-bound polysomes (10, 11). About half of these mRNAs contain a binding site for the Puf3 RBP in their 3′-UTR (12), and the loss of PUF3 gene expression influences mRNA association with mitochondria (11). Because the 3′-UTR sequences of certain yeast and human mitochondrial genes (i.e., OXA1) are functionally conserved and important for mRNA localization (13), it is likely that the machinery for targeting mRNA to mitochondria evolved from simple eukaryotes.
Yet despite advances in understanding the importance of mRNA trafficking, an overall picture of genomewide mRNA localization (the “mRNA localizome”) is lacking. To better understand the extent of mRNA localization in yeast, we developed a novel gene-tagging strategy to visualize mRNAs in vivo (14). This technique inserts binding sites [(e.g., the MS2 aptamer/loop sequence (MS2L)] for the MS2 bacteriophage coat protein (MS2-CP) into any gene of interest in the yeast genome. On coexpression of MS2-CP fused with GFP(x3), endogenously expressed mRNAs can be observed in vivo for the first time. This technique, called m-TAG, has allowed us to localized endogenous ASH1 and SRO7 mRNA to the bud tip, PEX3 mRNA to the ER, and OXA1 mRNA to the mitochondria (14). In the present study, we used m-TAG to localize mRNAs coding for proteins involved in peroxisome biogenesis and function.
Peroxisomes are found in all eukaryotic cells and facilitate functions related to the β-oxidation of fatty acids and synthesis of cholesterol, bile acids, and plasmogens (15). The existence of heritable disorders related to peroxisome dysfunction underscores the importance of this organelle in lipid metabolism in humans (16). Importantly, some features of peroxisomes resemble those of mitochondria and chloroplasts, including the posttranslational importation of proteins into preexisting organelles. However, peroxisomes differ in that they are surrounded by a single lipid bilayer, do not contain DNA or ribosomes, and import all of their protein content from the cytoplasm. Many peroxisomal proteins contain a peroxisomal targeting signal (PTS) that is sufficient for targeting to the peroxisome matrix. PTS1 is a tripeptide consensus sequence at the C terminus of some proteins (15, 17), while others use a signal at the N terminus called PTS2 (15, 18).
By using fluorescence imaging and subcellular fractionation experiments, we show 3 localization patterns for mRNAs encoding peroxisomal proteins (mPPs). One set of mPPs associates with peroxisomes, a finding that hints at the cotranslational importation of proteins via membrane-bound polysomes. A second set, comprising PEX3 mRNA, associates with ER and is consistent with the fact that Pex3 translocates to the ER (19). Finally, a third set of mRNAs does not localize to peroxisomes. Thus, at least 3 mRNA targeting paths are involved in the importation of proteins into this organelle. These may define distinct import routes as a consequence of protein synthesis on ribosomes associated with peroxisomes, ER-bound ribosomes, or free ribosomes in the cytoplasm.
Results
mRNAs Coding for Specific Peroxins Localize to the Peroxisome.
To examine endogenous mPP localization, we used m-TAG to create strains tagged with the MS2L sequence (Table S1). We first localized mRNAs encoding proteins involved in peroxisome biogenesis, called peroxins (PEX1–3, 5–8, 10–15, 17, 19, 21, 22, 27–30, and 32). On the expression of MS2-CP-GFP(x3) in cells bearing the tagged genes, we observed that few had fluorescent RNA granules when grown on glucose-containing medium. But when grown under conditions that induce peroxisome proliferation (i.e., media containing oleate), we saw a large increase in the number of cells bearing fluorescent granules. We then determined that between 56% and 80% of granules seen in the MS2L-tagged PEX1, 5, 8, 11, 12, 13, 14, or 15 strains colocalized with peroxisomes labeled with a peroxisomal matrix marker, RFP-PTS1 (68%, 56%, 66%, 80%, 58%, 78%, 60%, and 78% colocalization, respectively; Fig. 1A and Table S2). Thus, mPPs associate with peroxisomes, although we noted that the number of RFP-labeled peroxisomes observed per cell (≈2–6) was usually greater than the number of granules (≈1–3). This may indicate that mPPs are in transient/intermittent association with peroxisomes, or that there are distinct (i.e., mature) peroxisomes that do not associate with mRNA.
Fig. 1.
Localization of endogenous mRNAs encoding peroxins. (A) Representative fluorescence microscopy images of cells bearing the MS2L sequence integrated into different genes (as indicated; see the ORFINTstrains in Table S2) and transformed with plasmids expressing MS2-CP fused with 3 GFP molecules [MS2-CP-GFP(x3)] and RFP-PTS1, as a marker for the peroxisomes, are shown. Cells were grown overnight on medium containing oleate and induced with the same medium lacking methionine for 1 h before visualization. The percentage of GFP-labeled RNA granules that colocalize with peroxisomes is shown. mRNA indicates labeled RNA granules; RFP-PTS1 indicates peroxisomes. (Scale bar: 2 μm). (B) Integration of the MS2 loops does not alter protein function. MS2L-integrated yeast strains (as indicated) were grown to log phase on glucose-containing medium, normalized for cell number, diluted serially, and plated by drops onto solid medium containing oleate.
In contrast, other peroxin mRNAs, such as PEX6, 10, and 27–29, showed a low level of colocalization with peroxisomes (33%, 30%, 32%, 40%, and 32%, respectively), while some (i.e., PEX2, 7, 17, 18, 22, 30, and 32) showed little to no colocalization (6%, 5%, 24%, 18%, 18%, 16%, and 20%, respectively; Fig. 1A and Table S2). These results indicate significant variability in the extent of peroxin mRNA localization to peroxisomes. As a control, we examined the ability of mRNAs known to localize to the bud (e.g., ASH1) or mitochondria (e.g., ATP2, OXA1) to colocalize with peroxisomes. We found that tagged ASH1, ATP2, and OXA1 mRNAs showed a very low level of colocalization with peroxisomes (18%, 18%, and 12%, respectively; Fig. S1A). Thus, background mRNA colocalization with peroxisomes (probably due to the compact nature of yeast cells) is on the order of <20%. mPP localization to the peroxisome was considered significant when colocalization values were on the order of ≈50%, although lower values (i.e., 30%–40%) might indicate a transient or intermittent association.
Although m-TAG can visualize granules containing as few as 2 copies of mRNA (14), we did not observe fluorescent granules in some strains (i.e., PEX10) using MS2-CP-GFP(x3) (Table S2). This indicates that these mPPs either are present in single copy or are poorly expressed. Thus, we used MS2-CP fused to 4 GFP molecules [MS2-CP-GFP(x4)] to increase their fluorescence signature. We observed fluorescent granules in a high percentage (≈30%) of cells expressing MS2-CP-GFP(x4); however, only a few mPPs (i.e., PEX10 and 27–29) had granules that colocalized with peroxisomes to any degree (30%, 32%, 40%, and 32%, respectively), while others (i.e., PEX18 and 22) showed no colocalization (18% and 18%, respectively; Table S2). Unlike MS2-CP-GFP(x3), MS2-CP-GFP(x4) yielded large fluorescent granules in ≈10% of cells, which might have been protein–RNA aggregates that were unable to localize properly. Thus, mPPs visualized with MS2-CP-GFP(x4) could register lower than actual values of colocalization. However, we have found no differences in mRNA localization using either MS2-CP-GFP(x3) or MS2-CP-GFP(x4) when localizing mRNAs encoding secreted or mitochondrial proteins in other ongoing studies.
Functional peroxisomes are required for yeast to be able to use oleate as a carbon source. To verify that the MS2L sequence inserted between the ORF and 3′-UTR does not alter protein function, we examined the ability of the tagged strains to grow on oleate-containing plates (Fig. 1B). Yeast expressing tagged PEX3 and PEX14 mRNAs grew like wild-type cells, whereas strains lacking these genes were unable to grow (Fig. 1B). Thus, MS2L insertion does not alter protein function, as shown previously (14).
mRNAs Coding for Specific Peroxisomal Matrix Proteins Localize to the Peroxisome.
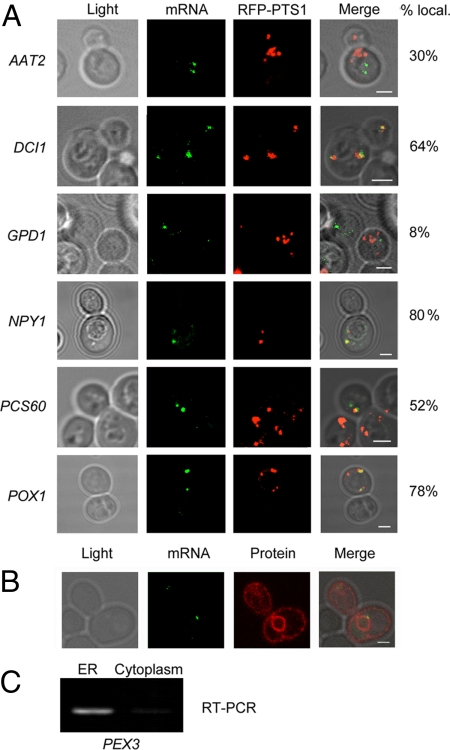
Another group of genes encodes peroxisomal matrix proteins, many of which have PTSs to ensure importation into peroxisomes. In some cases (i.e., PCD1, POX1, and TES1), however, no known PTS has been identified, and the targeting mechanism is unclear. We examined the localization of mRNAs encoding matrix proteins and found that some (i.e., DCI1, NPY1, PCS60, and POX1) colocalized with peroxisomes to a high degree (64%, 80%, 52%, and 78%, respectively; Fig. 2A and Table S2). In contrast, others (i.e., AAT2, CIT2, and MDH3) showed a low level of colocalization (30%, 40%, and 42%, respectively), while some (i.e., PCD1 and POT1) showed none (8% and 24%, respectively; Fig. 2A and Table S2). Thus, the ability of matrix protein mRNAs to localize with peroxisomes varies as well. We performed quantitative fluorescence analysis (14) to determine the transcript number in POX1 granules and found an average of 3.2 copies per cell (range, 1.1–7.1 copies; n = 14 granules). This number matches microarray studies that predict ≈3 copies of POX1 mRNA on oleate-containing medium, according to the Saccharomyces Genome Database. Thus, m-TAG appears to detect the available mPPs, although we cannot rule out the possibility that some are missed using MS2-CP-GFP(x3).
Fig. 2.
Localization of endogenous mRNAs encoding matrix proteins. (A) Representative fluorescence microscopy images of cells bearing the MS2L sequence integrated into different genes (as indicated; see the ORFINTstrains in Table S2) and transformed with plasmids expressing MS2-CP-GFP(x3) and RFP-PTS1. Cells were grown on medium containing oleate and induced with the same medium lacking methionine for 1 h before visualization. The percentage of GFP-labeled RNA granules that colocalize with peroxisomes is shown. mRNA indicates labeled RNA granules; RFP-PTS1 indicates peroxisomes. (Scale bar: 2 μm). (B) PEX3 mRNA localizes to the ER. Representative fluorescence microscopy images of MS2L-tagged PEX3 cells transformed with plasmids expressing MS2-CP-GFP(x3) and Sec63-RFP (an ER marker). mRNA indicates labeled RNA granules; protein indicates ER labeled with Sec63-RFP. The percentage of GFP-labeled granules that colocalize with ER is shown. (C) Subcellular fractionation. Yeast expressing Sec63-GFP was fractionated by density gradient centrifugation into ER and cytoplasmic fractions (same fractions as shown in fig. 9b in ref. 7). RT-PCR was performed on DNase-treated RNA derived from these fractions using PEX3-specific primers. PCR samples were then electrophoresed and visualized on a 1% agarose gel. (See fig. 9b in ref. 7 for other RNAs detected using the same conditions.)
Several mRNAs encoding matrix proteins did not form fluorescent granules using MS2-CP-GFP(x3), as observed with some peroxin mRNAs (Table S2). We used MS2-CP-GFP(x4) to visualize these mPPs (i.e., ANT1, CTA1, FAA2, FAT1, FOX2, GPD1, and PXA1), but found that only a few (i.e., CTA1, FAT1, and PXA1) had significant colocalization with peroxisomes (36%, 32%, and 38%, respectively; Table S2).
PEX3 mRNA Localizes to the ER.
While some mPPs (e.g., POX1) colocalize with peroxisomes and others (e.g., GPD1) do not, we determined previously that PEX3 mRNA localizes to ER (14). We reexamined this association using Sec63-RFP as an ER marker (Fig. 2B) and found 80% colocalization between PEX3 mRNA and ER, as reported previously (14). We next determined whether PEX3 mRNA colocalizes with peroxisomes and found 30% colocalization (Table S2). Subcellular fractionation was used to verify the association of PEX3 mRNA with ER, using a nonlinear sucrose density gradient to separate the postnuclear supernatant (PNS) into ER and cytosolic fractions, respectively (7). PEX3 mRNA was observed only in the ER fraction (Fig. 2C), a finding identical to that for other yeast mRNAs (i.e., SEC4, CDC42, and ASH1) that associate with ER membranes (7). This contrasts with the RDN18 ribosomal RNA, which associates with both the ER and cytosolic fractions (7). Thus, PEX3 mRNA preferentially associates with ER.
To further analyze the specificity of mPP targeting, we examined the localization of the POX1 and GPD1 mRNAs in cells expressing Sec63-RFP (Fig. S1 B and C and Table S3). Interestingly, a high percentage of both RNAs colocalized with Sec63-RFP (59% and 71%, respectively; Table S3 and Fig. S1B and C). While mRNA localization to the ER could be due to the fact that ER fills much of the cell volume (see, e.g., Fig. S1B and C), we also demonstrated that peroxisomes decorate both nuclear and cortical ER (Fig. S1D). Thus, discerning whether mPP localization to the ER results from a direct association with ER membranes or is a consequence of binding to ER-associated peroxisomes is difficult. Alternatively, GPD1 mRNA, which encodes a glycerol-3-phosphate dehydrogenase found in the cytosol and peroxisomes, may preferentially localize to ER-like mRNAs encoding cytoplasmic proteins (13). This could allow for protein distribution to either compartment.
We next examined the localization of POX1 mRNA in cells expressing Oxa1-RFP, a mitochondrial marker, and found that 44% of POX1 granules colocalized with mitochondria (Table S4). In contrast, 82% of OXA1 mRNA granules [which localize to mitochondria (13, 14)] colocalized with Oxa1-RFP. The large number of cells having POX1 mRNA localized to mitochondria might stem from the fact that Oxa1-RFP-labeled mitochondria fill a substantial volume of the cell. Alternatively, evidence for cargo-selective transport between mitochondria and peroxisomes has been reported (20), indicating that these organelles are in close contact. Thus, mPPs may appear juxtaposed to mitochondria, without necessarily being associated with the mitochondrial membrane.
Subcellular Fractionation of Peroxisomes and the Detection of mPPs.
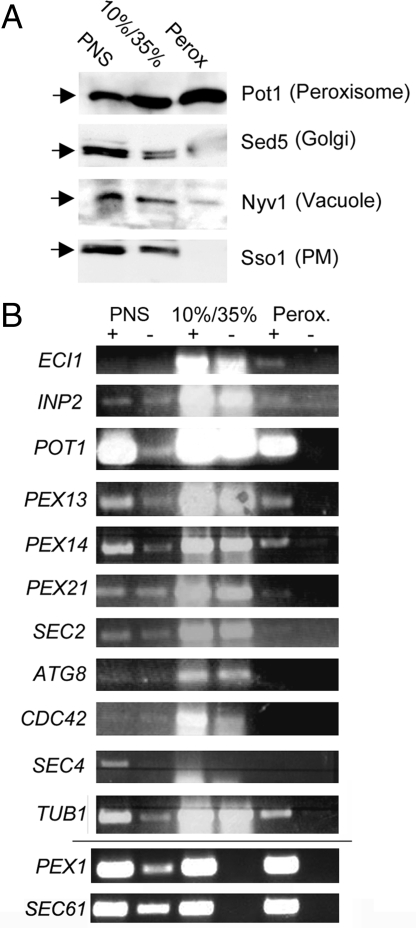
To verify mRNA localization to peroxisomes, we used subcellular fractionation and RT-PCR, which was used to demonstrate the association of polarized mRNAs (i.e., CDC42, SEC4, ASH1, and SRO7) with ER (7). We used a yeast peroxisome purification procedure and added an affinity purification step that uses anti-HA epitope-conjugated beads to pull down peroxisomes decorated with HA-tagged Pex30 expressed from the PEX30 locus. Yeast expressing PEX30-HA were grown on oleate and processed to obtain a purified peroxisomal fraction. The different fractions (e.g., PNS; 10%/35% Nycodenz interface that contains mitochondria/membranes; and peroxisome layer) were analyzed using antibodies against peroxisomal (e.g., Pot1), Golgi (e.g., Sed5), vacuolar (e.g., Nyv1), and plasma membrane (e.g., Sso1) proteins. We observed an enrichment of Pot1 in the peroxisome fraction, along with a concomitant reduction in the presence of other organellar markers (Fig. 3A).
Fig. 3.
Examination of mPP localization using cell fractionation and RT-PCR. (A) Western blot analysis of peroxisome purification. Peroxisomes were purified using density gradient centrifugation followed by affinity purification. Samples (40 μg) from the 3 fractions (PNS, postnuclear supernatant; 10%/35%, mitochondria-enriched membrane fraction; Perox, purified peroxisomes) were electrophoresed on SDS/PAGE gels and analyzed by immunoblotting. Antibodies (1:5,000) against Pot1 (peroxisome), Sed5 (Golgi), Nyv1 (Vacuole), and Sso1 (PM) were used for the chemiluminescent detection of proteins. (B) RT-PCR analysis of purified peroxisomes. Yeast cells were fractionated, and samples from the PNS, 10%/35%, and peroxisomal fractions were collected, from which RNA was purified. After DNase treatment and reverse transcription (RT; +), PCR was performed using gene-specific primers (as indicated). Samples without RT (-) were used as controls for DNA contamination. After PCR, samples were electrophoresed on agarose gels and documented. PEX1 and SEC61 mRNAs were detected in a parallel experiment.
In parallel, RNA isolated from these fractions was subjected to RT-PCR to identify mRNAs (Fig. 3B). Samples without RT also were used in the PCR as controls for DNA contamination and although amplification was observed in a few cases (i.e., with the 10%/35% interface), we saw a significant enrichment of mPPs in the peroxisome fraction. We could identify (Fig. 3B) mRNAs observed to localize with peroxisomes using microscopy, including PEX1 and PEX14 (Fig. 1A), as well as those that were not visualized (i.e., ECI1, INP2, and PEX21) (Table S2). Interestingly, POT1 mRNA was abundant in the peroxisome fraction (Fig. 3B), although only a low percentage of POT1 granules localized to peroxisomes with fluorescence microscopy (Fig. 2A). The reason for this is unclear, but it could be due to the elevated levels of POT1 mRNA in the cell lysate or because the MS2L sequence interferes with POT1 mRNA localization. Importantly, mRNAs known to localize with cortical ER (i.e., CDC42 and SEC4) (7) were not observed in the peroxisome fraction, although SEC61 mRNA was. This may represent nuclear ER contamination; studies have shown that peroxisomes are derived from ER (19). Moreover, we noted a close link between the ER and peroxisomes (Fig. S1D), which might explain the presence of ER-associated mRNAs (Fig. 3B). Finally, TUB1 mRNA also was detected (Fig. 3B), which may be related to findings showing that peroxisomes move along microtubules (21). Together, these results confirm that mPPs may associate with peroxisomes.
PEX14 mRNA Associates with Peroxisomes Throughout the Cell Cycle.
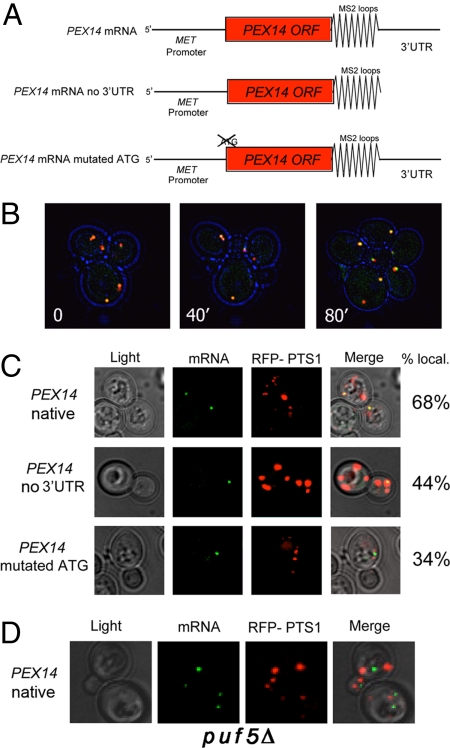
Pex14 is a peroxisomal membrane protein involved in the importation of PTS1- and PTS2-containing matrix proteins (22). The mechanism by which Pex14 is targeted and inserted into the peroxisome membrane is not clear, however. Because endogenous PEX14 mRNA associates with peroxisomes (Figs. 1A and 3B), we examined an mRNA–organelle association using time-lapse microscopy. To monitor PEX14 mRNA localization, we expressed PEX14 bearing the MS2 loops and 3′-UTR under the control of an inducible promoter (Fig. 4A), along with RFP-PTS1 and MS2-CP-GFP(x1) in wild-type cells. As shown in Fig. 4B and Movie S1, GFP-labeled mRNA is always in close proximity to the RFP-labeled peroxisomes throughout the cell division cycle. To help determine how PEX14 mRNA localizes with peroxisomes, we compared the localization of full-length PEX14 mRNA with PEX14 mRNA lacking its 3′-UTR and with PEX14 mRNA lacking its initial ATG to prevent translation (Fig. 4A). Plasmid-based expression of native PEX14 or PEX14 lacking its 3′-UTR was found to confer robust growth to pex14Δ cells on oleate-containing medium, whereas PEX14 lacking its ATG did not (Fig. S2A). Correspondingly, the expression of native PEX14 and PEX14 lacking its 3′-UTR but not its ATG could be verified by Western blot analysis (Fig. S2B). We then examined localization of the native and mutant PEX14 mRNAs, and found that 68% of native PEX14 granules colocalized with RFP-PTS1 (Fig. 4C), which was similar to endogenous PEX14 mRNA (Fig. 1A and Table S2). This was reduced to 44% when the 3′-UTR was removed and to 34% when the start codon was removed (Fig. 4C). Thus, both the 3′-UTR and translation initiation may contribute to PEX14 mRNA localization. While their removal does not block colocalization altogether, the differences observed are significant if background colocalization [up to 18%, as suggested from the mPP- and control mRNA-peroxisome localization experiments (Figs. 1A, 2A, and S1A)] is subtracted.
Fig. 4.
Localization of PEX14 mRNA. (A) Illustration of the different MS2L-tagged PEX14 mRNAs. Three different MS2L-tagged PEX14 constructs— native, no 3′-UTR, and mutated ATG—all expressed under the MET25-inducible promoter, were used. (B) PEX14 mRNA associates with peroxisomes throughout the cell cycle. Representative time-lapse microscopy images of cells expressing tagged native PEX14 mRNA, as well as MS2-CP fused with 1 molecule of GFP (MS2-CP-GFP) and RFP-PTS1, are shown. Cells were grown on glucose-containing medium, and images were obtained with a DeltaVision imaging system at different times (minutes). For a series of deconvoluted images, see Movie S1. (C) Removal of the 3′-UTR or ATG diminishes PEX14 mRNA localization to peroxisomes. Representative images of cells expressing the tagged PEX14 mRNAs (i.e., native, no 3′-UTR, and mutated ATG) cotransformed with plasmids expressing MS2-CP-GFP(x3) and RFP-PTS1 are shown. Cells were grown overnight on medium containing oleate and shifted to the same medium lacking methionine for 1.5 h. The percentage of cells showing mRNA colocalization with peroxisomes is indicated. (D) Loss of PUF5 expression diminishes PEX14 mRNA localization to peroxisomes. Representative images show cells expressing tagged native PEX14 mRNA, MS2-CP-GFP(x3), and RFP-PTS1 in GAL1-PUF5 cells grown overnight on medium containing oleate and shifted to the same medium lacking methionine for 1.5 h. mRNA indicates GFP-labeled RNA granules; RFP-PTS1 indicates peroxisomes.
We then examined the localization of PEX14 mRNA in pex3Δ cells (Fig. S1E). Cells lacking PEX3 are devoid of peroxisomes (19), and the expression of RFP-PTS1 led to cytosolic staining, as expected. Although fluorescent RNA granules were observed in these cells, they showed no specific pattern of localization. This indicates that the absence of either Pex3 or peroxisomes does not affect PEX14 mRNA granule formation.
Loss of Puf5 Expression Affects PEX14 mRNA Localization.
Puf5 belongs to the conserved Pumilio family (PUF) of RBPs. PUF proteins bind specific sequences in the 3′-UTR of target transcripts and inhibit the stability or translational control of these mRNAs (23). A systematic identification of mRNA targets for the PUF proteins has revealed that PEX14 mRNA binds to Puf5 (12). Thus, we examined whether the loss of PUF5 expression affects PEX14 mRNA localization. To control Puf5 levels, we inserted a GAL1 promoter upstream of the PUF5 gene (GAL1-PUF5) and grew both wild-type and GAL1-PUF5 yeast on either galactose-containing medium (to induce PUF5) or oleate-containing medium (to induce peroxisomes and turn off PUF5). Fig. S3 shows PUF5 mRNA detection under the different conditions. We then examined colocalization between PEX14 mRNA and RFP-labeled peroxisomes in GAL1-PUF5 cells grown on oleate. The amount of mRNA granules colocalized with peroxisomes was reduced from 68% (in wild-type cells) to 50% (Fig. 4D). While this difference is small, we did observe a fair number of cells that exhibited no colocalization (see, e.g., the example in Fig. 4D). Thus, Puf5 may regulate the amount of time that PEX14 mRNA interacts with peroxisomes, which could account for these differences.
Discussion
Our examination of the localization of mPPs revealed 3 basic patterns: peroxisome-associated, ER-associated, and nonperoxisomal. Beause mRNA localization and local translation represent an efficient means of targeting proteins to subcellular compartments (3, 24), it seems likely that peroxins and matrix proteins gain better access to the protein import machinery via mRNA targeting. Transcript localization to subcellular compartments, such as mitochondria (10, 13) or peroxisomes, is expected to result in a high concentration of newly translated protein at the organellar surface.
How peroxins target to peroxisomes is unclear, although several, such as Pex13 and Pex11, are targeted by binding to Pex19 and Pex3 (25). Yet a Pex13 mutant unable to bind Pex19 is imported into peroxisomes (26), suggesting the existence of multiple importation pathways. We found that many peroxin-encoded mRNAs colocalize with peroxisomes (Fig. 1A and Table S2), which may help their translation products to target the importation machinery. To visualize endogenous mPPs, we needed to induce gene expression by substituting oleic acid for glucose as the carbon source. Thus, mPP localization in cells grown on glucose, which have a low number of peroxisomes, is an open question. Yet PEX14 mRNA expressed from plasmids in these cells clearly colocalized with peroxisomes (Fig. 4B and Movie S1). Most, if not all, peroxisomes were labeled with fluorescent RNA granules under these mild overexpression conditions (e.g., CEN plasmid, MET25 promoter) indicating that the RNA-binding machinery on the peroxisome surface is not saturated, and that all peroxisomes have the propensity to bind mRNA. Although we do not know whether the mRNAs are present in RNPs, polysomes, or both, it seems likely that bound mPPs allow for the local translation and importation of functional proteins into peroxisomes. This is supported by the fact that plasmid-expressed MS2L-tagged PEX14 mRNA confers growth to pex14Δ cells on oleate-containing medium (Fig. S2A). Interestingly, mutation of the translation initiation codon in PEX14 diminished PEX14 mRNA localization (Fig. 4C), suggesting that PEX14 mRNA localization depends in part on translation. This also implies that translational control affects the association of polysome with peroxisomes, because the PTS1 signal is exposed to the importation machinery only at the end of translation. This makes it unlikely that mPPs localize in a PTS1-dependent fashion. PEX14 mRNA that lacks its 3′-UTR also showed diminished localization to peroxisomes but, ironically, conferred slightly better growth to pex14Δ cells than native PEX14 (Fig. S2). While studies have shown that sequences in the 3′-UTR affect mRNA localization (1, 3, 4, 27), these sequences are also known to control RNA stability (28). Thus, loss of the PEX14 3′-UTR might promote RNA stability and yield higher levels of translation.
Unlike other peroxin mRNAs, PEX3 mRNA showed little colocalization with peroxisomes (Table S2), but colocalized with ER in vivo (Fig. 2B) and copurified with ER membranes (Fig. 2C). PEX3 mRNA localization to ER membranes likely guarantees Pex3 access to the secretory pathway, which is necessary for peroxisome biogenesis de novo (19, 29). Yet other mPPs, such as POX1 and GPD1, also colocalized to some degree with the ER (Table S3). The association of mPP with the ER could result from direct targeting (i.e., as in the case of PEX3 mRNA) or indirectly due to the known ER–peroxisome connection (19). While our own observations indicate that peroxisomes are tightly juxtaposed with either cortical or nuclear ER (Fig. S1D), cell fractionation experiments reveal that mPPs copurify with peroxisomes (Fig. 3B). Thus, it seems likely that peroxin or matrix protein mRNAs interact with peroxisomal membranes rather than with ER. Due to the nature of peroxisome biogenesis and placement, some degree of interaction with the ER cannot be ruled out, however.
The second set of mPPs examined coded for matrix proteins, most harboring either the PTS1 or PTS2 signals. A few (e.g, Pox1, Inp1) lack a known PTS, however, and how they access peroxisomes is unclear. We found that many matrix protein mRNAs (e.g., POX1, DCI1) localize to peroxisomes (Figs. 2A and 3; Table S2), yet some (e.g., GPD1, PCD1) did not. The reason for the differences in mPP (either peroxin or matrix) localization is unclear; we found no correlation between the presence/absence of a known PTS and transcript localization. One possibility is that some proteins need to access the peroxisome immediately on translation, perhaps in consideration of protein folding issues. Likewise, assemblies of peroxisomal proteins might necessitate translational coordination for coimportation.
mRNA localization depends on interactions between cis-acting elements in the mRNA sequence and RBPs. PEX14 mRNA has been shown to be a target of Puf5 (12), and we found that the level of PUF5 expression had some effect on PEX14 mRNA localization (Fig. 4D). But because puf5Δ cells grow on oleate-containing medium, it is unlikely that this RBP is the only trans-acting factor involved in PEX14 mRNA localization. Together, these results demonstrate the localization of mPPs for the first time and suggest that multiple independent pathways are used for mRNA targeting. Moreover, they imply that localized translation and possibly cotranslational translocation may be involved in protein importation into peroxisomes.
Methods
Supporting Information.
See SI Text for a full description of the materials and methods used, including strains, growth conditions, plasmids, mRNA visualization, and peroxisome purification/mRNA analysis.
Peroxisome Isolation and mRNA Detection using RT-PCR.
Yeast expressing HA-Pex30 was grown at 30 °C to an OD600 of 1 in rich medium. Cells were transferred to a peroxisome induction medium containing 0.3% yeast extract, 0.5% peptone, 0.5% KH2PO4 (pH 6), and 0.225% oleate. After growth overnight, cells were spheroplasted, washed, and resuspended in peroxisome suspension buffer [PSB; 5 mM Mes, 1 mM KCl, 0.5 mM EDTA (pH 6), 12% PEG1500, and 160 mM sucrose] containing protease inhibitors. Spheroplasts were homogenized using a Dounce homogenizer and pelleted for 5 min at 1500 × g. The postnuclear supernatant was diluted with 50% Nycodenz (in PSB) to a final concentration of 10% and placed on a discontinuous Nycodenz gradient (35%; 50% Nycodenz in PSB). Gradients were centrifuged at 80,000 × g for 1.5 h at 4 °C and the peroxisome fraction (visible at the 35% and 50% interface) was removed, diluted with peroxisome dilution buffer [PDB; 50 mM Tris HCl (pH 7.5), 1 mM DTT, 12% PEG 1500, 160 mM sucrose, protein inhibitors, and Rnasin], and centrifuged at 80,500 × g for 20 min. Then the peroxisome fraction was resuspended in PDB and incubated with 40 μL HA-conjugated agarose beads overnight at 4 °C. Beads were washed with PDB, and samples of purified peroxisomes were obtained for Western blot analysis and mRNA purification. RNA was purified using an RNA purification kit and subjected to reverse- transcription and then PCR with specific oligonucleotides using standard procedures. PCR samples were electrophoresed on agarose gels and documented. Protein samples were electrophoresed on 8% SDS/PAGE gels, blotted, and detected in immunoblots using polyclonal anti-Pot1, anti-Sso1, anti-Nyv1, and anti-Sed5 antibodies.
Supplementary Material
Acknowledgments.
We thank K. Bloom, S. Keranen, A. Mayer, S. Michaelis, H. Pelham, and R. Singer for reagents. This study was supported by grants to J.E.G. from the Minerva Foundation, Y. Leon Benoziyo Institute for Molecular Medicine, Center for Scientific Excellence, Kahn Fund for Systems Biology, and Josef Cohn-Minerva Center for Biomembrane Research at the Weizmann Institute of Science, Israel. J.E.G. holds the Henry Kaplan Chair in Cancer Research. C.B. is supported by the Elise-Richter Program of the Austrian Science Fund.
Footnotes
The authors declare no conflicts of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910754106/DCSupplemental.
References
- 1.St Johnston D. Moving messages: The intracellular localization of mRNAs. Nat Rev Mol Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- 2.Kloc M, Zearfoss NR, Etkin LD. Mechanisms of subcellular mRNA localization. Cell. 2002;108:533–544. doi: 10.1016/s0092-8674(02)00651-7. [DOI] [PubMed] [Google Scholar]
- 3.Du TG, Schmid M, Jansen RP. Why cells move messages: The biological functions of mRNA localization. Semin Cell Dev Biol. 2007;18:171–177. doi: 10.1016/j.semcdb.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 4.Muller M, Heuck A, Niessing D. Directional mRNA transport in eukaryotes: Lessons from yeast. Cell Mol Life Sci. 2007;64:171–180. doi: 10.1007/s00018-006-6286-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takizawa PA, Vale RD. The myosin motor, Myo4p, binds Ash1 mRNA via the adapter protein, She3p. Proc Natl Acad Sci USA. 2000;97:5273–5278. doi: 10.1073/pnas.080585897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohl F, Kruse C, Frank A, Ferring D, Jansen RP. She2p, a novel RNA-binding protein, tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 2000;19:5514–5524. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aronov S, et al. mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:3441–3455. doi: 10.1128/MCB.01643-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerst JE. Message on the web: mRNA and ER co-trafficking. Trends Cell Biol. 2008;18:68–76. doi: 10.1016/j.tcb.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Margeot A, et al. In Saccharomyces cerevisiae, ATP2 mRNA sorting to the vicinity of mitochondria is essential for respiratory function. EMBO J. 2002;21:6893–6904. doi: 10.1093/emboj/cdf690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marc P, et al. Genome-wide analysis of mRNAs targeted to yeast mitochondria. EMBO Rep. 2002;3:159–164. doi: 10.1093/embo-reports/kvf025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saint-Georges Y, et al. Yeast mitochondrial biogenesis: A role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS ONE. 2008;3:e2293. doi: 10.1371/journal.pone.0002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerber AP, Herschlag D, Brown PO. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2004;2:e79. doi: 10.1371/journal.pbio.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sylvestre J, Margeot A, Jacq C, Dujardin G, Corral-Debrinski M. The role of the 3′-untranslated region in mRNA sorting to the vicinity of mitochondria is conserved from yeast to human cells. Mol Biol Cell. 2003;14:3848–3856. doi: 10.1091/mbc.E03-02-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haim L, Zipor G, Aronov S, Gerst JE. A genomic integration method to visualize localization of endogenous mRNAs in living yeast. Nat Methods. 2007;4:409–412. doi: 10.1038/nmeth1040. [DOI] [PubMed] [Google Scholar]
- 15.Platta HW, Erdmann R. The peroxisomal protein import machinery. FEBS Lett. 2007;581:2811–2819. doi: 10.1016/j.febslet.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 16.Steinberg SJ, et al. Peroxisome biogenesis disorders. Biochim Biophys Acta. 2006;1763:1733–1748. doi: 10.1016/j.bbamcr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 17.Brocard C, Hartig A. Peroxisome targeting signal 1: Is it really a simple tripeptide? Biochim Biophys Acta. 2006;1763:1565–1573. doi: 10.1016/j.bbamcr.2006.08.022. [DOI] [PubMed] [Google Scholar]
- 18.Lazarow PB. The import receptor Pex7p and the PTS2 targeting sequence. Biochim Biophys Acta. 2006;1763:1599–1604. doi: 10.1016/j.bbamcr.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 19.Tabak HF, van der Zand A, Braakman I. Peroxisomes: Minted by the ER. Curr Opin Cell Biol. 2008;20:393–400. doi: 10.1016/j.ceb.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Neuspiel M, et al. Cargo-selected transport from the mitochondria to peroxisomes is mediated by vesicular carriers. Curr Biol. 2008;18:102–108. doi: 10.1016/j.cub.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 21.Kulic IM, et al. The role of microtubule movement in bidirectional organelle transport. Proc Natl Acad Sci USA. 2008;105:10011–10016. doi: 10.1073/pnas.0800031105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Azevedo JE, Schliebs W. Pex14p, more than just a docking protein. Biochim Biophys Acta. 2006;1763:1574–1584. doi: 10.1016/j.bbamcr.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Wharton RP, Aggarwal AK. mRNA regulation by Puf domain proteins. Sci STKE. 2006;2006:pe37. doi: 10.1126/stke.3542006pe37. [DOI] [PubMed] [Google Scholar]
- 24.Paquin N, Chartrand P. Local regulation of mRNA translation: New insights from the bud. Trends Cell Biol. 2008;18:105–111. doi: 10.1016/j.tcb.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Rottensteiner H, et al. Peroxisomal membrane proteins contain common Pex19p-binding sites that are an integral part of their targeting signals. Mol Biol Cell. 2004;15:3406–3417. doi: 10.1091/mbc.E04-03-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fransen M, et al. Potential role for Pex19p in assembly of PTS-receptor docking complexes. J Biol Chem. 2004;279:12615–12624. doi: 10.1074/jbc.M304941200. [DOI] [PubMed] [Google Scholar]
- 27.Chabanon H, Mickleburgh I, Hesketh J. Zipcodes and postage stamps: mRNA localisation signals and their trans-acting binding proteins. Brief Funct Genomic Proteomic. 2004;3:240–256. doi: 10.1093/bfgp/3.3.240. [DOI] [PubMed] [Google Scholar]
- 28.Shalgi R, Lapidot M, Shamir R, Pilpel Y. A catalog of stability-associated sequence elements in 3′-UTRs of yeast mRNAs. Genome Biol. 2005;6:R86. doi: 10.1186/gb-2005-6-10-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.