Abstract
White adipocytes have a unique structure in which nearly the entire cell volume is occupied by one large lipid droplet. However, the molecular and cellular processes involved in the cytoplasmic remodeling necessary to create this structure are poorly defined. Autophagy is a membrane trafficking process leading to lysosomal degradation. Here, we investigated the effect of the deletion of an essential autophagy gene, autophagy-related gene 7 (atg7), on adipogenesis. A mouse model with a targeted deletion of atg7 in adipose tissue was generated. The mutant mice were slim and contained only 20% of the mass of white adipose tissue (WAT) found in wild-type mice. Interestingly, ≈50% of the mutant white adipocytes were multilocular. The mutant white adipocytes were smaller with a larger volume of cytosol and contained more mitochondria. These cells exhibited altered fatty acid metabolism with increased rates of β-oxidation and reduced rates of hormone-induced lipolysis. Consistently, the mutant mice had lower fed plasma concentrations of fatty acids and the levels decreased at faster rates upon insulin stimuli. These mutant mice exhibited increased insulin sensitivity. The mutant mice also exhibited markedly decreased plasma concentrations of leptin but not adiponectin, lower plasma concentrations of triglyceride and cholesterol, and they had higher levels of basal physical activity. Strikingly, these mutant mice were resistant to high-fat-diet-induced obesity. Taken together, our results indicate that atg7, and by inference autophagy, plays an important role in normal adipogenesis and that inhibition of autophagy by disrupting the atg7 gene has a unique anti-obesity and insulin sensitization effect.
Keywords: diabetes, differentiation, knockout, metabolism, obesity
Obesity, which is associated with type II diabetes, atherosclerosis, hypertension, and cancer, is reaching a pandemic level in the developed world. Essentially, obesity results from an imbalance between energy intake and energy expenditure (1, 2). Clinically, obesity is characterized by increased mass of white adipose tissue (WAT), which stores excess energy in the form of triglyceride (TG) and serves as a major energy reservoir in mammals. In addition, WAT is an endocrine organ that is central to energy homeostasis regulation. It integrates metabolic signals and in turn regulates systemic energy balance by secreting adipokines, including leptin, adiponectin, and tumor necrosis factor (TNF)-α (1, 3).
Consistent with its energy storage function, a mature white adipocyte has a unique structure in which almost the entire cytoplasm is occupied by one large (10–200 μm in diameter) unilocular TG-rich lipid droplet, while the rest of cytoplasm occupies negligible space. Extensive studies have elegantly revealed that a transcriptional network involving PPARγ plays a central role in orchestrating adipogenesis, the differentiation process that generates mature adipocytes from fibroblast-like preadipocytes (4–7). The development of the highly specialized cellular structure of white adipocytes requires massive cytoplasmic remodeling. This aspect of adipogenesis, however, has not been well-studied, and the cellular and molecular mechanisms underlying this remodeling process remain unclear.
Autophagy is a major cellular degradation process involving intracellular membrane trafficking toward the lysosome (8, 9). Autophagy is initiated by the emergence of double-membrane vesicles, known as autophagosomes, which engulf a portion of the cytoplasm. The autophagosome then delivers its cargo to the lysosome for degradation. Over the last decade, the molecular machinery of autophagy has been identified in both yeast and mammals. Most of the genes encoding components of autophagy machinery, named autophagy-related genes (atg), have been characterized (10, 11). Targeted deletion of essential autophagy genes in mice has revealed the various important functions of autophagy, including tumor suppression, neuronal protection, neonatal survival [(8–10) and references therein], as well as differentiation of erythrocytes (12, 13), lipid droplet formation, and lipid metabolism (14, 15) in hepatocytes.
The ability of autophagy to facilitate massive cytoplasmic degradation prompted us to investigate its possible involvement in adipogenesis. More than 25 years ago, an increased level of autophagosomes was observed when differentiating 3T3-L1 cells were analyzed morphologically with electron microscopy by Novikoff et al. (16). Recently, we showed that targeted deletion of atg5, an essential autophagy gene, markedly reduced white adipogenesis efficiency of the primary mouse embryonic fibroblasts (MEFs) (17). However, the atg5−/− mice die at birth (18), and white adipogenesis in mammals mainly occurs postnatally. Thus, the in vivo role of autophagy in white adipogenesis remains undefined.
The atg7 gene encodes an E1-like enzyme that is specifically involved in autophagosome formation and is essential for autophagy (11). To rule out the possibility that an autophagy-independent function of atg5 might be required for adipogenesis, and more importantly, to establish that autophagy is involved in normal adipogenesis in vivo, in the present study, we have investigated the requirement of atg7 in adipogenesis in the primary MEF model and in a mouse model with a targeted deletion of atg7 in adipose tissue. The atg7−/− primary MEFs exhibit drastically reduced adipogenesis efficiency. Moreover, the adipose-specific atg7 conditional knockout mice show striking phenotypes in the structure of adipocytes and exhibit an interesting combination of anti-obesity/anti-diabetic metabolic features. This study provides strong in vivo evidence that atg7, and by inference autophagy, is critical for normal adipogenesis.
Results
Atg7−/− Primary MEFs Exhibited Drastically Reduced Adipogenesis Efficiency and Adipose-Specific atg7 Knockout Mice Had Drastically Reduced White Fat Mass and Reduced Body Weight.
The wild-type and atg7−/− primary MEFs were induced for adipocyte differentiation, and the efficiency of adipogenesis was compared by microscopic analysis as well as by quantification of lipid droplet formation. As shown in Fig. S1, the atg7−/− primary MEFs mirrored the phenotypes of atg5−/− MEFs and exhibited a drastically reduced efficiency of adipogenesis. These results support that an autophagy defect is responsible for the adipogenesis defect observed in these cells.
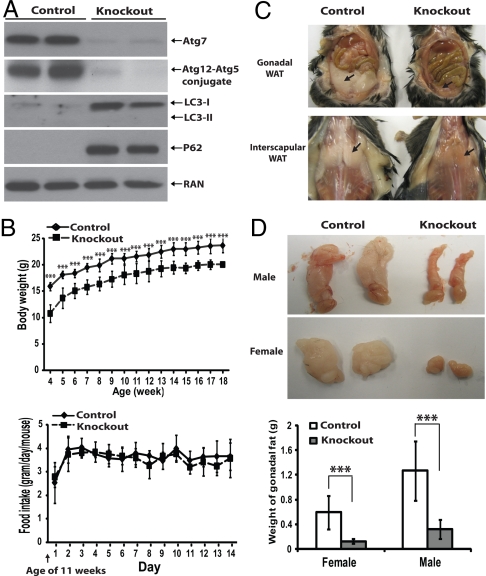
White adipogenesis in mammals mainly occurs postnatally; however, both the straight atg5−/− and atg7−/− mice die at birth (18, 19). To study the in vivo role of autophagy in adipogenesis, we generated an adipose-specific atg7 knockout mouse model by crossing flox-atg7 mice (19) with the aP2-cre mice, in which CRE expression is under the control of an adipose tissue-specific aP2 (fatty acid binding protein 4, FABP4) promoter (20). The homozygous atg7flox/flox/aP2-cre F2 mice were born in normal Mendelian ratios. The ablation of atg7 expression in white fat tissue was nearly complete (Fig. 1A). The deficiency of autophagy in adipose tissue was confirmed by three independent parameters (21) (Fig. 1A): (1) absence of LC3-II even in the presence of high levels of LC3-I, indicating an absence of steady-state levels of autophagosome formation; (2) accumulation of a common substrate of autophagic degradation, p62, indicating a lack of functional autophagic degradation (autophagy flux); and (3) undetectable levels of Atg5-Atg12 conjugate, a complex required for autophagosome formation. As the atg7 conditional knockout mice grew, they were visibly slim, seemingly more active, and shivered more frequently than their control wild-type littermates, but otherwise appeared normal. Both the male and female homozygous atg7 conditional knockout mice appeared to be infertile and failed to produce any offspring.
Fig. 1.
Adipose- specific atg7 knockout mice exhibited reduced body weight and WAT mass. (A) Immunoblotting analyses of lysates of white adipose tissue (female, uterine WAT) from control (atg7 flox/flox) and adipose-specific atg7 conditional knockout (atg7flox/flox; ap2-Cre) mice using indicated antibodies (Atg12-Atg5 conjugate was detected with an anti-Atg12 antibody). (B) Upper, body weight chart for control (female, n = 12) and atg7 conditional knockout (female, n = 11) mice from 4–18 weeks. Lower, a 2-week food intake chart for control (female, n = 6) and atg7 conditional knockout (female, n = 6) mice starting from week 11. (C) Representative pictures of control and atg7 conditional knockout mice at the age of 20 weeks, showing gonadal (Upper) and interscapular (Lower) white adipose tissue (WAT) as indicated by arrows. (D) Representative pictures of gonadal fat pad (uterine fat in female and epididymal fat in male) and quantification (Lower) from control (male, n = 10; female, n = 12) and atg7 conditional knockout (male, n = 5; female, n = 6) mice at the age of 18–20 weeks. ***, P < 0.001, Student's t test.
Body weight was compared between the atg7 knockout mice and their littermates after weaning (at 3 weeks of age). As shown in Fig. 1B upper panel, the average body weight of the atg7 adipose-specific knockout mice was ≈12 g at the age of 4 weeks vs. ≈16 g in the control atg7 wild-type mice. The difference in body weight was maintained and found to be statistically significant through 18 weeks of age when the experiment was stopped. Similar results were obtained with the male mice. Interestingly, the total food intake rates (per animal) were almost identical between the atg7 knockout and control mice, as shown in Fig. 1B (Lower), suggesting either a reduced efficiency in energy storage or an increased energy expenditure rate, or both, in the atg7 conditional knockout mice.
The fat tissue in the mice was analyzed. Fig. 1C shows the gross appearance of gonadal fat pads as well as white fat tissue in the scapular region, in which a striking reduction of fat mass in the atg7 conditional knockout mice was evident. The white adipose tissue in other regions of the mutant mice, including retroperitoneal fat and inguinal fat deposits, showed a similar extent of reduction in mass. Fig. 1D shows that the gonadal fat pads of the atg7 conditional knockout mice (uterine fat in female and epididymal fat in male) were typically 20% of the mass of those found in the control atg7 wild-type littermates. Importantly, other organs in the atg7 conditional knockout mice did not appear to have any defects and the weight of liver, heart, lungs, kidneys, and brain did not exhibit any significant difference from those in the control atg7 wild-type mice. Together, these results reveal that deletion of the atg7 gene in adipose tissue has a profound impact on the mass of WAT deposits in adult mice.
Atg7 Knockout WAT Contained Smaller Adipocytes and Had Large Populations of Multilocular Cells with Significant Amounts of Cytoplasm, But Exhibited No Apparent Changes in Adipocyte-Specific Gene Expression.
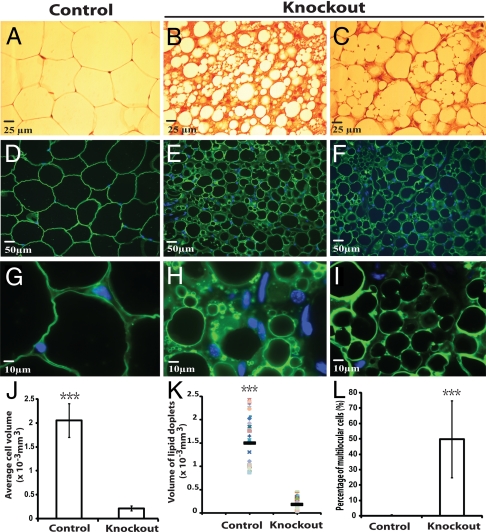
Histological analysis of gonadal fat was performed. Fig. 2 shows the results of uterine fat pad analysis. Hematoxylin and eosin (H&E) staining of tissues showed that wild-type WAT (Fig. 2A) was morphologically homogeneous and exhibited a typical structure in which almost the whole cell was occupied by one large lipid droplet, while cytoplasm was essentially undetectable. In contrast, the atg7 knockout white adipocytes were heterogeneous (Fig. 2 B and C). The mutant cells were smaller (Fig. 2J) and a large population of the cells contained a significant amount of cytoplasm (Fig. 2 B and C, stained in red). Immunofluorescence microscopy was performed with perilipin antibody, which labels the membrane of the lipid droplets in the cells (Fig. 2 D–I). While all of the wild-type adipocytes were unilocular (containing only one lipid droplet) (Fig. 2 D and G), ≈50% of the atg7 knockout adipocytes were multilocular (containing multiple lipid droplets) (Fig. 2 E, F, H, I, and L). On average, the size of the lipid droplets in the mutant adipocytes was smaller (Fig. 2 E, F, H, I, and K). Similar results were obtained from the epididymal fat pad analysis of male mice.
Fig. 2.
Histological and immunofluorescence analysis of gonadal WAT from control and atg7 conditional knockout mice. (A–C) Representative microscopic pictures of H&E stained sections of uterine WAT from control (atg7flox/flox, A) and adipose-specific atg7 conditional knockout mice (atg7flox/flox; aP2-Cre, B and C). (D–I) Representative microscopic pictures of immunofluorescence assays of uterine WAT from control (D and G) and atg7 conditional knockout mice (E, F, H, and I) with Perilipin A antibody. D–F were pictures of low magnification and G–I were pictures of high magnification. (J–L) Quantification of average cell volume, lipid droplet volume, and percentage of multilocular cells, as indicated, of uterine WAT from control and atg7 conditional knockout mice. ***, P < 0.001, Student's t test. The data show representative results of tissues from six pairs of female mice (control and atg7 knockout).
Quantitative PCR was performed to compare the mRNA levels of the important adipocyte-related genes, including gpam, cebpa, pparg, fabp4, ucp1, agpat2, and plin. As shown in Table S1, there is no significant change in the expression pattern of these genes between the atg7 knockout white fat and the wild-type control. It was noteworthy that although the atg7 knockout WAT gained a number of phenotypical features of brown fat, including multilocular lipid droplets, increased cytoplasmic volume, and enriched mitochondria content (Fig. 3), it did not show a significant increase in expression of the brown fat-specific genes including ucp1, cidea, elovl3, and prdm16 (Table S1). In addition, no change was observed in mRNA of enzymes involved in lipolysis, lipe and pnpla2 (Table S1).
Fig. 3.
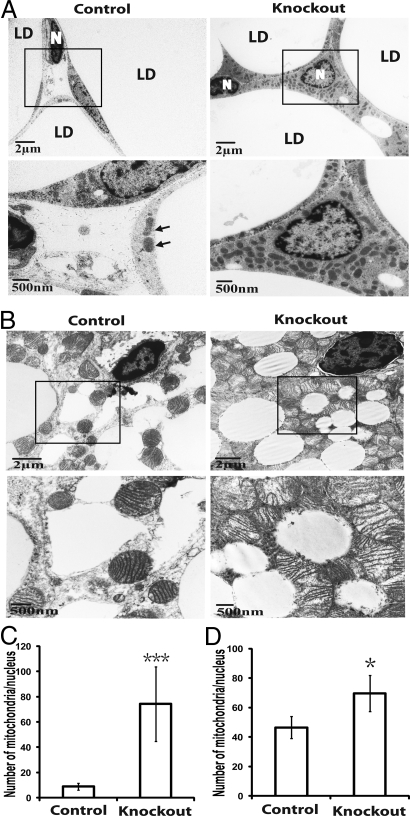
Adipose-specific atg7 knockout mice accumulated more mitochondria in WAT and BAT (brown adipose tissue). (A) Electron microscopic pictures of adipocytes from uterine WAT of control and adipose-specific atg7 knockout mice. Selected regions in low magnification images (within the squares) are shown below in high magnification. LD, lipid droplet; N, nucleus; arrows indicate mitochondria in the control tissue sample. (B) Representative electron microscopic pictures of interscapular BAT from control and atg7 conditional knockout mice. Selected regions in low magnification images (within the squares) are shown below in high magnification. (C and D) Quantification of mitochondria in WAT and BAT of control and adipose-specific atg7 knockout mice, respectively. Mitochondria number was counted in 25 random cells and expressed as the number of mitochondria per nucleus. *, P < 0.05; ***, P < 0.001, Student's t test.
Atg7 Knockout Adipose Tissues Had Increased Mitochondria Content.
Mitochondria play an important role in lipid metabolism and autophagy is critical for mitochondria elimination (22). We therefore analyzed mitochondria levels in adipose tissues of wild-type and atg7 knockout mice. Fig. 3 A and B show electron microscopic pictures of the white and brown adipocytes, respectively. The wild-type white adipocytes contained a limited volume of cytoplasm and relatively few mitochondria (Fig. 3A, Left). In contrast, the atg7 knockout adipocytes contained a larger cytoplasmic volume and a dramatically increased number of mitochondria (>8-fold increase, Fig. 3 A and C). High levels of mitochondria were also observed in the mutant brown adipocytes (Fig. 3 B and D). The cytoplasm of almost every single atg7 knockout brown fat cell was densely packed with mitochondria to such an extreme extent that, except for the lipid droplets, no mitochondria-free area, or other cellular structures could be easily identified (Fig. 3B, Right).
Atg7 Knockout Adipocytes Had Increased β-Oxidation Rates, Reduced Lipolysis Rates, and the Adipose-Specific atg7 Knockout Mice Exhibited Lower Fed Plasma Levels of Fatty Acids and Accelerated Rates of Fatty Acid Reduction in Response to Insulin.
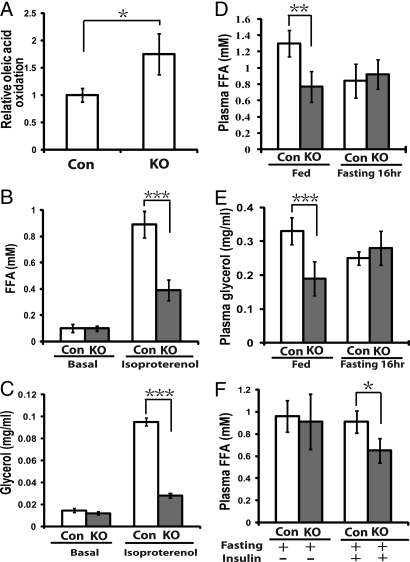
Mitochondria are the site of β-oxidation and mitochondria play an important role in lipid metabolism. We compared free fatty acid (FFA) catabolism in the wild-type and atg7 deleted adipocytes isolated from mice by measuring the rates of β-oxidation. The 1-14C- labeled oleic acid was added into the culture medium of adipocytes and the release of 14C- labeled CO2 was quantified. As shown in Fig. 4A, the primary adipocytes from the mutant mice exhibited almost double the rates of oleic acid oxidation found in wild-type cells. We then analyzed lipolysis rates in the isolated adipocytes (Fig. 4 B and C). As shown in Fig. 4 B and C, while no difference was observed between the basal lipolysis rates of the isolated mutant and wild-type adipocytes, the hormone induced lipolysis rate of the mutant cells was lower than that of the wild-type cells. We further measured the plasma levels of fatty acids in the wild-type and mutant mice. As shown in Fig. 4 D and E, the mutant mice exhibited significantly lower fed levels of plasma fatty acids and glycerol, even though no significant difference in fasting FFA levels was observed. Furthermore we compared the responsiveness of plasma FFA concentrations toward insulin stimuli. As shown in Fig. 4F, 30-min post insulin injection, the plasma concentration of free fatty acids in the mutant mice declined to a much lower level than in the wild-type mice.
Fig. 4.
Fatty acid oxidation and lipolysis analysis of the adipose-specific atg7 conditional knockout mice. (A) β-oxidation analysis of white adipocytes isolated from control (CON, n = 3) and atg7 conditional knockout mice (KO, n = 4). 1-14C labeled oleic acid was added to the medium containing isolated adipocytes (5 × 105), and 14CO2 trapped by the filter paper soaked with hyamine hydroxide was measured after a 3 h- incubation by a scintillation counter. (B and C) Lipolysis analysis of white adipocytes from control (CON, n = 3) and atg7 conditional knockout (KO, n = 4) mice. Adipocytes (5 × 105) from control and atg7 conditional knockout mice were incubated in medium for 2 h in the absence or presence of 10 μM isoproterenol. Free fatty acid (FFA) (B) and glycerol (C) levels were then measured. (D) Plasma FFA levels of fed and fasting (16 h) mice (Con, n = 6; KO, n = 6). (E) Plasma glycerol levels of the fed and fasting (16 h) mice (Con, n = 6; KO, n = 6). (F) Plasma FFA levels of the mice (fasted for 5 h) before and 30 min after i.p. injection of insulin (0.75 U/kg, Con, n = 6; KO, n = 6). *, P < 0.05, **, P < 0.01, ***, P < 0.001. Student's t test. These data are representative results from two independent experiments.
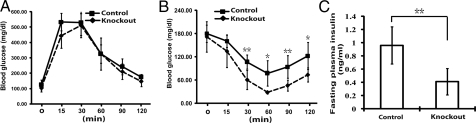
Adipose-Specific atg7 Knockout Mice Exhibited Increased Insulin Sensitivity.
Lipoatrophy in mice typically leads to the abnormal deposition of triglyceride in liver and muscles instead, which in turn causes insulin resistance (20). Surprisingly, unlike other lipoatrophy mouse models, the atg7 conditional knockout mice did not exhibit abnormal fat deposition in liver and muscles. We then measured insulin sensitivity in these mice by performing glucose tolerance and insulin tolerance tests. As shown in Fig. 5A (basal levels), as well as Table 1, no significant difference in fasting and fed plasma glucose levels was observed between the control atg7 wild-type and the atg7 conditional knockout mice. In addition, the mutant mice exhibited no significant difference in response to glucose in glucose tolerance tests compared to the wild-type mice (Fig. 5A), suggesting that the insulin secretion function of the pancreatic B cells of the mutant mice in response to glucose elevation was normal. Importantly, the mutant mice exhibited significantly increased sensitivity to insulin as measured by insulin tolerance tests (Fig. 5B). Consistent with increased insulin sensitivity, the fasting plasma insulin levels were lower in the mutant mice (Fig. 5C). These results show that the atg7 conditional knockout mice had significantly increased insulin sensitivity.
Fig. 5.
Adipose- specific atg7 conditional knockout mice exhibited increased insulin sensitivity. (A) Glucose tolerance tests. Control mice (male, n = 6) and atg7 conditional knockout mice (male, n = 6) were fasted overnight before receiving an i.p. injection of 2 g/kg glucose and blood glucose concentration was measured at indicated time points. (B) Insulin tolerance tests. Control mice (male, n = 6) and atg7 conditional knockout (male, n = 6) mice were fasted for 5 h before receiving an i.p. injection of 0.75 U/kg insulin and blood glucose concentration were measured at indicated time points. (C) Plasma insulin levels of the control (male, n = 6) and atg7 conditional knockout mice (male, n = 6) fasted for 5 h. *, P < 0.05; **, P < 0.01, Student's t test. These data are representative results from two independent experiments.
Table 1.
Metabolic parameters of control and atg7 conditional knockout mice
| Parameters | Control | Knockout | P value* |
|---|---|---|---|
| Glucose (mg/dl), fed | 146.25 ± 12.66 | 150.40 ± 18.31 | 0.13 |
| Glucose (mg/dl), fasting 16 hr | 90.17 ± 27.02 | 96.67 ± 11.89 | 0.60 |
| Lactate (mM), fed | 2.28 ± 0.25 | 2.00 ± 0.25 | 0.10 |
| Lactate (mM), fasting 16 hr | 1.48 ± 0.42 | 1.67 ± 0.21 | 0.34 |
| Cholesterol (mg/ml), fed | 2.62 ± 0.59 | 2.80 ± 0.74 | 0.68 |
| Cholesterol (mg/ml), fasting 16 hr | 2.64 ± 0.47 | 1.51 ± 0.46 | 0.008 |
| Triglyceride (mg/ml), fed | 0.85 ± 0.32 | 0.42 ± 0.13 | 0.01 |
| Triglyceride (mg/ml), fasting 16 hr | 0.33 ± 0.05 | 0.19 ± 0.04 | 0.0005 |
*Control: n = 6; Knockout: n = 6. Student's t test.
Adipose-Specific atg7 Knockout Mice Exhibited Markedly Decreased Leptin Levels, Reduced Plasma Concentrations of Triglycerides and Cholesterol, Increased Basal Physical Activities, and Were Resistant to High-Fat-Diet-Induced Obesity.
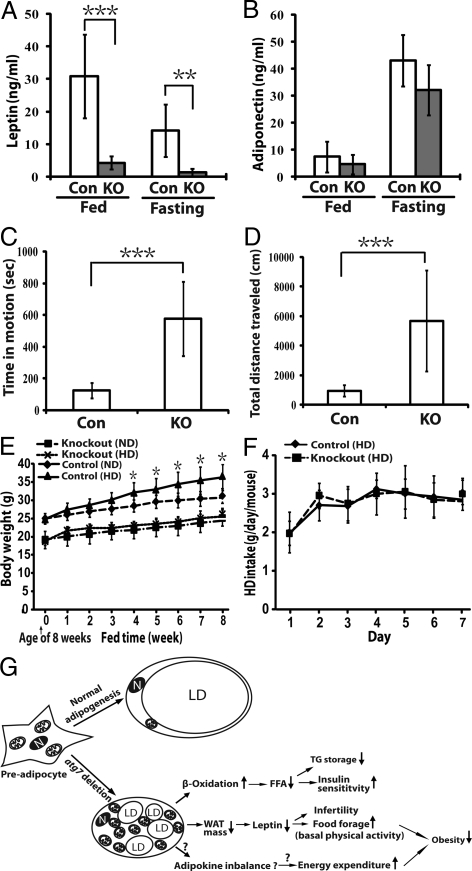
The plasma levels of leptin and adiponectin were measured. As shown in Fig. 6A, the fed and fasting plasma levels of leptin in the mutant mice were only ≈10% the levels found in the wild-type mice. Interestingly, no significant difference was observed in the plasma adiponectin levels between the mutant and wild-type mice (Fig. 6B). The plasma concentrations of other metabolites were also measured (Table 1). Significantly, the adipose-specific atg7 knockout mice also had reduced plasma concentrations of triglycerides and fasting levels of total cholesterol.
Fig. 6.
The adipose-specific atg7 conditional knockout mice exhibited altered adipokine secretion, increased basal physical activity, and were resistant to high-fat diet induced obesity. (A) Leptin levels of the fed and fasted (5 h) control (n = 6) and atg7 conditional knockout mice (n = 6). (B) Adiponectin levels of the fed and fasted (5 h) control (n = 6) and atg7 conditional knockout mice (n = 6). The data from (A) and (B) are representative results from two independent experiments. (C and D) Open Field Test of mouse activity. 12 wild-type and 12 atg7 conditional knockout mice were analyzed with VersaMax animal activity monitor (Accuscan) for a period of 20 min. Time in motion (C) and total distance traveled (D) were recorded. (E) Body weight chart of control (atg7flox/flox, male, n = 9) and adipose- specific atg7 conditional knockout mice (atg7flox/flox;aP2-cre, male, n = 6) fed with normal diet (ND) or high fat diet (HD) from the age of 8–16 weeks. (F) One-week HD food intake chart of control (male, n = 6) and atg7 conditional knockout (male, n = 6) mice starting from week 14. (G) A working model summarizing the phenotypes of the adipose-specific atg7 knockout mice and the proposed underlying mechanisms. Detail was described in Discussion. *, P < 0.05, **, P < 0.01, ***, P < 0.001, Student's t test. LD, lipid droplet; N: nucleus; TG, triglyceride; ↑, upregulation; ↓, downregulation.
The mutant mice had a similar rate of food intake compared to the wild-type mice (or even an increased rate if normalized against body weight), yet they accumulated less WAT mass, suggesting an increased rate of energy expenditure. We directly compared the basal physical activities of the wild-type and the atg7 conditional knockout mice using an animal activity monitor equipped with computer-linked infrared sensors that detect the movement of small animals in a defined open field (open-field test). As shown in Fig. 6 C and D, the mutant mice exhibited dramatically increased basal physical activities, as manifested by increased time in motion and increased total distance traveled during the testing period.
We further investigated if the adipose- specific atg7 knockout mice were more resistant to high-fat diet induced obesity. The age-matched control and atg7 conditional knockout mice were provided with a high-fat diet starting at the age of 8 weeks and continuing for 2 months. The body weight of each mouse was measured weekly. Fig. 6E shows the body weight chart. As expected, the wild-type mice gained ≈20% more body weight when fed with the high-fat diet during this 2-month period as compared to mice fed with a normal diet. In contrast, the mutant mice fed with the high-fat diet gained almost no additional weight compared to those fed a normal diet. Again, little difference in food intake was observed between the control atg7 wild-type mice and the atg7 conditional knockout mice (Fig. 6F).
Discussion
White adipocytes possess a unique cellular structure which contains a unilocular lipid droplet occupying almost the whole cell volume. In this report, we have investigated the functional role of autophagy in adipogenesis in vivo by generating and characterizing a mouse model in which an essential autophagy gene, atg7, is deleted in adipose tissue. Our study has demonstrated that atg7 deletion has a profound impact on the structure of white adipocytes in mice. The mutant mice developed white adipose mass that is only about one-fifth that found in the wild-type mice by the age of 18 weeks. While the total number of white adipocytes in the mutant mice appears similar to the wild-type mice, these adipocytes suffer severe morphological abnormalities, including reduced cell size, drastically increased cytoplasmic volume with higher mitochondria content, and a strikingly high percentage of cells (50%) containing multilocular lipid droplets. Despite the fact that the atg7 knockout white adipocytes possess a number of characteristics of normal brown adipocytes, including containing multiple smaller lipid droplets instead of one large one, they do not express significant levels of Ucp1 and other brown fat genes such as elovl3, cidea, and prdm16. In addition, the multilocularity of the mutant cells is not caused by an increase in lipolysis, since the mutant adipocytes exhibited no change in basal lipolysis rate and even a reduction in hormone induced lipolysis (Fig. 4 B and C). These phenotypes thus underscore the importance of the function of Atg7 in the formation of the highly specialized cellular structure during maturation of white adipocytes.
Most mouse models with lipoatrophy exhibit hyperlipidemia, hyperglycemia, and insulin tolerance as a result of defects in lipid storage and also show a consequent misdeposition of lipid in liver and muscles. Surprisingly, the adipose- specific atg7 knockout mice are euglycemic and are more sensitive to insulin. It appears that the structural changes of the mutant white adipocytes have led to interesting functional changes. Consistent with higher mitochondria content, the mutant adipocytes have increased rates of fatty acid β-oxidation. Moreover, the mutant adipocytes exhibit reduced hormone-induced lipolysis rates. Together, the changes of lipid metabolism in the mutant WAT may have led to the altered lipid homeostasis observed in the mutant mice, which exhibit decreased fed plasma levels of FFA and accelerated rates of FFA reduction upon insulin stimulus. In turn, the systemic changes of FFA homeostasis likely contribute to the enhanced insulin sensitivity (illustrated in Fig. 6G).
Another striking phenotype of the adipose-specific atg7 knockout mice is resistance to high-fat diet induced obesity. This must be either a result of increased energy expenditure or reduced energy utilization/storage efficiency or both. Indeed, the mutant mice are hyperactive [increased basal physical activity (Fig. 6 C and D)]. The adipose-specific atg7 knockout mice have drastically reduced plasma leptin levels, likely a result of the reduced WAT mass. It is likely that the reduction of leptin contributes to the hyperactivity phenotype (23) as well as infertility (3). However, the reduction of leptin alone cannot account for the phenotype of resistance to high-fat diet induced obesity, since a lack of leptin expression in mice with normal adipogenesis (the ob/ob mice) results in obesity (24). The exact mechanism underlying this phenotype remains unclear, but the answers may lie in the uniqueness of the atg7-deleted white adipocytes, which may have a reduced efficiency of energy storage (leakage due to increased basal free fatty acid oxidation levels) and an imbalanced adipokine secretion profile exemplified by a markedly reduced plasma leptin concentration but only a marginally decreased adiponectin concentration. The working model is illustrated in Fig. 6G.
atg7 encodes a E1- like enzyme that is specifically involved in autophagosome formation. The only known function of Atg7 is its involvement in autophagy and atg7 deletion in adipocytes leads to a total ablation of autophagy activity (19, 25) (Fig. 1). It is likely that an autophagy defect as a result of atg7 deletion has caused the phenotypes observed in the conditional knockout mice rather than some unknown function of Atg7. This conclusion is supported by the fact that the major structural alterations in the mutant adipocytes result from defects in degradation of cytosol and mitochondria, a major and specific function of autophagy. Further evidence supporting this conclusion comes from a separate study in which we have investigated the adipogenesis properties of primary MEFs derived from a mouse model with deletion of an independent autophagy gene, atg5 (17). These primary atg5−/− MEFs exhibit almost identical phenotypes as the atg7−/− MEFs and are reminiscent of the in vivo defect of the adipocytes with atg7 deletion as described in this study.
Phenotypically, obesity is caused by the excess accumulation and expansion of WAT. It is now becoming clear in humans that adipocytes are turned over at the rate of 10% of total adipocytes per year and that new adipogenesis occurs throughout adulthood (26). Our study here has presented in vivo evidence that inactivation of autophagy by ablation of Atg7 could alter adipogenesis, leading to the production of adipose tissue with a number of favorable anti-obesity and anti-diabetes features. This study thus has not only established a functional role of autophagy in adipogenesis, but may potentially open a venue for obesity and diabetes control.
Materials and Methods
Generation and Characterization of Adipose-Specific atg7 Knockout Mice.
The atg7flox/flox mice (25) and aP2 (Fabp4)-Cre transgenic mice ((20), obtained from The Jackson Laboratory) were crossed to produce the adipose tissue-specific atg7 conditional knockout mice atg7flox/flox; aP2-Cre. The genotypes of the mice were determined and deletion of atg7 in adipose tissue were confirmed with PCR using primers described (20, 25). The body weights of mutant and control mice were measured once every week after week 4. For food intake experiments, sets of mice at 11 weeks of age were housed individually in metabolic cages (Nalgene) with free access to food and water. Food cups and food scattered in the runway to the cups were weighed daily to determine food intake. For the high-fat diet (HD) experiment, six pairs of mice were fed with HD (60 kcal% fat, Research Diets) for 8 weeks at the age of 8 weeks, and body weights were measured once every week.
Western Blotting and Tissue Analyses.
Western blot analysis was carried out according to standard protocol. The sources of the antibodies are: Atg12 antibody (Cell Signaling Technology), Atg7 antibody (Cell Signaling Technology), MAP-LC3 antibody is made by Cocalico Biologicals using a recombinant rat MAP-LC3 protein as antigen; guinea pig p62 C-terminal specific antibody (American Research Products, Inc.); Ran antibody (C-20, Santa Cruz), Perilipin A antibody (Sigma).
At the age of 18–20 weeks, a set of control and mutant mice were killed, fat tissues were processed and analyzed by microscopy and electronic microscopy. Immunofluorescence analyses were performed on paraffin-embedded sections according to standard protocol with Perilipin A antibody (Sigma, 1:50 dilution) at 4 °C overnight. In some experiments, 100 ng/mL DAPI was added to the secondary antibody solution to co-stain the nuclei. A detailed method was described in SI Text.
Blood and Plasma Measurement.
Blood tests were performed with kits following the manufacturer's instruction: glucose, OneTouch UltraSmart blood glucose monitoring system (Lifescan); lactate, LACTATE PLUS lactate meter (Nova Biomedical Corporation). For plasma measurement, blood samples were collected from mice tail with heparinized microhematocrit capillary tubes (Fisher), and plasma was obtained by centrifuge the blood samples with Readacrit centrifuge (Clay Adams) for 3 min. Plasma glycerol and triglycerides were measured with a serum triglyceride determination kit (Sigma). Total cholesterols in plasma were measured with a total cholesterol/cholesteryl ester quantification kit (BioVision). Plasma FFA were measured with free fatty acids, half-micro test kit (Roche). Insulin, leptin, adiponectin levels were measured by ultra sensitive mouse insulin ELISA kit (Crystal Chem Inc.), mouse leptin ELISA kit (Crystal Chem Inc.), mouse adiponectin ELISA kit (Millipore), respectively.
Glucose and Insulin Tolerance Test.
For glucose tolerance tests, mice were starved overnight and i.p. injected with 20% glucose at a dose of 2 g/kg body weight. Blood was obtained from the tail at time points 0, 15, 30, 60, 90, and 120 min for glucose measurement. For insulin tolerance tests, mice were starved for 5 h and i.p. injected with 0.75U/kg body weight recombinant human insulin (Eli Lilly). Blood was obtained from the tail at time points 0, 15, 30, 60, 90, and 120 min for glucose measurement.
Lipolysis Assay, FFA Oxidation Measurement, and Open Field Test.
Primary white adipocytes were isolated, and lipolysis and FFA oxidation were measured as previously reported with some modification (27). Detailed method was described in SI Text. The open field test was conducted using a VersaMax animal activity monitor (Accuscan). All testing was conducted between the hours of 10:00 AM and 2:00 PM during the daylight phase of the vivarium light cycle. Mutant and wild-type mice were randomly selected and placed two at a time into diagonally opposite detection chambers and were allowed to acclimate. Following acclimation, movement data were recorded for a 20-min period. Data were collected using VersaMax software version 4.00–127E (2004) (Accuscan).
Supplementary Material
Acknowledgments.
We thank Drs. Loren Runnels and Li-Ting Su for assistance with the microscopy, Dr. Dawn L. Brasaemle for help with adipocyte differentiation, Raj Patel for technical assistance with the electron microscopy, Dr. Andrew Brooks for real-time PCR analyses, and Dr. Maral Mouradian and Dr. Kang-Woo Lee for the open-field test. This work was supported by National Institutes of Health Grants 5 F31 GM078857–02 (to R.B.) and 1R01 CA116088–01A1 and 1R01AG030081–01A1 (to Y.Z. and S.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906048106/DCSupplemental.
References
- 1.Rosen ED, Spiegelman BM. Adipocytes as regulators of energy balance and glucose homeostasis. Nature. 2006;444:847–853. doi: 10.1038/nature05483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shulman GI. Cellular mechanisms of insulin resistance. J Clin Invest. 2000;106:171–176. doi: 10.1172/JCI10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman JM. Leptin at 14 y of age: An ongoing story. Am J Clin Nutr. 2009;89:973S–979S. doi: 10.3945/ajcn.2008.26788B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gesta S, Tseng YH, Kahn CR. Developmental origin of fat: Tracking obesity to its source. Cell. 2007;131:242–256. doi: 10.1016/j.cell.2007.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006;7:885–896. doi: 10.1038/nrm2066. [DOI] [PubMed] [Google Scholar]
- 7.Rosen ED, Spiegelman BM. Molecular regulation of adipogenesis. Annu Rev Cell Dev Biol. 2000;16:145–171. doi: 10.1146/annurev.cellbio.16.1.145. [DOI] [PubMed] [Google Scholar]
- 8.Klionsky DJ. Autophagy: From phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8:931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]
- 9.Levine B, Klionsky DJ. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- 10.Mizushima N. Autophagy: Process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 11.Ohsumi Y, Mizushima N. Two ubiquitin-like conjugation systems essential for autophagy. Semin Cell Dev Biol. 2004;15:231–236. doi: 10.1016/j.semcdb.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Kundu M, et al. Ulk1 plays a critical role in the autophagic clearance of mitochondria and ribosomes during reticulocyte maturation. Blood. 2008;112:1493–1502. doi: 10.1182/blood-2008-02-137398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sandoval H, et al. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 2008;454:232–235. doi: 10.1038/nature07006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shibata M, et al. The MAP1-LC3 conjugation system is involved in lipid droplet formation. Biochem Biophys Res Commun. 2009;382:419–423. doi: 10.1016/j.bbrc.2009.03.039. [DOI] [PubMed] [Google Scholar]
- 15.Singh R, et al. Autophagy regulates lipid metabolism. Nature. 2009;458:1131–1135. doi: 10.1038/nature07976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novikoff AB, Novikoff PM, Rosen OM, Rubin CS. Organelle relationships in cultured 3T3–L1 preadipocytes. J Cell Biol. 1980;87:180–196. doi: 10.1083/jcb.87.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baerga R, Zhang Y, Chen P-H, Goldman S, Jin S. Targeted deletion of autophagy-related 5 (atg5) impairs adipogenesis in a cellular model and in mice. Autophagy. 2009 Nov 16;5(8) doi: 10.4161/auto.5.8.9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hara T, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 19.Komatsu M, et al. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He W, et al. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci USA. 2003;100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, et al. The role of autophagy in mitochondria maintenance: Characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy. 2007;3:337–346. doi: 10.4161/auto.4127. [DOI] [PubMed] [Google Scholar]
- 23.Hillebrand JJ, Kas MJ, van Elburg AA, Hoek HW, Adan RA. Leptin's effect on hyperactivity: Potential downstream effector mechanisms. Physiol Behav. 2008;94:689–695. doi: 10.1016/j.physbeh.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, et al. Positional cloning of the mouse obese gene and its human homologue. Nature. 1994;372:425–432. doi: 10.1038/372425a0. [DOI] [PubMed] [Google Scholar]
- 25.Komatsu M, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 26.Spalding KL, et al. Dynamics of fat cell turnover in humans. Nature. 2008;453:783–787. doi: 10.1038/nature06902. [DOI] [PubMed] [Google Scholar]
- 27.Nishino N, et al. FSP27 contributes to efficient energy storage in murine white adipocytes by promoting the formation of unilocular lipid droplets. J Clin Invest. 2008;118:2808–2821. doi: 10.1172/JCI34090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.