Abstract
Urodele amphibians and teleost fish regenerate amputated body parts via a process called epimorphic regeneration. A hallmark of this phenomenon is the reactivation of silenced developmental regulatory genes that previously functioned during embryonic patterning. We demonstrate that histone modifications silence promoters of numerous genes involved in zebrafish caudal fin regeneration. Silenced developmental regulatory genes contain bivalent me3K4/me3K27 H3 histone modifications created by the concerted action of Polycomb (PcG) and Trithorax histone methyltransferases. During regeneration, this silent, bivalent chromatin is converted to an active state by loss of repressive me3K27 H3 modifications, occurring at numerous genes that appear to function during regeneration. Loss-of-function studies demonstrate a requirement for a me3K27 H3 demethylase during fin regeneration. These results indicate that histone modifications at discreet genomic positions may serve as a crucial regulatory event in the initiation of fin regeneration.
Keywords: chromatin, polycomb
Mammals are unable to regenerate amputated appendages, such as limbs, yet it has long been known that salamanders and zebrafish perform such a feat and can completely restore lost body parts and repair severely damaged organs, including limbs, heart, and jaw, by a process known as epimorphic regeneration (1–4). This phenomena is typically broken down into three steps (1–4). First, a wound epidermis is formed at the site of damage by migrating epithelial cells that seals the wound from the environment. Next, disorganization and dedifferentiation of tissue near the wound results in the creation of a mass of undifferentiated cells, known as the blastema. Then, proliferation of blastema cells, concomitant with patterning and differentiation, results in the regeneration of the amputated portions of the damaged tissue. The defining characteristic of epimorphic regeneration is the formation of the blastema at the site of amputation. A fundamental question in the field is how amputation instructs certain cells near the wound site to dedifferentiate and take part in the re-growth and development of the amputated body part (1–4). A recent report has suggested that epigenetic mechanisms target a distal enhancer to silence the sonic hedgehog loci before regeneration in Xenopus (5).
The zebrafish exhibits an outstanding ability to regenerate different parts of its anatomy, including any of the paired and unpaired fins, the heart ventricle, and the spinal cord. Zebrafish is particularly useful for studies on regeneration since it has short generation times that make experiments requiring large number of animals feasible, and it has a fully sequenced and annotated genome. The zebrafish caudal fin is an established model of regeneration of a complex tissue that is easy to amputate, is not required for viability, and completely regenerates in a short time frame (7–10 days).
A gene silencing mechanism that may poise loci for reactivation has been described in ES cells (6–8). In ES cells, the transcription factors Sox2, Oct4, and nanog, serve as master regulators of pluripotency (9–11). They do so, in part, by concerted binding to the promoters of genes needed for pluripotency. Sox2, Oct4, and nanog also occupy the promoters of developmental regulatory genes that are silent in ES cells, but whose expression is associated with lineage commitment and differentiation. Most of these silent loci in ES cells are also bound by polycomb group (PcG) proteins (9–13). Nucleosomes near the transcription start sites of these silenced loci are decorated with trimethyl lysine 27 histone H3 (me3K27 H3) catalyzed by the polycomb repressive complex 2, PRC2 (8, 14). In some cases both trimethyl lysine 4 histone H3 (me3K4 H3), a product of TrX activity, and me3K27 H3 are observed at transcription start sites of developmental regulatory genes, thus creating a “bivalent” histone code that appears to poise these loci for activation in ES cells (6, 10, 15, 16). However, it has been suggested that the presence of bivalent chromatin is not indicative of cell multipotency or “stemness” (6, 17–20).
Upon ES cell differentiation, these so-called bivalent loci typically lose the repressive me3K27 H3 while at the same time maintaining the me3K4 H3 modification, a process that favors gene activation. A recent study has further delineated bivalent genes into two categories: bivalent positions associated with both PRC1 and PRC2, as opposed to those that are bound by PRC2 only (16). One mechanism by which me3K27 H3 is thought to be removed from bivalent loci is by the activity of a class of me3K27 H3-specific histone demethylases represented by mammalian UTX and Jmjd3 (21–28). These demethylases, therefore, function as positive regulators of transcription by relieving the negative effect of me3K27 H3 modifications on local chromatin structure.
Results and Discussion
Bivalent Chromatin at Genes Induced during Fin Regeneration.
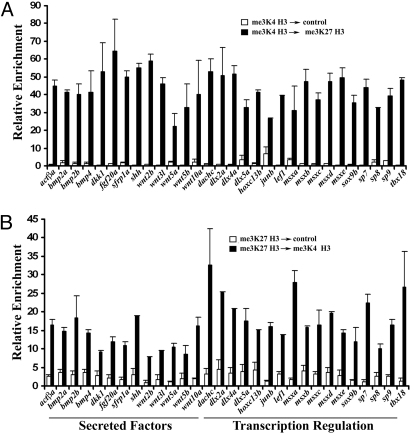
We used chromatin immunoprecipitation (ChIP) experiments to examine whether bivalent me3K4/me3K27 H3 chromatin could be detected in zebrafish caudal fin. Extracts prepared from normal adult zebrafish were subjected to a first ChIP with either me3K4 H3 or me3K27 H3 antibodies. These ChIP eluates were then re-ChIPed with control or me3K27 H3 (me3K4 H3) antibodies. We tested these sequential ChIP eluates by qPCR for enrichment of transcription start sites of genes expressed upon fin amputation. We focused on genes whose expression is induced upon fin amputation and code for putative developmental regulators (3, 29). We detected robust enrichment, indicative of bivalent me3K4/me3K27 H3 chromatin, at genes induced during regeneration. These genes code for secreted growth factors, morphogens, as well as regulators of transcription regulation (Fig. 1 A and B). Single me3K4 H3 and me3K27 H3 ChIP experiments with the same extract revealed strong enrichment of these histone modifications at unique genomic positions (Figs. S1 a and b and S2d) and peptide competition confirmed the specificity of the antibodies used in these studies (Fig. S2 b and c). These data indicate that many of the genes coding for developmental regulatory factors, which are induced in the regenerating fin (Fig. 2C), contain a bivalent me3K4/me3K27 H3 chromatin modification. This is consistent with the notion that this chromatin landmark is an indicator of genes that are poised for activation in response to developmental or environmental cues (6, 10, 15–20).
Fig. 1.
Bivalent me3K4/me3K27 H3 at promoters of genes implicated in regeneration. (A) Sequential ChIP first performed with me3K4 H3 followed by control or me3K27 H3 antibody. (B) Sequential ChIP first performed with me3K27 H3 followed by control or me3K4 H3antibody. Eluates from the second ChIP were examined by qPCR using primers near the transcription start sites of the indicated genes. Shown is the average of two independent sequential ChIPs and error bars represent deviation from the mean. Similar results were obtained from extracts from several cohorts.
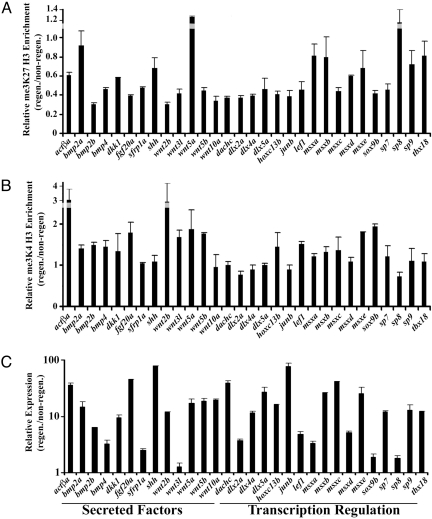
Fig. 2.
Resolution of bivalent chromatin during caudal fin regeneration. (A) me3K27 H3 ChIP in regenerating vs. non-regenerating fins. (B) me3K4 H3 ChIP in regenerating vs. non-regenerating fins. Eluates from the ChIPs were examined by qPCR using primers near the transcription start sites of the indicated genes. Data indicate the relative enrichment of ChIPs from regenerating tissue compared to non-regenerating tissue. Error bars indicate deviation from the mean from two experiments. Similar results were obtained from extracts from several cohorts. (C) Upregulation of genes during regeneration. The expression of the loci tested above was examined in non-regenerating and regenerating (48 hpa) adult caudal fins by qPCR and normalized to Ribosomal Protein L18 expression. Data are expressed as average expression levels relative to non-regenerating fins from two different cohorts (12 animals each) and error bars represent the deviation from the means derived from each cohort. Similar results were obtained in several independent experiments.
We tested for the presence of bivalent chromatin in other zebrafish tissues. Tissue residing at the base of the caudal fin is not able to regenerate in response to injury (Fig. S3a). However, we detected the bivalent me3K4/me3K27 H3 at a number of loci (Fig. S3 b and c). The zebrafish myocardium is capable of regeneration after injury (30) and we tested whether this myocardium contains bivalent chromatin at loci relevant for heart regeneration, such as hand2, tbx5, tbx18, tbx20, isl-1, and nkx2.5 (31). me3K27 H3 ChIPs from myocardium revealed that this set of genes were enriched for this histone modification at their promoters (Fig. S1c). Sequential ChIP experiments indicated that each of these genes contained a bivalent me3K4/me3K27 H3 promoter (Fig. S1 d and e). These results indicate that, in zebrafish, this chromatin modification exists in both regenerating and non-regenerating tissue. Our observations reinforce the idea that the presence of bivalent me3K4/me3K27 H3 is not ES cell or stem cell specific. As such, the modification may act to maintain loci in a dormant state with the potential for activation and this may occur in a array of cell types (15, 17, 20, 32–36).
Resolution of Bivalent Chromatin during Caudal Fin Regeneration.
Studies in both mouse and human ES cells, demonstrate that bivalent chromatin at silent genes usually resolves to a monovalent me3K4 H3 structure when the gene becomes active upon cell differentiation. Thus, we next asked whether repressive me3K27 H3 modifications residing in the putative bivalent me3K4/me3K27 H3 domains in zebrafish caudal fins are reduced during regeneration, since at this time these genes are highly expressed (Fig. 2C). We compared me3K4 H3 and me3K27 H3 ChIPs from non-regenerating tissue versus regenerating tissue at 48 h post amputation (hpa). A 48-hpa time point (Fig. S2a) was chosen because it allows the rapid identification and dissection of regenerating tissue.
We detected a significant reduction in repressive me3K27 H3 modifications at the start sites in a majority of genes examined, including fgf20a, lef1, and wnt genes in regenerating fins (Fig. 2A). Notably, several loci, including bmp2a, wnt5a, and sp8, maintained relatively high levels of me3K27 H3 during regeneration, and, as such represent cases in which the bivalent domain fails to resolve to monovalent me3K4 H3. This phenomena has also been reported in select cases in ES cells (6, 15), and is likely to represent an additional layer of complexity with respect to chromatin structure and the regulation of transcription. As opposed to silencing me3K27 H3 modifications, the presence of me3K4 H3 at transcription start sites typically correlates with transcription activation (6–8). me3K4 H3 ChIPs were generally increased or unchanged during regeneration (Fig. 2B). Genes we examined by ChIP are expressed in a subset of cells in the regenerate (2, 29, 37–42) (Fig. S4 d–f) and these ChIP data presented here should be interpreted in light of this. The relatively modest changes in me3K27 H3 during regeneration may reflect larger changes in me3K7 H3 at specific loci on a cell by cell basis. Collectively, our results indicate that the majority of bivalent loci we examined are resolved to monovalent me3K4 H3 after fin amputation, which coincides with gene activity. These data are consistent with the model that bivalent chromatin bearing genes are poised for activation, and they indicate that the balance between these two functionally opposing histone modifications is important for gene regulation during regeneration.
We turned to a small molecule inhibitor to test how altering histone methylation at bivalent positions would affect regeneration. 3-Deazaplanocin A (DZNEP) is a direct inhibitor of S-adenosyl methionine homocysteine hydrolase, an enzyme with roles in both epigenetic silencing and cellular senescence (43–47). We treated animals with this compound (15 μM) and tested them for the ability to regenerate. DZNEP greatly reduced the ability of fish to regenerate caudal fins and these animals displayed relatively few blastema cells (Fig. S5 a and b). Although global levels of me3K27 H3 were not affected by DZNEP, we detected a modest decrease in me3K27 H3 at the promoters of msxe and tbx18 in the presence of the drug. This correlated with over-expression of both of these genes in DZNEP treated animals, including that perturbation of histone methylation at bivalent positions is a potential mechanism by which DZNEP is able to inhibit regeneration.
me3K27 H3 Demethylases in Regenerating Caudal Fin.
We next asked how bivalent chromatin is resolved to active chromatin during regeneration. First we examined PcG expression and activity. We detected expression of all of the subunits of PRC1 and PRC2 in both the regenerating and non-regenerating fin, as well as PRC1 and PRC2 activity (Fig. 3A and Figs. S4 and S6). These studies do not support a model in which de-repression of genes during regeneration is accomplished by a global down regulation of PcG. Active histone demethylation of me3K27 H3 may also account for the resolution of dormant, bivalent me3K4/me3K27 H3 modifications to the active me3K4 H3 modification, resulting in the de-repression of select genes during regeneration. Two subclasses of jumonji-domain histone demethylases have been shown to specifically demethylate me3K27 H3 (trimethyl), me2K27 H3 (dimethyl) and meK27 H3 (monomethyl), namely UTX and Jmjd3 (21, 22, 24, 25). Sequence homology revealed four predicted zebrafish polypeptides (zf Kdm6a.1 or zf UTX, zf Kdm6a.2 or zf UTX-l, zf Kdm6b.1 or zf JmjD3, and zf Kdm6b.2 or zf JmjD3-like) that fall into this family (24, 48).
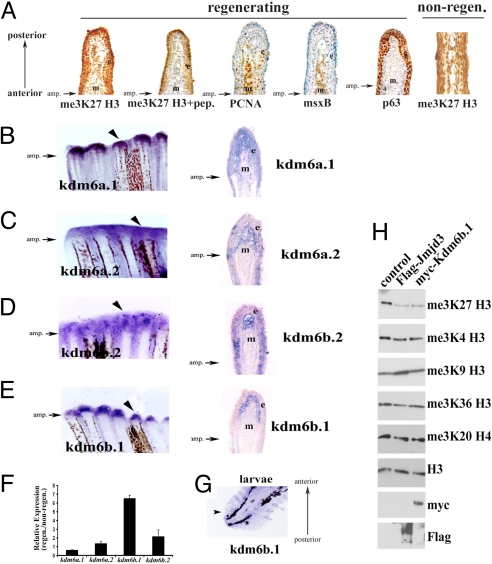
Fig. 3.
me3K27 H3 demethylases in caudal fin. (A) Global levels of me3K27 H3 remain intact during regeneration. Immunohistochemistry of longitudinal sections of 48 hpa regenerating caudal fins. From left to right, me3K27 H3 antibody; me3K27 H3 antibody with peptide competition; msxB; PCNA; p63. The right-most panel is a non-regenerating caudal fin stained for me3K27 H3. Antibody signal (brown) and hematoxylin counterstain (blue) are shown. (B–E) Expression of putative me3K27 H3 demethylases in regenerating caudal fins. Whole mount in situ hybridization of regenerating caudal fins at 48 h post amputation (B) kdm6a.1, (C) kdm6a.2, (D) kdm6b.2, and (E) kdm6b.1. Longitudinal sections of the fins in (B–E) reveal unique expression patterns and are shown in the right panels. The epidermis (e) and mesenchyme (m) are indicated in each panel. (F) Upregulation of kdm6b.1 during regeneration. The expression of putative me3K27 H3 demethylases was examined in non-regenerating and regenerating (48 hpa) adult caudal fins by qPCR and normalized to Ribosomal Protein L18 expression. Data are expressed as average expression levels relative to non-regenerating fins from two different cohorts (12 animals each) and error bars represent the deviation from the means derived from each cohort. Similar results were obtained in several independent experiments. (G) kdm6b.1 expression in larvae caudal fin at 24 hpa [72 h post fertilization (hpf)] by in situ hybridization. (H) Kdm6b.1 demethylates me3K27 H3. myc-tagged Kdm6b.1 was immunoprecipitated from transfected 293T cells and subjected to a histone demethylase assay, followed by immunoblotting. Some panels contain blots that were stripped and re-probed with different antibodies.
We isolated cDNA clones for each of these (Fig. S7) and tested their expression in regenerating caudal fins by in situ hybridization. Expression of the demethylases kdm6a.1, kdm6a.2 and kdm6b.2 was detected both in the epidermis and the blastema (Fig. 3 B–D). However, the expression of one of these demethylases, kdm6b.1, was detected only in the blastema in regenerating fins (Fig. 3E, left and right panels), and was strongly upregulated during regeneration compared to the other putative me3K27 H3 demethylases (Fig. 3F). kdm6b.1 expression was also detected in larvae caudal fins after amputation (Fig. 3G). Kdm6b.1 displayed me3K27 H3-specific demethylase activity comparable to that of mouse Jmjd3.
Kdm6b.1 Is Required for Regeneration in Zebrafish.
Given its activity, robust expression in the blastema, and its upregulation during regeneration, we undertook additional experiments to implicate the function of Kdm6b.1 demethylase during regeneration. Loss-of-function studies were performed to examine whether its function is needed for regeneration. We focused our efforts on zebrafish larvae, as opposed to adult animals, for the following reasons. First, studies in larvae using morpholinos oligonucleotides afford the ability to knock-down a gene of interest. Second, knockdown studies in adult caudal fin using morpholinos are most useful for later stages (48–72 hpa; after blastema formation) of regeneration since the injection of the morpholino can only be reliably performed in the relatively bulky tissue of the newly formed regenerate (49). In contrast, knockdown studies in larvae allow the observation of early regeneration phenotypes. Finally, caudal fins of larvae are capable of regenerating after amputation via a mechanism that appears very similar to that used by adult fish (37, 50–53).
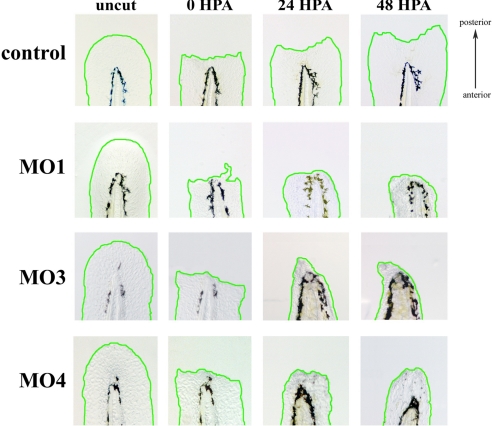
One-cell stage embryos were injected with control or Kdm6b.1-specific morpholinos (Fig. S8). At 48–72 h post injection, caudal fins were amputated just distal to the notochord (Fig. 4, first two columns of images). Care was taken to ensure that only injected animals that appeared grossly normal were used for regeneration analyses (Fig. S8). At 24 hpa, a defect in regeneration was visible in Kdm6 b.1 knockdown animals (Fig. 4, comparing the third column of images). By 48 hpa, animals injected with control morpholinos had regenerated normally (150 out of 150 animals regenerated normally). In contrast, Kdm6b.1 MO1 morphants exhibited a pronounced inability to regenerate caudal fins with only 67 out of 175 (33%) regenerating normally (Fig. 4, comparing the fourth column of images). Injection of three other independent Kdm6b.1 morpholinos also resulted in reduced regeneration (compare the fourth column of images in Fig. 4; see also Fig. S9). Hematoxylin staining of Kdm6b.1 knockdown animals revealed that caudal fins, after amputation, comprise densely packed cells (Fig. 5A), many of which were PCNA+ cells (Fig. 5B). In control animals, as previously reported (37), by 48 hpa the majority of proliferating, PCNA+ cells were found surrounding the notochord (Fig. 4B), indicating that a gross loss in cell proliferation was not the cause of the inhibition of regeneration in the morphants.
Fig. 4.
Kdm6b.1 is needed for fin regeneration. Analysis of regeneration in control versus Kdm6b.1 MO1, MO3, or MO4 injected animals was followed over time in individual animals. Representative examples of a control (upper panels) and Kdm6b.1 MO (lower panels) phenotypes are shown before amputation, immediately after amputation, and 24 and 48 h after amputation. The amputation planes are similar to that shown above and the same animal is shown before and after amputation up to 48 h.
Fig. 5.
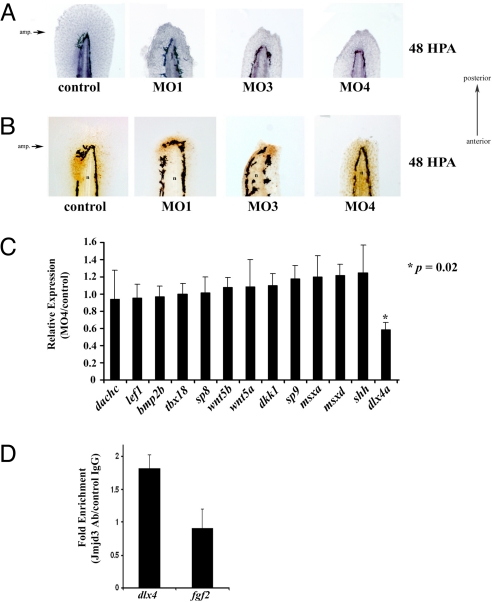
Identification of dlx4a as putative target of Kdm6b.1. (A) Control or Kdm6b.1 morpholino injected animals were amputated and allowed to regenerate for 48 h at which point they were fixed and stained with hematoxylin. The amputation planes are similar to that shown above. (B) Proliferating cells in control and Kdm6b.1 morphants. Control (left panel) or Kdm6b.1 MO (right panels) for 48 h, at which point they were stained with PCNA antibodies (PCNA+ cells are stained brown). n, notochord. The amputation planes are similar to that shown above. (C) dlx4a is downregulated in Kdm6b.1 MO animals. The expression levels of various candidate genes were tested in Kdm6 b.1 MO4 and control animals at 96 hpf (24 hpa) and normalized to Ribosomal Protein L18 expression. Data are expressed as average expression levels in the morphants relative to control animals. Error bars, the standard deviation and an asterisk (*) indicates a P value of 0.02 (n = 6 animals for each). (D) Jmjd3 associates with the dlx4 promoter in developing limb buds. Limb buds from approximately 20 pups were harvested at day e10.5 and processed for ChIP with control or Jmjd3 antibodies, followed by qPCR using primers at the transcription start sites of dlx4 or fgf2. Shown are the data from one experiment; error bars indicate the deviation from the mean duplicate ChIPs. Similar data were obtained in another experiment from different limb buds.
We reasoned that as a positive regulator of transcription, loss of Kdm6b.1 would lead to down regulation of target genes. We used a candidate gene approach to identify genes positively regulated by of Kdm6b.1. One gene that was reproducibly downregulated in Kdm6b.1 morphants was the distal homeobox gene, dlx4a (Fig. 5C and Fig. S10). This gene was an attractive candidate for being a Kdm6b.1 target gene during regeneration since dlx genes function during both fin and limb development, throughout evolution (54). Additionally, dlx4a is expressed in both the median fin folds, the pectoral fin buds in developing zebrafish (55), and, in the regenerating fin (29). Importantly, bivalent me3K4/me3K27 H3 domains at the dlx4a promoter is resolved to monovalent me3K4 H3 after fin amputation. We turned our attention to the developing mouse limb as a means to bolster our contention that Kdm6b.1 positively regulates expression dlx4a. In mouse, dlx4 (the zebrafish dlx4a ortholog) is expressed in the mesenchyme of the developing limb (56, 57) which provided us an opportunity to test the hypothesis that the mouse homolog of Kdm6b.1, Jmjd3 (Fig. S7) targets dlx4 in a similar physiological setting. Thus, we performed ChIP experiments to demonstrate a physical association between mouse Jmjd3 and the mouse dlx4 locus during limb development. Chromatin from e10.5 limb buds was immunoprecipitated with Jmjd3 antibodies (58) and eluates were tested for enrichment of dlx4 sequences at its transcription start site. We detected significant enrichment of dlx4 in Jmjd3 ChIPs, indicating an association between Jmjd3 and the dlx4 gene (Fig. 5D). In contrast, no enrichment was observed for the fgf2 promoter (Fig. 5D). These data argue in favor of our contention that dlx4a is regulated by Kdm6b.1 in zebrafish, as well as in mouse limb. Ultimately, identification of loci that are direct targets of Kdm6b.1 using whole genome ChIP approaches will illuminate the function of this histone demethylase during development and regeneration. Still, we should not rule out additional mechanisms which contribute to the loss of me3K27 H3 at some promoters, such as transcription coupled histone exchange, dilution of this histone modification during cell division, or due to the action of demethylases other than Kdm6b.1.
It is noteworthy that the expression of a number of bivalent genes remained unchanged in the Kdm6b.1 knockdown animals (Fig. S10a). This may be attributed to either redundancy or to demethylase specificity. It is still premature to implicate either as is not clear how me3K27 H3 demethylases are recruited to discreet genomic positions (24). In mouse ES cells, UTX knockdown does not alter global me3K27 H3 levels, indicative of redundancy (21). On the other hand, modest changes in me3K27 H3 levels in HeLa and 293T cells were seen upon UTX knock down (28). This is an important consideration for future investigations given our gene expression analysis and our finding that Kdm6a.2 associates with lef1 promoter in caudal fins (Figs. S7 and S10 c and d).
In the case of the zebrafish caudal fin, amputation leads to the reactivation of genes whose expression normally diminishes after development. We find that many of these same genes contain silent promoters decorated with a bivalent me3K4/me3K27 H3 domain, an epigenetic mark thought to represent a poised state of gene inactivity. During regeneration of the zebrafish caudal fin, transcription activation of these promoters occurs in part by removal of repressive PcG-dependent me3K27 H3, which is likely mediated the by histone demethylases, such as Kdm6b.1.
Collectively, the observations presented here support a model in which the zebrafish maintains a normal, non-regenerating gene expression program in the caudal fin which contributes to tissue homeostasis. Unlike non-regenerating animals, this gene expression program is switched after injury to the animal through the actions of plastic epigenetic mechanisms, including reversible histone modifications. Our results suggest that demethylation of me3K27 H3 contributes to gene expression in regenerating caudal fin. It is not yet clear how signals generated by fin amputation alter chromatin remodeling and uncovering these signal transduction pathways that converge on histone demethylase complexes after tissue damage will deepen our understanding of how a regenerative response can be achieved in response to tissue injury.
Bivalent me3K4/me3K27 H3 can be detected in various tissues and cell types (15, 17, 20, 33–36), suggesting that developmental regulatory genes may be poised for activation in a variety of cell types and animals. It will be interesting to examine bivalent chromatin modifications in non-regenerating tissue, for example, a mouse limb, to determine whether they contain poised, bivalent genes.
Experimental Procedures
Zebrafish.
Wild-type AB zebrafish were maintained at 28.5 °C. For fin amputations, fish were sedated in tricaine and the caudal fins were amputated with a razor blade and the fins were collected on ice in isotonic buffer. Fluoroscein-labeled (MO1) or unlabeled (MO2–MO4) morpholinos directed against zebrafish Kdm6b.1 (MO1: 5′-TGTACGGCCCATCTCGCTGTTACTG-3′, MO2: 5′ MO3: 5′-TTTCCAGCCATGCTGTGCTGTGTTG-3′, MO4) or control morpholino (5′-CCTCTTACCTCAGTTACAATTTAT-3′) were purchased from Gene Tools and injected in 1–2 cell stage embryos at a needle concentration of 0.2–0.75 mM. MO1, 2, and 3 are Kdm6b.1 translation blocking morpholinos; MO4 targets exon 8 of Kdm6b.1 that codes for a portion of the catalytic domain. For morpholino injections, eggs at the 1-cell stage were injected with 1 nL of the indicated morpholino oligonucleotide and were allowed to develop at 28.5 °C until 48–72 h, at which point the larvae were sedated and the caudal fin fold was amputated under a dissecting scope with a fine scalpel blade. Animals were returned to 28.5 °C and regeneration was observed. Details can be found in the Supporting Information.
cDNA Clone Isolation.
Zebrafish PcG and Kdmases cDNA were identified by blast searches and the corresponding full-length cDNAs were isolated by PCR using cDNA synthesized from caudal fins at 48 hpa.
In Situ Hybridization and Histology.
Fins were fixed in PFA and processed for in situ hybridization using antisense RNA probes generated from the indicated cDNA clones. Some fins were cryosectioned and stained with fast red after development of the in situ hybridization. Details can be found in the Supporting Information.
Chromatin Immunoprecipitation.
ChIP was performed using standard protocols (Millipore) with modifications in the extract preparation. ChIP was quantified by qPCR (ABI) by fitting to standard curves generated using dilutions of genomic DNA. For sequential ChIP experiments, immune complexes were eluted as above in the presence of 10 mM DTT, diluted 50-fold in IP buffer and immunoprecipitated with control IgG or histone antibodies as indicated. Subsequent wash steps and elutions were performed as for single ChIP experiments. Details can be found in the Supporting Information.
Demethylation Assay.
For in vitro demethylation assays, transfected 293 cells were lysed in buffer (10 mM HEPES-NaOH, pH 7.6, 185 mM NaCl, and 0.5% Nonidet P-40) containing 1 mM DTT, 0.2 mM PMSF, and a protease inhibitor mixture (Complete EDTA-free, Roche) and extracts were immunoprecipitated with Flag or myc antibodies. Immune-complexes were washed and reactions were initiated by adding 5 μg core histones from calf (Sigma) to the immune-complexes in 1x KDM reaction buffer (50 mM HEPES-KOH (pH 8.0), 100 μM Fe(NH4)2(SO4)2, 1 mM α-ketoglutarate, and 2 mM ascorbate) at 37 °C for 5 h with mixing followed by SDS/PAGE and immunoblotting with the indicated antibodies.
Supplementary Material
Acknowledgments.
We thank M. G. Rosenfeld (University of California, San Diego) for the mouse Jmjd3 expression vector; E. Canaani (Weizmann Institute of Science) and K. Helin (University of Copenhagen) for providing antisera; C. Murawsky and B. Emerson for suggestions for the demethylase assay; K. Stankunas and C. Kintner for comments on the manuscript; and the National Cancer Institute for DZNEP. This work was supported by a training grant from the California Institute for Regenerative Medicine (S.S.), work in the laboratory of J.C.I.B. was supported by grants from Fundacion Cellex, the G. Harold and Leila Y. Mathers Charitable Foundation, the Ipsen Foundation and the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904132106/DCSupplemental.
References
- 1.Brockes JP, Kumar A. Appendage regeneration in adult vertebrates and implications for regenerative medicine. Science. 2005;310:1919–1923. doi: 10.1126/science.1115200. [DOI] [PubMed] [Google Scholar]
- 2.Poss KD, Keating MT, Nechiporuk A. Tales of regeneration in zebrafish. Dev Dyn. 2003;226:202–210. doi: 10.1002/dvdy.10220. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez Alvarado A, Tsonis PA. Bridging the regeneration gap: Genetic insights from diverse animal models. Nat Rev Genet. 2006;7:873–884. doi: 10.1038/nrg1923. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka EM. Regeneration: If they can do it, why can't we? Cell. 2003;113:559–562. doi: 10.1016/s0092-8674(03)00395-7. [DOI] [PubMed] [Google Scholar]
- 5.Yakushiji N, et al. Correlation between Shh expression and DNA methylation status of the limb-specific Shh enhancer region during limb regeneration in amphibians. Dev Biol. 2007;312:171–182. doi: 10.1016/j.ydbio.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 7.Mikkelsen TS, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 9.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 12.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 13.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pietersen AM, van Lohuizen M. Stem cell regulation by polycomb repressors: Postponing commitment. Curr Opin Cell Biol. 2008;20:201–207. doi: 10.1016/j.ceb.2008.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Mendenhall EM, Bernstein BE. Chromatin state maps: New technologies, new insights. Curr Opin Genet Dev. 2008;18:109–115. doi: 10.1016/j.gde.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ku M, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genetics. 2008;4:e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Araki Y, et al. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity. 2009;30:912–925. doi: 10.1016/j.immuni.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyazaki M, et al. Thymocyte proliferation induced by pre-T cell receptor signaling is maintained through polycomb gene product Bmi-1-mediated Cdkn2a repression. Immunity. 2008;28:231–245. doi: 10.1016/j.immuni.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 19.Pan G, et al. Whole-genome analysis of histone H3 lysine 4 and lysine 27 methylation in human embryonic stem cells. Cell Stem Cell. 2007;1:299–312. doi: 10.1016/j.stem.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 20.Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci USA. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 22.De Santa F, et al. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 23.Jepsen K, et al. SMRT-mediated repression of an H3K27 demethylase in progression from neural stem cell to neuron. Nature. 2007;450:415–419. doi: 10.1038/nature06270. [DOI] [PubMed] [Google Scholar]
- 24.Lan F, et al. A histone H3 lysine 27 demethylase regulates animal posterior development. Nature. 2007;449:689–694. doi: 10.1038/nature06192. [DOI] [PubMed] [Google Scholar]
- 25.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 26.Smith ER, et al. Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol Cell Biol. 2008;28:1041–1046. doi: 10.1128/MCB.01504-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swigut T, Wysocka J. H3K27 demethylases, at long last. Cell. 2007;131:29–32. doi: 10.1016/j.cell.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 28.Xiang Y, et al. JMJD3 is a histone H3K27 demethylase. Cell Res. 2007;17:850–857. doi: 10.1038/cr.2007.83. [DOI] [PubMed] [Google Scholar]
- 29.Schebesta M, Lien CL, Engel FB, Keating MT. Transcriptional profiling of caudal fin regeneration in zebrafish. Sci World J. 2006;1:38–54. doi: 10.1100/tsw.2006.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 31.Lepilina A, et al. A dynamic epicardial injury response supports progenitor cell activity during zebrafish heart regeneration. Cell. 2006;127:607–619. doi: 10.1016/j.cell.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 32.Ohm JE, et al. A stem cell-like chromatin pattern may predispose tumor suppressor genes to DNA hypermethylation and heritable silencing. Nat Genet. 2007;39:237–242. doi: 10.1038/ng1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 34.Sanz LA, et al. A mono-allelic bivalent chromatin domain controls tissue-specific imprinting at Grb10. EMBO J. 2008;27:2523–2532. doi: 10.1038/emboj.2008.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Attema JL, et al. Epigenetic characterization of hematopoietic stem cell differentiation using miniChIP and bisulfite sequencing analysis. Proc Natl Acad Sci USA. 2007;104:12371–12376. doi: 10.1073/pnas.0704468104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taverna SD, et al. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc Natl Acad Sci USA. 2007;104:2086–2091. doi: 10.1073/pnas.0610993104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawakami A, Fukazawa T, Takeda H. Early fin primordia of zebrafish larvae regenerate by a similar growth control mechanism with adult regeneration. Dev Dyn. 2004;231:693–699. doi: 10.1002/dvdy.20181. [DOI] [PubMed] [Google Scholar]
- 38.Kawakami Y, et al. Wnt/beta-catenin signaling regulates vertebrate limb regeneration. Genes Dev. 2006;20:3232–3237. doi: 10.1101/gad.1475106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lien CL, Schebesta M, Makino S, Weber GJ, Keating MT. Gene expression analysis of zebrafish heart regeneration. PLoS Biol. 2006;4:e260. doi: 10.1371/journal.pbio.0040260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoick-Cooper CL, et al. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- 41.Thummel R, Ju M, Sarras MP, Jr, Godwin AR. Both Hoxc13 orthologs are functionally important for zebrafish tail fin regeneration. Dev Genes Evol. 2007;217:413–420. doi: 10.1007/s00427-007-0154-3. [DOI] [PubMed] [Google Scholar]
- 42.Whitehead GG, Makino S, Lien CL, Keating MT. fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310:1957–1960. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]
- 43.Tan J, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21:1050–1063. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rocha PS, et al. The Arabidopsis HOMOLOGY-DEPENDENT GENE SILENCING1 gene codes for an S-adenosyl-L-homocysteine hydrolase required for DNA methylation-dependent gene silencing. Plant Cell. 2005;17:404–417. doi: 10.1105/tpc.104.028332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mull L, Ebbs ML, Bender J. A histone methylation-dependent DNA methylation pathway is uniquely impaired by deficiency in Arabidopsis S-adenosylhomocysteine hydrolase. Genetics. 2006;174:1161–1171. doi: 10.1534/genetics.106.063974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leal JF, et al. Cellular senescence bypass screen identifies new putative tumor suppressor genes. Oncogene. 2008;27:1961–1970. doi: 10.1038/sj.onc.1210846. [DOI] [PubMed] [Google Scholar]
- 47.Berns K, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 48.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 49.Thummel R, et al. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev Dyn. 2006;235:336–346. doi: 10.1002/dvdy.20630. [DOI] [PubMed] [Google Scholar]
- 50.Mathew LK, et al. Unraveling tissue regeneration pathways using chemical genetics. J Biol Chem. 2007;282:35202–35210. doi: 10.1074/jbc.M706640200. [DOI] [PubMed] [Google Scholar]
- 51.Mathew LK, Sengupta SS, Ladu J, Andreasen EA, Tanguay RL. Crosstalk between AHR and Wnt signaling through R-Spondin1 impairs tissue regeneration in zebrafish. FASEB J. 2008;22:3087–3096. doi: 10.1096/fj.08-109009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rojas-Munoz A, et al. ErbB2 and ErbB3 regulate amputation-induced proliferation and migration during vertebrate regeneration. Dev Biol. 2009;327:177–190. doi: 10.1016/j.ydbio.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 53.Yoshinari N, Ishida T, Kudo A, Kawakami A. Gene expression and functional analysis of zebrafish larval fin fold regeneration. Dev Biol. 2009;325:71–81. doi: 10.1016/j.ydbio.2008.09.028. [DOI] [PubMed] [Google Scholar]
- 54.Panganiban G, et al. The origin and evolution of animal appendages. Proc Natl Acad Sci USA. 1997;94:5162–5166. doi: 10.1073/pnas.94.10.5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akimenko MA, Ekker M, Wegner J, Lin W, Westerfield M. Combinatorial expression of three zebrafish genes related to distal-less: Part of a homeobox gene code for the head. J Neurosci. 1994;14:3475–3486. doi: 10.1523/JNEUROSCI.14-06-03475.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gray PA, et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science. 2004;306:2255–2257. doi: 10.1126/science.1104935. [DOI] [PubMed] [Google Scholar]
- 57.Shou S, Scott V, Reed C, Hitzemann R, Stadler HS. Transcriptome analysis of the murine forelimb and hindlimb autopod. Dev Dyn. 2005;234:74–89. doi: 10.1002/dvdy.20514. [DOI] [PubMed] [Google Scholar]
- 58.Agger K, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23:1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.