Abstract
NL63 coronavirus (NL63-CoV), a prevalent human respiratory virus, is the only group I coronavirus known to use angiotensin-converting enzyme 2 (ACE2) as its receptor. Incidentally, ACE2 is also used by group II SARS coronavirus (SARS-CoV). We investigated how different groups of coronaviruses recognize the same receptor, whereas homologous group I coronaviruses recognize different receptors. We determined the crystal structure of NL63-CoV spike protein receptor-binding domain (RBD) complexed with human ACE2. NL63-CoV RBD has a novel β-sandwich core structure consisting of 2 layers of β-sheets, presenting 3 discontinuous receptor-binding motifs (RBMs) to bind ACE2. NL63-CoV and SARS-CoV have no structural homology in RBD cores or RBMs; yet the 2 viruses recognize common ACE2 regions, largely because of a “virus-binding hotspot” on ACE2. Among group I coronaviruses, RBD cores are conserved but RBMs are variable, explaining how these viruses recognize different receptors. These results provide a structural basis for understanding viral evolution and virus–receptor interactions.
Keywords: receptor protein, SARS coronavirus, spike protein receptor-binding domain, virus-binding hotspots
A fundamental yet unresolved puzzle in virology is how viruses evolve to recognize their receptor proteins (1). Specifically, how do different viruses recognize the same receptor protein, and how do similar viruses recognize different receptor proteins? Do viruses select their receptor proteins by chance, or do they target specific virus-binding hotspots on these receptor proteins? Structural information of virus–receptor interfaces can potentially answer these questions. To date, although a few studies have obtained structural information for a single virus–receptor interface (2–6), no study has provided structural information for the interfaces between different viruses and their common receptor protein. Here we provide such structural information, by showing that nonhomologous receptor-binding proteins of 2 coronaviruses bind to the same “virus-binding hotspot” on their common protein receptor.
A recently identified human coronavirus, NL63 (NL63-CoV), is associated with common colds, croup, and other respiratory diseases (7, 8). Potent neutralizing antibodies against NL63-CoV are detected in sera from nearly all humans older than 8 years, suggesting that NL63-CoV infection is common in childhood (7, 9). NL63-CoV belongs to the coronavirus family, a group of enveloped, positive-stranded RNA viruses that infect many mammalian and avian species. Coronaviruses are classified into 3 serologic and genetic groups: mammalian group I, mammalian group II, and avian group III (10). NL63-CoV is the only group I coronavirus known to use angiotensin-converting enzyme 2 (ACE2) as its receptor (9), whereas the others use aminopeptidase-N (APN) (10–12). Curiously, ACE2 is also the receptor for the severe acute respiratory syndrome (SARS) coronavirus (SARS-CoV) (13), a group II coronavirus responsible for SARS (14, 15).
Coronaviruses enter cells through a large spike protein on their envelopes (10). The coronavirus spike protein is a membrane-anchored trimer and contains 2 subunits, receptor-binding subunit S1 and membrane-fusion subunit S2 (Fig. 1A). The S2 subunits from group I and group II coronaviruses share both sequence and structural homology (16); they contain homologous heptad-repeat segments that fold into a conserved trimers-of-hairpin structure, which is essential for membrane fusion (Fig. 1A) (16, 17). Surprisingly, the S1 subunits from group I and group II coronaviruses have no obvious sequence homology. Nevertheless, they can both be divided approximately into N-terminal region, central region, and C-terminal region (Fig. 2 A and B). The S1 central regions of both NL63-CoV and SARS-CoV are defined receptor-binding domains (RBDs) that are sufficient for high-affinity binding to their common receptor ACE2 (18–22).
Fig. 1.
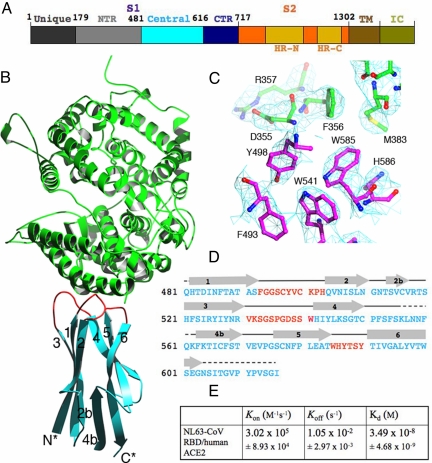
Structure of NL63-CoV RBD complexed with human ACE2. (A) Domain structure of the NL63-CoV spike protein. Unique, unique region; NTR, N-terminal region; Central, central region; CTR, C-terminal region; HR-N, heptad-repeat N; HR-C, heptad-repeat C; TM, transmembrane anchor; IC, intracellular tail. The unique domain only exists in NL63-CoV and is not involved in receptor binding (19, 20). (B) Overall structure of NL63-CoV RBD complexed with human ACE2. The RBD core is in cyan, RBMs in red, and ACE2 in green. (C) Averaged electron density map contoured at 1.0 σ and covering a portion of the NL63-CoV–ACE2 interface. (D) Sequence and secondary structures of NL63-CoV RBD. Beta-strands are drawn as arrows. RBMs are in red; the remainder of the RBD is in cyan. Disordered regions are shown as dashed lines. (E) Kinetics and binding affinity of NL63-CoV RBD and human ACE2 by surface plasmon resonance using Biacore. Structural illustrations were made using Povscript (31).
Fig. 2.
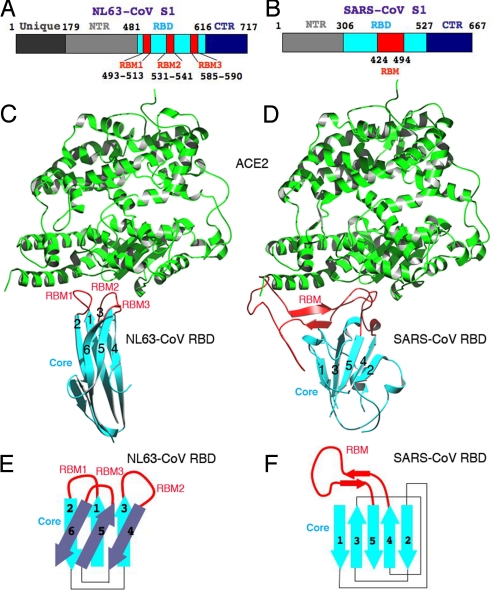
Structural comparison of NL63-CoV and SARS-CoV RBDs. (A) Domain structure of NL63-CoV S1. The boundaries of the RBD were determined by limited proteolysis of longer S1 fragments, followed by N-terminal sequencing and mass spectrometric analysis of the digestion fragments. The RBMs were identified from the crystal structure of the NL63-CoV RBD in complex with ACE2. (B) Domain structure of SARS-CoV S1 (5). (C) Another view of the structure of the NL63-CoV-RBD–ACE2 complex, which is derived by rotating the one in Fig. 1B by 90° clockwise along a vertical axis. (D) Structure of the SARS-CoV-RBD–ACE2 complex (PDB 2AJF) (5), from the same orientation as in C. (E) Schematic illustration of the topology of NL63-CoV RBD. Strands are drawn as arrows. (F) Schematic illustration of the topology of SARS-CoV RBD.
To date, the crystal structure of SARS-CoV RBD complexed with ACE2 is the only atomic structure available for any coronavirus S1 (Fig. 2D) (5). SARS-CoV RBD contains 2 subdomains, a core and a receptor-binding motif (RBM); RBM exclusively contacts ACE2. ACE2 contains a claw-like peptidase domain, with 2 lobes encircling the active site. SARS-CoV binds to the outer surface of the N-terminal lobe of the ACE2 peptidase domain. Structural information has been lacking for group I coronavirus S1, either alone or in complex with its receptor.
How do NL63-CoV and SARS-CoV both use ACE2 as their receptor, despite no obvious sequence homology in their S1 subunits? One hypothesis is that NL63-CoV and SARS-CoV share homologous RBMs, and that through RNA recombination, SARS-CoV acquired its RBM from NL63-CoV or an NL63-CoV–related group I coronavirus, gaining binding affinity for ACE2 and infectivity for human cells (20, 23). Extensive mutagenesis studies have been done to characterize the interactions between NL63-CoV and ACE2 but have failed to yield consistent results regarding the NL63-CoV binding site on ACE2 or the domain boundaries of NL63-CoV RBD (19–21). It is also intriguing that NL63-CoV and other group I coronaviruses recognize different receptors despite obvious sequence homology in their S1 subunits. Here we report the crystal structure of NL63-CoV RBD complexed with its human receptor ACE2, revealing structural mechanisms whereby NL63-CoV recognizes the same receptor as SARS-CoV but a different receptor from other group I coronaviruses.
Results and Discussion
Structure Determination.
To prepare NL63-CoV RBD for crystallization, we designed 36 RBD constructs with different N- and C-termini, on the basis of biochemical studies (19–21) and secondary structure prediction of the S1 sequence. We expressed and purified each of these fragments in insect cells. Alpha-chymotrypsin treatment of one of these fragments (residues 461–616) yielded a smaller but more stable fragment (residues 481–616), which was subsequently crystallized in complex with human ACE2 peptidase domain. We determined the structure by molecular replacement, using ACE2 as the search model (Figs. 1B and 2C). A 4-fold noncrystallographic symmetry averaging within the crystal and multiple cross-crystal averaging of the ACE2 region with 3 other structures (SARS-CoV-RBD–ACE2 complex, ACE2, and ACE2-inhibitor complex) (5, 24) dramatically improved the electron density in the NL63-CoV RBD region (Fig. 1C). We refined the structure at 3.3-Å resolution (Table S1). The final model contains residues 19–614 of human ACE2 and residues 482–602 (except for a disordered loop 555–565) of NL63-CoV RBD. The model also contains glycans N-linked to viral residues 486 and 512 and to ACE2 residues 90 and 546.
Structure of NL63-CoV RBD.
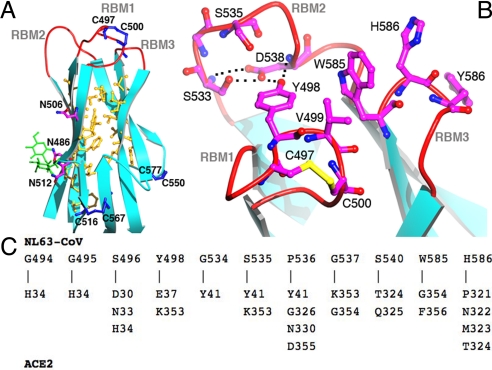
The core of NL63-CoV RBD can be best described as a β-sandwich. It consists of 2 layers of 3-stranded β-sheets, one with mixed polarity (strands 3–1-2) and the other antiparallel (strands 4–5–6) (Figs. 1 B and D and 2 C and E). The 2 layers stack tightly against each other through extensive hydrophobic interactions (Fig. 3A). A short 3-stranded, antiparallel β-sheet (strands 2b-4b-6) stabilizes the distal end of the RBD (opposite the receptor-binding interface); this end contains both the N- and C-termini of the RBD and hence likely interacts with the rest of S1 and is not as ordered as the receptor-binding interface (Fig. 1B). Three disulfide bonds are present in the RBD (Fig. 3A). The Cys-497–Cys-500 disulfide bond strengthens a critical receptor-binding loop (see below). Two other disulfide bonds, connecting Cys-516–Cys-567 and Cys-550–Cys-577, strengthen the region at the distal end. Alanine substitutions for Cys-516, Cys-550, Cys-567, or Cys-577 decrease protein stability and abolish protein expression (Table S2) (21). Overall, the β-sandwich structure of NL63-CoV RBD is a unique protein fold; a search of the protein database using DALI (25) did not reveal any related structures.
Fig. 3.
Structural features of NL63-CoV RBD and RBMs. (A) Beta-sandwich core structure of NL63-CoV RBD. Hydrophobic residues between β-sheet layers are in yellow, cysteines in blue, glycans in green, and glycosylated asparagines in magenta. There exist 3 predicted N-linked glycosylation sites in the RBD (Asn-486, Asn-506, and Asn-512), and 2 of them are confirmed in the structure (Asn-486 and Asn-512). (B) NL63-CoV RBMs. (C) Residues on NL63-CoV RBMs that directly contact ACE2.
Three discontinuous receptor-binding sites on NL63-CoV RBD, which we term receptor-binding motifs (RBMs), are presented by the β-sandwich core to bind ACE2. On the basis of their order in the primary structure, we define the 3 RBMs as RBM1 (residues 493–513), RBM2 (residues 531–541), and RBM3 (residues 585–590) (Figs. 1D and 3B). All 3 RBMs are connected to β-strands and are thus β-loops. RBM3 is more compact than RBM1 and RBM2. Nevertheless, RBM1 is strengthened by the Cys-497–Cys-500 disulfide bond, and RBM2 is reinforced by key hydrogen bonds involving Asp-538 (Fig. 3B). Alanine substitutions for Cys-497, Cys-500, or Asp-538 abolish receptor binding without significantly affecting protein expression (Table S2) (21). At the top of the RBD, the 3 protruding RBMs surround a shallow bowl-shaped cavity, which is important for receptor binding (Fig. 4A).
Fig. 4.
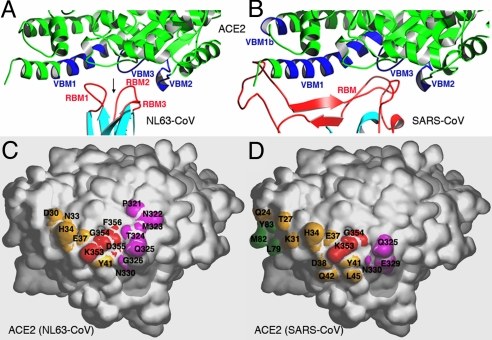
Structural comparison of NL63-CoV–ACE2 and SARS-CoV–ACE2 interfaces. (A) Enlarged view of the NL63-CoV–ACE2 interface, from the same orientation as in Fig. 2C. VBMs on ACE2 are in blue, and RBMs on NL63-CoV are in red. Arrow indicates the bowl-shaped cavity surrounded by 3 viral RBMs. (B) Enlarged view of the SARS-CoV–ACE2 interface, from the same orientation as in A. (C) Footprint of NL63-CoV on the surface of ACE2. The view is derived from the one in A by rotating ACE2 by 90° along a horizontal axis, in such a way that the edge facing the viewer moves up. VBM1 residues are in orange, VBM2 residues in magenta, and VBM3 residues in red. (D) Footprint of SARS-CoV on the surface of ACE2, from the same orientation as in C. VBM1b residues are in green.
NL63-CoV and SARS-CoV RBDs have no structural homology in either cores or RBMs, although they are both located in the S1 central regions (Fig. 2 A and B). The core of SARS-CoV RBD is a single-layer, 5-stranded antiparallel β-sheet, stabilized by 3 short α-helices (Fig. 2 D and F); the core of NL63-CoV RBD is a β-sandwich with novel topology (Fig. 2 C and E). The number of β-sheet layers, the number of β-strands, and most notably, the topologies of the β-sheets all differ in the 2 RBD cores. The RBMs of SARS-CoV and NL63-CoV are also completely different. The SARS-CoV RBM is a continuous 70-residue-long subdomain that lies on one edge of the core (Fig. 2 D and F). The NL63-CoV RBMs are 3 discontinuous β-loops (Fig. 2 C and E). The lack of structural homology in RBD cores and, more importantly, the lack of structural homology in RBMs, indicate 2 independent ways in which NL63-CoV and SARS-CoV recognize their common receptor protein.
Structure of NL63-CoV–Receptor Interface.
NL63-CoV RBD binds to the outer surface of the N-terminal lobe of the ACE2 peptidase domain (Figs. 1B and 2C). Three discontinuous virus-binding sites on ACE2, which we term virus-binding motifs (VBMs), are directly involved in virus binding (Fig. 4 A and C). On the basis of their order in the primary structure, we define the 3 VBMs as VBM1 (residues 30–41 on the N-terminal helix), VBM2 (loop containing residues 321–330), and VBM3 (loop containing residues 353–356). The binding surfaces of ACE2 and RBD are complementary, with receptor VBM3 inserting into the bowl-shaped cavity on RBD (Figs. 4A and 5A). All of the predicted and identified glycosylation sites on RBD and ACE2 are located away from the virus–receptor interface (Fig. 3A) (24) and thus do not interfere with virus–receptor interactions. A total of 11 viral residues directly contact 16 receptor residues (Figs. 3C and 4C and Table S3), and the binding buries approximately 1,300 Å2 total surface area. This binding interface is slightly smaller than other virus–receptor binding interfaces that typically range between 1,400 and 1,800 Å2 (2–6). Nevertheless, the binding affinity between NL63-CoV RBD and ACE2 is high. By surface plasmon resonance using Biacore, we measured the dissociation constant between the 2 proteins as 34.9 nM (Figs. 1E and Fig. S1).
Fig. 5.
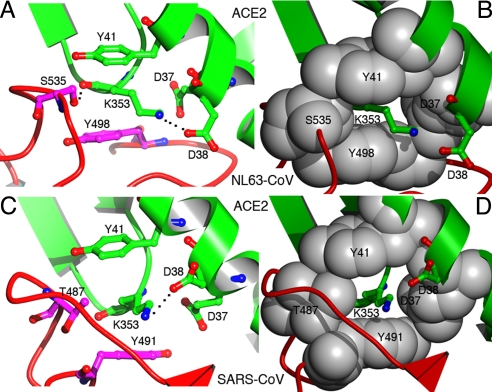
A common virus-binding hotspot on ACE2 for the binding of both NL63-CoV and SARS-CoV. (A) Stick-and-ball representation of the hotspot at the NL63-CoV–ACE2 interface. (B) Corey-Pauling-Koltun representation of the hotspot at the NL63-CoV–ACE2 interface. (C) Stick-and-ball representation of the hotspot at the SARS-CoV–ACE2 interface. (D) Corey-Pauling-Koltun representation of the hotspot at the SARS-CoV–ACE2 interface.
NL63-CoV and SARS-CoV bind to common regions on ACE2, even though they have no structural homology in RBD cores or RBMs. Both viruses recognize the same 3 VBMs on the outer surface of the N-terminal lobe of ACE2 (SARS-CoV recognizes an additional short VBM1b) (Fig. 4 A and B). Compared with SARS-CoV RBD, NL63-CoV RBD has less extensive contact with VBM1 but more extensive contact with VBM2 and VBM3 (Fig. 4 C and D). In addition, nearly the same number of ACE2 residues directly contacts each of the 2 viruses. Among these ACE2 residues, 7 directly contact both of the viruses (Table S3). Overall, although SARS-CoV forms a larger binding interface with ACE2 than NL63-CoV does, the 2 viral RBDs bind to ACE2 with similar affinity (Figs. 1E and Fig. S1) (26).
A Common Virus-Binding Hotspot on ACE2.
A virus-binding hotspot on ACE2 lies at the center of the NL63-CoV–receptor interface. In the structure of unbound ACE2, Lys-353 projects into solution (24). Upon NL63-CoV binding, Lys-353 becomes embedded in a hydrophobic tunnel surrounded by 2 aromatic rings of ACE2 Tyr-41 and NL63-CoV Tyr-498 and by 2 alkyl chains of ACE2 Asp-37 and NL63-CoV Ser-535 (Fig. 5 A and B). At the end of the tunnel, ACE2 Asp-38 forms a salt bridge with Lys-353, neutralizing its charge. Because of the hydrophobic environment, this salt bridge is energetically stabilizing (27) and critical for virus–receptor interactions. Alanine substitutions for Lys-353 or any other residues involved in the hotspot structure abolish NL63-CoV binding (20, 21). The same virus-binding hotspot on ACE2 is also key to the binding of SARS-CoV. The structures of the hotspots at the 2 different virus–receptor interfaces are strikingly similar (Fig. 5). The receptor parts are nearly identical, with subtle changes in protein side chain conformations. In the viral parts, Thr-487 and Tyr-491 on SARS-CoV replace Ser-535 and Tyr-498 on NL63-CoV, respectively, as 2 of the 4 tunnel walls. Adaptation to the hotspot on ACE2 is critical for SARS-CoV pathogenesis. A single T487S mutation that disturbs the hotspot decreases the binding affinity between SARS-CoV RBD and human ACE2 by more than 30-fold and may explain why the SARS infections in 2003–2004 were mild (23, 26, 28). Therefore, virus-binding hotspots on receptor proteins likely dictate virus–receptor interactions, viral pathogenesis, and viral transmissibility and thus are potentially major binding targets for viruses.
Receptor-Recognition Mechanisms of Other Group I Coronaviruses.
Despite their homologous spike proteins, NL63-CoV and other group I coronaviruses recognize different receptors. The S1 central regions of porcine transmissible gastroenteritis virus (TGEV), feline infectious peritonitis virus (FIPV), porcine respiratory coronavirus (PRCV), and canine coronavirus (CCoV) are highly homologous to each other, and they all use APN from their respective host as receptors (29). On the basis of structural data for NL63-CoV RBD and sequence alignment of the S1 central domains of group I coronaviruses (Fig. S2), we make the following predictions. First, the β-sandwich core structure of NL63-CoV RBD is likely conserved among group I coronaviruses. The sequences of all of the β-strands are conserved, so are all of the 4 cysteines in the core that form 2 disulfide bonds and strengthen the β-sandwich structure. Second, we predict that the 3 ACE2-binding RBMs in NL63-CoV are APN-binding sites in TGEV, FIPV, PRCV, and CCoV; structural changes in these RBMs result in the recognition of different receptors by these homologous viruses. Compared with NL63-CoV RBMs, the predicted RBM1 of other group I coronaviruses has a 2-residue insertion and loses a critical disulfide bond, the predicted RBM2 has a 9-residue insertion and 2 additional cysteines that likely form a disulfide bond, and the predicted RBM3 contains a 1-residue deletion. These structural changes are expected to abolish ACE2-binding capabilities of NL63-CoV and may help confer APN-binding capabilities of other group I coronaviruses. Structural studies of these RBDs and RBMs will further elucidate the receptor-recognition mechanisms of these important animal coronaviruses.
Evolution of Coronaviruses.
Coronaviruses are believed to have common ancestors because they share similar replication mechanisms, genomic structures, and overall gene sequences (10). Among all of the coronavirus genes, the one encoding the spike protein is the most variable. Between the spike protein subunits, S1 is more variable than S2. The current structural divergences of the S1 subunits reveal the tremendous evolutionary pressure that coronaviruses face to adapt to different host receptors, and they also reflect on the evolutionary history of coronaviruses and their receptor selections.
Our study clarifies the baffling evolutionary relationships between NL63-CoV and SARS-CoV. First, because of the lack of structural homology in RBD cores and more importantly, the lack of structural homology in RBMs, NL63-CoV and SARS-CoV must have evolved their ACE2-binding capabilities independently, disproving the previous hypothesis that the newly emerged SARS-CoV acquired its RBM from the more ancient NL63-CoV or an NL63-CoV-related group I coronavirus (20, 23). Second, binding to the same virus-binding hotspot on their common receptor protein, the 2 viruses present a remarkable case of functionally convergent evolution that occurred after divergent evolution. Whereas differences in ancient host environments likely caused the 2 viruses to initially diverge, the virus-binding hotspot on their common receptor protein was the probable driving force for the subsequent convergent evolution.
Materials and Methods
Protein Purification and Crystallization.
The NL63-CoV spike protein receptor-binding region (residues 481–616) containing an N-terminal honey bee melittin signal peptide and a C-terminal His tag was expressed in Sf9 insect cells using the Bac-to-Bac system (Invitrogen) and then purified as previously described (5, 30). In brief, the protein was harvested from Sf9 cell supernatants and loaded onto a Ni-NTA column. The protein was eluted from Ni-NTA column with imidazole and further purified by gel filtration chromatography on Superdex 200 (GE Healthcare). The protein was concentrated to 10 mg/mL. The peptidase domain of human ACE2 was expressed and purified using the same protocol as above. To purify the RBD–ACE2 complex, ACE2 was incubated with excess NL63-CoV RBD for 15 min at room temperature, and the complex was purified by gel filtration chromatography.
Crystals of the RBD–ACE2 complex were grown in sitting drops at 12 °C, over wells containing 20% PEG6000 and 100 mM Na citrate pH 5.5. Crystals were harvested in 1 week, stabilized in 25% PEG6000, 100 mM Na citrate pH5.5, and 30% ethylene glycol, and flash frozen in liquid nitrogen.
Structure Determination and Refinement.
X-ray diffraction data were collected at Advanced Photon Source (Illinois) beamline 19 ID. The crystal contains 4 complexes per asymmetric unit. The structure was determined by molecular replacement using ACE2 peptidase domain structure as the search model (5). To overcome the model bias problem common to the molecular replacement method, we performed 4-fold noncrystallographic symmetry averaging within the crystal and multiple cross-crystal averaging of the ACE2 region with 3 other structures (SARS-CoV-RBD–ACE2 complex, ACE2 by itself, and in complex with an inhibitor) (5, 24). This procedure dramatically improved electron density in the NL63-CoV RBD region. The structure of the NL63-CoV-RBD–ACE2 complex was refined to a final Rfree of 30.8% and Rwork of 27.6%. Data and refinement statistics are shown in Table S1. Software used for data processing, structure determination, and refinement is also listed in Table S1.
Kinetics and Binding Affinity of NL63-CoV RBD and Human ACE2 by Surface Plasmon Resonance Using Biacore.
The binding reactions between NL63-CoV RBD and human ACE2 were assayed by surface plasmon resonance using a Biacore 3000. ACE2 was immobilized on a C5 sensor chip directly. The surface of the sensor chip was first activated with N-hydroxysuccinimide; ACE2 was then injected and immobilized to the surface of the chip; last, the remaining activated surface of the chip was blocked with ethanolamine. Soluble NL63-CoV RBD was introduced at a flow rate of 20 μL/min at different concentrations. Kinetic parameters were determined with BIAevaluation software (Biacore) and are shown in Fig. 1E.
Supplementary Material
Acknowledgments.
We thank Drs. Stephen Harrison, Kathryn Holmes, Robert Geraghty, Doug Ohlendorf, Carrie Wilmot, and Yuhong Jiang for comments; Dr. Michael Farzan for NL63-CoV spike gene; Matthew Wilken for technical support; and staff at Advanced Photon Source beamline 19 ID for assistance in data collection. This work was supported by a University of Minnesota Academic Health Cencer (AHC) Seed Grant and an AHC Faculty Research Development Grant (to F.L.), and by Minnesota Partnership for Biotechnology and Medical Genomics Grant (to University of Minnesota). Computer resources were provided by the Basic Sciences Computing Laboratory of the University of Minnesota Supercomputing Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.J. is a guest editor invited by the Editorial Board.
Data deposition: Coordinate and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3KBH).
This article contains supporting information online at www.pnas.org/cgi/content/full/0908837106/DCSupplemental.
References
- 1.Baranowski E, Ruiz-Jarabo CM, Domingo E. Evolution of cell recognition by viruses. Science. 2001;292:1102–1105. doi: 10.1126/science.1058613. [DOI] [PubMed] [Google Scholar]
- 2.Bewley MC, Springer K, Zhang YB, Freimuth P, Flanagan JM. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science. 1999;286:1579–1583. doi: 10.1126/science.286.5444.1579. [DOI] [PubMed] [Google Scholar]
- 3.Carfi A, et al. Herpes simplex virus glycoprotein D bound to the human receptor HveA. Mol Cell. 2001;8:169–179. doi: 10.1016/s1097-2765(01)00298-2. [DOI] [PubMed] [Google Scholar]
- 4.Kwong PD, et al. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li F, Li WH, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 6.Xu K, et al. Host cell recognition by the henipaviruses: Crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc Natl Acad Sci USA. 2008;105:9953–9958. doi: 10.1073/pnas.0804797105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Hoek L, et al. Identification of a new human coronavirus. Nature Medicine. 2004;10:368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fouchier RAM, et al. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci USA. 2004;101:6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofmann H, et al. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc Natl Acad Sci USA. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lai MMC, Holmes KV. Coronaviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields' Virology. Philadelphia: Lippincott, Williams & Wilkins; 2001. pp. 1163–1186. [Google Scholar]
- 11.Delmas B, et al. Aminopeptidase-N is a major receptor for the enteropathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeager CL, et al. Human aminopeptidase-N is a receptor for human coronavirus-229e. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li WH, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ksiazek TG, et al. A novel coronavirus associated with severe acute respiratory syndrome. N Engl J Med. 2003;348:1953–1966. doi: 10.1056/NEJMoa030781. [DOI] [PubMed] [Google Scholar]
- 15.Peiris JSM, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Q, et al. Core structure of S2 from the human coronavirus NL63 spike glycoprotein. Biochemistry. 2006;45:15205–15215. doi: 10.1021/bi061686w. [DOI] [PubMed] [Google Scholar]
- 17.Xu YH, et al. Crystal structure of severe acute respiratory syndrome coronavirus spike protein fusion core. J Biol Chem. 2004;279:49414–49419. doi: 10.1074/jbc.M408782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Babcock GJ, Esshaki DJ, Thomas WD, Ambrosino DM. Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. J Virol. 2004;78:4552–4560. doi: 10.1128/JVI.78.9.4552-4560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hofmann H, et al. Highly conserved regions within the spike proteins of human coronaviruses 229E and NL63 determine recognition of their respective cellular receptors. J Virol. 2006;80:8639–8652. doi: 10.1128/JVI.00560-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li WH, et al. The S proteins of human coronavirus NL63 and severe acute respiratory syndrome coronavirus bind overlapping regions of ACE2. Virology. 2007;367:367–374. doi: 10.1016/j.virol.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin HX, et al. Identification of residues in the receptor-binding domain (RBD) of the spike protein of human coronavirus NL63 that are critical for the RBD-ACE2 receptor interaction. J Gen Virol. 2008;89:1015–1024. doi: 10.1099/vir.0.83331-0. [DOI] [PubMed] [Google Scholar]
- 22.Wong SK, Li WH, Moore MJ, Choe H, Farzan M. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J Biol Chem. 2004;279:3197–3201. doi: 10.1074/jbc.C300520200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li WH, et al. Animal origins of the severe acute respiratory syndrome coronavirus: Insight from ACE2-S-protein interactions. J Virol. 2006;80:4211–4219. doi: 10.1128/JVI.80.9.4211-4219.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Towler P, et al. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J Biol Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Holm L, Sander C. Touring protein fold space with Dali/FSSP. Nucleic Acids Res. 1998;26:316–319. doi: 10.1093/nar/26.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li WH, et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Daopin S, Anderson DE, Baase WA, Dahlquist FW, Matthews BW. Structural and thermodynamic consequences of burying a charged residue within the hydrophobic core of T4 lysozyme. Biochemistry. 1991;30:11521–11529. doi: 10.1021/bi00113a006. [DOI] [PubMed] [Google Scholar]
- 28.Li F. Structural analysis of major species barriers between humans and palm civets for severe acute respiratory syndrome coronavirus infections. J Virol. 2008;82:6984–6991. doi: 10.1128/JVI.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tusell SM, Schittone SA, Holmes KV. Mutational analysis of aminopeptidase N, a receptor for several group 1 coronaviruses, identifies key determinants of viral host range. J Virol. 2007;81:1261–1273. doi: 10.1128/JVI.01510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li F, Li WH, Farzan M, Harrison SC. Interactions between SARS coronavirus and its receptor. Adv Exp Med Biol. 2006;581:229–234. doi: 10.1007/978-0-387-33012-9_38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fenn TD, Ringe D, Petsko GA. POVScript+: A program for model and data visualization using persistence of vision ray-tracing. J Appl Crystallogr. 2003;36:944–947. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.