Abstract
Gram-positive bacteria elaborate pili and do so without the participation of folding chaperones or disulfide bond catalysts. Sortases, enzymes that cut pilin precursors, form covalent bonds that link pilin subunits and assemble pili on the bacterial surface. We determined the x-ray structure of BcpA, the major pilin subunit of Bacillus cereus. The BcpA precursor encompasses 2 Ig folds (CNA2 and CNA3) and one jelly-roll domain (XNA) each of which synthesizes a single intramolecular amide bond. A fourth amide bond, derived from the Ig fold of CNA1, is formed only after pilin subunits have been incorporated into pili. We report that the domains of pilin precursors have evolved to synthesize a discrete sequence of intramolecular amide bonds, thereby conferring structural stability and protease resistance to pili.
Keywords: CNA B domain, jelly roll domain, protease resistance, sortase
The cell wall envelope of Gram-positive bacteria can be thought of as a surface organelle that encompasses anchored proteins and pili, which enable microbes to adhere to and invade host tissues or escape innate immune responses (1). In contrast to eukaryotic cells and Gram-negative microbes, Gram-positive bacteria do not fold secreted proteins in a designated subcellular compartment with chaperones or oxidoreductases that generate disulfide bonds (2). In Gram-positive bacteria, signal peptide carrying precursors of surface proteins are secreted via the Sec pathway and require C-terminal sorting signals for their incorporation into the envelope (1). Sortases function as transpeptidases that cleave precursor proteins at short motif sequences of their sorting signals, generating thioester linked intermediates with their active site cysteine residues (3). For sortase A, B, or C, these intermediates are resolved by the nucleophilic attack of the amino group of peptidoglycan crossbridges, a reaction that immobilizes surface proteins in the bacterial cell wall envelope (1). In Bacillus cereus, a pilin-specific sortase (sortase D) recognizes the side chain ε-amino group of lysine within the YPKN pilin motif of the major pilus subunit, BcpA, as a nucleophile (4). The product of this reaction is an intermolecular amide bond that covalently links each subunit within the pilus fiber. The LPXTG sorting signal of BcpA precursors can be cut by sortase D or A (5). The latter reaction prevents the incorporation of additional pilin subunits and promotes anchoring of BcpA pili to peptidoglycan crossbridges. BcpA pili contain a minor pilin, BcpB, which is positioned at the tip of each fiber (6). Analogously to the tip proteins of other pili, BcpB is thought to function as an adhesin for host tissues (7, 8). The sorting signal of BcpB is cleaved by sortase D, but not by sortase A. As the resulting acyl intermediate between BcpB and sortase D can only be resolved by the nucleophilic attack of the YPKN motif, the minor subunit is deposited at the tip of BcpA pili (6).
In previous studies, we engineered BcpA variants with strategically placed polyhistidine (His6) affinity tags and methionine (Met) residues that were assembled into pili. Cyanogen bromide (CNBr) treatment of purified pili generated homoserine-lactone breaks at Met residues and peptide fragments that could be analyzed by mass spectrometry and Edman degradation, thereby revealing the presence of 2 amide bonds, Lys162-Thr522, the sortase D-derived intermolecular linkage, as well as Lys37-Asn163, an intramolecular amide bond positioned in the first CNA-B domain of BcpA (designated CNA1) (4). Purification and mass spectrometry of the recombinant major pilin, rBcpA, which lacks the N-terminal signal peptide and C-terminal sorting signal, suggested the presence of intramolecular amide bonds in 2 additional predicted Ig folds, CNA2 and CNA3 (4). Nevertheless, the contributions of intramolecular bonds toward pilus structure and function have not yet been revealed.
Here we report the 3D x-ray structure of BcpA. The pilin precursor encompasses 3 reverse-Ig (Ig) fold domains (CNA1–3) and one jelly-roll domain (designated XNA). CNA2, CNA3 and XNA, but not CNA1, synthesize intramolecular amide bonds within the precursor, whereas the intramolecular amide of CNA1 is formed only in mature, assembled pili. Informed by these results, we examined the contribution of intramolecular amide bonds toward pilus structure and function.
Results
Intramolecular Amide Bonds in the BcpA Precursor.
Recombinant rBcpAHis-6, lacking the N-terminal signal peptide and the C-terminal LPVTG sorting signal of the pilin precursor, was purified by affinity chromatography from cleared lysates of Escherichia coli (SI Appendix, Fig. S1). The predicted mass of the primary rBcpAHis-6 translational product is 56,697.27 (SI Appendix, Table S1). We observed an average mass of 56,648.60, a measurement that is in agreement with the formation of 3 intramolecular amide bonds, each resulting in the loss of 17 Da (NH3) (SI Appendix, Table S1). Analysis of the amino acid sequence of BcpA identified 3 CNA-B domains (Pfam05738), Ig-like structures proposed to form a single intramolecular amide bond (9). Each of the 3 CNA-B domains of rBcpAHis-6 is predicted to position a glutamic acid residue in close proximity to lysine and asparagine residues; the glutamic acid is thought to enable the formation of an intramolecular amide bond between the side chains of lysine-asparagine (9). As expected, alanine substitution of the conserved glutamic acid within CNA2 (Glu223Ala) or CNA3 (Glu472Ala) abrogated the formation of the corresponding amide bonds, as the average mass of each variant rBcpAHis-6 increased by 17 Da (4) (SI Appendix, Table S1). Similarly, alanine substitution of residues engaged in the intramolecular amide (Lys174 or Asn256 for CNA2, as well as Lys417 or Asn512 for CNA3) abolished the formation of the corresponding amide bond; these rBcpAHis-6 mutants displayed only a mass difference of 34 Da as compared to the predicted average mass of their primary translational products (SI Appendix, Table S1). Surprisingly, alanine substitutions of Lys37 or Asn163 in CNA1 did not affect the average mass of purified rBcpAHis-6. Thus, although Lys37 and Asn163 engage in an intramolecular amide bond immediately adjacent to the sortase D-derived intermolecular link of assembled pili (4), this bond is not formed in rBcpA.
The CNA1 Domain of the BcpA Precursor Is Sensitive to Protease.
Purified rBcpAHis-6 failed to produce diffracting crystals and was subjected to limited protease cleavage. Treatment with trypsin generated a protease resistant product, rBcpA*, that migrated farther on Coomassie-stained SDS/PAGE than rBcpAHis-6 without protease treatment (SI Appendix, Fig. S1). Edman degradation identified Asn163, of the YPKN pilin motif, as the N-terminal residue of BcpA*, as all subsequently released residues agreed with the predicted downstream sequence of BcpA (SI Appendix, Table S2). Edman degradation released a single amino acid per sequencing cycle, confirming that the CNA1 domain within rBcpAHis-6, unlike its counterpart within assembled pili (vide infra), lacks the amide bond between Lys37 and Asn163. BcpA*, when examined by electrospray ionization Fourier transform mass-spectrometry (ESI-FTMS), generated an observed average mass of 40,377.2 (SI Appendix, Fig. S1). The predicted average mass of BcpA* without intramolecular amide bonds is 40,427.96. These data suggest that BcpA* spans the remaining length of rBcpAHis-6 beginning at Asn163 and forms 3 intramolecular amide bonds. BcpA* was expressed as a fusion to the C-terminal end of GST (GST, pJB171) (SI Appendix, Fig. S1). GST-BcpA* was isolated by affinity chromatography and the GST tag removed with thrombin and BcpA* purified by gel filtration chromatography (SI Appendix, Fig. S1). An average mass of 39,097.48 was observed for BcpA*. The predicted average mass for BcpA* is 39,150.54, which is in agreement with the presence of 3 intramolecular amide bonds (SI Appendix, Table S1).
Crystal Structure of BcpA*.
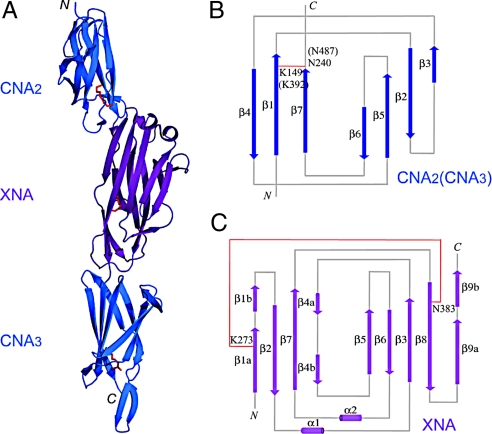
BcpA* was crystallized (SI Appendix, Fig. S1) and its structure solved using multiple-wavelength anomalous dispersion (MAD). BcpA* has an elongated, 3-domain structure with a length of 134 Å and a width of 15 to 30 Å (Fig. 1). Each of the 3 domains in BcpA*—CNA2, XNA and CNA3—adopts β-sandwich structures (Fig. 1A). CNA2 and CNA3 possess a reverse Ig-fold, comprised of one 4-stranded β-sheet and one 3-stranded β-sheet (Fig. 1B). XNA assumes a jelly-roll structure with 9 β-strands (Fig. 1C). A single intramolecular amide bond was identified in CNA2 and CNA3 (Fig. 1A). Continuous electron density demonstrated the existence of amide bonds between the ε-amino group of lysine and the carboxamide group of asparagine, i.e., Lys174-Asn265 in CNA2 and Lys417-Asn512 in CNA3. In each case, a catalytic glutamic acid (Glu223 or Glu472) is positioned in hydrogen bonding distance of the amide bond.
Fig. 1.
Crystal structure of BcpA* and topology diagrams of the CNA and XNA domains. (A) The crystal structure of BcpA* was solved using MAD. The CNA2 and CNA3 domains are shown as blue ribbons and the XNA domain purple. The Lys and Asn residues forming the intramolecular amide bonds in each of the 3 crystallized domains are shown as red sticks. Topology diagrams of the CNA (B) and XNA (C) domains show the arrangement of β-strands in the reverse Ig-fold (CNA) and jelly-roll fold (XNA). Both CNA2 and CNA3 share the same overall topology. β-strands are shown as arrows and α-helices as cylinders. Intramolecular amide bonds are shown as red lines.
A previously unrecognized amide bond between Lys273-Asn383 was identified in XNA (Fig. 1C and SI Appendix, Fig. S2). In contrast to CNA2 and CNA3, where amide bond formation is thought to be catalyzed by the side chain of glutamic acid, Asp312 is positioned in close proximity to Lys273 and Asn383 (SI Appendix, Fig. S2). The side chain carboxylic acid of aspartic acid forms hydrogen bonds with both the carbonyl oxygen and amide hydrogen of the intramolecular amide bond (SI Appendix, Fig. S2).
In the folded structure of BcpA*, the amide bonds of CNA2 and CNA3 are located toward the C-terminal end of each domain, near the interdomain connections between CNA2 and XNA as well as CNA3 and the LPVTG sorting signal (Fig. 1A and SI Appendix, Fig. S2). The CNA2/CNA3 amide bonds link the first and last β-strands of each domain, with participating lysine and asparagine residues located on adjacent, parallel β-strands of the 3-stranded β-sheet (SI Appendix, Fig. S2). The XNA structure includes a β-sandwich and the intramolecular amide bond covalently joins the first and penultimate strands of the domain. The β-strands on which the lysine and asparagine residues reside assume antiparallel polarity, are separated by one β-strand, and are located on opposite sides of the β-sandwich. This arrangement provides a tether between 2 β-sheets as the core structural unit of the XNA domain.
In mature polymerized BcpA, an intermolecular amide bond between Lys162 of the YPKN motif, positioned immediately adjacent to the N-terminal Asn163 of BcpA*, is linked end-to-end with the carboxyl group of Thr522, which is located 7 residues downstream of the C-terminal Ser515 in Fig. 1A. Both termini of BcpA* extend onto the surface of the structure. We presume that these parts of the pilin protein are reorganized during pilus assembly via the sortase D-catalyzed linkage as well as the folding of the CNA1 domain.
Catalytic Requirements for the Intramolecular Amide Bonds of BcpA.
rBcpAHis-6 (primary translation product mass 56,697.27; observed mass 56,644.26) or its variants were purified and their ability to synthesize the intramolecular amide bonds in 3 domains—CNA2, XNA and CNA3—was examined by a combination of molecular biology and mass spectrometry. As reported above, alanine substitution of the catalytic residue of CNA2 (Glu223) or CNA3 (Glu472) abrogated formation of the corresponding amide bond (SI Appendix, Table S1). Similarly, alanine substitution of any one of the 6 residues engaged in the intramolecular amide bonds (Lys174-Asn265 for CNA2, Lys273-Asn383 for XNA, Lys417-Asn512 for CNA3) prevented formation of the corresponding amide linkage (SI Appendix, Table S1). Consistent with the proposed role of Asp312 as the catalytic residue for amide bond formation in XNA, alanine substitution of this residue also abolished amide bond formation in this domain (SI Appendix, Table S1). We constructed a triple alanine substitution variant in all 3 catalytic residues, Glu223, Asp312, and Glu472 (pJB217). This mutant rBcpAHis-6 did not form any amide bonds, as its observed mass was within the experimental error for the predicted mass of the primary translational product (SI Appendix, Table S1).
We constructed 3 plasmids (pJB150, pJB187 and pJB200) to isolate branched peptides with intramolecular amide bonds in CNA2, XNA, and CNA3 from rBcpAHis-6 (SI Appendix, Table S1). Intramolecular amide bonds were demonstrated by mass spectrometry and Edman degradation (SI Appendix, Fig. S3 and Tables S3–S8). In summary, synthesis of intramolecular amide bonds occurs at Lys-Asn residues that are positioned within electron bonding distance of each folded domain of BcpA and must be in close proximity to their catalytic carboxylic residue, Glu or Asp. All mutations that altered catalytic or participating residues also prevented amide bond formation, indicating that the contributions of participating Asn or Lys residues, as well as their Glu or Asp catalytic residues, are highly specific.
Intramolecular Amide Bonds in BcpA Pili.
To identify intramolecular amide bonds in pili, we used plasmid pJB230, encoding BcpA with a Met-His6 tag at Gly150, i.e., within the CNA1 domain. To avoid generation of complex mass spectra caused by multiple intramolecular linkages, the BcpA variant of pJB230 carries an alanine substitution at Asn163, which prevents synthesis of the CNA1 amide bond (Lys37-Asn163). BcpA pili were isolated from Bacillus anthracis srtA::ermC (pJB230) and cleaved with CNBr (SI Appendix, Fig. S4). Met-His6 tagged fragments were purified by affinity chromatography and analyzed by Coomassie Blue stained SDS/PAGE (SI Appendix, Fig. S4). Edman degradation identified compound A as a branched peptide with 2 NH2-terminal residues (SI Appendix, Fig. S4 and Table S9). Compound A harbors the intermolecular amide bond of BcpA (Lys162-Thr522, SI Appendix, Fig. S4). Its observed average mass (14,624.64) is in agreement with the intramolecular amide of CNA2 (SI Appendix, Fig. S4). To identify the residues involved in the CNA2 amide link, compound A was cleaved with trypsin. The structure of a peptide with the average mass of 2,130.07 [NH2-GAVDLIKTGVNEK-(NNEEPTM*)-CO2H)], was characterized by CAD fragmentation of the doubly charged ion at m/z 1065.54 (SI Appendix, Fig. S4). The a1′ and b1′ fragment ions demonstrate the amide linkage between Lys174-Asn265 (SI Appendix, Table S10).
BcpA expressed from plasmid pJB232 also harbors the Met-His6 tag at Gly150; further, this BcpA variant includes Met residues in the vicinity of Asn265 (Met-Glu-Val-Lys-Asn265 in CNA2), Asn383 (Met-Leu-Asn383 in XNA) and Asn512 (Met-Glu-Asn512 in CNA3), i.e., of residues engaged in intramolecular amide bonds of the pilin precursor (Fig. 2A). Pili were isolated from B. anthracis srtA::ermC (pJB232) (10), cleaved with CNBr, and purified by affinity chromatography (Fig. 2BC). The linear translational BcpA product of pJB232, predicted mass 55,834, harbors 6 Met residues. CNBr would be predicted to generate 7 daughter fragments with mass 14,178 (Leu382-Met510), 13,376 (Glu262-Met381), 13,119 [(His6) Gly150-Met261], 7,681 (Asp26-Met96, the NH2-terminal fragment following signal peptidase cleavage of BcpA), 5,955 [Gly97-Glu149 (Ala-Ser-Met)], 917 (Glu511-Met518), and 428.53 (Leu-Pro-Val-Thr522)]. CNBr treatment of BcpA pili released a compound with a mass of 49,728.86 as determined by ESI-FTMS (Fig. 2D). This measurement can be explained by the presence of 4 intramolecular amide bonds in BcpA pili and one intermolecular amide bond connecting 6 CNBr fragments of BcpA (calculated mass 49,735.28). The seventh CNBr fragment, m/z 5,955, is located between Gly97-Glu149 within CNA1 [Gly97-Glu149 (Ala-Ser-Met)]. Because Gly97-Glu149 is not linked by intra- or intermolecular amide bonds to the remainder of the pilin, this compound does not copurify with the His6 tagged BcpA from isolated pili (Fig. 2CE). Edman degradation of the 49,728.86 compound released 6 residues per cycle, confirming the presence of 5 amide bonds within each BcpA subunit of assembled pili (Fig. 2E, SI Appendix, Table S11). Together, these data indicate that BcpA pili are assembled from one intermolecular (Lys162-Thr522) as well as 4 intramolecular amide bonds involving Lys37-Asn163, Lys174-Asn265, Lys273-Asn383, and Lys417-Asn512.
Fig. 2.
Four intramolecular and one intermolecular amide bond are formed in BcpA pili. (A) Schematic to illustrate the structure or BcpA pili assembled from the CNA1, CNA2, XNA, and CNA3 domains, the side chain residues (Lys and Asn) engaged in the formation of intramolecular amide bonds as well as the intermolecular amide bond between Lys162 (YPKN motif) and Thr522 (in the LPVT remnant of the cleaved sorting signal). BcpA pilin variants encoded by pJB232 harbor 6 internal methionine residues that can be cleaved with cyanogen bromide (CNBr) to generate homoserine lactone breaks. (B) B. anthracis srtA::ermC (pJB232) was stained with α-BcpA antibodies, 10 nm gold anti-rabbit conjugate, and uranyl acetate. Images were viewed at 8,260× and the bar represents a distance of 1 μm. (C) Pili were isolated from bacilli, cleaved with CNBr, purified by Ni-NTA chromatography and eluate analyzed by Coomassie stained SDS/PAGE. (D) The mass of compound B (arrow in C) was determined by LC-ESI-FTMS. (E) The structure of compound B as derived by Edman degradation and mass spectrometry. Diagram displays the peptide sequence of 6 branched peptides, generated by CNBr cleavage at 6 internal methionine (M) residues, and the intra- and intermolecular amide bonds that connect all 6 peptides.
Intramolecular Amide Bonds Confer Structural Stability on Pili.
Pili are known to be resistant to proteases, a prerequisite for their functional attributes as microbial adhesins on host tissues (11). Microbial adhesion occurs on mucosal surfaces, a host milieu endowed with innate defenses including proteases (11). BcpA pili were purified from B. anthracis srtA::ermC harboring pJB12 and the wild-type pilus operon bcpA-srtD-bcpB. Pili assembled with wild-type BcpA were resistant to trypsin treatment, which is used here as an indicator for the structural integrity of these fibers (Table 1). We constructed a series of BcpA variants with alanine substitutions at Asn163, Lys174, Asp312, or Glu472, thereby disrupting each of the 4 intramolecular amide bonds in CNA1, CNA2, XNA, or CNA3, respectively. When expressed in bacilli and analyzed by immunoblotting, these variants assembled BcpA pili (SI Appendix, Fig. S5). Purified pili were identified by labeling with immunogold particles and viewed by electron microscopy (Fig. 3). Alanine substitutions that abrogated the intramolecular amide bonds of CNA1, CNA2, or CNA3 did not affect pilus assembly or length (Table 1). Alanine substitution of Asp312, the catalytic residue of XNA, reduced the length of pili. However, Lys273Ala, a mutation that also abrogates formation of the XNA intramolecular amide, did not affect pilus length (Fig. 3, Table 1). Similarly, alanine substitution of Glu223, the catalytic residue of CNA2, dramatically reduced pilus assembly and length (SI Appendix, Fig. S5). Alanine substitution at Lys174, which abrogates the intramolecular amide between Lys174-Asn265 in the CNA2 domain, did not display such phenotype. Thus, the CNA2 and XNA derived intramolecular amide bonds are not essential for pilus polymerization (Table 1). Nevertheless, 2 catalytic residues for amide bond formation, Glu223 and Asp312, contribute in a discrete manner to BcpA substrate recognition and sortase D-catalyzed polymerization of pili.
Table 1.
Intramolecular amide bonds determine length and protease resistance of BcpA pili
| BcpA mutant | Plasmid | Pili without trypsin |
Pili after 2.2 × 10−7 M trypsin |
||
|---|---|---|---|---|---|
| Length,* μm | P value† | Length,* μm | P value† | ||
| Wild type | pJB12 | 1.45 (0.52) | — | 1.41 (0.57) | — |
| Asn163Ala | pJB109 | 1.57 (1.02) | 0.63 | 1.00 (0.68) | 0.02 |
| Lys174Ala | pJB61 | 1.33 (0.50) | 0.47 | 0.18 (0.13) | 1.3 × 10−14 |
| Lys273Ala | pJB210 | 1.21 (0.37) | 0.10 | 0.38 (0.29) | 2.0 × 10−11 |
| Asp312Ala | pJB236 | 0.75 (0.31) | 1.2 × 10−5 | 0.07 (0.03) | 3.9 × 10−14 |
| Glu472Ala | pJB107 | 1.17 (0.51) | 0.07 | 0.45 (0.24) | 1.4 × 10−9 |
| Asn163Ala; Lys174Ala | pJB240 | 1.29 (0.89) | 0.60 | 0.19 (0.06) | 2.1 × 10−18 |
| Asn163Ala; Asp312Ala | pJB238 | 0.79 (0.19) | 8.0 × 10−6 | 0.06 (0.04) | 3.5 × 10−14 |
| Asn163Ala; Glu472Ala | pJB235 | 1.00 (0.36) | 0.002 | 0.29 (0.14) | 3.6 × 10−14 |
| Lys174Ala; Glu472Ala | pJB243 | 1.26 (0.58) | 0.25 | 0.16 (0.05) | 8.9 × 10−15 |
| Asn163Ala; Asp312Ala; Glu472Ala | pJB239 | 1.02 (0.33) | 2.7 × 10−3 | 0.10 (0.04) | 8.9 × 10−14 |
| Asn163Ala; Lys174Ala; Glu472Ala | pJB245 | 1.04 (0.49) | 0.09 | 0.19 (0.05) | 8.6 × 10−13 |
*Average pilus length measurement (n ≥ 20 immunoreactive particles) with the standard error of the mean in parentheses.
†Statistical significance was evaluated with the two-tailed homoscedastic Student's t test, and P values were recorded.
Fig. 3.
Intramolecular amide bonds are required for the functional assembly of BcpA pili. (A) Diagram of polymerized BcpA pili with 4 intramolecular and one intermolecular amide bonds. (B–K) Wild-type BcpA pili and their variants were purified from B. anthracis srtA::ermC. Isolated pili were either mock treated or incubated with trypsin. The integrity of isolated pili was analyzed by immunogold labeling and transmission electron microscopy. See Table 1 for quantification of pilus length.
To test whether BcpA mutant pili resist proteases, we incubated isolated fibers with trypsin, an enzyme that cleaves polypeptides at lysine, arginine, and histidine. Positively charged residues are present in great abundance in BcpA—16% of all residues in the mature pilin. When viewed by immunogold labeling and electron microscopy, pili assembled from BcpA variants that cannot form intramolecular amide bonds in CNA2 (Lys174Ala), XNA (Lys273Ala), or CNA3 (Glu472Ala) were all exquisitely sensitive to trypsin, which cleaved these fibers into very small fragments (Fig. 3 and Table 1). Alanine substitution at Asn163, which abrogates synthesis of the CNA1 derived intramolecular amide, caused a moderate but significant increase in protease sensitivity. Taken together our data suggest that the intramolecular amide bonds of CNA2 and XNA display the largest contributions toward structural stability of BcpA pili. Furthermore, pili assembled from Asp312Ala variants appeared thinner (15 nm diameter) than those harboring the intramolecular amide bond of XNA or the wild-type residue Asp312, but lacking the Lys273-Asn383 bond (30 nm diameter) (Fig. 3 B and H). Of note, our pilus detection method requires antibody labeling and therefore cannot distinguish between the possibilities that the diameter of the pilus structure may be diminished in XNA variants or that the reactivity of pili with BcpA-specific antibodies has been affected by the Asp312Ala substitution.
Discussion
Pili, filamentous fibers often more than 1 μm in length, are comprised of protein subunits (pilins) and endowed with adhesive subunits at their tips (12). Pili enable adherence and tissue invasion of bacterial pathogens. Even a single proteolytic cut along the pilus shaft, which may occur during assembly or in the mature structure, would disable these adhesive factors and confer host resistance to infection. If so, most bacterial pathogens are confronted with a similar challenge—how to prevent cleavage of their pili by host proteases.
Assembly of protease resistant pili in the envelope of Gram-negative bacteria involves folding catalysts as well as the oxidizing environment of the periplasm (13, 14), a subcellular compartment between the cytoplasmic and outer membranes of these microbes (13, 14). Gram-negative pilin subunits, for example GafD, PapG, or FimH (15–17), are synthesized as precursors with jelly-roll domains (18). The location of disulfide bonds in these domains varies from structure to structure; however, they generally connect β-strands or loops to a β-strand in the same β-sheet. Stabilizing disulfide bonds are acquired via the dsb pathway in the periplasm (19). The oxidized products are subsequently assembled by an usher protein, the central catalyst of pilus formation, which translocates nascent pili across the outer membrane (20–22). Homologues of pilins, their folding chaperones, and outer membrane ushers are found in the genome sequences of many Gram-negative pathogens, implying that pilus assembly occurs via a conserved pathway (23).
The envelope of Gram-positive bacteria is comprised of thick cell wall sacculi; however, these structures lack the folding catalysts and oxidizing environment of the periplasm (1). Nevertheless, assembly of pili in Gram-positive bacteria also proceeds by a universal mechanism, involving major pilin subunits with YPKN pilin motifs and LPXTG sorting signals, as well as pilin-specific sortases (24). Previous work solved the crystal structures of 2 pilin proteins from Gram-positive bacteria—the major subunit, Spy0128, of T1 pili from Streptococcus pyogenes (9), and the minor subunit, GBS52, from Streptococcus agalactiae (25). Both Spy0128 and GBS52 possess 2 intramolecular amide (isopeptide) bonds, and both structures encompass tandem Ig-like folds, i.e., 2 CNA-B domains. Intramolecular amide (isopeptide) bonds exist in other cell-surface proteins structurally characterized from Gram-positive bacteria (9). Specifically, 2 domains, CNA-A and CNA-B, of the collagen-binding adhesin CNA from S. aureus harbor intramolecular amide bonds (26, 27).
The CNA2 and CNA3 domains in the current BcpA structure display the same reverse IgG fold of CNA-B and intramolecular amide bonds that reside on adjacent and parallel β-strands, belonging to the same β-sheet (27). Intra-sheet bonds covalently connect the first and last strands of the domain and are buried within the hydrophobic core of a β-sandwich. XNA is the first CNA-A-like domain characterized in pili of Gram-positive bacteria. XNA assumes the same jelly-roll domain fold as Gram-negative pilins, albeit that XNA generates an amide bond joining opposite faces of the β-sandwich domain. This unique feature suggests that pili derived from XNA domains may be more stable to proteases and resistant to thermal denaturing than pili assembled only from CNA-B domains. The residue essential for forming the intramolecular amide bond of XNA is Asp, in contrast to Glu from the CNA-B domains.
Most pilin subunits in Gram-positive bacteria appear to be built around 2 β-sheets with each sheet containing a number of β-strands (28). Further, most pilin subunits appear to be comprised of only 2 CNA-B domains (28). Lancefield and Dole reported Group A streptococcal T antigens to be resistant to trypsin treatment (29). T antigens are now appreciated as the major subunit of Group A streptococcal pili that also include minor pilin subunits (30). The recombinant precursor of the major pilin of T1 antigens harbors 2 CNA-B domains, each endowed with an intramolecular Asn-Lys amide bond (9) of what appear to be the structural equivalents of B. cereus BcpA CNA2 and CNA3 domains. In the T1 pilin precursor, these intramolecular bonds are required for resistance of the recombinant protein to protease (31). Nevertheless, the contribution of these bonds toward structural stability of T1 pili in the streptococcal envelope is not yet known.
Here we examined BcpA pili of B. cereus with molecular biology and structural analyses and derive a new model for the formation of pilus fibers (Fig. 4). Assembly of fully functional BcpA pili requires intramolecular amide bonds in each of 4 domains—CNA1, CNA2, XNA, and CNA3. Amide bonds in 3 BcpA domains, CNA2, XNA, and CNA3, are found in the recombinant pilin precursor as well as in assembled pili. We presume all 3 intramolecular amide linkages are formed before the BcpA precursor is cleaved by sortase D. Such a mechanism may not only confer protease resistance to the precursor, but it could also enable proper packing of BcpA intermediates en route to the final pilus structure. A fourth intramolecular amide, derived from the CNA1 domain, is synthesized only after the BcpA pilin precursor has been polymerized via the Lys162-Thr522 bond. In the absence of the CNA1 amide bond, pili remain partially sensitive to protease cleavage. As CNA1 includes the YPKN pilin motif, we presume the physiological role of its amide bond may be to protect the peptide sequence surrounding the sortase D-catalyzed intermolecular amide bond by stabilizing the reverse Ig-fold of this domain. Pili assembled without the XNA amide bond, but not pili lacking the intramolecular linkages of CNA1, CNA2, or CNA3, appear thinner than their wild-type counterparts. If so, a second important function of the XNA domain may be to ensure the proper packing of BcpA pilin subunits into the mature structure of the pilus fiber.
Fig. 4.
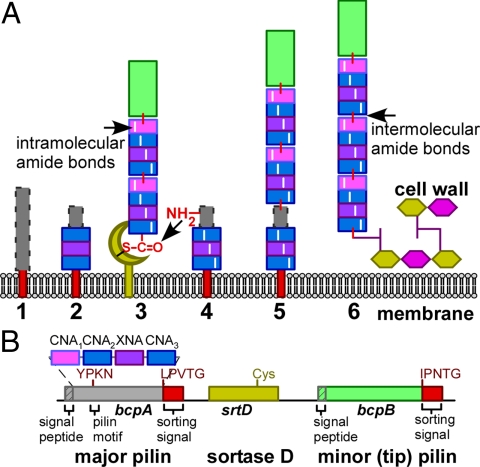
Model for pilus assembly in Bacillus cereus. (A) The unfolded BcpA precursor (1) is secreted across the plasma membrane and its N-terminal signal peptide removed. The CNA2, CNA3 and XNA domains assume reverse Ig or jelly-roll folds (2), form intramolecular amide bonds and enable presentation of the YPKN amino group (4) for nucleophilic attack at the sortase D-acyl intermediate with nascent pili, including the BcpB tip pilin (3). The product of this reaction initially links the incoming BcpA subunit to the LPXT motif of the growing pilus. At this stage, the CNA1 domain of the incoming BcpA subunit is still unfolded (gray box with dashed borders) (5). Following folding and intramolecular amide bond formation by the CNA1 domain, the mature, protease resistant pilus is immobilized in the cell wall envelope of bacilli (6). (B) Diagram displays the bcpA-srtD-bcpB gene cluster of B. cereus and the relevant features of the encoded products.
Materials and Methods
Bacterial Plasmids and Strains.
Coding sequences of the bcpA-srtD-bcpB operon were PCR amplified and cloned into pLM5, to generate IPTG-inducible expression via the Pspac promoter in B. anthracis strains. Oligonucleotides were used to create single amino acid substitutions in bcpA by site directed mutagenesis (SI Appendix, Table S12); pJB7 (BcpAHis-6 in pET24-b) was used as template to derive substitution mutations via Quick-Change mutagenesis.
BcpA for Crystallization.
Purified BcpAHis-6 was digested with trypsin and the resistant core analyzed by mass spectrometry and Edman degradation. E. coli BL21 (pJB215–6 expressing GST-BcpA*) was grown in 1.5 L of M9 minimal media to OD600 0.6, 100 ml of cell culture was discarded, and amino acids were added (lysine 0.1 mg/L, threonine 0.1 mg/L, phenylalanine 0.1 mg/L, leucine 0.05 mg/L, isoleucine 0.05 mg/L, valine 0.05 mg/L, and selenomethionine 0.06 mg/L). GST-BcpA was purified by GST-affinity chromatography, and the GST tag was removed by on-column cleavage with 55 U of thrombin (GE). BcpA was concentrated (Amicon Ultra–15 centrifugal filter) and purified by gel filtration chromatography (Superose 12 column). Selenomethionine-substituted BcpA crystallized at room temperature with a reservoir solution of 20% PEG 3350 and 0.2 M calcium acetate hydrate. Crystals were frozen in N2 (l) following cryoprotection with the reservoir solution containing 20% glycerol. Data were collected at the Life Sciences Collaborative Access Team (LS-CAT) beamline 21-ID-D at the Advanced Photon Source at Argonne National Laboratory (SI Appendix, Table S13). The structure was determined by the multiple-wavelength anomalous dispersion method with PHENIX (32).
Purification and Cleavage of Pili.
B. anthracis srtA::ermC harboring plasmids were grown for 16–24 h at 30 °C on LB agar supplemented with 1.5 mM IPTG and 20 μg/ml kanamycin. Bacteria and pili were scooped into water or 50 mM ammonium bicarbonate, cells removed by centrifugation, and aliquots of pili were digested with CNBr, mixed with loading buffer for SDS/PAGE and immunoblots or subjected to immunogold electron microscopy. CNBr-cleaved material was purified by Ni-NTA affinity chromatography (4). Eluate was subjected to SDS/PAGE and Coomassie Blue R-250 staining, transferred to PVDF membrane and stained with amido black for Edman degradation, or analyzed by LC-ESI-FTMS for molecular weight determination. RP-HPLC fractions from bacilli harboring pJB230 were digested with trypsin and analyzed by mass spectrometry.
Trypsin Cleavage of Pili.
Bacilli were suspended in 1.75 ml of 50 mM ammonium bicarbonate and OD600 normalized to 1.1. Cells were removed by centrifugation twice at 6,000 × g for 5 min, and 0.5 ml of pili were incubated with 2.68 μg of sequencing grade modified trypsin (Promega) overnight at 37 °C for 19 h. Aliquots were stained with α-BcpA antisera and goat anti-rabbit IgG 10 nm gold conjugate before electron microscopy.
Bacterial Strains, Pilus Purification, Structure Determinations.
Detailed methods are described in SI Appendix.
Supplementary Material
Acknowledgments.
We thank Dr. Xiaojing Yang and staff at Argonne National Laboratory-Advanced Photon Source for crystallography assistance. This work was supported by US Public Health Service Grants AI38897 (to O.S.) and AI074658 (to C.H.). J.M.B is a trainee of National Institutes of Health Medical Scientist Training Program Grant GM07281. C.B.P. is a National Science Foundation Graduate Research Fellow. O.S. and C.H. are members of and supported by the Region V “Great Lakes” Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium (NIAID, NIH Award 1-U54-AI-057153). Data collection was performed at the Life Sciences Collaborative Access Team (LS-CAT) beamline 21-ID-D and the Structural Biology Center beamline 19-BM at the Advanced Photon Source at Argonne National Laboratory, which are supported by the National Institutes of Health and the United States Department of Energy.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910887106/DCSupplemental.
References
- 1.Marraffini LA, DeDent AC, Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol Mol Biol Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarvas M, Harwood CR, Bron S, van Dijl JM. Post-translational folding of secretory proteins in Gram-positive bacteria. Biochim Biophys Acta. 2004;1694:311–327. doi: 10.1016/j.bbamcr.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 3.Ton-That H, Liu G, Mazmanian SK, Faull KF, Schneewind O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc Natl Acad Sci USA. 1999;96:12424–12429. doi: 10.1073/pnas.96.22.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budzik JM, et al. Amide bonds assemble pili on the surface of bacilli. Proc Natl Acad Sci USA. 2008;105:10215–10220. doi: 10.1073/pnas.0803565105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Budzik JM, Oh SY, Schneewind O. Cell wall anchor structure of BcpA pili in Bacillus anthracis. J Biol Chem. 2008;283:36676–36686. doi: 10.1074/jbc.M806796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budzik JM, Oh SY, Schneewind O. Sortase D forms the covalent bond that links BcpB to the tip of Bacillus cereus pili. J Biol Chem. 2009;284:12989–12997. doi: 10.1074/jbc.M900927200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dramsi S, et al. Assembly and role of pili in group B streptococci. Mol Microbiol. 2006;60:1401–1413. doi: 10.1111/j.1365-2958.2006.05190.x. [DOI] [PubMed] [Google Scholar]
- 8.Nelson AL, et al. RrgA is a pilus-associated adhesin in Streptococcus pneumoniae. Mol Microbiol. 2007;66:329–340. doi: 10.1111/j.1365-2958.2007.05908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- 10.Gaspar AH, et al. Bacillus anthracis sortase A (SrtA) anchors LPXTG motif-containing surface proteins to the cell wall envelope. J Bacteriol. 2005;187:4646–4655. doi: 10.1128/JB.187.13.4646-4655.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sauer FG, Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. Bacterial pili: Molecular mechnaisms of pathogenesis. Curr Opin Microbiol. 2000;3:65–72. doi: 10.1016/s1369-5274(99)00053-3. [DOI] [PubMed] [Google Scholar]
- 12.Sauer FG, Remaut H, Hultgren SJ, Waksman G. Fiber assembly by the chaperone-usher pathway. Biochim Biophys Acta. 2004;1694:259–267. doi: 10.1016/j.bbamcr.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 13.Kuehn MJ, Normark S, Hultgren SJ. Immunoglobulin-like PapD chaperone caps and uncaps interactive surfaces of nascently translocated pilus subunits. Proc Natl Acad Sci USA. 1991;88:10586–10590. doi: 10.1073/pnas.88.23.10586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacob-Dubuisson F, et al. PapD chaperone function in pilus biogenesis depends on oxidant and chaperone-like activities of DsbA. Proc Natl Acad Sci USA. 1994;91:11552–11556. doi: 10.1073/pnas.91.24.11552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merckel MC, et al. The structural basis of receptor-binding by Escherichia coli associated with diarrhea and septicemia. J Mol Biol. 2003;331:897–905. doi: 10.1016/s0022-2836(03)00841-6. [DOI] [PubMed] [Google Scholar]
- 16.Dodson KW, et al. Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor. Cell. 2001;105:733–743. doi: 10.1016/s0092-8674(01)00388-9. [DOI] [PubMed] [Google Scholar]
- 17.Choudhury D, et al. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichica coli. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 18.Remaut H, Waksman G. Structural biology of bacterial pathogenesis. Curr Opin Struct Biol. 2004;14:161–170. doi: 10.1016/j.sbi.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Bardwell JC, McGovern K, Beckwith J. Identification of a protein required for disulfide bond formation in vivo. Cell. 1991;67:581–589. doi: 10.1016/0092-8674(91)90532-4. [DOI] [PubMed] [Google Scholar]
- 20.Remaut H, et al. Fiber formation across the bacterial outer membrane by the chaperone/usher pathway. Cell. 2008;133:640–652. doi: 10.1016/j.cell.2008.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishiyama M, Ishikawa T, Rechsteiner H, Glockshuber R. Reconstitution of pilus assembly reveals a bacterial outer membrane catalyst. Science. 2008;320:376–379. doi: 10.1126/science.1154994. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y, Smith BS, Chen LX, Baxter RH, Deisenhofer J. Insights into pilus assembly and secretion from the structure and functional characterization of usher PapC. Proc Natl Acad Sci USA. 2009;106:7403–7407. doi: 10.1073/pnas.0902789106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sauer FG, et al. Chaperone-assisted pilus assembly and bacterial attachment. Curr Opin Struct Biol. 2000;10:548–556. doi: 10.1016/s0959-440x(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 24.Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 25.Krishnan V, et al. An IgG-like domain in the minor pilin GBS52 of Streptococcus agalactiae mediates lung epithelial cell adhesion. Structure. 2007;15:893–903. doi: 10.1016/j.str.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Symersky J, et al. Structure of the collagen-binding domain from a Staphylococcus aureus adhesin. Nat Struct Biol. 1997;4:833–838. doi: 10.1038/nsb1097-833. [DOI] [PubMed] [Google Scholar]
- 27.Deivanayagam CC, et al. Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure Fold Des. 2000;8:67–78. doi: 10.1016/s0969-2126(00)00081-2. [DOI] [PubMed] [Google Scholar]
- 28.Kang HJ, Middleditch M, Proft T, Baker EN. Isopeptide bonds in bacterial pili and their characterization by X-ray crystallography and mass spectrometry. Biopolymers. 2009;91:1126–1134. doi: 10.1002/bip.21170. [DOI] [PubMed] [Google Scholar]
- 29.Lancefield RC, Dole VP. The properties of T antigens extracted from group A hemolytic streptococci. J Exp Med. 1946;84:449–471. doi: 10.1084/jem.84.5.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mora M, et al. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc Natl Acad Sci USA. 2005;102:15641–15646. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kang HJ, Baker EN. Intramolecular isopeptide bonds give thermodynamic and proteolytic stability to the major pilin protein of Streptococcus pyogenes. J Biol Chem. 2009;284:20729–20737. doi: 10.1074/jbc.M109.014514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.