Abstract
Modification of eukaryotic proteins is a powerful strategy used by pathogenic bacteria to modulate host cells during infection. Previously, we demonstrated that Helicobacter pylori modify an unidentified protein within mammalian cell lysates in a manner consistent with the action of a bacterial ADP-ribosylating toxin. Here, we identified the modified eukaryotic factor as the abundant nuclear factor poly(ADP-ribose) polymerase-1 (PARP-1), which is important in the pathologies of several disease states typically associated with chronic H. pylori infection. However, rather than being ADP-ribosylated by an H. pylori toxin, the intrinsic poly(ADP-ribosyl) polymerase activity of PARP-1 is activated by a heat- and protease-sensitive H. pylori factor, resulting in automodification of PARP-1 with polymers of poly(ADP-ribose) (PAR). Moreover, during infection of gastric epithelial cells, H. pylori induce intracellular PAR-production by a PARP-1-dependent mechanism. Activation of PARP-1 by a pathogenic bacterium represents a previously unrecognized strategy for modulating host cell signaling during infection.
Keywords: apoptosis, infection, PARP-1, toxin
Chronic infection with Helicobacter pylori is a significant risk for the development of peptic ulcer disease and gastric cancer in humans (1–3). During infection, H. pylori generate several protein factors that modulate host cells and tissues in a manner that contributes to pathogen colonization and persistence, as well as the pathophysiological changes associated with gastric disease (4). Previous work indicated that a soluble factor within mammalian cell lysates was modified in the presence of NAD and H. pylori culture filtrate (HPCF) (5). Initial characterization suggested that the eukaryotic factor was ADP-ribosylated (5). Notably, the ADP-ribosylation of eukaryotic targets has been identified as the action of multiple protein toxins and effectors important for bacterial virulence (6). Because the modified eukaryotic factor demonstrated an apparent molecular weight greater than any of the currently identified ADP-ribose acceptors, we hypothesized that this factor is a member of the growing list of eukaryotic proteins targeted for ADP-ribosylation by pathogens (5).
The objective of this study was to identify the mammalian protein previously demonstrated to be modified in an H. pylori-dependent manner (5). Here, we demonstrate that the modified protein is the nuclear factor poly(ADP-ribose) polymerase-1 (PARP-1), which is involved in the pathogenesis of several cancers and inflammatory disorders (7, 8). Unexpectedly, rather than being modified by a bacterial ADP-ribosylating toxin, the intrinsic catalytic activity of PARP-1 is activated by a heat- and protease-sensitive factor(s) within the HPCF, resulting in automodification with polymers of poly(ADP-ribose) (PAR). Moreover, we demonstrated that PARP-1-dependent PAR production occurs in gastric epithelial cells infected with H. pylori. H. pylori activation of PARP-1 may represent a strategy for pathogen modulation of host signaling.
Results
Identification of Modified Proteins.
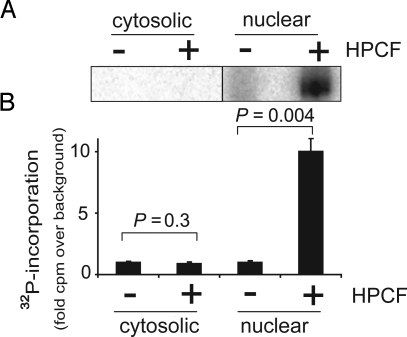
To identify the eukaryotic protein previously reported to be modified in an HPCF-dependent manner (5), lysate prepared from HeLa cells was incubated with [32P-adenylate] NAD in the absence or presence of culture filtrate prepared from H. pylori 26695 (HPCF). Two-dimensional gel electrophoresis of the samples (SI Materials and Methods) revealed a dominant 32P-radiolabeled species of approximately 135 kDa only in the presence of HPCF (Fig. S1). Three proteins were identified by LC-MS analysis of the extracted radiolabeled band, ATP: citrate lyase, the kinesin heavy chain, and poly(ADP-ribose) polymerase-1 (PARP-1) (Table S1). Further analysis revealed that 32P-radiolabel was incorporated only into the nuclear fraction of HeLa lysates, but not into the non-nuclear fraction (Fig. 1 A and B). Because only PARP-1 (9), but neither ATP:citrate lyase (10) nor the kinesin heavy chain (11), is considered to be a nuclear protein, these results suggested that PARP-1 may be the eukaryotic factor previously shown to modified in an HPCF-dependent manner (5).
Fig. 1.
HPCF-dependent 32P-radiolabeling of a nuclear protein. The cytosolic or nuclear fraction (5 mg/mL) of HeLa cells were incubated with [32P-adenylate] NAD (50 mM) for 15 min at 25 °C in the absence (-) or presence (+) of HPCF (100 μg/mL). (A) The samples were resolved by SDS/PAGE, and radiolabeled bands were visualized by phosphorimaging of the dried gels. (B) 32P-radiolabel incorporated into TCA precipitable material from cytosolic or nuclear fractions was quantified by scintillation counting. The data are presented as the fold-32P-radiolabel incorporated over background (cell lysate fractions in the absence of HPCF Statistical significance was calculated for differences in 32P-incorporation between samples containing HPCF and controls lacking HPCF.
PARP-1 Is Required for HPCF-Dependent 32P-Radiolabeling.
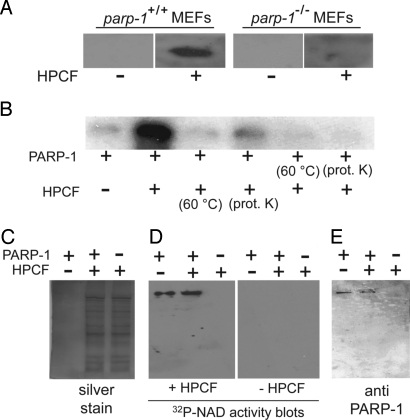
To validate that PARP-1 is required for HPCF-dependent modification of cellular lysates, an activity blot assay was used to compare the incorporation of 32P-radiolabel into lysates prepared from immortalized wild-type mouse embryonic fibroblasts (MEFs) to lysates from MEFs lacking PARP-1 (parp-1−/− MEFs) (12). The proteins within wild-type or parp-1−/− MEF lysates were resolved by SDS/PAGE, transferred to nitrocellulose blots, and then incubated with 32P-NAD in the absence or presence of HPCF. In the presence of HPCF, a single 32P-radiolabeled band was visible on blots prepared from wild-type but not parp-1−/− MEF lysates (Fig. 2A). This result indicated that PARP-1 is required for the HPCF-dependent incorporation of 32P-radiolabel into mammalian cell lysates and, moreover, strongly suggested that PARP-1 is the protein that is modified.
Fig. 2.
PARP-1 is an HPCF-dependent 32P-radiolabel acceptor. (A) Activity blots of wild-type or parp-1−/− MEF lysates incubated with 32P-NAD in the absence or presence of HPCF. (B) Purified, recombinant PARP-1 (50 pM) that had been pretreated by heating (60 °C for 15 min), incubation with proteinase K (100 μg/mL for 1 h, followed by PMSF addition), or, incubation on ice (1 h; indicated by “-”), and [32P-adenylate] NAD (50 μM), was incubated at 25 °C in the absence or presence of HPCF (50 μg/mL) that had been pretreated by heating (60 °C for 15 min), incubation with proteinase K (100 μg/mL for 1 h, followed by PMSF addition), or, incubation on ice (1 h; indicated by “-”). After 15 min, the samples were evaluated for 32P-incorporation by resolving the samples using SDS/PAGE, and then visualized after autoradiography of the dried gels. (C–E) HPCF alone (50 μg/mL) (as indicated by those lanes labeled as (-) PARP-1/(+) HPCF), purified PARP-1 (50 pM) (as indicated by those lanes labeled as (+) PARP-1/(-) HPCF), or mixtures of HPCF (50 μg/mL) and purified PARP-1 (50 pM) (as indicated by those lanes labeled as (+) PARP-1/(+) HPCF) were resolved by SDS/PAGE. Single gels were divided, and evaluated for the profile of total protein by silver staining (C), 32P-radiolabel incorporation by activity blotting in the absence or presence of HPCF (D), or, the presence of PARP-1 by Western blotting (E). Note that in (C), silver stained PARP-1 was not visible at the 50 pM concentration used.
PARP-1 Is Sufficient for HPCF-Dependent 32P-Radiolabeling.
To directly test whether PARP-1 functions as an HPCF-dependent 32P-radiolabel acceptor in the absence of additional eukaryotic factors, 32P-radiolabel incorporation was evaluated using a recombinant form of PARP-1 (13) expressed in E. coli, and purified to homogeneity. Visibly more 32P-radiolabel was associated with single protein band (at the molecular weight of PARP-1) than in the absence of HPCF (Fig. 2B). Notably, 32P-radiolabel incorporation was nearly decreased to background levels when HPCF was pretreated either by preheating at 60 °C, or by incubating with proteinase K, which strongly supports the notion that the factor(s) within HPCF required for 32P-radiolabeling is a protein. 32P-radiolabeling was visibly reduced when PARP-1 was pretreated either by preheating at 60 °C, or incubation with proteinase K. These experiments indicated that in the absence of additional eukaryotic factors, PARP-1 alone is sufficient for HPCF-dependent 32P-radiolabel incorporation.
To further define the requirements and specificity of HPCF-dependent PARP-1 modification, SDS/PAGE was used to resolve mixtures of HPCF and purified PARP-1, as well as HPCF alone or PARP-1 alone. Gels were divided, and evaluated for total protein by silver staining (Fig. 2C), 32P-radiolabel incorporation by the activity blot assay (Fig. 2D), and the presence of PARP-1 by Western blotting (Fig. 2E). These studies revealed, despite the presence of many visible silver-stained proteins in lanes containing HPCF (Fig. 2C), the presence of only a single, dominant 32P-radiolabeled band on activity blots, but only on those activity blots incubated with HPCF, and only in those lanes containing purified recombinant PARP-1 (Fig. 2D). Moreover, the 32P-radiolabeled band exhibited similar electrophoretic migration as PARP-1 cross-reacting material (Fig. 2E), but only in lanes of the activity blots containing PARP-1, and only when incubated with HPCF. These results further confirmed PARP-1 as an acceptor of HPCF-dependent 32P-radiolabel transfer.
A Functional PARP-1 Catalytic Domain Is Required for HPCF-Dependent 32P-Radiolabeling.
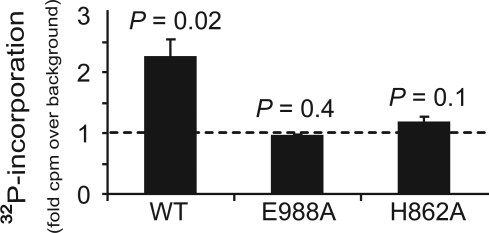
Although PARP-1 is required for HPCF-dependent incorporation of 32P-radiolabel, it was not clear which intrinsic properties of full-length PARP-1 are important for HPCF-dependent modification. Notably, in the presence of NAD and an activating co-factor, full-length PARP-1 is automodified by an intermolecular reaction (e.g., in trans) with polymers of ADP-ribose due to the poly(ADP-ribosyl) polymerase activity of the catalytic domain (CD) (14, 15). To evaluate the potential importance of a functional CD for HPCF-dependent modification of PARP-1, two catalytically attenuated mutants, PARP-1 (E988A) or PARP-1 (H862A) (16), each of which contains a point mutation in the CD active site, were incubated with [32P-adenylate] NAD in presence of HPCF. These experiments revealed that neither PARP-1 (E988A) or PARP-1 (H862A) was significantly radiolabeled in the presence of HPCF (Fig. 3). These results indicated that a functional catalytic domain of PARP-1 is required for HPCF-dependent 32P-radiolabel incorporation.
Fig. 3.
PARP-1 catalytic activity is required for PARP-1 to be modified with 32P-radiolabel in the presence of HPCF. PARP-1, PARP-1 (H862A), or PARP-1 (E988A) (each at 50 pM) and [32P-adenylate] NAD (50 μM) were incubated 25 °C in the absence or presence of HPCF (50 μg/ml). After 15 min, the samples were scored for 32P-incorporation by scintillation counting of TCA-precipitated material. The data are presented as fold-increase in 32P-radiolabel incorporation versus a control conducted with wild-type PARP-1 and [32P-adenylate] NAD in the absence of HPCF. Statistical significance was calculated for differences in 32P-incorporation between samples containing HPCF and the control lacking HPCF.
Poly(ADP-Ribosyl) Polymerase Activity Is Important for HPCF-Dependent 32P-Radiolabeling of PARP-1.
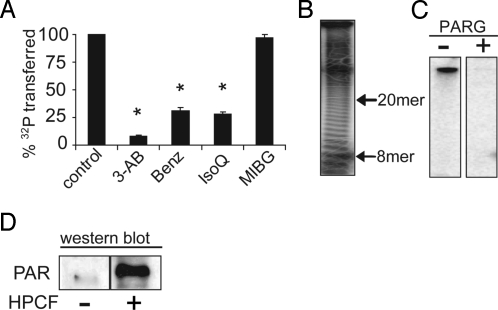
The requirement for a functional PARP-1 catalytic domain suggested that HPCF-dependent 32P-radiolabeling of PARP-1 may be a result of PARP-1 poly(ADP-ribosylation) activity, rather than modification by a bacterial ADP-ribosylating toxin. To explore the relationship between poly(ADP-ribosylation) activity and HPCF-dependent 32P-radiolabeling of PARP-1, we tested the effects of several known poly(ADP-ribosylation) inhibitors on the HPCF-mediated transfer of 32P-radiolabel to PARP-1. These experiments revealed that poly(ADP-ribosylation) inhibitors (Benz, 3-AB, IsoQ) significantly reduced 32P-radiolabel incorporation into PARP-1 (Fig. 4A). In contrast, a mono(ADP-ribosylation) inhibitor [MIBG, (17)] did not reduce 32P-radiolabel incorporation into PARP-1. These data support the importance of poly(ADP-ribosyl) polymerase activity for PARP-1 modification.
Fig. 4.
PARP-1 is activated in an HPCF-dependent manner to catalyze self-modification with PAR. PARP-1 (50 pM) along with either [32P-adenylate] NAD (50 μM) (A–C) or non-radiolabeled NAD (D) were incubated at 25 °C in the absence or presence of HPCF (50 μg/mL). (A) The reactions were carried out in the absence (control) or presence of 3-aminobenzamide (3-AB, 3 mM), benzamide (Benz, 3 μM), isoquinolinone (IsoQ, 1 μM), or m-iodobenzylguanidine (MIBG, 100 μM). * indicates that differences in % 32P transferred relative to the control lacking inhibitor are statistically significant (P < 0.05). (B) The completed reactions were incubated at 60 °C in the absence or presence of NaOH (100 mM). After 60 min, released PAR was resolved by high percentage TBE acrylamide gels (19% acrylamide, 1% bis-acrylamide), and visualized after autoradiography. The 8-mer and 20-mer PAR chain, which co-migrate with the bromophenol blue and xylene cyanol loading dyes respectively (35, 36) are indicated next to the autoradiographs. (C) The reactions were further incubated at 25 °C in the absence or presence of PARG (100 ng/mL). After 15 min, the samples were fractionated by SDS/PAGE, and scored for 32P-incorporation by SDS/PAGE followed by autoradiography of the dried gels. (D) After 15 min, the samples were probed for PAR by Western blot analysis using anti-PAR antibodies.
PARP-1 Is Automodified with PAR in an HPCF-Dependent Manner.
PARP-1 catalyzed poly(ADP-ribosylation) typically results in the synthesis of PAR polymers ranging in length from several to approximately 200 individual ADP-ribose units (18). To explore whether HPCF-dependent modification of PARP-1 is associated with the generation of PAR polymers, we exposed 32P-radiolabeled PARP-1 to alkaline pH, which releases intact PAR polymers from PARP-1. Incubation of modified PARP-1 with NaOH resulted in the loss of 32P-radiolabel from PARP-1, indicating that the PARP-1 modification is sensitive to alkaline pH. The NaOH-treated reactions were resolved by high-resolution polyacrylamide electrophoresis, and visualized by autoradiography, revealing that alkaline treatment of 32P-radiolabeled PARP-1 yielded a laddered pattern of bands (Fig. 4B), characteristic of PAR modification (19). These data suggested that PARP-1 modification is associated with the production of PAR chains of variable lengths.
To validate that PARP-1 is modified with PAR, we tested whether 32P-radiolabeled PARP-1 is sensitive to poly(ADP-ribose)glycohydrolase (PARG), which specifically hydrolyzes the ribose-ribose linkages of PAR, but not the ribose-amino acid acceptor linkages of mono-ADP-ribose modification. These experiments revealed that PARG treatment with PARG resulted in almost a complete loss of detectable 32P-radiolabel (Fig. 4C). These data indicate that HPCF-dependent 32P-incorporation into PARP-1 is sensitive to PARG, and support the idea that PARP-1 is modified by PAR. PAR specific antibodies were used to confirm that purified recombinant PARP-1 is modified with PAR in an HPCF-dependent manner. Western blot analysis of modified PARP-1 revealed PAR cross-reacting material (Fig. 4D). Our results support a model that HPCF promotes PARP-1 automodification with PAR by activating the intrinsic poly(ADP-ribosylation) activity of PARP-1.
H. pylori Infection of Gastric Epithelial Cells Results in the 3-AB-Sensitive Incorporation of 32P-Radiolabel into TCA-Precipitable Material.
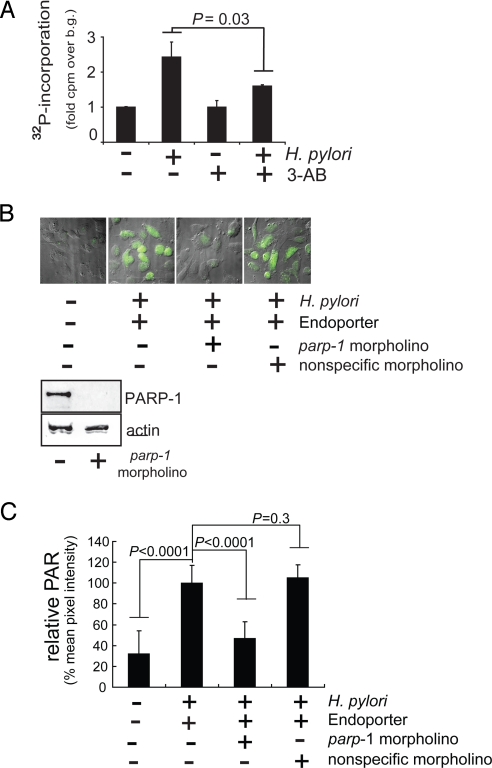
Although PARP-1 is activated in an HPCF-dependent manner within a cell-free system, it was not clear whether PARP-1 activation occurs as a consequence of H. pylori infection of gastric epithelial cells. To evaluate this possibility, human-derived adenocarcinoma gastric (AGS) cells were infected with H. pylori 26695 in the absence or presence of 3-AB (5 mM). After 8 h, the cells were washed, permeabilized, and incubated with [32P-adenylate] NAD. The cells were lysed, and the amount of 32P-radiolabel incorporated into TCA-precipitable material was quantified. These experiments revealed a significantly higher level of 32P-radiolabel incorporated into the TCA-precipitated fraction in H. pylori-infected cells than in non-infected cells (Fig. 5A). Moreover, 32P-radiolabel incorporation was significantly decreased when the infections were conducted in the presence of the PARP-1 inhibitor, 3-AB (Fig. 5A). These results indicated that H. pylori infection of gastric epithelial cells resulted in the incorporation of 32P-radiolabel into TCA-precipitable material in a manner that is sensitive to the presence of the PARP-1 inhibitor, 3-AB.
Fig. 5.
H. pylori infection induces PARP-1 activation within gastric epithelial cells. AGS cells preincubated in the absence or presence of 3-AB (5 mM) (A), or, in the absence or presence of Endoporter transfection reagent with or without parp-1 or non-specific morpholinos (B and C), were further incubated in the absence or presence of H. pylori 26695 (MOI 100). (A) The cells were analyzed for the incorporation of 32P-radiolabel. Statistical significance was calculated for the differences in 32P-radiolabel incorporation between H. pylori infected cells with or without 3-AB. (B) The cells were analyzed for the production of intracellular PAR by immunostaining using mouse anti-PAR. Each panel in is an overlay of images collected using DIC/fluorescence microscopy. Immediately below the microscopy images are Western blots demonstrating the relative PARP-1 (top panels) and actin (lower panels) cross-reacting material from lysates of cells pretreated in the absence or presence of parp-1 morpholinos. (C) Quantitative analysis of the fluorescent intensities associated with PAR staining was carried out using ImageJ 1.42i software (Wayne Rasband, National Institutes of Health). For each cell analyzed, the mean pixel intensity of Alexa Fluor 488 fluorescence within the nuclear region (as indicated by DAPI staining), minus the background mean pixel intensity of arbitrary areas in each field void of cell, was determined. Data were collected and averaged from at least 10 cells in each of three independent experiments (>30 cells total), and normalized against cells that had been pretreated with Endoporter transfection agent alone before H. pylori infection. Statistical significance was calculated for the differences in relative mean pixel intensity between untreated cells or H. pylori-infected cells pretreated with Endoporter in the presence of parp-1 or non-specific morpholinos and those H. pylori-infected cells pretreated with Endoporter alone.
PAR Is Synthesized with H. pylori-Infected Gastric Epithelial Cells.
Although 32P-radiolabel is incorporated into the TCA-precipitated fraction in H. pylori-infected cells, it was not clear whether or not intracellular PAR had been synthesized. To evaluate whether H. pylori infection results in the synthesis of intracellular PAR, AGS cells that had been infected with H. pylori 26695 (MOI 100) were probed for PAR by indirect immunofluorescence microscopy, using anti-PAR antibodies. These experiments revealed visibly higher levels of PAR within the nuclei of infected AGS cells than in uninfected cells (Fig. 5B). These results indicate that PAR is produced in mammalian cells in response to H. pylori infection.
PARP-1 Is Required for PAR Synthesis Within H. pylori-Infected Gastric Epithelial Cells.
Although PAR is synthesized within H. pylori infected cells, it was not clear whether or not PARP-1 was required for generation of this polymer. To evaluate the requirement for PARP-1, AGS cells, in which cellular PARP-1 had been knocked down using a parp-1-specific morpholino (Fig. 5B), were infected with H. pylori 26695 (MOI 100). Infected cells were probed for PAR by indirect immunofluorescence microscopy, using antibodies specific for PAR. These experiments revealed visibly lower levels of PAR within infected PARP-1 knockdown cells than in either untreated cells or cells pretreated with a non-specific morpholino (Fig. 5 B and C). The decrease in intracellular PAR is unlikely to be due to alterations in bacterial interactions with AGS cells, as approximately the same number of intracellular H. pylori were recovered (SI Materials and Methods) from cells with parp-1 knocked down as untreated cells or those treated with non-specific morpholinos (Fig. S2). These data indicate that PARP-1 is required for PAR synthesis within H. pylori-infected AGS cells.
Discussion
During infection of the gastric mucosa, H. pylori modulate the properties of host cells and tissues to generate a more suitable niche for colonization and persistence. In this study, we demonstrated H. pylori-dependent modification of the abundant and important nuclear protein PARP-1. However, contrary to our original hypothesis, PARP-1 is not mono-ADP-ribosylated by the action of an H. pylori toxin or effector. Rather, PARP-1 undergoes automodification with PAR as a result of H. pylori-mediated activation of the intrinsic poly(ADP-ribosylation) activity of PARP-1. PARP-1 activation by a bacterial factor in a cell-free system has not been previously reported.
PARP-1 was originally characterized as an abundant nuclear factor that facilitates DNA base excision repair. However, recent work has expanded the physiologic functions of PARP-1, and it is now clear that this important protein is involved in many disease pathologies, including cancer (7, 8). Interestingly, PARP-1 synthesized PAR was recently identified as an important trigger of apoptosis, which is a hallmark of H. pylori infection (1, 20). PARP-1 generated PAR exits the nucleus and induces the release of mitochondria-associated apoptosis inducing factor (AIF), which, in turn, translocates into the nucleus and promotes caspase-independent apoptosis (21, 22). Notably, H. pylori infection was recently demonstrated to induce the release of AIF from mitochondria (23, 24). While beginning to explore the consequences of H. pylori PARP-1 activation, we validated that H. pylori 26695 infection of AGS cells with H. pylori 26695 resulted in the release of AIF from mitochondria (Fig. S3). However, H. pylori-mediated AIF release was significantly inhibited in the presence of the PARP-1 inhibitor, 3-AB (Fig. S3). These results suggest the possibility, which is currently being investigated, that one consequence of H. pylori-mediated PARP-1 activation may be the induction of apoptosis via the PAR-mediated release of AIF from mitochondria. Notably, PARP-1 activation occurs considerably before PARP-1 cleavage, a commonly used marker for late stage apoptosis, reported at 24 h in H. pylori-infected cells (24).
Several non-bacterial proteins have now been identified as PARP-1 activators. In a cell-free system, mammalian phosphorylated extracellular signal-regulated kinase-2 (ERK2) (25), and the chromatin insulating CCCTC-binding factor (CTCF) (26) have been recently demonstrated to bind and activate PARP-1. Simian virus 40 (SV40) capsid protein VP3 (27) and the polyomavirus capsid protein viral protein 1 (VP1) (28), have all been demonstrated to activate PAR synthesis by interacting directly with PARP-1, suggesting that PARP-1 activation may be a consequence of the infectious cycle of these viruses. Analogous to known protein activators of PARP-1, we hypothesize that HPCF-dependent PAR synthesis may involve direct interactions of a HPCF protein factor(s) with PARP-1. In support of the involvement of a protein factor(s), we demonstrated that HPCF PARP-1 activating factor(s) is sensitive to either heat (60 °C) or protease treatment (Fig. 2B). Consistent with the notion that HPCF must access intracellular PARP-1 for activation to occur, incubation of AGS cells with HPCF premixed with the Chariot transfection reagent, which promotes protein entry into cells, resulted in significantly more PAR production than cells incubated with HPCF alone (Fig. S4). Although H. pylori are taken up into gastric epithelial cells in vitro (Fig. S2), it is not clear at this time whether the PARP-1 activating factor accesses PARP-1 within the nuclei of intact cells following release from intracellular bacteria, or by an alternate mechanism. Finally, when incubated in the presence of 100-fold molar excess of each of the recombinant forms of the three isolated PARP-1 functional domains [the DNA binding domain (DBD), the automodification domain (AD), or the catalytic domain (CD)], only the DBD inhibited HPCF-dependent PARP-1 activation (Fig. S5), suggesting that the DBD may directly interact with the HPCF factor.
We are currently working to identify the factor(s) within HPCF responsible for activating PARP-1. In preliminary studies to evaluate the possible role of the known H. pylori virulence factors, CagA and VacA, in PARP-1 activation, we established that HPCF prepared from H. pylori G27 lacking the cag pathogenicity island (G27 ΔPAI) activated PARP-1 to the same extent as wild-type H. pylori G27, while purified vacuolating toxin (VacA) (29) did not induce detectable PARP-1 activation. Bioinformatic analyses of the genomes of H. pylori 26695 and J99 have not revealed genes encoding proteins with amino acid sequence homologies to known PARP-1-activating proteins. Moreover, our attempts to purify the PARP-1 activating factor(s) from HPCF using standard biochemical approaches have not yet been successful; we speculate that the H. pylori PARP-1 activating factor is present in low abundance under the conditions that we cultivate the bacterium in liquid medium.
In summary, our data demonstrate PARP-1 activation by a pathogenic bacterium, which has not been previously reported. Because PARP-1 is emerging as an important factor in the development of several pathologies that are typically associated with chronic H. pylori infection (7, 8), future work will focus on the importance and potential roles that direct PARP-1 activation by H. pylori may have in the development of gastric disease.
Materials and Methods
Bacterial Culture Filtrates and Mammalian Lysates.
Mammalian lysates were prepared from HeLa cells (ATCC no. CCL-2), as described previously (5). The nuclear fraction of HeLa cells was prepared using the Nuclear Extract Kit (Active Motif).
In Vitro Modification Experiments.
Culture filtrates were prepared as described (5) from H. pylori TIGR strain 26695 (ATCC no. 700392; ATCC, J99 (ATCC no. 700824) and 60190 (ATCC no. 49503), G27, or G27 lacking the cag pathogenicity island (30, 31) (ΔPAI; gift from K. Guilleman). In vitro modification of mammalian lysates was carried out as described previously (5).
To score 32P-incorporation into PARP-1, purified recombinant PARP-1 (50 pM) was incubated in reaction buffer (50 mM Tris, pH 7.6, 10 mM NaCl, 10 mM MgCl2, and 1 mM DTT) with 32P NAD (50 μM) (1,000 Ci/mmol; GE Healthcare BioSciences) at 25 °C in the absence or presence of H. pylori culture filtrate (50 μg/mL). 32P-radiolabel incorporation was quantified by scintillation counting of TCA precipitable material, as previously described (5). Alternatively, the reactions were resolved by SDS/PAGE, and visualized after autoradiography of the dried gels. In preliminary studies, we established that culture filtrates prepared from H. pylori strains 26695, 60190, J99, or G27 all induced detectable 32P-radiolabel incorporation into PARP-1, whereas culture filtrates prepared from several unrelated species, E. coli XL1-Blue (Invitrogen, Inc.), Campylobacter jejuni 81–176 (gift from P. Guerry), and Serratia marcescens MB1911 (32), were unable to induce detectable 32P-radiolabel incorporation into PARP-1. In some studies, the following PARP inhibitors were included: benzamide (Benz), 3-aminobenzamide (3-AB), isoquinolinone (IsoQ), or [N-(6-Oxo-5,6-dihydrophenanthridin-2-yl)-N,N-dimethylacetamide.HCl] (PJ34) (all from Sigma). The mono(ADP-ribosylation) inhibitor m-Iodobenzylguanidine (MIBG; Sigma), was also included in some experiments. Alkaline hydrolysis to release polymers of PAR from modified PARP-1, and visualization of PAR polymers were conducted as described previously (19). Modified PARP-1 was incubated at 25 °C in the absence or presence of pure, recombinant PARG (100 ng/mL; Trevigen Inc.). After 15 min, the reactions were stopped by the addition of SDS (SDS)-polyacrylamide gel electrophoresis (PAGE) sample buffer and boiling.
Expression and Purification of Recombinant Forms of PARP-1.
Wild-type and mutant forms of PARP-1 were expressed as recombinant proteins with amino-terminal hexa-histidine fusion peptides in E. coli SG13009 transformed with plasmid pSD6.3, and purified as described previously (13). Full-length PARP-1 comprised residues 18–1,014, and contained the D214A mutation, which was previously demonstrated to express at higher levels while retaining activity (13). Using Ni-CAM affinity resin (Sigma), each recombinant protein was purified from the soluble fraction of bacterial lysate to approximately 99% homogeneity, as estimated by Coomassie Brilliant Blue stained SDS-PA gels.
Mutant forms of PARP-1 were generated using the Quick-change site directed mutagenesis kit (Stratagene). Oligonucleotide synthesis (Table S2) and DNA sequencing was performed at the University of Illinois Carver Biotechnology Center (www.biotech.uiuc.edu/). For studies using smaller, mutant forms of PARP-1, the DBD comprised PARP-1 residues 18–372, the AD comprised residues 372–536, and the CD comprised residues 524–1,014.
Activity Blots.
PARP-1 activity blots were performed as described previously (19). Wild-type or parp-1−/− MEF lysates, or purified recombinant PARP-1, were resolved by SDS/PAGE, and electrotransferred to nitrocellose. Premixing of HPCF with PARP-1 before SDS/PAGE, as indicated by the middle lanes of each blot in Fig. 2D (labeled as (+) PARP-1/(+) HPCF), was neither sufficient for (in the blot labeled -HPCF) nor interfered with (in the blot labeled +HPCF) 32P-radiolabeling of PARP-1. The transferred proteins were incubated at 25 °C in renaturation buffer (50 mM Tris, pH 8, 100 mM NaCl, 1 mM DTT, 0.3% Tween-20, 20 μM Zn(II) acetate, and 2 mM MgCl2) with 32P-NAD (50 μM) in the absence or presence of HPCF (5 mL of 250 μg/mL). After 15 min, the blots were washed. When 32P-NAD was used, activity blots were visualized following autoradiography. For detection of PAR production, the blots were probed with PAR monoclonal antibodies (clone 10HA; Trevigen). PARP-1 or actin within lysates was visualized by Western blot analysis, where the blots were probed, respectively, with PARP-1 monoclonal antibody (Trevigen) or pan Ab-5 (clone ACTN05) mouse monoclonal antibody (Thermo Scientific). Following incubation with primary antibodies, all blots were incubated with horseradish peroxidase-conjugated rabbit, anti-mouse secondary antibodies. Cross-reacting material was visualized by exposing the blots to x-ray film in the presence of the Enhanced Chemiluminescence Immunoblotting Reagent (Pierce).
In Vitro H. pylori Infections.
AGS cells (ATCC no. CRL-1739; plated at 0.5 × 105 cells per well), were infected with H. pylori 26695 or G27 at MOI 100, which is a MOI typically used in studies of H. pylori infection of AGS cells (33), at 37 °C within a humidified environment under 5% CO2 and 10% O2. Alternatively, AGS cells were incubated with HPCF (50 μg/mL) in the absence or presence of Chariot protein transfection reagent (Active Motif), according to manufacturer's instructions. After 8 or 4 h, respectively, the monolayers were washed to remove H. pylori or HPCF, and the cells were incubated with PARP-1 cellular activity buffer [50 mM, pH 7.5, 2 mM MgCl2, 25 mM NaCl, 25 mM KCl, 0.01% digitonin, and 32P NAD (10 μM, 1 μCi/mL)] at 37 °C within a humidified environment and under 5% CO2 and 10% O2. After 15 min, the PARP-1 activity buffer was removed, the reactions were stopped and the cells were lysed with 1× SDS/PAGE loading dye and 10% glycerol. The incorporation of 32P-radiolabel into TCA-precipitable material was quantified in Scintiverse BD mixture (Fisher) using a LS-1500 scintillation counter (Beckman). In preliminary studies, we established that 32P-radiolabelling occurred only when cells were exposed to either H. pylori 26695 or G27, but not with heat-killed organisms (60 °C for 30 min).
Alternatively, the cells were fixed by incubation in ice-cold 1:1 methanol:acetone. After 30 min, the cells were permeabilized by incubating three times for 5 min with Tween 20 (0.1% in PBS pH 7.2) at room temperature. The cells were incubated with anti-PAR (1:500 dilution of commercial stock, 1 h, 25 °C), and after washing, with rabbit anti-mouse IgG conjugated to Alexa Fluor 488 (Molecular Probes; 1:1,000 dilution of commercial stock, 1 h, 25 °C). After washing, the samples were exposed to SlowFade reagent (Molecular Probes), and imaged with a Delta Vision RT microscope (Applied Precision), EX 490/20 and EM 528/38, using an Olympus Plan Apo 40× oil objective with NA 1.42 and working distance of 0.15 mm. Differential interference contrast (DIC) images were collected for all fields. Images were processed using SoftWoRX Explorer Suite (Applied Precision). Analysis of PAR staining in the fluorescent microscopy images was carried out using ImageJ 1.43f software (Wayne Rasband, NIH) and relative PAR staining in control (untreated) and morpholino (parp-1 and non-specific) treated cells was calculated by measuring the % mean pixel intensity values. Data were collected and averaged from at least 10 cells in each of three independent experiments (>30 cells total), and in each case subtracting the average background values in each field. In preliminary studies, we established that visible PAR was detected in AGS cells infected with either H. pylori 26695 or G27, but not with heat-killed organisms. For PARP-1 silencing experiments, monolayers of AGS were incubated with Endo-Porter delivery reagent (Gene Tools) at 37 °C and either the parp-1 specific morpholino antisense oligonucleotide (AGAGCTTATCCGAAGACTCCGCCAT) or a non-specific control nucleotide (CCTCTTACCTCAGTTACAATTTATA) (each at 20 μM; Gene Tools). After 24 h, PARP-1 silencing was confirmed by Western blot analysis using anti-PARP-1 or anti-actin, as described above under Activity Blots.
AIF Release.
Release of mitochondria-associated AIF into the cytosol was quantified using a previously described method (34).
Statistics.
Unless otherwise indicated, all experiments were performed at least three independent times in triplicate. Error bars represent standard deviations. P values were calculated with the Student's t-test using paired, one-tailed distribution. P < 0.05 indicates statistical significance. All statistics, including means, standard deviations, and Student's t-tests, were calculated using Microsoft Excel (version 11.0).
Supplementary Material
Acknowledgments.
We thank Dr. Gilbert de Murcia (University of Strasbourg, France) for the generous gift of wild-type and parp-1−/− mouse embryonic fibroblasts and Mr. Ian Gut and Mr. Amandeep Gargi for critical reading of the manuscript. This work was supported from a grant from the National Institutes of Health R01 AI045928 (to S.R.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0906753106/DCSupplemental.
References
- 1.Kusters JG, van Vliet AH, Kuipers EJ. Pathogenesis of Helicobacter pylori infection. Clin Microbiol Rev. 2006;19:449–490. doi: 10.1128/CMR.00054-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McNamara D, El-Omar E. Helicobacter pylori infection and the pathogenesis of gastric cancer: A paradigm for host-bacterial interactions. Dig Liver Dis. 2008;40:504–509. doi: 10.1016/j.dld.2008.02.031. [DOI] [PubMed] [Google Scholar]
- 3.Suerbaum S, Michetti P. Helicobacter pylori infection. N Engl J Med. 2002;347:1175–1186. doi: 10.1056/NEJMra020542. [DOI] [PubMed] [Google Scholar]
- 4.Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136:1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nossa CW, Blanke SR. Modification of a mammalian cell protein in the presence of [32P-adenylate]NAD: Evidence for ADP ribosylation activity associated with Helicobacter pylori. Infect Immun. 2006;74:3071–3076. doi: 10.1128/IAI.74.5.3071-3076.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krueger KM, Barbieri JT. The family of bacterial ADP-ribosylating exotoxins. Clin Microbiol Rev. 1995;8:34–47. doi: 10.1128/cmr.8.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hakme A, Wong HK, Dantzer F, Schreiber V. The expanding field of poly(ADP-ribosyl)ation reactions. 'Protein Modifications: Beyond the Usual Suspects' Review Series. EMBO Rep. 2008;9:1094–1100. doi: 10.1038/embor.2008.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13:3046–3082. doi: 10.2741/2909. [DOI] [PubMed] [Google Scholar]
- 9.Smith S. The world according to PARP. Trends Biochem Sci. 2001;26:174–179. doi: 10.1016/s0968-0004(00)01780-1. [DOI] [PubMed] [Google Scholar]
- 10.Tucek S. Subcellular distribution of acetyl-CoA synthetase, ATP citrate lyase, citrate synthase, choline acetyltransferase, fumarate hydratase and lactate dehydrogenase in mammalian brain tissue. J Neurochem. 1967;14:531–545. doi: 10.1111/j.1471-4159.1967.tb09552.x. [DOI] [PubMed] [Google Scholar]
- 11.DeLuca JG, et al. Purification and characterization of native conventional kinesin, HSET, and CENP-E from mitotic hela cells. J Biol Chem. 2001;276:28014–28021. doi: 10.1074/jbc.M102801200. [DOI] [PubMed] [Google Scholar]
- 12.de Murcia JM, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci USA. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagnon SN, Desnoyers S. Single amino acid substitution enhances bacterial expression of PARP-4D214A. Mol Cell Biochem. 2003;243:15–22. doi: 10.1023/a:1021645327079. [DOI] [PubMed] [Google Scholar]
- 14.Altmeyer M, et al. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mendoza-Alvarez H, Alvarez-Gonzalez R. Poly(ADP-ribose) polymerase is a catalytic dimer and the automodification reaction is intermolecular. J Biol Chem. 1993;268:22575–22580. [PubMed] [Google Scholar]
- 16.Marsischky GT, Wilson BA, Collier RJ. Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation. Evidence for active site similarities to the ADP-ribosylating toxins. J Biol Chem. 1995;270:3247–3254. doi: 10.1074/jbc.270.7.3247. [DOI] [PubMed] [Google Scholar]
- 17.Loesberg C, van Rooij H, Smets LA. Meta-iodobenzylguanidine (MIBG), a novel high-affinity substrate for cholera toxin that interferes with cellular mono(ADP-ribosylation) Biochim Biophys Acta. 1990;1037:92–99. doi: 10.1016/0167-4838(90)90106-p. [DOI] [PubMed] [Google Scholar]
- 18.Burkle A. Poly(ADP-ribose). The most elaborate metabolite of NAD+ Febs J. 2005;272:4576–4589. doi: 10.1111/j.1742-4658.2005.04864.x. [DOI] [PubMed] [Google Scholar]
- 19.Shah GM, et al. Methods for biochemical study of poly(ADP-ribose) metabolism in vitro and in vivo. Anal Biochem. 1995;227:1–13. doi: 10.1006/abio.1995.1245. [DOI] [PubMed] [Google Scholar]
- 20.Maeda S, Mentis AF. Pathogenesis of Helicobacter pylori infection. Helicobacter. 2007;12(1 Suppl):10–14. doi: 10.1111/j.1523-5378.2007.00529.x. [DOI] [PubMed] [Google Scholar]
- 21.David KK, Andrabi SA, Dawson TM, Dawson VL. Parthanatos, a messenger of death. Front Biosci. 2009;14:1116–1128. doi: 10.2741/3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Dawson VL, Dawson TM. Poly(ADP-ribose) signals to mitochondrial AIF: A key event in parthanatos. Exp Neurol. 2009 doi: 10.1016/j.expneurol.2009.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh M, Prasad KN, Saxena A, Yachha SK. Helicobacter pylori induces apoptosis of T- and B-cell lines and translocates mitochondrial apoptosis-inducing factor to nucleus. Curr Microbiol. 2006;52:254–260. doi: 10.1007/s00284-005-0103-1. [DOI] [PubMed] [Google Scholar]
- 24.Ashktorab H, et al. H. pylori-induced apoptosis in human gastric cancer cells mediated via the release of apoptosis-inducing factor from mitochondria. Helicobacter. 2008;13:506–517. doi: 10.1111/j.1523-5378.2008.00646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohen-Armon M, et al. DNA-independent PARP-1 activation by phosphorylated ERK2 increases Elk1 activity: A link to histone acetylation. Mol Cell. 2007;25:297–308. doi: 10.1016/j.molcel.2006.12.012. [DOI] [PubMed] [Google Scholar]
- 26.Guastafierro T, et al. CCCTC-binding factor activates PARP-1 affecting DNA methylation machinery. J Biol Chem. 2008;283:21873–21880. doi: 10.1074/jbc.M801170200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon-Shaag A, Yosef Y, Abd El-Latif M, Oppenheim A. The abundant nuclear enzyme PARP participates in the life cycle of simian virus 40 and is stimulated by minor capsid protein VP3. J Virol. 2003;77:4273–4282. doi: 10.1128/JVI.77.7.4273-4282.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carbone M, et al. PARP-1 interaction with VP1 capsid protein regulates polyomavirus early gene expression. J Mol Biol. 2006;363:773–785. doi: 10.1016/j.jmb.2006.05.077. [DOI] [PubMed] [Google Scholar]
- 29.Gupta VR, et al. Sphingomyelin functions as a novel receptor for Helicobacter pylori VacA. PLoS Pathog. 2008;4:e1000073. doi: 10.1371/journal.ppat.1000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guillemin K, Salama NR, Tompkins LS, Falkow S. Cag pathogenicity island-specific responses of gastric epithelial cells to Helicobacter pylori infection. Proc Natl Acad Sci USA. 2002;99:15136–15141. doi: 10.1073/pnas.182558799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Covacci A, Rappuoli R. Tyrosine-phosphorylated bacterial proteins: Trojan horses for the host cell. J Exp Med. 2000;191:587–592. doi: 10.1084/jem.191.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marty KB, et al. Characterization of a cytotoxic factor in culture filtrates of Serratia marcescens. Infect Immun. 2002;70:1121–1128. doi: 10.1128/IAI.70.3.1121-1128.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki M, et al. Helicobacter pylori CagA phosphorylation-independent function in epithelial proliferation and inflammation. Cell Host Microbe. 2009;5:23–34. doi: 10.1016/j.chom.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 34.Yilmaz O, Jungas T, Verbeke P, Ojcius DM. Activation of the phosphatidylinositol 3-kinase/Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infect Immun. 2004;72:3743–3751. doi: 10.1128/IAI.72.7.3743-3751.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alvarez-Gonzalez R, Jacobson MK. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry. 1987;26:3218–3224. doi: 10.1021/bi00385a042. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka M, et al. Demonstration of high molecular weight poly (adenosine diphosphate ribose) Nucleic Acids Res. 1978;5:3183–3194. doi: 10.1093/nar/5.9.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.