Abstract
The emergence of multidrug-resistant (MDR) tuberculosis (TB) highlights the urgent need to understand the mechanisms of resistance to the drugs used to treat this disease. The aminoglycosides kanamycin and amikacin are important bactericidal drugs used to treat MDR TB, and resistance to one or both of these drugs is a defining characteristic of extensively drug-resistant TB. We identified mutations in the −10 and −35 promoter region of the eis gene, which encodes a previously uncharacterized aminoglycoside acetyltransferase. These mutations led to a 20–180-fold increase in the amount of eis leaderless mRNA transcript, with a corresponding increase in protein expression. Importantly, these promoter mutations conferred resistance to kanamycin [5 μg/mL < minimum inhibitory concentration (MIC) ≤40 μg/mL] but not to amikacin (MIC <4 μg/mL). Additionally, 80% of clinical isolates examined in this study that exhibited low-level kanamycin resistance harbored eis promoter mutations. These results have important clinical implications in that clinical isolates determined to be resistant to kanamycin may not be cross-resistant to amikacin, as is often assumed. Molecular detection of eis mutations should distinguish strains resistant to kanamycin and those resistant to kanamycin and amikacin. This may help avoid excluding a potentially effective drug from a treatment regimen for drug-resistant TB.
The World Health Organization estimates that 9.2 million new cases of tuberculosis (TB) occur each year (1). Despite intensive efforts to ensure proper drug dosages and patient compliance with drug regimens, multidrug-resistant (MDR) and extensively drug-resistant (XDR) strains of Mycobacterium tuberculosis have emerged (2). These strains cause extensive mortality in immunocompromised individuals (3) and hinder the control and prevention of the disease. XDR and MDR TB infections cannot be adequately treated with the first-line anti-TB drugs and require expensive, prolonged treatment with second-line anti-TB drugs. The rapid determination of the resistance profile of an isolate can facilitate selection of an appropriate drug regimen and preclude development of additional drug resistances. Rapid detection of resistances is best achieved with molecular diagnostic approaches, particularly in developing countries where access to culture facilities is limited. Such strategies require a detailed understanding of the molecular basis for drug resistance. Although the mechanisms of resistance to first-line drugs such as isoniazid and rifampin are well characterized, much less is known about such mechanisms for the second-line drugs (4).
An important second-line anti-TB drug is the aminoglycoside kanamycin (KAN), which binds to the 16S rRNA in the 30S ribosomal subunit and inhibits protein synthesis (5). In other bacteria, characterized mechanisms of KAN resistance include altered efflux or influx of the drug, inactivation of the drug by enzymatic modification, and mutation or methylation of rRNA, which disrupts binding of the drug to the ribosome (5). In contrast, our understanding of the mechanism of KAN resistance in M. tuberculosis is limited. Mutations in the 16S rRNA gene, rrs, can cause high-level resistance to KAN [minimum inhibitory concentration (MIC) ≥80 μg/mL], and some mutations can confer cross-resistance to other second-line drugs, including amikacin (AMK) and capreomycin (CAP) (6). However, up to 80% of KAN-resistant (KANR) clinical isolates display low-level resistance to KAN (5 μg/mL < MIC < 80 μg/mL), do not contain rrs mutations, and do not exhibit cross-resistance (7–10). The molecular basis of this low-level resistance is unclear. We report here the discovery and characterization of unique mutations, common in clinical isolates of M. tuberculosis, which confer low-level resistance to KAN by causing overexpression of the enhanced intracellular survival protein, Eis.
Results
C-14T Mutation in eis (Rv2416c) Confers KAN Resistance and Increases Expression of eis.
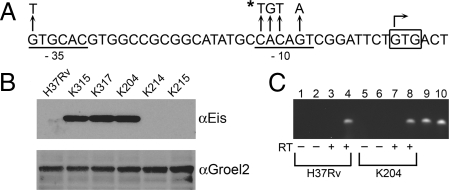
In a previous study (6), M. tuberculosis K204 [supporting information (SI) Table S1] was isolated as a spontaneous KANR mutant of M. tuberculosis H37Rv. Strain K204 is resistant to low levels of KAN (MIC of 25 μg/mL); susceptible to AMK (MIC ≤4 μg/mL), CAP (MIC ≤10 μg/mL), and viomycin (MIC ≤10 μg/mL); and harbors a WT rrs gene. To identify the mutation that confers KAN resistance in this strain, a cosmid library constructed from K204 genomic DNA was introduced into the pansusceptible H37Rv strain, and 5 KANR transformants were isolated. Rapid amplification of transposon ends (RATE) and sequence analysis of the transformants identified a common C-to-T transition located 14 bp upstream of the start codon of the eis gene (Rv2416c) encoding the enhanced intracellular survival protein (Fig. 1A).
Fig. 1.
Characterization of eis promoter and expression. (A) eis promoter sequence and predicted promoter elements in M. tuberculosis. Mutations identified in clinical isolates are denoted by arrows. The mutation identified in the K204 eis promoter region is noted by the asterisk. The eis transcription start site is denoted by a bent arrow. The −10 and −35 regions are underlined, and the start codon is boxed. The extended −10 region (TGn motif) is located directly upstream of the −10 region. (B) Immunoblot analysis of cell lysates generated from strains harboring either WT (H37Rv, K214, K215) or the C-14T eis allele (K204, K315, K317). The lysates were probed with either anti-Eis or anti-Groel2 serum. (C) RNA from H37Rv (lanes 1–4) and K204 (lanes 5–8) was used in 1-step RT-PCR reactions. Odd numbered lanes contain primer pair AZ198 (anneals at −20 to −1 of the eis start codon) and AZ87; even lanes contain primers AZ199 (anneals at +1 to +20) and AZ87. H37Rv genomic DNA (lanes 9 and 10) was used as a positive control for each primer set. The + and − signs indicate whether reverse transcriptase was added to the reaction and serve as negative controls for each primer set and RNA sample.
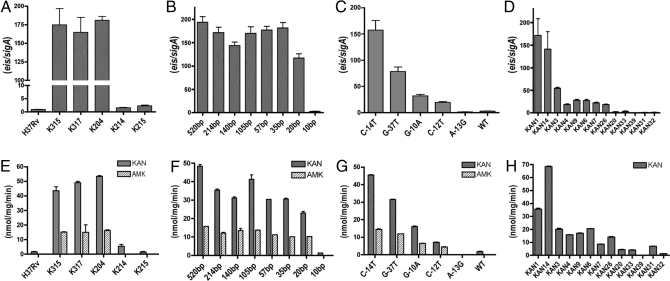
To evaluate the contribution of this gene to KAN resistance, we performed quantitative (q)RT-PCR assays using RNA isolated from H37Rv and K204 and compared the level of eis transcripts with that of the housekeeping gene sigA in each strain. The ratio of eis to sigA transcripts in K204 was ≈180-fold higher than the ratio in H37Rv (Fig. 2A). Comparison of sigA mRNA transcripts relative to 16S rRNA revealed no significant difference in sigA expression between strains, indicating that the increased amount of eis transcript measured in K204 was not due to differential expression of sigA (Fig. S1).
Fig. 2.
Analysis of eis expression and acetyltransferase activity. The ratio of eis/sigA transcripts (A–D) was determined by qRT-PCR and normalized to H37Rv. (A) H37Rv, K204, and allelic exchange derivatives. (B) H37RvΔeis complemented with eis promoter truncation constructs; bp (base pairs) indicates the length of sequence upstream from the eis start codon present on the complementing plasmid. (C) H37RvΔeis complemented with eis promoter alleles. (D) Clinical isolates. Acetyltransferase activity (E–H) was measured from crude cell lysates and is expressed in nmol per mg per min. (E) H37Rv, K204, and allelic exchange derivatives. (F) H37RvΔeis complemented with eis promoter truncation constructs. (G) H37RvΔeis complemented with different eis promoter alleles (H) Clinical isolates. In all panels, error bars represent 3 independent experiments.
To determine whether the C-14T mutation confers resistance to KAN and is responsible for the increased level of eis transcript in K204, we introduced the point mutation into the parent strain H37Rv by allelic exchange using the specialized-linkage transducing phage, phAlexC-14T (Table S2). We also used the specialized-linkage transduction system to revert the mutant eis allele in K204 to the WT allele. Two representative strains containing either the C-14T eis allele (K315 and K317; Table S1) or WT eis allele (K214 and K215; Table S1) were chosen for further analysis. The introduction of the C-14T allele into H37Rv (K315 and K317) was sufficient to confer at least a 10-fold increase in resistance to KAN (MIC of 20–25 μg/mL; Table 1) and ≈180-fold increase in eis transcript levels (Fig. 2A). Conversely, the reversion of the C-14T mutant eis allele to the WT allele (K214 and K215) caused a decrease in the MIC of KAN (2 μg/mL; Table 1) and the amount of eis transcript to levels similar to that observed for H37Rv (Fig. 2A). The increase in eis transcripts correlated with an increase in Eis protein, as determined by immunoblots on cell lysates generated from each strain. Eis was detected in cell lysates generated from strains harboring the C-14T allele (K204, K315, and K317) but could not be detected in cell lysates generated from strains harboring the WT allele (H37Rv, K214, and K215; Fig. 1B). Similarly, Eis was detected in immunoblots of culture supernatants from strains harboring the C-14T allele (K204 and K315) but not the WT allele (H37Rv and K214; Fig. S2). Thus, the C-14T allele is responsible for the increased eis transcripts, KAN MICs, and protein levels in the strains harboring this mutation.
Table 1.
Eis mutations and aminoglycoside resistance levels
| Strain | eis | MIC (μg/mL) |
|
|---|---|---|---|
| KAN | AMK | ||
| H37Rv | WT | 2 | 0.5 |
| K214 | WT | 2 | 0.5 |
| K215 | WT | 2 | 0.5 |
| K204 | C-14T | 25 | 3 |
| K315 | C-14T | 25 | 3 |
| K317 | C-14T | 20 | 3 |
| Plasmid complementing H37RvΔeis | |||
| pAZ38 | WT | 2 | 0.5 |
| pAZ29 | C-14T | 20 | 2 |
| pAZ40 | G-37T | 20 | 1 |
| pAZ39 | G-10A | 10 | 1 |
| pAZ41 | C-12T | 10 | 1 |
| pAZ42 | A-13G | 5 | 0.5 |
| Clinical isolate | |||
| KAN1 | C-14T | 40 | <4 |
| KAN14 | C-14T | 40 | <4 |
| KAN3 | G-37T | 20 | <4 |
| KAN4 | G-37T | 20 | <4 |
| KAN9 | G-37T | 10 | <4 |
| KAN6 | G-10A | 10 | <4 |
| KAN7 | G-10A | 10 | <4 |
| KAN26 | G-10A | 80 | <4 |
| KAN20 | C-12T | 10 | <4 |
| KAN33 | C-12T | <5 | <4 |
| KAN39 | A-13G | <5 | <4 |
| KAN31 | WT | <5 | <4 |
| KAN32 | WT | <5 | <4 |
Because single point mutations in another gene, rrs, can confer resistance to both AMK and KAN (6), we measured the MICs of the parental strains and allelic exchange derivatives to determine whether the eis C-14T point mutation affected the resistance level of AMK. Strains harboring the C-14T eis allele exhibited a 6-fold increase in their MIC (from 0.5 μg/mL to 3 μg/mL; Table 1). Although the MIC for AMK increased, these strains are defined as susceptible to AMK in current testing and treatment guidelines (11, 12).
Transcription from the eis Promoter Produces a Leaderless mRNA.
Using RACE (rapid amplification of cDNA ends) and primer extension analysis, the transcription start site in H37Rv and K204 was mapped to the first G of the start codon of eis (Fig. 1B), indicating that transcription from both the WT and mutant eis promoter produces a leaderless mRNA transcript. We confirmed the location of the transcription start site with RT-PCR assays using primers that annealed either directly upstream (annealing at −20 to −1) or downstream of the GTG start codon (annealing at +1 to +20) of eis in conjunction with a reverse primer. When cDNA generated either from H37Rv or K204 was used as template, a PCR product was only detected when the primer overlapping the GTG start codon was present in the reaction (Fig. 1C). Identification of the transcription start site allowed us to define the −10 and −35 regions of the eis promoter and map the C-14T mutation to the −10 promoter region (Fig. 1A).
To identify any cis-acting elements present in the promoter region, we complemented an eis deletion strain, H37RvΔeis (Table S1), with constructs containing the entire ORF of eis and various derivatives of the mutant eis promoter (Fig. S3). Constructs containing between 520 bp and 35 bp of the upstream promoter region exhibited similar levels of eis transcripts, regardless of the amount of upstream sequence (Fig. 2B). A promoter construct containing 20 bp of upstream sequence demonstrated somewhat decreased eis transcript levels when compared with the longer upstream regions but produced >100-fold higher transcript levels than the WT promoter in H37Rv. Because the 20-bp promoter construct lacks the −35 region of the eis promoter, the data indicate that the −10 region of the promoter is able to initiate transcription. Finally, only minimal transcript levels were detected with a construct harboring only 10 bp of upstream sequence, which lacks both the −10 and −35 promoter regions (Fig. 2B). On the basis of sequence homology and these results, the eis promoter seems to use an extended −10 region (Figs. 1A and 2B).
Eis Is an Aminoglycoside Acetyltransferase That Acetylates and Inactivates KAN and AMK.
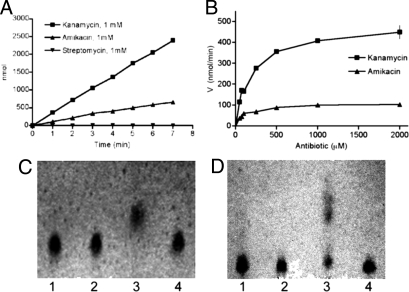
Eis shares sequence homology and secondary structural characteristics with the GNAT superfamily of acetyltransferases, which includes the aminoglycoside N-acetyltransferases (13, 14). To determine whether recombinant Eis protein exhibited acetyltransferase activity, we tested KAN, AMK, and streptomycin (SM) individually as substrates using a colorimetric assay that quantifies the conversion of acetyl-CoA to CoA-SH. Reactions that lacked either Eis protein, acetyl-CoA, or any of the 3 substrates exhibited no acetyltransferase activity. Acetyltransferase activity was detected with KAN (specific activity of 447 nmol per min per mg) and AMK (98.4 nmol per min per mg), but not with SM (Fig. 3A). Eis acetylated KAN at a rate 3.3-fold higher than AMK (366 nmol/min and 111 nmol/min, respectively) and exhibited estimated Km values for KAN at 154 μM and AMK at 112 μM (Fig. 3B). Additionally, Eis demonstrated a 4-fold higher maximum acetylation rate of KAN (0.47 μmol/min) compared with AMK (0.12 μmol/min). Finally, the Vmax/Km ratio for KAN was 3-fold higher than AMK, indicating that Eis acetylated KAN more efficiently than AMK, even though Eis seems to bind AMK with a higher affinity. This difference in substrate utilization by Eis likely explains the observed lack of cross-resistance to KAN and AMK for isolates harboring eis mutations.
Fig. 3.
Eis acetylates KAN and AMK. (A) Acetylation rates for KAN, AMK, and SM. (B) Michaelis-Menton plots for KAN and AMK. Acetyltransferase reactions were performed using either (C) KAN or (D) AMK as substrate, and the products were developed on TLC plates. Lane 1: unmodified antibiotic; lane 2: no acetyl-CoA control; lane 3: complete acetyltransferase reaction; lane 4: no Eis control. Rf values under these solvent conditions: unmodified KAN, 0.31; acetyl-KAN, 0.44; unmodified AMK, 0.09; acetyl-AMK, 0.46.
To determine whether Eis modifies KAN and AMK, we analyzed the acetyltransferase assay products using TLC. An increase of the migration of the acetylated products on a TLC plate confirmed that KAN (100% of substrate by densitometric analysis) and AMK (56% of substrate) were modified by Eis (Fig. 3 C and D). When 20 μL of acetyltransferase reactions were spotted onto a lawn of Mycobacterium smegmatis cells, reactions that lacked the Eis protein or acetyl-CoA produced zones of clearing of approximately 5.0–5.8 cm (Fig. S4). Conversely, when complete reactions with KAN or AMK were spotted, no zone of inhibition was detected (Fig. S4, lane 4), indicating inactivation of both antibiotics. Similar zones of inhibition were detected for SM in the presence or absence of Eis, indicating that SM is not inactivated by Eis (Fig. S4). If 40-μL reactions were spotted onto M. smegmatis, a zone of clearing was detected for AMK (4.0 cm) but not for KAN, consistent with the observation that AMK is acetylated less efficiently than KAN.
A colorimetric assay was used to quantify acetyltransferase activity in cell lysates of eis mutants. Activity was detected in cell lysates from strains harboring the C-14T allele but not from strains harboring the WT eis allele (Fig. 2E). The lysates exhibited approximately 3 times more activity when KAN was used as the substrate as compared with AMK. We also used the in vitro acetyltransferase assay to analyze activity detected from cell lysates harboring the eis promoter truncation constructs and found that the activity generally correlated with the amount of eis transcript produced in each strain (Fig. 2F).
Clinical Isolate Analysis.
We sequenced the eis and rrs alleles of KANR clinical isolates that did not exhibit any cross-resistance to AMK or CAP. Of the 42 isolates analyzed, 33 (79%) harbored a mutation within the promoter region of eis (Table 2). We observed the mutation identified in K204 (C-14T) and discovered 2 unique mutations (G-37T and G-10A) that mapped within the −35 or −10 region of the eis promoter (Fig. 1A and Table 2). Nine (21%) of the KANR clinical isolates did not contain a mutation within eis or rrs. We also analyzed 16 KANS clinical isolates and did not find a mutation in either locus in 12 (75%) of the isolates (Table 2). However, 4 KANS isolates did contain a mutation (A-13G or C-12T) within the promoter region of eis.
Table 2.
Distribution of eis mutations in clinical isolates
| eis allele | Isolates, n (%) | KAN MIC* (μg/mL) |
|---|---|---|
| C-14T | 10 (24) | 25–40 |
| G-10A | 14 (33) | 10–20 |
| G-37T | 8 (19) | 20 |
| C-12T | 1 (2) | 10 |
| WT | 9 (21) | 10–40 |
| Total | 42 (100) | |
| C-12T | 3 (19) | <5 |
| A-13G | 1 (6) | <5 |
| WT | 12 (75) | <5 |
| Total | 16 (100) |
*Isolates with MICs >5 μg/mL are considered resistant to KAN.
Analysis of the level of eis expression in representative clinical isolates with each promoter mutation revealed a general correlation between eis transcript levels, KAN MICs, and acetyltransferase activity (Fig. 2 D and H and Table 1). Clinical isolates with the eis mutations C-14T, G-37T, and G-10A exhibited detectable acetyltransferase activity against KAN that correlated with what is considered clinically significant levels of KAN resistance (MIC ≥5 μg/mL) (11). Several clinical isolates (KAN1 and KAN14) also exhibited detectable activity against AMK. However, these strains have AMK MIC ≤4 μg/mL and therefore are considered to be susceptible to AMK (Table 1) (11). The clinical isolates harboring a C-12T mutation (KAN20 and KAN33) exhibited only slightly elevated levels of eis transcripts and a minimal amount of acetyltransferase activity (Fig. 2 D and H). Clinical isolates containing WT eis (KAN31 and KAN32) or the A-13G mutation (KAN39) demonstrated eis transcript levels similar to that of the pansusceptible H37Rv strain, had minimal or no detectable acetyltransferase activity, and were susceptible to KAN and AMK according to Clinical and Laboratory Standards Institute (CLSI) guidelines (11) (Fig. 2 D and H and Table 1).
There was some variation in observed MICs between strains with identical mutations. It is possible that clinical isolates harbor additional mutations at undefined genomic loci that may contribute to the KANR phenotype. To assess the contributions of individual eis mutations to KAN resistance in a clean genetic background, we complemented the knockout strain, H37RvΔeis, with each mutation individually. Generally, the transcript level, aminoglycoside resistance, and acetyltransferase activity were higher in complemented H37RvΔeis strains than clinical isolates with the corresponding mutation (Fig. 2 C and G). However, the same hierarchy of mutations remained. Complementation of H37RvΔeis with the C-14T promoter allele resulted in the highest eis transcript level, acetyltransferase activity, and MICs to KAN and AMK (Fig. 2 C and G and Table 1). Compared with the C-14T allele, the G-37T, G-10A, and C-12T promoter alleles demonstrate progressively lower eis transcript levels, acetyltransferase activity, and aminoglycoside resistance (Fig. 2 C and G and Table 1). Complementation of H37RvΔeis with either the WT or A-13G promoter allele produced eis transcript levels, acetyltransferase activity, and KAN and AMK MICs similar to levels observed for the parental strain H37Rv (Fig. 2 C and G and Table 1).
Discussion
Here we describe a previously unidentified mechanism of KANR in M. tuberculosis due to point mutations in the promoter region of the enhanced intracellular survival gene, eis. The KAN resistance conferred by eis promoter mutations is due to the significant increase in eis transcript levels and corresponding increase in the levels of an enzyme that acetylates and inactivates KAN. Importantly, although Eis was found to have some activity against AMK, eis mutant strains are susceptible to AMK (MIC ≤4 μg/mL) according to current testing guidelines (11) and treatment guidelines (12), suggesting the possibility that AMK could be used to treat strains displaying the resistance pattern of eis mutants.
The lack of cross-resistance to AMK is explained by the finding that Eis utilizes KAN as substrate 3-fold more efficiently than AMK. The difference in substrate utilization is likely due to differences in the structures of KAN and AMK. AMK contains an L-hydroxyaminobuteroyl amide group substitution in the N1 position of the deoxystreptamine ring (5), which may sterically hinder the acetylation by Eis.
Because the mutations in K204 and KANR clinical isolates cluster at the −10 region or −35 region of the eis promoter, it is likely that the increase in transcription is due to improving the strength of the existing promoter. Recently, it was shown that sigA binds to the promoter region and regulates expression of the eis gene (15). Indeed, the −10 regions of the eis mutant promoters (C-14T, TACAGT; G-10A, CACAAT; C-12T, CATAGT) are more homologous with the SigA consensus sequence (TA(G/T)(A/G)AT) (16) than the WT promoter (CACAGT). Conversely, the A-13G promoter (CGCACT) shares less homology with the SigA consensus sequence and does not increase eis transcript levels. Along similar lines, the −35 region of the WT eis (GTGCAC) promoter is similar to the consensus sequence proposed for group A mycobacterial promoters (TTGACn) (16), and the observed G-37T mutation (TTGCAC) improves the match with the SigA −35 consensus sequence. Although it seems likely that these mutations increase sigA binding to the eis promoter, studies are currently underway to clarify the mechanism of increased transcription at the mutant eis promoters and to identify any regulators that may be acting to facilitate the increase.
Our results demonstrate that the transcription start site of the eis promoter in K204 and H37Rv maps to the GTG start codon and produces a leaderless mRNA transcript (Fig. 1). This differs from the published transcription start site for eis reported to be 33 bp upstream of the GTG start codon (17). One possible reason for the discrepancy is that the transcription start site of eis was not mapped in M. tuberculosis but rather from a multicopy plasmid in M. smegmatis and the avirulent derivative of H37Rv, M. tuberculosis H37Ra, which both differ significantly from H37Rv. A growing number of mycobacterial genes have been shown to produce leaderless mRNAs (18–20), including the aminoglycoside acetyltransferase gene aac(2′-Id), in M. smegmatis (21). Interestingly, sequence analysis of the eis promoter indicated the presence of an extended −10 region motif (TGN) just upstream of the −10 region (Fig. 1A). Our observation that the 20 bp of sequence upstream of the GTG start codon are sufficient to initiate transcription suggests that this motif does indeed act as an extended −10 region; however, the presence of both the −10 and −35 region is required for optimal expression from the eis promoter. The extended −10 motif is a characteristic shared with other leaderless mRNA-producing mycobacterial promoters (18, 20, 21) and may imply a conserved regulatory mechanism for these poorly understood transcripts.
Analyses of the eis mutant strains revealed consistent correlations of eis mutations with eis transcription level, aminoglycoside MICs, and acetyltransferase activity. Our data suggest that M. tuberculosis strains harboring a C-14T, G-37T, or G-10A eis mutation are likely to be resistant to KAN but susceptible to AMK in currently used laboratory tests. The higher levels of eis transcript observed in the H37RvΔeis complemented strains when compared with clinical isolates with the same mutation are probably due to read-through transcription that can occur when genes are expressed at a nonnative locus. This is supported by the fact that H37RvΔeis complemented with the WT eis allele produces almost 3-fold greater eis transcript than the parental H37Rv strain (Fig. 2C). Read-through transcription may also account for the increased level of eis expression in H37RvΔeis complemented with the C-12T eis allele. Although the C-12T complement had an increased KAN MIC, no clinical isolates harboring the C-12T mutation were resistant to KAN (Table 1). This suggests that strains harboring a C-12T mutation may have an MIC close to the critical concentration used to test for KAN susceptibility. Our data also suggest that another mechanism of resistance to KAN is due to mutation(s) in as-yet-unidentified gene(s) because ≈20% of our clinical isolates exhibited KAN resistance that could not be explained by rrs or eis mutations. For example, the clinical isolate KAN26, which harbors a G-10A mutation, expresses low but detectable levels of acetyltransferase activity and has an MIC to KAN of 80 μg/mL that cannot be explained by the eis mutation alone.
The function of Eis in M. tuberculosis is unclear. The high Km value of Eis with either KAN (154 μM) or AMK (112 μM) argues that neither antibiotic is a natural substrate of the protein. Eis was originally reported for its role in intracellular survival of mycobacteria in macrophages (22). Recently, activation of the eis gene in a W-Beijing strain of M. tuberculosis due to increased sigA expression levels has been linked to an enhanced survival phenotype in monocytes (15). This observation suggests that a similar phenotype could be observed with eis promoter mutants and raises the possibility that treatment with KAN could select for strains with improved survival in macrophages. Additionally, purified recombinant Eis protein was shown to be sufficient in stimulating the expression of antiinflammatory cytokines in macrophages (17) and disrupting the cross-regulation of T cells (23). Therefore, it is possible that overexpression of eis could also lead to more robust immune-modulatory effects in the host. Although it is unclear how the acetylation function of Eis modulates both cytokine production and intracellular survival, because Eis is secreted into host cell cytoplasm (24), one may speculate that Eis could be acetylating host factors during infection that could affect these different phenotypes. Even though the natural substrate(s) for Eis are unknown, the protein could be also involved in metabolic activities of the cell. Aminoglycoside acetyltransferases have been hypothesized to play a role in metabolism (25), although the only clear example of a metabolic function for one of these proteins is the O-acetylation of peptidoglycan in Providencia stuartii (26, 27).
Our findings have significant clinical and diagnostic implications. For example, the current CLSI guidelines (11) recommend using KAN as the class representative for both KAN and AMK resistance. That is, strains found to be susceptible (or resistant) to KAN would be assumed to be susceptible (or resistant) to AMK. Although this seems to be mostly true for resistance due to mutations in the rrs gene, our data indicate that KANR strains due to eis mutations are AMK susceptible. Therefore, cross-resistance between KAN and AMK should not be assumed, and to avoid excluding a potentially effective drug from a regimen, susceptibility testing for KAN and AMK should be conducted individually.
In several research studies, low-level KANR strains differ from high-level KANR strains by both lacking rrs mutations and cross-resistance to other ribosome binding drugs, including AMK (6–10). The prevalence of mutations in the eis promoter in 79% of the low-level KANR clinical isolates studied and absence from the KANS isolates studied suggests that these mutations account for much of the observed low-level KAN resistance. Specifically, our data indicate that at the molecular level, KANR, AMKS isolates are likely to harbor an eis mutation, whereas KANR, AMKR strains are likely to harbor an rrs mutation. Over all, our findings suggest that these mutations could serve as a molecular marker to identify KAN-resistant strains and distinguish low-level resistant isolates from high-level, cross-resistant isolates, leading to more appropriate treatment regimens for those infected with drug-resistant strains.
Methods
Bacterial Strains, Media, and DNA Manipulations.
The strains and plasmids used in this study are listed in Table S1 and Table S2 and were cultivated as previously described (6). Details of culture conditions, cloning, and MIC procedures can be found in SI Text.
Expression and Purification of Eis.
The eis gene was amplified by PCR using primers AZ109 and AZ120 (Table S3), and the amplicon cloned into pET19b (Novagen) to create an N-terminal histidine-tagged fusion protein. The His-Eis fusion was purified from the lysate of an isopropyl-beta-D-thiogalactopyranoside–induced culture using a His-Bind NiNTA column (Novagen). The eluate containing the His-Eis protein was dialyzed against 50 mM Tris·HCl pH 8.0, and concentrated using a spin column (Millipore). Purity was assessed by SDS-PAGE, and the protein was quantified using the MicroBCA Protein Kit (Pierce).
Immunoblot Analysis.
Immunoblots were performed as previously described (13). Briefly, 10 μg of whole-cell lysate were separated on 12% Bis-Tris SDS-PAGE gels (Invitrogen) and transferred to PVDF membranes. Membranes were incubated with primary antibody specific to Eis (1:2,500) or GroEL2 (1:50) for 1 h in 1 mM PBS with Tween 20 (PBS-Tw20) containing 0.25% skim milk, washed with PBS-Tw20, and then probed for 1 h with anti-IgG secondary antibody (1:10,000) conjugated to HRP. The reactive bands were visualized using ECL detection reagents (Amersham). αEis antibodies were a gift from Richard Friedman of Arizona State University. αGroEL2 and αAg85 complex antibodies were obtained from the Colorado State University TB Research Materials Contract.
Cell Lysate Collection and Acetyltransferase Assays.
M. tuberculosis strains were grown to mid-log phase, washed, harvested, resuspended in 50 mM Tris·HCl pH 8.0, transferred to tubes containing glass beads (Biospec Products), and lysed using a Mini-Beadbeater (BioCold Scientific). Acetyltransferase assays were carried out as described previously (28) using either purified His-Eis protein or cell lysate. To quantify acetylation, the degree of colorimetric change associated with either cell lysate or purified protein was subtracted from that of a control mixture that lacked antibiotic substrate. The reaction mixture (1 mL) contained 0.1 mM acetyl-CoA (Sigma), 1 mM KAN or AMK, 1 mM 5,5′-dithio-bis(2-nitrobenzoate) (Sigma), 50 mM Tris-HCl pH 8.0, and either 10 μg purified His-Eis protein or 50 μg cell lysate. One unit of enzyme activity is defined as the amount of enzyme that catalyzes the formation of 1 nmol 2-nitro-5-benzoate (14,151 M−1cm−1) per min at room temperature at A412. For kinetic analysis, reaction conditions were the same as above except final KAN and AMK concentrations varied from 50 nM to 2 μM. Triplicate sets of reactions at each concentration were initiated with the addition of antibiotic substrate, and Lineweaver-Burk plots were used to estimate the kinetic constants for each antibiotic.
TLC and Disk-Diffusion Assays.
Acetyltransferase reactions for TLC analysis and disk-diffusion assays were performed in 20-μL reaction volumes; 5-μL portions of each reaction were spotted onto 0.25-mm silica gel TLC plates (Whatman) and developed with methanol/ammonium hydroxide (5:2). Aminoglycosides and acetylated products were detected by spraying the TLC plates with orcinol and charring at 110 °C for 10 min. For disk-diffusion assays, the entire 20-μL acetyltransferase reaction was applied to a Whatman filter disk (Millipore) and placed on a lawn of M. smegmatis cells on Mueller-Hinton agar and incubated for 3 days at 37 °C.
Quantitative RT-PCR.
M. tuberculosis strains were grown to mid-log phase (OD600 0.4–0.6) in Middlebrook 7H9 medium; bacteria were harvested and lysed using a FastPrep 120 (Bio 101 Savant); and RNA purified using an RNeasy kit (Qiagen). One microgram of DNase-treated RNA from each M. tuberculosis strain was used to generate cDNA using the Promega Reverse Transcription System. Quantification of transcripts from 2 μL cDNA was performed by real-time PCR using Probe Master Mix (Roche) in a LightCycler480 detection system. Reactions to detect the sigA transcript used primers AZ139 and AZ140 and probe UPL#133 (Roche). Primers AZ141 and AZ142 and UPL#62 probe (Roche) were used to detect the eis transcript. Primers AZ146 and AZ147 and UPL#38 probe (Roche) were used to detect the rrs transcript. The relative amounts of each PCR product were calculated from standard curves obtained from PCR with the same primers and probes and serially diluted K204 cDNA.
Primer Extension, RACE, and RT-PCR.
Fifty micrograms of RNA from M. tuberculosis H37Rv or K204 bacteria were incubated with 100 pmol of 5′FAM-labeled primer AZ143 at 70 °C for 10 min, followed by 10 min on ice and 20 min at 58 °C, and allowed to cool to room temperature for 15 min. cDNA was synthesized using the Promega Reverse Transcription System. The primer-extension products were purified using a Zymo DNA cleanup kit, spiked with 0.5 μL ROX markers (MapMarker 1000, BioVentures), and analyzed on an ABI 3130xl sequencer using the Fragment Analysis module and PeakScanner software. RACE was performed using a 5′/3′ RACE Kit, 2nd Generation (Roche). For K204 and H37Rv, 100 ng or 2 μg of RNA was used for cDNA synthesis, respectively, using primer AZ85 (12.5 μM) and 80 U transcriptor reverse transcriptase (Roche). cDNA was purified using a High Pure PCR Product Purification Kit (Roche). A homopolymeric A-tail was added to the 3′end of the purified cDNA by incubation of the cDNA with recombinant terminal transferase (80 U) and dATP (200 nM) for 20 min at 37 °C. Five microliters of polyadenylated cDNA was amplified using Promega PCR Master Mix and 0.5 μM of primers AZ87 and an oligo(dT) anchor primer (Roche). Amplified cDNA products were purified using a ZymoClean Gel DNA Recovery Kit (Zymo), cloned into pCR2.1-TOPO (Invitrogen), and sequenced with AZ143. For 2-step RT-PCR reactions, cDNA was synthesized using the Promega Reverse Transcription System, except random hexamers were replaced with primer AZ87. Either 200 ng of K204 RNA or 3 μg of H37Rv RNA was used as template. Two microliters of cDNA was added to PCR reactions containing the primers (AZ179 and AZ87) or (AZ180 and AZ87) using Promega PCR Master Mix. PCR conditions were as follows: initial denaturation at 97 °C for 5 min, 30 cycles of 97 °C for 20 sec, 60 °C for 20 sec, and 72 °C for 45 sec; followed by a final elongation step at 72 °C for 2 min. PCR products were analyzed on agarose gels.
Supplementary Material
Acknowledgments.
We thank Richard Friedman for generously providing the anti-Eis antibody and Daniel Kalman, Patrick M. Reeves, and Angela Starks for critical reading of the manuscript and helpful suggestions.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907925106/DCSupplemental.
References
- 1.World Health Organization. Antituberculosis Drug Resistance in the World Report No. 4. Geneva: World Health Organization; 2008. [Google Scholar]
- 2.U.S. Centers for Disease Control and Prevention. Emergence of Mycobacterium tuberculosis with extensive resistance to second line drugs-worldwide, 2000–2004. MMWR Morb Mortal Wkly Rep. 2006;55:301–305. [PubMed] [Google Scholar]
- 3.Gandhi NR, et al. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet. 2006;368:1575–1580. doi: 10.1016/S0140-6736(06)69573-1. [DOI] [PubMed] [Google Scholar]
- 4.Johnson R, et al. Drug resistance in Mycobacterium tuberculosis. Curr Issues Mol Biol. 2006;8:97–111. [PubMed] [Google Scholar]
- 5.Magnet S, Blanchard JS. Molecular insights into aminoglycoside action and resistance. Chem Rev. 2005;105:477–498. doi: 10.1021/cr0301088. [DOI] [PubMed] [Google Scholar]
- 6.Maus CE, Plikaytis BB, Shinnick TM. Molecular analysis of cross-resistance to capreomycin, kanamycin, amikacin, and viomycin in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2005;49:3192–3197. doi: 10.1128/AAC.49.8.3192-3197.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alangaden GJ, et al. Mechanism of resistance to amikacin and kanamycin in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1998;42:1295–1297. doi: 10.1128/aac.42.5.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kruuner A, Jureen P, Levina K, Ghebremichael S, Hoffner S. Discordant resistance to kanamycin and amikacin in drug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2003;47:2971–2973. doi: 10.1128/AAC.47.9.2971-2973.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki Y, et al. Detection of kanamycin-resistant Mycobacterium tuberculosis by identifying mutations in the 16S rRNA gene. J Clin Microbiol. 1998;36:1220–1225. doi: 10.1128/jcm.36.5.1220-1225.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taniguchi H, et al. Molecular analysis of kanamycin and viomycin resistance in Mycobacterium smegmatis by use of the conjugation system. J Bacteriol. 1997;179:4795–4801. doi: 10.1128/jb.179.15.4795-4801.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Committee for Clinical Laboratory Standards. Susceptibility testing of mycobacteria, norcardiae, and other aerobic actinomycetes. Wayne, PA: National Committee for Clinical Laboratory Standards; 2003. Approved Standard M24-A. [PubMed] [Google Scholar]
- 12.Tuberculosis Coalition for Technical Assistance. The Hague: Tuberculosis Coalition for Technical Assistance; 2006. [Google Scholar]
- 13.Samuel LP, et al. Expression, production and release of the Eis protein by Mycobacterium tuberculosis during infection of macrophages and its effect on cytokine secretion. Microbiology. 2007;153:529–540. doi: 10.1099/mic.0.2006/002642-0. [DOI] [PubMed] [Google Scholar]
- 14.Vetting MW, et al. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 15.Wu S, et al. Activation of the eis gene in a W-Beijing strain of Mycobacterium tuberculosis correlates with increased SigA levels and enhanced intracellular growth. Microbiology. 2009;155:1272–1281. doi: 10.1099/mic.0.024638-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez M, Smith I. Determinants of mycobacterial gene expression. In: Hatfull GF, Jacobs WR Jr, editors. Molecular Genetics of Mycobacteria. Washington, DC: American Society for Microbiology Press; 2000. pp. 111–129. [Google Scholar]
- 17.Roberts EA, Clark A, McBeth S, Friedman RL. Molecular characterization of the eis promoter of Mycobacterium tuberculosis. J Bacteriol. 2004;186:5410–5417. doi: 10.1128/JB.186.16.5410-5417.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson M, et al. The Mycobacterium tuberculosis purine biosynthetic pathway: Isolation and characterization of the purC and purL genes. Microbiology. 1996;142(Pt 9):2439–2447. doi: 10.1099/00221287-142-9-2439. [DOI] [PubMed] [Google Scholar]
- 19.Timm J, et al. Transcription and expression analysis, using lacZ and phoA gene fusions, of Mycobacterium fortuitum beta-lactamase genes cloned from a natural isolate and a high-level beta-lactamase producer. Mol Microbiol. 1994;12:491–504. doi: 10.1111/j.1365-2958.1994.tb01037.x. [DOI] [PubMed] [Google Scholar]
- 20.Dhandayuthapani S, Mudd M, Deretic V. Interactions of OxyR with the promoter region of the oxyR and ahpC genes from Mycobacterium leprae and Mycobacterium tuberculosis. J Bacteriol. 1997;179:2401–2409. doi: 10.1128/jb.179.7.2401-2409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mick V, et al. Transcriptional analysis of and resistance level conferred by the aminoglycoside acetyltransferase gene aac(2′)-Id from Mycobacterium smegmatis. J Antimicrob Chemother. 2008;61:39–45. doi: 10.1093/jac/dkm440. [DOI] [PubMed] [Google Scholar]
- 22.Wei J, et al. Identification of a Mycobacterium tuberculosis gene that enhances mycobacterial survival in macrophages. J Bacteriol. 2000;182:377–384. doi: 10.1128/jb.182.2.377-384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lella RK, Sharma C. Eis (enhanced intracellular survival) protein of Mycobacterium tuberculosis disturbs the cross regulation of T-cells. J Biol Chem. 2007;282:18671–18675. doi: 10.1074/jbc.C600280200. [DOI] [PubMed] [Google Scholar]
- 24.Dahl JL, Wei J, Moulder JW, Laal S, Friedman RL. Subcellular localization of the Intracellular survival-enhancing Eis protein of Mycobacterium tuberculosis. Infect Immun. 2001;69:4295–4302. doi: 10.1128/IAI.69.7.4295-4302.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Udou T, Mizuguchi Y, Wallace RJ., Jr Does aminoglycoside-acetyltransferase in rapidly growing mycobacteria have a metabolic function in addition to aminoglycoside inactivation? FEMS Microbiol Lett. 1989;48:227–230. doi: 10.1111/j.1574-6968.1989.tb03304.x. [DOI] [PubMed] [Google Scholar]
- 26.Payie KG, Clarke AJ. Characterization of gentamicin 2′-N-acetyltransferase from Providencia stuartii: Its use of peptidoglycan metabolites for acetylation of both aminoglycosides and peptidoglycan. J Bacteriol. 1997;179:4106–4114. doi: 10.1128/jb.179.13.4106-4114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Payie KG, Rather PN, Clarke AJ. Contribution of gentamicin 2′-N-acetyltransferase to the O acetylation of peptidoglycan in Providencia stuartii. J Bacteriol. 1995;177:4303–4310. doi: 10.1128/jb.177.15.4303-4310.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamano Y, Hoshino Y, Nakamori S, Takagi H. Overexpression and characterization of an aminoglycoside 6′-N-acetyltransferase with broad specificity from an epsilon-poly-L-lysine producer, Streptomyces albulus IFO14147. J Biochem. 2004;136:517–524. doi: 10.1093/jb/mvh146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.