Abstract
Phosphoinositide-specific phospholipase C (PLC) is a central effector for many biological responses regulated by G-protein–coupled receptors including Drosophila phototransduction where light sensitive channels are activated downstream of NORPA, a PLCβ homolog. Here we show that the sphingolipid biosynthetic enzyme, ceramide kinase, is a novel regulator of PLC signaling and photoreceptor homeostasis. A mutation in ceramide kinase specifically leads to proteolysis of NORPA, consequent loss of PLC activity, and failure in light signal transduction. The mutant photoreceptors also undergo activity-dependent degeneration. Furthermore, we show that a significant increase in ceramide, resulting from lack of ceramide kinase, perturbs the membrane microenvironment of phosphatidylinositol 4, 5, bisphosphate (PIP2), altering its distribution. Fluorescence image correlation spectroscopic studies on model membranes suggest that an increase in ceramide decreases clustering of PIP2 and its partitioning into ordered membrane domains. Thus ceramide kinase–mediated maintenance of ceramide level is important for the local regulation of PIP2 and PLC during phototransduction.
Signal transduction via G-protein–coupled receptors (GPCRs) is vital for many cellular processes including vision, olfaction, taste, and neurotransmission. Extensive studies on proteins constituting this family and their interactions reveal complex signaling networks regulated at multiple levels (1). Lipids play equally important roles in GPCR signaling, as most of the signal transduction machinery is membrane associated. How lipids regulate GPCR signaling is being addressed in recent years (2). Our current knowledge of how lipids influence each other to form membrane microenvironments and how this modulates proteins during signal transduction in a multicellular organism is limited. In this study, we address this issue in the context of Drosophila phototransduction, a prototypic G-protein–coupled phosphoinositide cascade, by genetically modulating the sphingolipid ceramide.
Analyses of Drosophila phototransduction have led to the identification, characterization, and regulation of many signaling components (3). Phototransduction begins with the absorption of light by rhodopsin, followed by the activation of a G protein (Gαq). Gαq activates the critical effector NORPA, a phospholipase C (PLC) that catalyzes the hydrolysis of phosphatidylinositol 4, 5-bisphosphate (PIP2) into two important second messengers, diacylglycerol and inositol 1, 4, 5-trisphosphate. Activation of PLC leads to gating of two transduction channels, transient receptor potential (TRP) and TRP-like. Although many of the proteins involved in phototransduction have been well characterized, we are only beginning to understand how lipids and enzymes involved in lipid metabolism regulate this cascade (4–7). Sphingolipids are integral components of all eukaryotic cell membranes and also act as second messengers in diverse signaling pathways (8). The sphingolipid biosynthetic pathway is an evolutionarily conserved route that generates and interconverts various sphingolipids such as ceramide, sphingosine, ceramide 1–phosphate and sphingosine 1–phosphate (9). We showed earlier that modulating this biosynthetic pathway by targeted overexpression of Drosophila neutral ceramidase (CDase), an enzyme that converts ceramide to sphingosine, rescues retinal degeneration in an arrestin mutant, and facilitates membrane turnover in a rhodopsin null mutant by modulating the endocytic machinery (10–12). Although these studies established that ceramide metabolism is important for survival of photoreceptors, they did not evaluate its role in signaling events during phototransduction.
Ceramide kinase (CERK), a recently cloned lipid kinase, phosphorylates ceramide to ceramide 1–phosphate (C-1-P), thereby decreasing ceramide levels (13, 14). Here we show that Drosophila ceramide kinase (DCERK) regulates PLC activity, function, and the local organization of PIP2 in GPCR signaling by controlling the ceramide level. Genetic, biochemical, and electrophysiological analyses of DCERK deficient flies reveal a severe down regulation of NORPA and failure in phototransduction. Increased ceramide levels in the mutant also alter the level and membrane microenvironment of PIP2 that correlates with a failure of NORPA to localize to the membranes. Using fluorescence image correlation spectroscopy in supported bilayers, we show that ceramide perturbs both the protein-dependent and -independent compartmentalization of PIP2, thus providing a biophysical basis for the effect of ceramide on PIP2. These findings show that sphingolipids and phospholipids cooperate in vivo to establish a suitable membrane microenvironment for signaling mediated by PLC.
Results
Identification and Characterization of Drosophila CERK (DCERK).
A BLAST search of the Drosophila melanogaster genome identified CG16708 as the Drosophila homolog of the CERK gene and was named DCERK. DCERK is on the right arm of the third chromosome at 82F11–83A1. It encodes a protein of 687 aa and is 35% identical to human CERK (Supporting Information (SI) Fig. S1). Western analyses using monoclonal antibodies raised against DCERK protein showed that it is expressed during all developmental stages (Fig. S2A). Membrane association analyses showed that DCERK is an integral membrane protein that is released from membranes by detergent treatment (Fig. S2B). Hydrophobicity analysis predicts two transmembrane helices (residues 258–282 and 515–533) in the protein. Immunofluorescence analyses of Schneider cells and third-instar larval wing discs using DCERK antibodies showed that the protein predominantly localized to the plasma membrane (Figs. 1A, S2 C and D). Like the mammalian enzyme, DCERK has a diacylglycerol kinase domain and a Ca2+/calmodulin binding domain. The N terminus of mammalian CERK contains a pleckstrin homology (PH) domain that binds PIP2 and targets CERK to the membrane. Although primary sequence comparisons of DCERK do not reveal a PH domain except for a conserved tryptophan residue, secondary structure predictions indicate that DCERK has a noncanonical PH domain. We confirmed that DCERK could phosphorylate ceramide by establishing stable cells expressing tagged DCERK or control vector and measuring enzyme activity in membrane fractions by a chemiluminescence assay that measures ATP depletion (15). DCERK used ceramide but not sphingosine or diacylglycerol as a substrate (Fig. S3 A and B). Sphingolipid enriched fractions were prepared from stable cells expressing DCERK and ceramide and C-1-P levels were measured by ultraperformance liquid chromatography in conjunction with mass spectrometry (UPLC-MS/MS). The analyses confirmed that ceramide kinase overexpression increased C-1-P and decreased ceramide level (Fig. S3C).
Fig. 1.
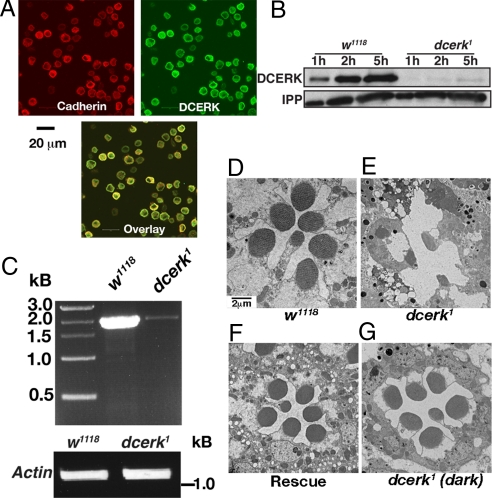
dcerk1 Is a severe hypomorph showing light-dependent photoreceptor degeneration. (A) Immunostaining of Schneider cells with antibodies to DCERK and N-cadherin, a plasma membrane marker. DCERK shows plasma membrane staining. (B) Western blot analysis of one, three, and five head extracts (1 h, 3 h, and 5 h) from w1118 and dcerk1 with DCERK antibody (3H7) reveals less than 2% DCERK protein in the mutant lanes. The blot is probed with an antibody to inositol polyphosphate 1–phosphatase (IPP) as a loading control. (C) RT-PCR analysis shows significant reduction in DCERK transcript in dcerk1. Reactions with actin serve as a control. Transmission electron micrographs (TEM) showing photoreceptors of 5-day-old (D) w1118 (E) dcerk1 with extensive degeneration, (F) DCERK transgene in dcerk1 showing degeneration is rescued, and (G) dark-raised dcerk1 showing no significant degeneration. Bars, 2 μm.
dcerk1 Mutant Undergoes Light-Dependent Photoreceptor Degeneration.
To understand how CERK regulates ceramide metabolism in vivo and, in particular, to analyze whether it regulates photoreceptor structure and function, we generated mutants in DCERK. Two P element lines (CG16708BG01100 and P{EPgy2}EY07850) inserted in the DCERK gene were identified in Fly Base. Because both lines expressed significant amounts of DCERK protein, we excised the P element CG16708BG01100 (inserted in the first intron) using standard techniques. The genetic scheme used to mobilize it is described in Fig. S4A. A total of 640 independent excision lines were established of which 57 were lethal. The lethal lines were analyzed by transgenic rescue using a wild-type copy of DCERK, whereas the viable lines were analyzed by western analysis for decrease or lack of DCERK protein. This led to the isolation of dcerk1 mutant, and western analysis of dcerk1 fly head extracts showed less than 2% intact protein compared with control level (Fig. 1B). Reverse transcription–polymerase chain reaction (RT-PCR) analysis of dcerk1 showed a significant reduction in the DCERK transcript level (Fig. 1C). PCR analysis suggested that ≈6 kb of the original P element left behind affected the DCERK transcript level (Fig. S4B). The above data show that dcerk1 is a severe hypomorphic mutant of DCERK. dcerk1 Flies are semilethal, and show fertility defects. All of the phenotypes were rescued by the introduction of a DCERK transgene into dcerk1.
Because our earlier published results indicated that ceramide is an important regulator of photoreceptor viability, we analyzed photoreceptors of dcerk1 for possible defects. As seen in Fig. 1E, dcerk1 showed severe photoreceptor degeneration, cells appeared vacuolated with almost no intact rhabdomeres. A time course depicting progressive degeneration of the photoreceptors is shown in Fig. S4C. The degenerative phenotype was rescued by introducing a transgene expressing DCERK (Fig. 1F). Degeneration of dcerk1 depended on light activation of phototransduction, as photoreceptors of dark raised flies did not show morphological signs of degeneration (Fig. 1G).
dcerk1 Mutant Does Not Respond to Light, and Its PLC Level and Activity Are Severely Downregulated.
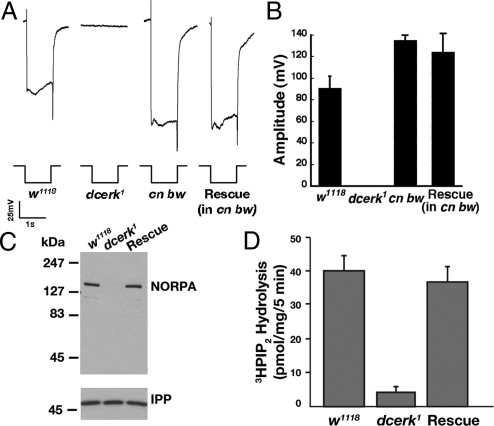
To test whether photoreceptor function was also defective in dcerk1, we performed electroretinogram recordings (ERGs) from control, mutant, and rescued flies raised in the dark. Surprisingly, despite their intact morphology, mutants did not respond to the light stimulus (Fig. 2A). However dcerk1 rescued with a DCERK transgene showed normal ERGs (Fig. 2 A and B). The lack of light response in dcerk1 retina suggested that CERK regulates a crucial step in phototransduction. Mutations in NORPA, the critical effector of phototransduction, led to defective light-induced electrical responses, with null mutants being blind (16–18). Because dcerk1 were blind, we checked the steady-state level of NORPA by immunoblotting using 1-day-old dcerk1 maintained in the dark. We observed that NORPA protein was absent in dcerk1 and that the expression of a DCERK transgene in the mutant rescued the NORPA level (Fig. 2C). In overexposed blots, a small amount of NORPA can be detected in 1-day-old flies (Fig. S5C). Real-time PCR analysis revealed that the amount of NORPA transcript was not different between w1118 and dcerk1, suggesting that DCERK primarily regulates NORPA posttranscriptionally (Fig. S5D). Although w1118 retinal extracts showed high PLC activity, mutant extracts lacked this activity, and it was restored in the rescued flies (Fig. 2D). Our results thus far show that the lack of DCERK downregulates NORPA and that this results in loss of its activity and function in photoreceptor cells.
Fig. 2.
Severe downregulation of NORPA expression in dcerk1 flies renders them unresponsive to light. (A) ERG recordings from dark-raised w1118, dcerk1, and rescued flies. The dcerk1 do not respond to light, whereas rescued flies exhibit robust response. Cn bw flies were also used as control, as the rescued flies were in cn bw background. (B) Quantitative analysis of ERG recordings shows that the amplitude in the rescued flies is comparable to that in control flies. (C) Western blot of head extracts probed with NORPA antibody shows that its expression is severely reduced in dcerk1 and rescued in dcerk1 expressing DCERK transgene. (D) Retinal extracts of dcerk1 do not show PLC activity, whereas activity of rescued flies is comparable to that of w1118. Error bars denote standard deviation (n = 3).
Localization and Levels of Other Phototransduction Components Are Not Affected in dcerk1 Mutant.
In Drosophila photoreceptors, INAD, a scaffolding protein, organizes signaling components including NORPA into a supramolecular complex to maximize the efficiency of phototransduction (19, 20). We tested whether DCERK specifically influenced NORPA or if it could affect the concentration or localization of other signaling components by western and immunofluorescence analyses. As seen in Fig. S6 A and B, no significant difference was observed between control and dcerk1 for any of the proteins tested in 1-day-old flies. Immunostaining with a NORPA antibody revealed that its level was too low to be detected (Fig. S6B). By day 14, dark-raised dcerk1 showed loss of TRP (Fig. S6C). Fig. S6D shows immunostaining of eyes with antibodies to DCERK and Rh1 in w1118 and dcerk1. In these sections, CERK is present in rhabdomeres and other membranous compartments of photoreceptors, overlying cone, and lens cells, whereas Rh1 shows specific rhabdomere staining of R1–R6 cells. That CERK can localize to rhabdomeres is also seen in thin sections of photoreceptors co-stained with antibodies to DCERK and INAD (Fig. S7A).
DCERK Regulates NORPA and Photoreceptor Function by Modulating the Ceramide Level.
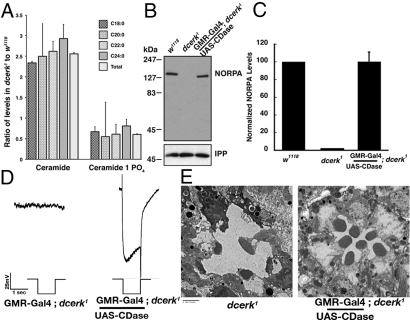
To test whether the substrate, ceramide, or the product C-1-P was responsible for regulating NORPA, we measured their levels in lipid extracts from control and dcerk1 by UPLC-MS/MS. The mutant flies showed a 250–300% increase in ceramide levels in all of the species measured and a modest decrease (40%) in C-1-P levels when compared with w1118 (Fig. 3A). The levels of other sphingolipids such as ceramide phosphoethanolamine and glucosyl ceramide did not change significantly (<30%) between w1118 and dcerk1 extracts. It is likely that other pathways or proteins could contribute to the synthesis of C-1-P, as it is decreased by only 40% in the mutant. To determine whether the significant increase in ceramide level in dcerk1 was crucial for downregulating NORPA, we overexpressed CDase in dcerk1. CDase overexpression decreases the ceramide level by catalyzing the hydrolysis of ceramide to sphingosine and a fatty acid (10). Targeted overexpression of CDase in dcerk1 photoreceptors rescued NORPA, suggesting that an increase in ceramide contributed to a decrease in NORPA level (Fig. 3 B and C). Lipid extracts from dcerk flies expressing CDase indeed showed a decrease in ceramide in all species measured except C-20 (Fig. S7B). ERGs carried out on dcerk1 expressing CDase showed that overexpression of CDase rescues the lack of light response observed in dcerk1 (Fig. 3D). CDase expression also rescued the retinal degeneration in dcerk1 (Fig. 3E).
Fig. 3.
dcerk mutant accumulates ceramide, and reducing the ceramide level restores NORPA expression and function. (A) Total lipids extracted from dcerk1 and w1118 flies are subjected to UPLC-MS/MS. Individual ceramide and ceramide 1–phosphate species (with tetradecasphinganine backbone to which 18–, 20–, 22–, or 24–carbon chain fatty acids are attached) are measured and values are calculated for dcerk1 relative to w1118. (B) Expression of ceramidase (CDase) transgene rescues NORPA expression in dcerk1. (C) Quantification of NORPA shows that the level is comparable in the CDase dcerk1 rescued flies compared with the control. (D) ERGs showing CDase transgene rescues light response in dcerk1. (E) CDase overexpression rescues retinal degeneration in dcerk1.
Ceramide Levels Regulate the Localization and Level of PIP2.
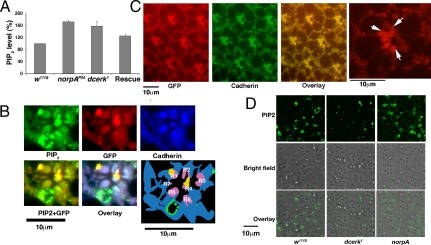
Because PLC acts on PIP2, its localization and function is modulated by the lipid bilayer (21). We evaluated whether an increase in ceramide could alter global membrane properties such as bilayer fluidity. We measured fluorescence polarization as a measure of membrane fluidity, and these experiments showed that membrane fluidity did not change significantly in dcerk1 (22, Fig. S7C). As bulk membrane properties like fluidity were not affected, we decided to test whether the membrane microenvironment was perturbed in dcerk1 due to an increase in ceramide concentration. Because conventional procedures that involve 3Hinositol labeling followed by HPLC separation of radiolabeled phosphoinositides could not be adapted to flies, we measured the PIP2 level in control and dcerk1 by standardizing a coupled nonradioactive TLC separation of phosphoinositides followed by an enzyme-linked immunosorbent assay (ELISA) using a monoclonal antibody that recognizes PIP2 with high specificity (23). dcerk1 showed a 60% increase in the PIP2 level compared with the control (Fig. 4A). NORPA null mutants lacking the major PLC used as a control showed an accumulation of PIP2. We then analyzed whether increased ceramide could affect the distribution of PIP2 using the PIP2 antibody in thin sections of control and dcerk1 mutant photoreceptor cells. As seen in Fig. 4B (labeled PIP2), PIP2 appeared to be enriched in doughnut-shaped clusters and spots at the rhabdomeres and their base. We then confirmed that these clusters recruit PLC by expressing PLCδ1PH-GFP in photoreceptors and staining thin sections of photoreceptors with a GFP antibody. In this widely used tool to visualize the distribution of PIP2, the PH domain of PLCδ1, which specifically binds to PIP2 at the plasma membrane, is fused to GFP (24). As seen in Fig. 4B (labeled GFP), in addition to rhabdomere staining, clusters were visible at the membrane. The plasma membrane was stained with cadherin in these sections (labeled Cadherin). Fig. 4B (labeled PIP2+GFP) shows the overlay of PIP2 and GFP, suggesting that PLCδ can co-localize to PIP2 enriched clusters at the membrane. To test whether clustering is unique to PIP2, we also expressed the PH domain of general receptor for phosphoinositides-1 (GRP1) fused to GFP which binds specifically to phosphatidylinositol-3,4,5 -P3(PIP3). As seen in Fig. 4C, punctate dots of PIP3 staining can be seen at the rhabdomeres and their base but big clusters seen with PIP2 are absent here. To analyze the distribution of PIP2 in dcerk1, thin sections of photoreceptors were stained with PIP2 antibody. Unlike w1118, PIP2 was not clustered but fragmented in dcerk1 and staining was diffuse (Fig. 4D and Fig. S7D). The diffuse staining in the mutant is better appreciated in experiments using quantum dot conjugated secondary antibody (Fig. S8A). norpA null mutant photoreceptors stained with PIP2 antibody showed PIP2 clusters (Fig. 4D and Fig. S8A). The ceramide level in norpA mutant would be unaltered and the microenvironment likely to be unperturbed. Morphometric analyses showed there were six to eight PIP2 clusters in w1118 and norpA null mutant ommatidium, whereas there were two to three in dcerk1 (Fig. S7E). Our results suggest an increase in the ceramide level disrupts the organization of PIP2-enriched areas at the plasma membrane.
Fig. 4.
Increased ceramide level alters PIP2 membrane microenvironment in dcerk1. (A) The PIP2 level is elevated in dcerk1 compared with w1118. (B) Immunostaining of photoreceptors expressing PLCδPH GFP in w1118 cn bw background with PIP2, GFP and cadherin antibodies. PIP2 clustering can be seen at the membranes, and the overlay shows that PLCδPH GFP localizes to these clusters. A cartoon of the staining shows PIP2 in green, GFP in red, and overlay in yellow. Purple rhabdomeres are from overlay of cadherin in blue and GFP in red. (C) Immunostaining of photoreceptors expressing GRP1PH GFP in w1118 cn bw background with GFP and cadherin antibodies. PIP3 staining, unlike PIP2, is not in large clusters. Last panel is a magnified view of one ommatidium; arrows show small dots of PIP3 in the rhabdomere vicinity. (D) PIP2 is disorganized in dcerk1. Photoreceptors of w1118, dcerk1 and norp A null are immunostained with PIP2 antibody. The w1118 and norp A null photoreceptors show PIP2 staining in clusters, which are less in dcerk1.
NORPA That Is Not Membrane Associated Is Not Functional and Is Targeted for Degradation.
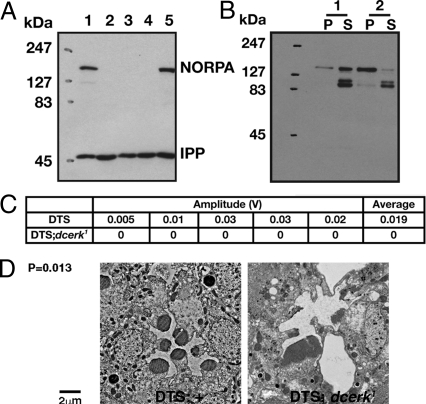
As the interaction of PLC with its substrate PIP2 requires membrane association, and as PIP2 organization is affected in dcerk1, we examined the membrane association of NORPA in dcerk1. We tested whether reducing ubiquitin-mediated proteosomal degradation in vivo would allow us to detect NORPA in dcerk1. We used a dominant temperature-sensitive (DTS) mutation, DTS5, that affects the β6 proteosomal subunit (25). If NORPA is targeted for degradation in dcerk1, then expressing DTS5 in dcerk1 photoreceptors should result in restoration of NORPA. As seen in Fig. 5A, this was indeed the case; quantitative analysis of NORPA levels is shown in Fig. S8B. We fractionated head extracts from control and dcerk1 expressing the DTS5 subunit into pellet and supernatant fractions and looked for NORPA by western analysis. These studies showed that in control almost all of the NORPA fractionates with the pellet underscoring its membrane localization, whereas most of the NORPA in dcerk expressing DTS5 was in the soluble fraction and hence cytosolic. To test whether this NORPA was functional, ERGs were carried out on dcerk1 expressing DTS5. Whereas DTS5 flies showed a light response, DTS5 flies in dcerk1 showed no response (Fig. 5C). Photoreceptor degeneration in dcerk1 could also not be rescued in these flies (Fig. 5D). Taken together, a likely explanation for our results is that NORPA that is not membrane associated because of the disorganization of PIP2 is unstable and is targeted for degradation.
Fig. 5.
NORPA is unable to bind efficiently to membranes in dcerk1 and is degraded. (A) NORPA expression is detected in western analysis of dcerk1 expressing DTS5. Lanes 1: w1118; lane 2: dcerk1; lane 3: UAS-DTS5;dcerk1; lane 4: GMR-Gal4;dcerk1; and lane 5: GMR-Gal4/UAS-DTS5;dcerk1. (B) NORPA is unable to bind to membranes in dcerk1. Fractionation experiments on head extracts from dcerk1 expressing DTS5 subunit in photoreceptors (1) and control DTS5 subunit (2) show that a significant fraction of NORPA is soluble in dcerk1 expressing DTS5, whereas it is in the pellet fraction in the control extract. (C) Amplitude values from ERGs show that DTS5 flies in dcerk1 show no light response, indicating NORPA is not functional. (D) Ultrastructure of photoreceptors expressing DTS5 in dcerk1 still degenerate.
Ceramide Affects PIP2 Clustering and Its Partitioning into Liquid Ordered Membrane Domains.
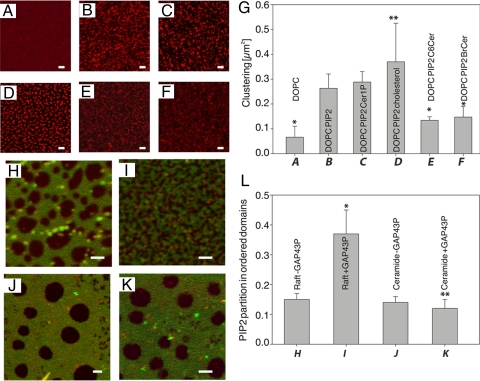
Cellular membranes are not homogeneous but compartmentalized into liquid ordered (Lo), liquid disordered (Ld), and gel (Lβ) phases. In the Ld phase, lipids diffuse freely, while in the Lβ phase they are immobile and the Lo phase is an intermediate stage. The existence of PIP2 compartmentalization in vivo has been rationalized by two hypotheses: (i) PIP2 molecules can cluster independent of proteins or other lipids, for example, through hydrogen bonding between their head groups; or (ii) PIP2 can be laterally sequestered by certain proteins such as MARCKS, GAP43 into cholesterol-rich membrane domains (26). We explored each of these scenarios in model membranes with defined components to understand how ceramide accumulation could influence PIP2. To address protein-independent clustering, we prepared Ld supported bilayers made of dioleoylphosphatidylcholine (DOPC), 3% molar brain PIP2, and a trace amount of fluorescent PIP2 (BodTMRPIP2) for visualization. Under these conditions, BodTMRPIP2 forms clusters that can be visualized using fluorescence laser scanning microscopy (Fig. 6). Fluorescence correlation spectroscopy was used to show that membrane fluidity is not affected by the presence of the clusters (Fig. S8C). It was recently shown that in free-standing membranes, PIP2 forms similar clusters that depend on the presence of divalent cations such as Ca2+ (27). Next, we evaluated the effects of incorporating ceramides in these preparations on the clustering and distribution of PIP2, and image correlation spectroscopic (ICS) analysis was performed to quantify the degree of PIP2 clustering (Fig. 6G). The spatial segregation of BodTMRPIP2 depends on the presence of brain PIP2 in the lipid mixture, as simple DOPC samples show no significant heterogeneities (Fig. 6A). The incorporation of either brain ceramide or short-chain C6 ceramide significantly decreases cluster formation (Fig. 6 E and F). C-1-P does not prevent PIP2 clustering, whereas cholesterol seems to promote it (Fig. 6 C and D). From these experiments, it appears that the presence of ceramide has a direct effect on the clustering of PIP2 in model membranes. A possible explanation for this phenomenon lies in the peculiar biophysical properties of the ceramide molecule. Ceramide has a small polar head (like cholesterol), and it is both donor and acceptor for hydrogen bonding (28). The former feature might explain why ceramide and cholesterol interact with PIP2: the energetically unfavorable contact between water and their hydrophobic moieties are avoided by the large PIP2 polar head (“umbrella effect”) (29). Nevertheless, the distinctive structural properties of ceramide (e.g., the amidic group that is absent in cholesterol and phospholipids) could influence the hydrogen bond network of surrounding molecules. Ceramide might act as a local chaotrope that perturbs the PIP2–water–hydrogen bond network, thus allowing the electrostatic repulsion between PIP2 molecules to disperse the clusters (30).
Fig. 6.
Ceramide affects protein-independent PIP2 clustering and GAP-43P–induced PIP2 partition in ordered domains. Fluorescence images of BodTMRPIP2 (0.005% mol) in supported bilayers containing (A) DOPC, (B) DOPC + 3% BrPIP2, (C) B + 5% Cer-1-P, (D) B + 5% cholesterol, (E) B + 5% C6 ceramide, and (F) B + 5% brain ceramide at room temperature. The presence of ceramide affects clustering of PIP2. (G) Degree of clustering of PIP2 for all samples described above. This quantity is defined as inverse of the cluster density, as calculated via ICS on an average of eight 23 × 23-μm2 images for each sample. *E and F are statistically distinguishable from B with P < 0.01. **D is statistically distinguishable from B with P < 0.1. (H) Fluorescence image of a supported bilayer showing Lo domains in Ld phase. Red color refers to BodTMRPIP2 and green to BodChol (0.01% mol). The Lo domains appear dark in both channels. (I) When the bilayer is treated with palmitoylated GAP-43P, the Lo domains are fragmented and show a significant degree of fluorescence in the red channel. (J and K) Analogous imaging of bilayers with ceramide-rich domains in Ld phase, without and with GAP-43P respectively. Ceramide-rich domains are dark in both channels. (L) Partition of BodTMRPIP2 in Lo or ceramide-rich domains. This is calculated as the ratio between the average red signal in Lo (or ceramide) domains—dark in the green channel and the signal in the Ld phase—bright in the green channel. *I is statistically distinguishable from H with P < 0.01 and **K is statistically indistinguishable from sample J with P = 0.3. (Scale bars, 2 μm.)
Protein/lipid dependent PIP2 clusters (scenario 2): A second line of thought regarding PIP2 compartmentalization is that certain proteins, for example, the diacylated protein GAP-43, can bind PIP2 and partition into small raft domains, thus offering a molecular basis for PIP2 segregation (31). In this context, ceramide might play an important role inducing structural alterations in the lateral organization of the membrane bilayer. Nonphosphorylated long-chain ceramides can form specific membrane microdomains that can exist in gel phase (Lβ) within raft domains of Lo phase (32). Such ceramide-rich domains are characterized by very tight lipid packing. To test whether such lateral segregation properties of ceramide within membrane bilayers could explain the loss of PIP2 aggregation seen in vivo in CERK mutants, we examined the effect of increasing ceramide on PIP2 organization in supported bilayer showing raft-like phase separation. BodTMRPIP2 was added to the lipid mixture in trace amounts to study the organization of PIP2, whereas a green fluorescent lipid analog (BodChol) was used as a marker for the Ld phase. In these preparations, PIP2 localizes to the Ld phase of the membranes (Fig. 6H) because of the multiple unsaturation in the sn-2 hydrocarbon chain that would make PIP2 partitioning into tightly packed domains energetically unfavorable. In this set of experiments, we monitored PIP2 partition into ordered domains, either raft-like or ceramide-rich, as this could be a more relevant mechanism for PIP2 segregation in vivo, and we did not focus on the Ca2+ dependent PIP2 clustering only partially observable in the Ld phase because of washing with 15 mM ethylenediaminetetraacetic acid (EDTA). Following a published protocol (31), we verified that the addition of doubly palmitoylated peptide GAP-43P (i.e., the N terminus plus the basic effector domains of GAP-43) relocates a portion of the PIP2 to the Lo (raft-like) phase of the membrane (Fig. 6I). Substitution of ≈50% of sphingomyelin with ceramide results in bilayers containing ceramide-rich domains in a matrix of Ld phase. Here again, PIP2 is localized to the Ld phase (Fig. 6J). Interestingly, the addition of the acylated GAP-43P showed no significant change in the dimension of the ordered domains (Fig. 6K). Furthermore, very little of the PIP2 relocates to these ceramide rich domains. Figure 6L shows quantification of the fluorescent intensity originating from PIP2 in the ordered domains compared with that from PIP2 in the disordered phase. These experiments suggest where PIP2 clustering in vivo is determined by the partition of this lipid into small protein/lipid domains, the accumulation of ceramide, and the formation of highly ordered, ceramide-rich domains could effectively decrease PIP2 segregation. The conclusions drawn from fluorescence experiments described above correlate well with the observed effects of increased ceramide in DCERK mutants on PIP2 organization in photoreceptor cells.
Discussion
In this study, we show that DCERK regulates the ceramide level to maintain PLC and membrane organization of PIP2 during phosphoinositide-mediated GPCR signaling in Drosophila. Our results are summarized in a model depicted in Fig. S9. Morphometric analysis of PIP2 clusters suggests that there are still some clusters in dcerk1; thus, the loss of NORPA in dcerk mutant may not be due only to the loss of PIP2 clustering. Although the effect of ceramide on PIP2 is central, increased ceramide could affect other membrane properties that can downregulate NORPA or other proteins. Also, INAD is required for localization of NORPA and TRP is required for localization of INAD (20). Because TRP protein is affected over time in dcerk1, additional interactions mediated by INAD and TRP could also contribute to NORPA stability. Photoreceptor degeneration seen in cerk mutants is also not simply caused by loss of NORPA protein, as it is more severe and sets in earlier than in norpA null mutants. There could be other effects of ceramide contributing to degeneration.
Recent clinical studies have identified mutations in the human ceramide kinase like (CERKL) gene in patients with autosomal recessive retinitis pigmentosa (33). No CERKL homolog has been identified in the Drosophila genome. Interestingly, DCERK shares 31% identity with human CERKL (Fig. S1), and possibly DCERK could perform some CERKL functions also. It would be worthwhile to test whether CERK regulates PLCβ4, the closest homolog of NORPA among mammalian PLCs, thereby participating in the mammalian visual process. Recent analyses of cerk−/− mice revealed that CERK could function in cerebellar Purkinje cells (which are also enriched in PLCβ4) and in neutrophil homeostasis (34, 35). Because the mechanism by which mammalian CERK regulates these processes is not known, it would be interesting to test whether these functions are also mediated through PLC. Although our current understanding limits a direct co-relationship between mammalian CERK and PLCs, it is likely that similar ceramide-regulated microenvironments could operate in other phosphoinositide-dependent signaling such as insulin signaling in adipocytes.
In summary, our data show that modulation of the ceramide level by CERK regulates PIP2 and PLCβ function in Drosophila. Because PIP2 and PLC are fundamental components of GPCR signaling, uncovering their regulation by ceramide through CERK should lead to a better understanding of lipid regulation in signaling.
Methods
Genetic Screen and Isolation of dcerk Mutants.
The genetic scheme and method to generate jump-out lines from DCERK P-element are outlined in SI Materials and Methods. Five hypomorphic mutants including dcerk1 were obtained from the screen.
Methods for electron microscopy and immunohistochemistry, assay for ceramide kinase activity, estimation of sphingolipids by mass spectrometry and PIP2 by ELISA, and model membrane experiments are described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank the Bloomington Stock Center, Indiana, Drs. Andy Zelhof, and Eric Baerahcke for fly stocks. We thank Dr. Charles Zuker and Ann Becker and Drs. Craig Montell, Hong-sheng Li, and John Sisson for their gifts of antibodies. We thank Drs. Mike Czech and David Lambright for helpful discussions. We thank Kathya Acharya for her work on the figures. Mr. Jun Yonekubo (Nihon Waters K.K) is gratefully acknowledged for helpful discussions and experimental support. This work is supported by National Institutes of Health grant R01EY16469 (to U.A.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911028106/DCSupplemental.
References
- 1.Marinissen MJ, Gutkind JS. G-protein-coupled receptors and signaling networks: Emerging paradigms. Trends Pharmacol Sci. 2001;22:368–376. doi: 10.1016/s0165-6147(00)01678-3. [DOI] [PubMed] [Google Scholar]
- 2.Escriba PV, Wedegaertner PB, Goni FM, Vogler O. Lipid-protein interactions in GPCR-associated signaling. Biochim Biophys Acta. 2007;1768:836–852. doi: 10.1016/j.bbamem.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Montell C. Visual transduction in Drosophila. Annu Rev Cell Dev Biol. 1999;15:231–268. doi: 10.1146/annurev.cellbio.15.1.231. [DOI] [PubMed] [Google Scholar]
- 4.Chyb S, Raghu P, Hardie RC. Polyunsaturated fatty acids activate the Drosophila light-sensitive channels TRP and TRPL. Nature. 1999;397:255–259. doi: 10.1038/16703. [DOI] [PubMed] [Google Scholar]
- 5.LaLonde MM, et al. Regulation of phototransduction responsiveness and retinal degeneration by a phospholipase D-generated signaling lipid. J Cell Biol. 2005;169:471–479. doi: 10.1083/jcb.200502122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang FD, Matthies HJ, Speese SD, Smith MA, Broadie K. Rolling blackout, a newly identified PIP2-DAG pathway lipase required for Drosophila phototransduction. Nat Neurosci. 2004;7:1070–1078. doi: 10.1038/nn1313. [DOI] [PubMed] [Google Scholar]
- 7.Garcia-Murillas I, et al. lazaro encodes a lipid phosphate phosphohydrolase that regulates phosphatidylinositol turnover during Drosophila phototransduction. Neuron. 2006;49:533–546. doi: 10.1016/j.neuron.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Hannun YA, Obeid LM. Principles of bioactive lipid signalling: Lessons from sphingolipids. Nat Rev Mol Cell Biol. 2008;9:139–150. doi: 10.1038/nrm2329. [DOI] [PubMed] [Google Scholar]
- 9.Acharya U, Acharya JK. Enzymes of sphingolipid metabolism in Drosophila melanogaster. Cell Mol Life Sci. 2005;62:128–142. doi: 10.1007/s00018-004-4254-1. [DOI] [PubMed] [Google Scholar]
- 10.Acharya U, et al. Modulating sphingolipid biosynthetic pathway rescues photoreceptor degeneration. Science. 2003;299:1740–1743. doi: 10.1126/science.1080549. [DOI] [PubMed] [Google Scholar]
- 11.Acharya U, Mowen MB, Nagashima K, Acharya JK. Ceramidase expression facilitates membrane turnover and endocytosis of rhodopsin in photoreceptors. Proc Natl Acad Sci USA. 2004;101:1922–1926. doi: 10.1073/pnas.0308693100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Acharya JK, et al. Cell-nonautonomous function of ceramidase in photoreceptor homeostasis. Neuron. 2008;57:69–79. doi: 10.1016/j.neuron.2007.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugiura M, et al. Ceramide kinase, a novel lipid kinase. Molecular cloning and functional characterization. J Biol Chem. 2002;277:23294–23300. doi: 10.1074/jbc.M201535200. [DOI] [PubMed] [Google Scholar]
- 14.Wijesinghe DS, Lamour NF, Gomez-Munoz A, Chalfant CE. Ceramide kinase and ceramide-1-phosphate. Methods Enzymol. 2007;434:265–292. doi: 10.1016/S0076-6879(07)34015-9. [DOI] [PubMed] [Google Scholar]
- 15.Munagala N, et al. Identification of small molecule ceramide kinase inhibitors using a homogeneous chemiluminescence high throughput assay. Assay Drug Dev Technol. 2007;5:65–73. doi: 10.1089/adt.2006.046. [DOI] [PubMed] [Google Scholar]
- 16.Bloomquist BT, et al. Isolation of a putative phospholipase C gene of Drosophila, norpA, and its role in phototransduction. Cell. 1988;54:723–733. doi: 10.1016/s0092-8674(88)80017-5. [DOI] [PubMed] [Google Scholar]
- 17.Pearn MT, Randall LL, Shortridge RD, Burg MG, Pak WL. Molecular, biochemical, and electrophysiological characterization of Drosophila norpA mutants. J Biol Chem. 1996;271:4937–4945. doi: 10.1074/jbc.271.9.4937. [DOI] [PubMed] [Google Scholar]
- 18.Zhu L, McKay RR, Shortridge RD. Tissue-specific expression of phospholipase C encoded by the norpA gene of Drosophila melanogaster. J Biol Chem. 1993;268:15994–16001. [PubMed] [Google Scholar]
- 19.Mishra P, et al. Dynamic scaffolding in a G protein-coupled signaling system. Cell. 2007;131:80–92. doi: 10.1016/j.cell.2007.07.037. [DOI] [PubMed] [Google Scholar]
- 20.Tsunoda S, Zuker CS. The organization of INAD-signaling complexes by a multivalent PDZ domain protein in Drosophila photoreceptor cells ensures sensitivity and speed of signaling. Cell Calcium. 1999;26:165–171. doi: 10.1054/ceca.1999.0070. [DOI] [PubMed] [Google Scholar]
- 21.Rebecchi MJ, Pentyala SN. Structure, function, and control of phosphoinositide-specific phospholipase C. Physiol Rev. 2000;80:1291–1335. doi: 10.1152/physrev.2000.80.4.1291. [DOI] [PubMed] [Google Scholar]
- 22.Rao RP, et al. Ceramide transfer protein function is essential for normal oxidative stress response and lifespan. Proc Natl Acad Sci USA. 2007;104:11364–11369. doi: 10.1073/pnas.0705049104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laux T, et al. GAP43, MARCKS, and CAP23 modulate PI(4,5)P(2) at plasmalemmal rafts, and regulate cell cortex actin dynamics through a common mechanism. J Cell Biol. 2000;149:1455–1472. doi: 10.1083/jcb.149.7.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varnai P, Balla T. Visualization of phosphoinositides that bind pleckstrin homology domains: Calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol. 1998;143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belote JM, Fortier E. Targeted expression of dominant negative proteasome mutants in Drosophila melanogaster. Genesis. 2002;34:80–82. doi: 10.1002/gene.10131. [DOI] [PubMed] [Google Scholar]
- 26.Epand RM. Proteins and cholesterol-rich domains. Biochim Biophys Acta. 2008;1778:1576–1582. doi: 10.1016/j.bbamem.2008.03.016. [DOI] [PubMed] [Google Scholar]
- 27.Carvalho K, Ramos L, Roy C, Picart C. Giant unilamellar vesicles containing phosphatidylinositol(4,5)bisphosphate: Characterization and functionality. Biophys J. 2008;95:4348–4360. doi: 10.1529/biophysj.107.126912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cremesti AE, Goni FM, Kolesnick R. Role of sphingomyelinase and ceramide in modulating rafts: Do biophysical properties determine biologic outcome? FEBS Lett. 2002;531:47–53. doi: 10.1016/s0014-5793(02)03489-0. [DOI] [PubMed] [Google Scholar]
- 29.Huang J, Feigenson GW. A microscopic interaction model of maximum solubility of cholesterol in lipid bilayers. Biophys J. 1999;76:2142–2157. doi: 10.1016/S0006-3495(99)77369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levental I, Janmey PA, Cebers A. Electrostatic contribution to the surface pressure of charged monolayers containing polyphosphoinositides. Biophys J. 2008;95:1199–1205. doi: 10.1529/biophysj.107.126615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong J, et al. Role of GAP-43 in sequestering phosphatidylinositol 4,5-bisphosphate to Raft bilayers. Biophys J. 2008;94:125–133. doi: 10.1529/biophysj.107.110536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiantia S, Kahya N, Ries J, Schwille P. Effects of ceramide on liquid-ordered domains investigated by simultaneous AFM and FCS. Biophys J. 2006;90:4500–4508. doi: 10.1529/biophysj.106.081026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tuson M, Marfany G, Gonzalez-Duarte R. Mutation of CERKL, a novel human ceramide kinase gene, causes autosomal recessive retinitis pigmentosa (RP26) Am J Hum Genet. 2004;74:128–138. doi: 10.1086/381055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graf C, et al. Neutropenia with impaired immune response to Streptococcus pneumoniae in ceramide kinase-deficient mice. J Immunol. 2008;180:3457–3466. doi: 10.4049/jimmunol.180.5.3457. [DOI] [PubMed] [Google Scholar]
- 35.Mitsutake S, et al. The generation and behavioral analysis of ceramide kinase-null mice, indicating a function in cerebellar Purkinje cells. Biochem Biophys Res Commun. 2007;363:519–524. doi: 10.1016/j.bbrc.2007.09.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.