Abstract
Seed development in plants involves the coordinated growth of the embryo, endosperm, and maternal tissue. Several genes have been identified that influence seed size by acting maternally, such as AUXIN RESPONSE FACTOR2, APETALA2, and DA1. However, given the lack of gain-of-function effects of these genes on seed size, it is unclear whether their activity levels are limiting in WT plants and whether they could thus be used to regulate seed size in development or evolution. Also, whether the altered seed sizes reflect local gene activity or global physiological changes is unknown. Here, we demonstrate that the cytochrome P450 KLUH (KLU) regulates seed size. KLU acts locally in developing flowers to promote seed growth, and its activity level is limiting for seed growth in WT. KLU is expressed in the inner integument of developing ovules, where it non-cell autonomously stimulates cell proliferation, thus determining the growth potential of the seed coat and seed. A KLU-induced increase in seed size leads to larger seedlings and higher relative oil content of the seeds. Genetic analyses indicate that KLU acts independently of other tested maternal factors that influence integument cell proliferation. Thus, the level of KLU-dependent growth factor signaling determines size in ovules and seeds, suggesting this pathway as a target for crop improvement.
Keywords: Arabidopsis, clonal analysis, cytochrome P450, seed growth
Seed size in higher plants is an important trait with respect to ecology and agriculture (1). For example, larger seeds are less easily dispersed, but offer the germinating seedling a larger supply of nutrients, thus increasing its competitiveness during seedling establishment and tolerance to adverse environmental conditions. At the same time, limited resources in the mother plant generally cause a tradeoff between the number and size of the seeds produced (2). As for agriculture, increasing seed size has been a crucial contributor to the yield increases in crop plants during domestication (3).
Seeds are formed by the coordinated growth of maternal sporophytic and zygotic tissues (4). The zygotic tissues are the result of double fertilization, with one sperm cell fertilizing the diploid central cell to yield the triploid endosperm and the other sperm cell fertilizing the haploid egg cell to give rise to the diploid embryo. These maternal gametes lie within the embryo sac that develops in the nucellus region of the ovule (5). The nucellus is surrounded by the integuments, protective organs that form the maternal component of the mature seed after fertilization, the seed coat (6).
The size of seeds is known to be influenced by parent-of-origin effects, with a paternal genome excess causing seed overgrowth, whereas a maternal genome excess reduces seed size (7). In addition, recent genetic studies in the model species Arabidopsis thaliana and rice have identified a number of factors affecting seed size by acting in the maternal and/or zygotic tissues. Among the zygotically acting factors, a small cascade of genes comprising the HAIKU1, HAIKU2, and MINISEED3 loci promote endosperm growth in Arabidopsis (8). On the maternal side, several factors are required to increase or limit final seed size, and natural variation in the activity of such factors contributes to seed size differences in Arabidopsis accessions (9). The Arabidopsis WRKY transcription factor TRANSPARENT TESTA GLABRA2 (TTG2) is necessary to promote cell expansion in the integuments and allow for normal seed growth (10). By contrast, the Arabidopsis transcription factors APETALA2 (AP2) and AUXIN RESPONSE FACTOR2 (ARF2, also known as MEGAINTEGUMENTA) and the ubiquitin interaction motif-containing DA1 protein limit seed size (11–14). ARF2 and DA1 act by restricting cell proliferation in the integuments, which has led to the suggestion that the cell number, and thus the size of the seed coat, physically limits seed size (14). This is supported by the reduced seed size that results from decreased cell proliferation in integuments, when the cell cycle inhibitor KIP-RELATED PROTEIN2 is overexpressed or the activity of the DNA methyltransferase MET1 is reduced (10, 15). In rice, four unique proteins have been identified through quantitative genetic studies of grain size (3, 16–18). The protein of unknown function encoded by the qSW5 gene, the RING-finger E3 ubiquitin ligase encoded by GW2, the transmembrane protein encoded by GS3, and the nuclear polyubiquitin-binding protein encoded by GW5 are all required to limit final grain size and weight. For at least some of the allelic variants, changes are already detected in developing flowers, suggesting that the respective genes act in maternal tissues.
Despite this progress, two important questions about the maternal control of seed size remain unanswered. One question has to do with the fact that, in most cases, it is not clear to what extent the altered seed sizes in the mutants mentioned above reflect a local requirement for the respective genes in developing flowers or whether seed size changes result from altered physiology and resource status of the mother plant. A positive correlation between maternal resource status and seed size has been amply demonstrated (2), and this question is particularly pertinent in cases such as arf2 or da1 mutants that pleiotropically increase overall plant size, and thus likely alter photosynthetic capacity and other physiological parameters. The other question has to do with the fact that, to our knowledge, no opposite gain-of-function phenotypes on seed size have been described for any of the maternally acting genes mentioned above, making it difficult to judge their regulatory potential.
Here, we identify the cytochrome P450 KLUH (KLU)/CYP78A5 (encoded by locus At1g13710) as a maternal regulator of seed size. KLU has been shown previously to promote growth of leaves and floral organs and to prolong the plastochron (19, 20). Based on its non-cell autonomous mode of action, which does not seem to involve any of the known phytohormones, the protein was suggested to be involved in the generation of a unique mobile growth stimulator. We show here that KLU function is required locally in developing flowers and that its activity level is limiting for seed growth in a WT background. Our findings highlight the presumed growth-signaling pathway defined by KLU as a potential target for evolutionary modification of seed size as well as for crop improvement.
Results
KLU Is Expressed in the Inner Integument Throughout Ovule Development.
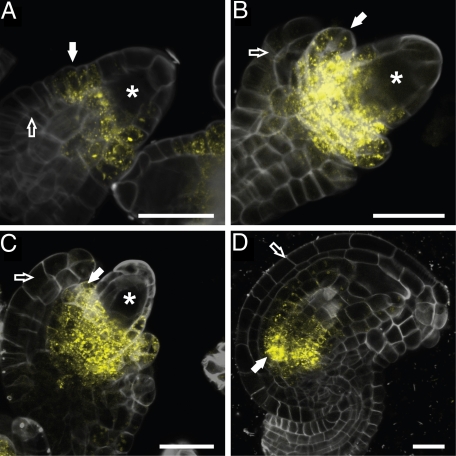
The KLU gene is expressed in a spatially restricted pattern in leaves and floral organs, where it plays an important role in promoting growth; in addition, KLU expression has been reported in ovules (19, 21). To determine the activity pattern of the KLU promoter during ovule development in more detail, we followed fluorescence in a pKLU::vYFPer reporter line. In this line, the expression of a cell-autonomous endoplasmic reticulum-localized version of VENUS-YFP is under the control of 4 kb of KLU upstream genomic sequence. YFP fluorescence was detected at the base of the nucellus in the region initiating the inner integument from as early as stage 2-II (Fig. 1A). The reporter gene continues to be active at the base of the nucellus and in the inner integument as the latter grows out (Fig. 1 B and C). Expression persists longest at the base of the inner integument in mature ovules (Fig. 1D). By contrast, no expression was detected in the outer integument at any stage of its growth. Thus, the KLU promoter, as assessed using the current reporter construct, is specifically active in the inner integument throughout ovule development.
Fig. 1.
Expression pattern of KLU in developing ovules. (A–D) Fluorescence of a vYFPer reporter protein expressed under the control of the KLU promoter during progressively later stages of ovule development. Reporter expression is detected at the base of the nucellus and in the inner integument (solid arrow), but is absent from the outer integument (open arrow). Asterisk indicates the megaspore mother cell (A and B) and developing embryo sac (C) within the nucellus. (A) Stage 2-II ovule. (B) Stage 2-III ovule. (C) Stage 2-V ovule. (D) Stage 3-VI ovule. Stages are in accordance with those described by Schneitz et al. (32). (Scale bars: 20 μm.)
KLU Is Required and Sufficient to Promote Seed Growth.
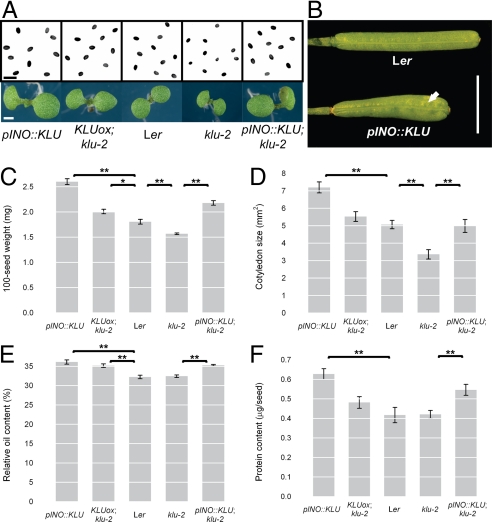
Given its role in controlling organ size in leaves and flowers (19, 20), we asked whether the expression of KLU in ovules is functionally relevant and regulates the size of the seeds originating from the ovules. To address this, we measured final seed size and weight of klu-2 loss-of-function mutants and plants overexpressing KLU in its endogenous expression pattern (KLUox, klu-2; see ref. 19 for details). Seeds from klu-2 mutants were 13% lighter and 16% smaller than seeds of WT plants, whereas KLU-overexpressing plants produced seeds that were 11% heavier and 10% larger than controls (Fig. 2 A and C and Fig. S1A). Thus, the KLU activity level is positively correlated with final seed size.
Fig. 2.
Effects of altered KLU activity in developing ovules. (A) Light micrographs of mature seeds (Upper) and 7-day old seedlings (Lower) of the genotypes indicated below. (B) Expression of KLU in ovules from the pINO::KLU construct (Bottom) leads to the formation of wider siliques with uneven growth of the silique walls (solid arrow) compared with the slender siliques of WT with their smooth surface (Top). (C–F) Quantification of seed and embryo characteristics in response to altered KLU activity. Ten plants per genotype were grown to maturity without any assisted pollination and harvested for measurements. The KLUox; klu-2 line has higher expression of KLU than WT in the endogenous pattern [same as klu-2 RLox2 in the article by Anastasiou et al. (19)], and is therefore most appropriately compared with Ler WT plants. Concerning KLU activity in ovules, the initial 4 genotypes represent a series of decreasing activity levels, whereas the fifth genotype represents an ovule-specific rescue of KLU function in an otherwise mutant background, and is therefore compared with nontransgenic klu-2 mutants. (C) Weight of 100 seeds. (D) Cotyledon area of 7-day-old seedlings. (E) Relative oil content of seeds as determined by NMR spectroscopy. (F) Absolute protein content per seed. Values shown are mean ± SEM. *Significantly different from control at P < 0.05, **significantly different from control at P < 0.01; both after Bonferroni correction. (Scale bars: A, 1 mm; B, 5 mm.)
KLU Acts Maternally to Promote Seed Growth.
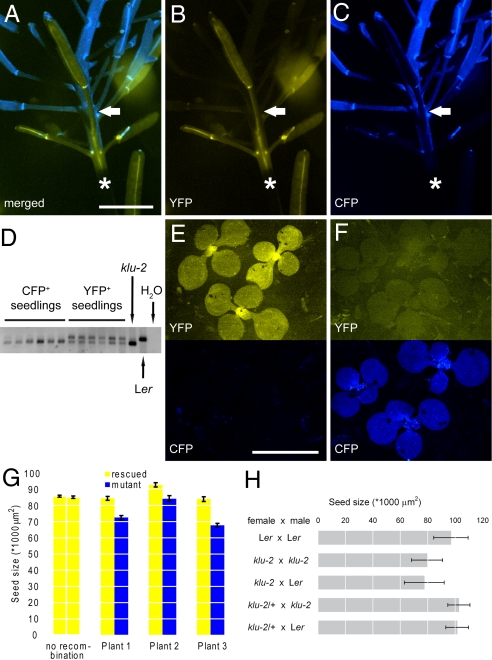
Expression of KLU has also been detected in developing embryos (21). Therefore, we asked whether KLU function is only required in maternal sporophytic tissue or whether KLU activity in the embryo also contributes to promoting seed growth. To answer this, we performed a set of reciprocal crosses (Fig. 3H). Mutations in KLU are fully recessive (19), allowing us to treat heterozygous plants/embryos as WT in the following. Pollinating klu-2 mutant plants with WT pollen leads to the development of WT embryos within a mutant seed coat. However, the size of the resulting seeds was not rescued and was indistinguishable from that of self-pollinated klu-2 mutants (Fig. 3H). To assess the reciprocal combination of genotypes, we pollinated klu-2/+ heterozygous plants with klu-2 mutant pollen. This gives rise to 50% of seeds with a homozygous mutant embryo developing within a phenotypically WT seed coat. The size of seeds from such crosses had the same mean and SD (as a measure of variability) as the size of seeds from the control cross (klu-2/+ × Ler WT) and from self-pollination of WT (Fig. 3H). This indicates that the embryo and endosperm genotype for KLU do not influence seed size. The indistinguishable size of seeds from the Ler × Ler and klu-2/+ × Ler crosses also formally rules out a female gametophytic requirement for KLU function. Thus, together, the results of our crosses indicate that KLU is solely required in the sporophytic tissue of the mother plant to promote seed growth.
Fig. 3.
KLU acts locally in the maternal tissue of developing flowers to promote seed growth. (A–C) Fluorescence micrographs of one of the three genetically grafted plants analyzed in G, showing the transition in the inflorescence from genotypically WT tissue marked by YFP fluorescence (yellow) to klu-2 mutant tissue marked by CFP expression (blue). The approximate point of transition is indicated by the solid arrow. The asterisk indicates a section of stem covered by the shadow of the silique at the bottom of the image. (A) Merged image. (B) YFP channel. (C) CFP channel. (D–F) Analysis of seedlings germinated from seeds that were harvested from WT (E) or mutant (F) siliques of the plant shown in A–C. (D) PCR analysis of seedlings shown in E and F to detect the presence of the WT KLU allele in the rescue construct. Whereas YFP-positive seedlings still contain the WT allele, it has been lost from the CFP-positive but YFP-negative seedlings. Amplifications on nontransgenic klu-2 mutant and Ler WT DNA are shown as controls. (E) Seedlings from YFP-positive WT silique. (F) Seedlings from CFP-positive klu-2 mutant silique. (E and F, Top) YFP channel. (E and F, Bottom) CFP channel. (G) Size of seeds from WT (yellow bars) and mutant (blue bars) siliques of three independent chimeras. The leftmost two bars show seed size from early (Left, flowers 1–6) and late (Right, flowers 20–25) siliques of a nonrecombined rescue plant for comparison. In all three recombined plants, seeds from mutant siliques were significantly smaller at P < 0.05 (two-tailed t test). (H) Size of seeds resulting from the indicated reciprocal crosses. Note that error bars in H show SD as a measure of variability in the seed populations. Values shown are mean ± SEM (G) and mean ± SD (H). (Scale bars: 5 mm.)
KLU Is Required Locally in Flowers to Promote Seed Growth.
As mentioned previously, any maternally acting gene could influence seed size by local effects in developing ovules or flowers or through more global changes in the plant's resource level. This issue is particularly relevant for genes with obvious effects on other aspects of plant growth or physiology. To answer this question for KLU, we compared the sizes of seeds produced by WT and mutant flowers developing on the same plant. To be able to do so, we generated plants with a genotypically split inflorescence, where the initial 10 to 15 flowers had WT KLU activity, whereas the flowers formed after this were klu mutant (Fig. 3 A–F). This was achieved by CRE/loxP-mediated excision of a rescue transgene in a homozygous klu-2 mutant background (Fig. S2 A and B). Temporal control over the excision event was afforded by expressing CRE under the control of the ethanol-inducible AlcR–AlcA system (22). Spatially, recombination was restricted to the stem cells of the shoot meristem by using the stem cell-specific CLAVATA3 (CLV3) promoter (pCLV3::AlcR–AlcA::CRE; Fig. S2A) (23). The presence of the rescue transgene in cells is indicated by expression of an adjacent 35S::vYFPer reporter, whereas after CRE/loxP-mediated excision, the YFP reporter is lost together with the rescue transgene and a 35S::CFPer reporter is generated instead (Fig. 3 A–F).
We grew the yellow-fluorescing rescued plants for 12 days before inducing CRE expression in the stem cells of the shoot meristem. This allowed for the formation of a phenotypically WT rosette and basal inflorescence, followed by the development of a blue-fluorescing upper part of the inflorescence, where the klu-2 mutation was uncovered (Fig. 3 A–C). Measuring the size of seeds produced by WT and klu-2 mutant flowers from three such chimeric plants indicated that seeds produced by mutant flowers were between 9 and 19% smaller than those from rescued flowers (Fig. 3G). This difference is not simply attributable to the position of the flowers on the inflorescence, because seed size in non-recombined plants was stable for at least the initial 25 siliques (Fig. 3G and Fig. S2C). Therefore, we conclude that the effects of KLU on seed size are not the result of global changes in plant resource status but, instead, that KLU function is required locally in developing flowers to ultimately promote seed growth.
KLU Acts Independently of Other Maternal Factors That Influence Integument Cell Proliferation.
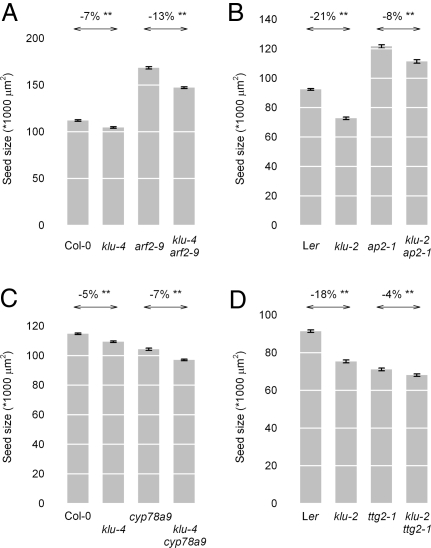
To determine whether KLU interacts genetically with previously identified maternal factors to regulate seed growth, we analyzed the size of double-mutant seeds compared with those from single mutants. For both ap2 and arf2, seeds from double mutants with klu had an intermediate size between those of the respective single mutants (Fig. 4 A and B), suggesting that KLU acts independently of AP2 and ARF2. Independent action of ARF2 and KLU was also supported by the additive phenotype of the double mutant in leaves and petals (Fig. S3 A–F).
Fig. 4.
Double-mutant analysis. (A–D) Seed sizes of the indicated single- and double-mutant combinations. Percentages above arrows indicate the reduction in seed size caused by eliminating KLU function in the respective backgrounds. (A) arf2–9 klu-4 double mutants. (B) ap2–1 klu-2 double mutants. (C) cyp78a9 klu-4 double mutants. (D) klu-2 ttg2–1 double mutants. **Difference indicated by the arrow is statistically significant at P < 0.01 (two-tailed t test).
The CYP78A9 gene encodes a cytochrome P450 that is closely related to KLU and is specifically expressed in the funiculus of developing ovules (24). Overexpression of CYP78A9 leads to enlarged and wider siliques, similar to the ones formed by the INNER NO OUTER promoter (pINO)::KLU-expressing plants (see below; Fig. 2B) (24), yet no loss-of-function phenotype has been described for cyp78a9 mutants. To test whether this gene functions in regulating seed size and may act redundantly with KLU, we analyzed seeds from cyp78a9 klu-4 double mutants relative to single-mutant seeds. Loss of CYP78A9 function indeed reduces seed size (Fig. 4C); however, we did not detect a genetic interaction between CYP78A9 and KLU, because double mutant seeds showed a purely additive phenotype.
The TTG2 gene acts maternally to promote cell expansion in integuments and growth of the endosperm (10). Seeds from klu-2 ttg2–1 double mutants were significantly smaller than those of either single mutant, yet the difference was comparatively minor. We tested whether the reduced requirement for KLU in a ttg2–1 mutant background reflected a downregulation of KLU expression; however, this was not the case, and both KLU and CYP78A9 showed normal expression levels in ttg2–1 mutant gynoecia (Fig. S4, Table S1). Thus, the phenotype of the double mutant suggests that klu mutant integument cells are less dependent on TTG2 function to promote expansion than are cells in WT integuments.
Elevated KLU Activity in Ovules Is Sufficient to Increase Seed Size.
To test whether increasing KLU activity only in ovules is sufficient to enhance seed growth, we expressed KLU in outer integuments using pINO (Fig. S5 A and B) (25), which leads to a considerable increase in total KLU mRNA in dissected gynoecia (Fig. S5C), and thus to both ectopic expression and overexpression. The pINO::KLU transgene in a WT background caused a strong increase in seed size and weight (30% and 44%, respectively; Fig. 2 A and C and Fig. S1A). Also, introducing the pINO::KLU construct into a klu-2 mutant background increased seed size and weight by a similar margin (33% in area and 40% in weight; Fig. 2 A and C and Fig. S1A). In addition to enlarged seeds, pINO::KLU-expressing plants formed wider siliques than WT with a more uneven valve surface, suggestive of local tissue overgrowth (Fig. 2B). These results indicate that KLU activity in WT ovules is limiting for seed growth and that KLU expression either in the inner integument, as in WT, or in the outer integument, as in pINO::KLU; klu-2 plants, is able to promote seed growth. Together with the effect of ovule-specific overexpression on increased silique width, these findings support the non-cell autonomy of KLU function that was previously concluded from its action in leaves and floral organs (19).
KLU Promotes Cell Proliferation in Integuments.
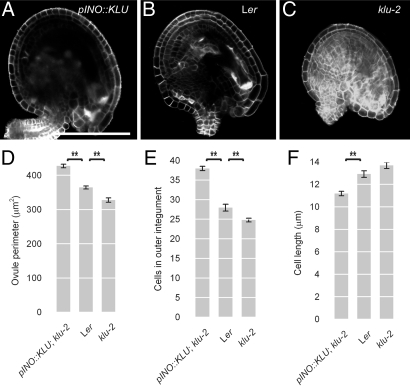
Having established that local KLU activity in the integuments of developing ovules is required and sufficient to promote seed growth, a plausible hypothesis is that its effect on final seed size is mediated by reduced or increased cell proliferation in integuments, giving rise to fewer or more cells, respectively, in the prospective seed coat. To test this, we characterized mature ovules from klu-2 mutants and pINO::KLU-expressing plants relative to WT at 2 days after emasculation (Fig. 5 A–C). Indeed, the outer integuments were longer and consisted of more and smaller cells in pINO::KLU-expressing plants than in WT, whereas in klu-2 mutant ovules, the outer integuments were shorter, consisting of fewer cells (Fig. 5 A–F). The magnitude of the changes in integument cell numbers closely parallels the differences in final seed size (compare Figs. 2C and 5E). This suggests that by controlling the extent of cell proliferation in developing integuments, KLU determines the growth potential of the seed coat, which ultimately appears to limit growth of the seeds.
Fig. 5.
KLU promotes cell proliferation in the integuments. (A–C) Confocal optical sections through Calcofluor White-stained ovules at 2 days after emasculation. (A) Ovule from a pINO::KLU-expressing plant. (B) WT ovule from a Ler plant. (C) klu-2 mutant ovule. (D–F) Quantification of ovule dimensions in response to changing KLU activity. (D) Length of the outer integument as measured from the insertion point at the funiculus to the tip at the micropyle. (E) Number of cells in the outer integument. (F) Average length of cells in the outer integument as calculated from outer integument length and cell number for individual ovules. Values shown are mean ± SEM. **Significantly different from control at P < 0.01 after Bonferroni correction. (Scale bar: 100 μm.)
KLU and Seed Yield.
We next characterized the effects of changes in KLU activity in developing ovules on overall seed yield and related parameters. To begin with, we asked whether the changes in seed size are reflected in the size of the embryos and resulting seedlings by measuring cotyledon area 7 days after germination. Cotyledon size closely paralleled the altered seed sizes (Fig. 2 A, C, and D). The changes in cotyledon size in klu-2 mutants and KLUox; klu-2 plants may represent the combined effects of altered seed size and different KLU activity in the developing embryo. However, the strong cotyledon enlargement in seedlings derived from pINO::KLU-expressing plants, which do not have altered KLU activity in developing embryos themselves, is most likely to reflect enhanced embryo growth attributable to the larger seed volume only.
Despite the strong effects of KLU on individual seed size, pINO::KLU overexpression in a WT background did not lead to a higher yield in terms of total seed weight (Fig. S1B), because the seed size effects at the whole-plant level were offset by a reduced number of seeds per silique and per plant (Fig. S1 C–E). These observations raise the possibility that the increased seed size in pINO::KLU plants might merely be an indirect effect of reduced within-plant competition for resources by a smaller total number of developing seeds. However, this explanation appears unlikely for the following reasons. In a klu-2 mutant background, pINO::KLU expression increased seed size to the same extent as in WT, yet the total seed number of the transgenic plants was the same as in nontransgenic klu-2 controls (Fig. 2C and Fig. S1C). As a consequence, the total seed yield in pINO::KLU; klu-2 plants relative to klu-2 mutants was proportionately increased by 34% (Fig. S1B). Thus, at least in this genetic background, an increased individual seed size is not correlated with a reduced overall number of seeds, and thus leads to an overall higher seed yield. Furthermore, under our growth conditions, even a severe limitation of the number of seeds that develop per plant did not increase seed size in either WT or pINO::KLU-expressing plants (Fig. S1G). Similarly, reduced competition within individual siliques cannot explain the difference in seed size between WT and pINO::KLU-expressing plants; when siliques were matched according to the number of seeds they contained, the average size of seeds from individual siliques was still significantly larger in KLU-overexpressing plants than in WT (Fig. S1H).
The reason for the reduced seed set per silique in pINO::KLU plants appears to be a defect on the maternal side, because pollinating WT flowers with pollen from the transgenic plants produced normal numbers of seeds per silique (50.8 ± 2.3, n = 6), whereas the reciprocal cross yielded only 27 ± 0.86 (n = 6) seeds per silique. Reduced fertility is known to delay overall plant senescence (26). Consistent with this, pINO::KLU-expressing plants formed more total aerial biomass (seeds plus nonseed tissue), resulting in a reduced harvest index (the proportion of total seed yield divided by total aerial biomass; Fig. S1F).
Increased Seed Size Correlates with Higher Relative Oil Content.
An increased size reduces the surface-to-volume ratio of a seed. Assuming a constant thickness of the seed coat, which contains little to no storage oil, and considering the apparently increased embryo (see above), which contains oil as a storage product in the Brassicaceae, this change in seed dimensions is predicted to result in a higher relative oil content in seeds from pINO::KLU-expressing plants (27). Indeed, their relative oil content was increased by 9–12% in both a WT and a klu-2 mutant background (Fig. 2E). The magnitude of this effect is comparable to the ≈5% increase in the estimated ratio of embryonic-to-total seed volume (see SI Text), suggesting that the higher oil content is at least partly attributable to geometric changes. The enlarged seeds from pINO::KLU-expressing plants also contained more total protein than respective control seeds (Fig. 2F). Here, however, the relative protein content was largely unchanged when correcting for the higher seed weight. Thus, even though the enlarged seeds in pINO::KLU-expressing plants do not increase yield in terms of total seed weight per plant, the increased relative oil content suggests that the transgenic plants produce more total seed oil than WT.
Discussion
In summary, our results suggest the following model for the control of integument growth and, ultimately, the determination of seed size. KLU expression in a restricted region at the base of the nucellus and in the inner integument leads to the generation of a mobile growth signal that moves throughout the inner and outer integuments, where it stimulates cell proliferation. The cell number in the integuments of the mature ovule, in turn, sets the growth potential of the seed coat after fertilization. As the embryo develops, the seed coat acts as a physical constraint on embryo growth, thus ultimately providing an upper limit to final seed size.
Based on the functional characterization of KLU in the control of leaf and petal growth, it was proposed that the KLU-dependent growth signal was mobile enough to equilibrate to a largely homogeneous concentration throughout the organ, despite only being generated in the periphery (19). If true, this would suggest that in developing ovules, both integuments are likely to be exposed to very similar levels of this signal. A likely advantage of using such a mobile growth regulator to control cell proliferation in the integuments is that growth of both integuments would proceed in a coordinated manner, ensuring the right proportions among the different components that make up the mature and functionally integrated ovule.
We demonstrate a previously undiscovered requirement for the CYP78A9 protein in promoting seed growth. CYP78A9 is closely related to KLU, belonging to the same subfamily of cytochrome P450 enzymes. This close relation raises the possibility that both proteins may be active on the same or very similar substrate compounds; yet, no genetic interaction between the two factors was found in our double-mutant analysis. It will be interesting to determine whether combined overexpression of these two genes could be used to bring about even stronger increases in seed size than those observed in pINO::KLU-expressing plants.
The above model supports the conclusion from the analysis of arf2 mutants that integument growth limits final seed size (14). It also unifies the roles of KLU in regulating growth of leaves, floral organs, and integuments by the generation of a growth signal that is limiting in a WT background (19). Using clonal analysis, we have shown unambiguously that KLU is required locally in developing flowers to promote seed growth and that its loss and gain of function lead to opposite seed size phenotypes, indicating that the level of KLU activity is limiting for seed growth in WT. Therefore, this presumed growth factor signaling pathway has the potential to be used in the adjustment of seed size during individual plant development and, at the same time, represents a plausible target for the modification of seed size in plant evolution. Because an orthologous growth-signaling pathway appears to exist in rice (19, 20, 28), KLU-dependent maternal control of seed size may also be conserved in monocotyledonous species.
Materials and Methods
Plant Lines and Growth Conditions.
All mutants used have been described before: klu-2, klu-4, and the endogenous KLU overexpression line (19); arf2–9/mnt (14); ap2–1 (29); and ttg2–1 (30). The cyp78a9 insertion allele was isolated from the Sainsbury Laboratory Arabidopsis Transposants collection of En/Spm insertion mutants (SM_3_556, Nottingham Arabidopsis Stock Centre ID code: N56511).
Plants were grown under long-day conditions (16 h of light/8 h of dark) at 20 °C, with a light intensity of 120 μmol photons m−2·s−1. Plant transformation was performed using “floral dip.” Ethanol induction was performed by treating plants with ethanol vapor within sealed plastic bags for 40–60 min.
Construction of Transgenes, PCR Genotyping, and qRT-PCR.
Details of transgene construction, PCR genotyping, and qRT-PCR are provided in SI Text.
Seed Size Measurements.
To measure seed size, dried seeds were photographed on white paper using a Zeiss SteREO Lumar stereomicroscope fitted with a Zeiss AxioCam MRm digital camera. After thresholding and converting the photographs to binary images, seed size was measured using the “Analyze particles” function of ImageJ (National Institutes of Health). At least 100 seeds from 10 different siliques were analyzed per genotype.
To determine seed weight, batches of 100 seeds were weighed using a Sartorius ME5–0CE microbalance.
Confocal Microscopy and Measurement of Ovule Dimensions.
To measure ovule dimensions using confocal microscopy, flowers just before anthesis were emasculated to prevent self-pollination. Two days later, ovules from the emasculated flowers were dissected, stained using a 0.1% solution of Calcofluor White, and imaged using a Leica SP2 confocal microscope. The length of the outer integuments was measured using Zeiss AxioVision software.
Measurement of Relative Oil and Protein Content.
Relative oil content was determined as described by Hobbs et al. (31), using an MQC NMR instrument (Oxford Instruments).
To determine the protein content per seed, 100 seeds were ground in a total of 1 mL of extraction buffer [200 mM Tris-HCl (pH 7.5), 250 mM NaCl, 25 mM EDTA (pH 8), and 0.5% SDS], and protein concentration was determined using the Bio-Rad Protein assay.
Supplementary Material
Acknowledgments.
We thank David Smyth, Rod Scott, Thomas Laux, and the Nottingham Arabidopsis Stock Centre for providing seeds; Charles Gasser for the pINO; and Patrick Laufs for the CLV3::AlcR construct. We are grateful to Timothy Wells and Lesley Phillips for excellent plant care. We thank Isabel Bäurle and members of the Lenhard laboratory for critical reading and helpful comments on the manuscript. This work was supported by a David Phillips Fellowship from the Biotechnology and Biological Sciences Research Council (to M.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907024106/DCSupplemental.
References
- 1.Harper JL, Lovell PH, Moore KG. The shapes and sizes of seeds. Annu Rev Ecol Syst. 1970;1:327–356. [Google Scholar]
- 2.Venable DL. Size-number trade-offs and the variation of seed size with plant resource status. Am Nat. 1992;140:287–304. [Google Scholar]
- 3.Shomura A, et al. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet. 2008;40:1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- 4.Berger F, Grini PE, Schnittger A. Endosperm: An integrator of seed growth and development. Curr Opin Plant Biol. 2006;9:664–670. doi: 10.1016/j.pbi.2006.09.015. [DOI] [PubMed] [Google Scholar]
- 5.Colombo L, Battaglia R, Kater MM. Arabidopsis ovule development and its evolutionary conservation. Trends Plants Sci. 2008;13:444–450. doi: 10.1016/j.tplants.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 6.Haughn G, Chaudhury A. Genetic analysis of seed coat development in Arabidopsis. Trends Plants Sci. 2005;10:472–477. doi: 10.1016/j.tplants.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Scott RJ, Spielman M, Bailey J, Dickinson HG. Parent-of-origin effects on seed development in Arabidopsis thaliana. Development. 1998;125:3329–3341. doi: 10.1242/dev.125.17.3329. [DOI] [PubMed] [Google Scholar]
- 8.Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:17531–17536. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alonso-Blanco C, Blankestijn-de Vries H, Hanhart CJ, Koornneef M. Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc Natl Acad Sci USA. 1999;96:4710–4717. doi: 10.1073/pnas.96.8.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia D, Fitz Gerald JN, Berger F. Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell. 2005;17:52–60. doi: 10.1105/tpc.104.027136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jofuku KD, Omidyar PK, Gee Z, Okamuro JK. Control of seed mass and seed yield by the floral homeotic gene APETALA2. Proc Natl Acad Sci USA. 2005;102:3117–3122. doi: 10.1073/pnas.0409893102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, Zheng L, Corke F, Smith C, Bevan MW. Control of final seed and organ size by the DA1 gene family in Arabidopsis thaliana. Genes Dev. 2008;22:1331–1336. doi: 10.1101/gad.463608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohto MA, Fischer RL, Goldberg RB, Nakamura K, Harada JJ. Control of seed mass by APETALA2. Proc Natl Acad Sci USA. 2005;102:3123–3128. doi: 10.1073/pnas.0409858102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schruff MC, et al. The AUXIN RESPONSE FACTOR 2 gene of Arabidopsis links auxin signalling, cell division, and the size of seeds and other organs. Development. 2006;133:251–261. doi: 10.1242/dev.02194. [DOI] [PubMed] [Google Scholar]
- 15.FitzGerald J, Luo M, Chaudhury A, Berger F. DNA methylation causes predominant maternal controls of plant embryo growth. PLoS One. 2008;3:e2298. doi: 10.1371/journal.pone.0002298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan C, et al. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor Appl Genet. 2006;112:1164–1171. doi: 10.1007/s00122-006-0218-1. [DOI] [PubMed] [Google Scholar]
- 17.Song XJ, Huang W, Shi M, Zhu MZ, Lin HX. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- 18.Weng J, et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008;18:1199–1209. doi: 10.1038/cr.2008.307. [DOI] [PubMed] [Google Scholar]
- 19.Anastasiou E, et al. Control of plant organ size by KLUH/CYP78A5-dependent intercellular signalling. Dev Cell. 2007;13:843–856. doi: 10.1016/j.devcel.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 20.Wang JW, Schwab R, Czech B, Mica E, Weigel D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell. 2008;20:1231–1243. doi: 10.1105/tpc.108.058180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zondlo SC, Irish VF. CYP78A5 encodes a cytochrome P450 that marks the shoot apical meristem boundary in Arabidopsis. Plant J. 1999;19:259–268. doi: 10.1046/j.1365-313x.1999.00523.x. [DOI] [PubMed] [Google Scholar]
- 22.Deveaux Y, et al. The ethanol switch: A tool for tissue-specific gene induction during plant development. Plant J. 2003;36:918–930. doi: 10.1046/j.1365-313x.2003.01922.x. [DOI] [PubMed] [Google Scholar]
- 23.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 24.Ito T, Meyerowitz EM. Overexpression of a gene encoding a cytochrome P450, CYP78A9, induces large and seedless fruit in Arabidopsis. Plant Cell. 2000;12:1541–1550. doi: 10.1105/tpc.12.9.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meister RJ, Kotow LM, Gasser CS. SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integuments. Development. 2002;129:4281–4289. doi: 10.1242/dev.129.18.4281. [DOI] [PubMed] [Google Scholar]
- 26.Nooden LD, Penney JP. Correlative controls of senescence and plant death in Arabidopsis thaliana (Brassicaceae) J Exp Bot. 2001;52:2151–2159. doi: 10.1093/jexbot/52.364.2151. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Beisson F, Pollard M, Ohlrogge J. Oil content of Arabidopsis seeds: The influence of seed anatomy, light and plant-to-plant variation. Phytochemistry. 2006;67:904–915. doi: 10.1016/j.phytochem.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 28.Miyoshi K, et al. PLASTOCHRON1, a timekeeper of leaf initiation in rice, encodes cytochrome P450. Proc Natl Acad Sci USA. 2004;101:875–880. doi: 10.1073/pnas.2636936100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jofuku KD, den Boer BG, Van Montagu M, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002;14:1359–1375. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hobbs DH, Flintham JE, Hills MJ. Genetic control of storage oil synthesis in seeds of Arabidopsis. Plant Physiol. 2004;136:3341–3349. doi: 10.1104/pp.104.049486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schneitz K, Hülskamp M, Pruitt RE. Wild-type ovule development in Arabidopsis thaliana: A light microscope study of cleared whole-mount tissue. Plant J. 1995;7:731–749. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.