Abstract
A quantitative linear model accurately (R2 = 0.88) describes the thermostabilities of 54 characterized members of a family of fungal cellobiohydrolase class II (CBH II) cellulase chimeras made by SCHEMA recombination of three fungal enzymes, demonstrating that the contributions of SCHEMA sequence blocks to stability are predominantly additive. Thirty-one of 31 predicted thermostable CBH II chimeras have thermal inactivation temperatures higher than the most thermostable parent CBH II, from Humicola insolens, and the model predicts that hundreds more CBH II chimeras share this superior thermostability. Eight of eight thermostable chimeras assayed hydrolyze the solid cellulosic substrate Avicel at temperatures at least 5 °C above the most stable parent, and seven of these showed superior activity in 16-h Avicel hydrolysis assays. The sequence-stability model identified a single block of sequence that adds 8.5 °C to chimera thermostability. Mutating individual residues in this block identified the C313S substitution as responsible for the entire thermostabilizing effect. Introducing this mutation into the two recombination parent CBH IIs not featuring it (Hypocrea jecorina and H. insolens) decreased inactivation, increased maximum Avicel hydrolysis temperature, and improved long time hydrolysis performance. This mutation also stabilized and improved Avicel hydrolysis by Phanerochaete chrysosporium CBH II, which is only 55–56% identical to recombination parent CBH IIs. Furthermore, the C313S mutation increased total H. jecorina CBH II activity secreted by the Saccharomyces cerevisiae expression host more than 10-fold. Our results show that SCHEMA structure-guided recombination enables quantitative prediction of cellulase chimera thermostability and efficient identification of stabilizing mutations.
SCHEMA is a computational approach to identifying blocks of sequence that minimize structural disruption when they are recombined in chimeric proteins (1). SCHEMA recombination of eight blocks from three fungal cellobiohydrolase class II (CBH II)2 genes was used in our previous work to create a library of 38 = 6,561 chimeric sequences, all having the native Hypocrea jecorina cellulose binding module and linker and observed to feature a degree of glycosylation similar to that found in native CBH IIs secreted by fungi (2). Synthesis and characterization of selected CBH II chimeras expressed in Saccharomyces cerevisiae revealed enzymes with thermostabilities and cellulose hydrolysis performance superior to those of the parent enzymes from Humicola insolens, H. jecorina, and Chaetomium thermophilum.
Our prior analysis showed that a qualitative model based on sequence-stability data from 23 functional chimeras (categorizing blocks as destabilizing, stabilizing, or neutral) could identify highly stable chimeras in the SCHEMA library (2). When studying SCHEMA recombination of a bacterial cytochrome P450, we previously estimated that building a quantitative regression model would require stability measurements for at least 35 representative sequences (3). We therefore synthesized an additional 18 CBH II chimeras to further explore the sequences that the qualitative model predicted would encode the most thermostable chimeras. If sequence blocks contribute additively and independently of their context, as was found for SCHEMA chimeras of cytochrome P450 (3), then quantitative stability prediction would be possible based on stability data from a very limited sampling of the thousands of possible chimeras. Here we show that a quantitative CBH II chimera stability model can in fact be constructed and also that it was possible, using site-directed mutagenesis experiments, to pinpoint a single amino acid substitution that is responsible for the large stabilizing contribution of one of the SCHEMA blocks.
Highly thermostable fungal CBH IIs are potentially useful for the degradation of cellulosic substrates in biofuels, textile, and other applications (4). High thermostability translates to longer half-lives at elevated hydrolysis temperatures, where viscosity and microbial contamination are reduced (5). We therefore investigated how selected thermostable CBH II chimeras perform in the hydrolysis of crystalline cellulose (Avicel) at elevated temperatures (up to 70 °C). All of the thermostable chimeras tested have specific activities on phosphoric acid swollen cellulose (PASC) at 50 °C that are comparable with the most active parent (H. jecorina CBH II) and hydrolyze Avicel at temperatures higher than any of the three parent enzymes, including the CBH II from the thermophilic fungus H. insolens.
EXPERIMENTAL PROCEDURES
Parent and chimeric genes encoding CBH II enzymes were cloned into yeast expression vector YEp352/PGK91-1-αss, and expression in synthetic dextrose casamino acids (SDCAA) medium was carried out as described previously (2). For Avicel activity assays, yeast peptone dextrose (YPD) culture supernatants were brought to 1 mm phenylmethylsulfonyl fluoride and 0.02% NaN3 and used without concentration. CBH II enzyme activity was measured by adding dilutions of concentrated culture supernatant to 37.5 μl of PASC and 225 μl of 50 mm sodium acetate, pH 4.8, and incubating for 2 h at 50 °C. Reducing sugar equivalents formed were determined via Nelson-Somogyi assay, as described (2).
CBH II enzyme T50 values were measured by adding concentrated CBH II SDCAA expression culture supernatant to 50 mm sodium acetate, pH 4.8, at a concentration giving A520 of 0.5 as measured in the Nelson-Somogyi reducing sugar assay after incubation with endoglucanase-treated PASC (2). 200 μl of CBH II enzyme/buffer mixtures were incubated in a water bath at the temperature of interest for 10 min. After incubation, 37.5 μl of endoglucanase-treated PASC and 62.5 μl of 50 mm sodium acetate were added, and hydrolysis was carried out for 2 h at 50 °C. The incubation temperature at which the enzyme lost one-half of its activity was determined by linear interpolation of the Nelson-Somogyi assay A520 values plotted versus temperature.
For 16-h Avicel PH101 (Fluka) hydrolysis measurements, 0.3 μg of purified CBH II was incubated with 3 mg of Avicel in 270 μl of 50 mm sodium acetate, pH 4.8, in PCR tubes placed in a water bath for 16 h. Tubes were cooled in a room temperature water bath for 10 min and centrifuged at 1000 × g for 10 min, and supernatants were withdrawn for reducing sugar analysis.
For estimation of CBH II activity in YPD expression culture supernatants, supernatant volumes ranging from 2 to 40 ml were added to 800 μl of 33 mg/ml Avicel suspended in 50 mm sodium acetate, pH 4.8, in conical tubes. CBH IIs were allowed to bind Avicel at 4 °C for 1 h, centrifuged at 2000 × g for 2 min, and washed twice with 50 mm sodium acetate, pH 4.8. After the second wash, CBH II-bound Avicel was resuspended in 2.75 ml of sodium acetate buffer, split into 270-μl aliquots, and incubated at 50 °C for 2.5 h. Centrifugation and supernatant reducing sugar analysis were carried out as above.
The LinearRegression package in Mathematica was used to fit CBH II chimera T50 data to a 17-parameter, block additive model and was also used for cross-validation analysis. Block effects are reported relative to a parent 1 (H. insolens CBH II) reference state with 16 parameters representing substitution of each of the eight blocks from parents 2 and 3.
RESULTS
Values of T50, defined here as the temperature at which an enzyme loses 50% of its activity during a 10-min incubation, were determined for the three parent cellobiohydrolases, 33 active CBH II chimeras from our prior work and 18 additional chimeras that qualitative stability modeling predicted to be among the most thermostable, i.e. containing none of the seven predicted destabilizing blocks and either three or four of the four predicted stabilizing blocks (2). All 51 chimera sequences are listed in supplemental Table 1. Reculturing and reconcentrating all of the predicted thermostable chimeras previously classified as not secreted (2) allowed us to obtain sufficient amounts of 12112132, 13111132, and 13322332 CBH IIs for T50 determination. Because T50 measurements require more enzyme in each assay than the half-life (t½) measurements made in our prior work, we were unable to obtain T50 values for three poorly expressed, relatively unstable chimeras (21223122, 23231222, and 33123313) from the initial t½ data set. The complete set of T50 values for the chimeras and parent CBH IIs is provided in supplemental Table 1. The amino acid sequences for all these CBH IIs appear in supplemental Table 2. All 31 predicted thermostable chimeras tested in this and previous work have T50 values more than two degrees higher than that of the most thermostable parent enzyme (64.8 °C).
Applying linear regression to the sequence-stability data resulted in a 10-parameter model that fit the observed T50 values with R2 = 0.88 (Fig. 1). To better estimate the predictive capacity of the regression model outside the training set, we performed 11-fold cross-validation resulting in a R2 of 0.57, where removal of two outliers (11313121 and 22222222) increases the cross-validation R2 to 0.76. The regression model uses the most stable parent 1 (H. insolens) as the reference state T50 and includes nine additional terms having p values ≤0.1. The model parameters (supplemental Table 3) show that a single block, block 7 from parent 3 (B7P3), is by far the strongest contributor to chimera thermostability relative to H. insolens CBH II. This block from C. thermophilum CBH II contributes ∼8.5 °C to the stability of chimeras that contain it. Two of the eight remaining blocks with p values ≤0.1 were found to make smaller stability contributions, of 1.2 and 2.7 °C, whereas the other six decrease stability.
FIGURE 1.
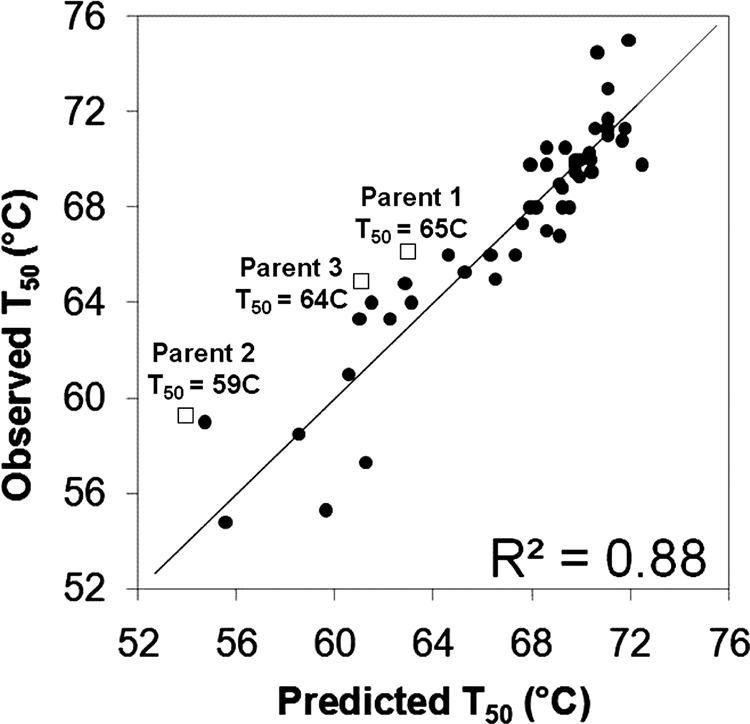
Observed and predicted T50 values for CBH II parents and 51 CBH II chimeras. The line denotes the linear regression model equation (parameters in supplemental Table 3). Parent CBH II T50 values are denoted as squares.
Alignment of the B7P1 and B7P3 sequences (supplemental Fig. 1) shows that block 7 differs at 10 out of 56 amino acid positions in the H. insolens and C. thermophilum enzymes. In the background of the chimera with the highest T50 value, 21311131, we individually mutated each residue in B7P3 to the corresponding residue in B7P1 and determined T50 values for each. We found that only one mutation, S313C, markedly altered the thermostability of the chimera; this single mutation reduced the T50 of 21311131 by ∼10 °C (supplemental Fig. 2).
To study the effect of the reverse mutation in different backgrounds, genes for the H. insolens and H. jecorina parent CBH IIs encoding the C313S substitution (C314S in H. insolens and C311S in H. jecorina) were constructed and expressed, and the T50 values of the enzymes were determined. We also quantified the stabilities of chimeras 11111131 and 22222232, in which the stabilizing B7P3 is substituted into the wild type H. insolens and H. jecorina enzymes. Both the B7P3 block substitution and the Cys-Ser single mutation markedly stabilized the parent CBH IIs; the largest effect was an ∼8 °C increase in T50 for H. jecorina CBH II containing the C311S substitution (supplemental Fig. 3). The Cys-Ser mutation was also tested in two chimeras, 31311112 and 13231111, that did not contain B7P3 as well as in a homologous CBH II (from Phanerochaete chrysosporium), which was not in the recombination parent set. The P. chrysosporium CBH II catalytic domain is only 55–56% identical to the parent CBH II catalytic domains. All of these enzymes were stabilized by the Cys-Ser substitution; the P. chrysosporium CBH II was stabilized by a remarkable 10 °C (supplemental Fig. 4).
Eight of the thermostable CBH II chimeras and the parent enzymes containing the equivalent C313S mutation were His6-tagged and purified so that their specific activities could be determined. As shown in supplemental Table 4, the specific activities, as measured on amorphous cellulose (PASC) at 50 °C, for these chimeras and native enzymes containing the Cys-Ser mutation are similar to those of the wild type parents. Thus the increased thermostability does not come at the expense of specific activity.
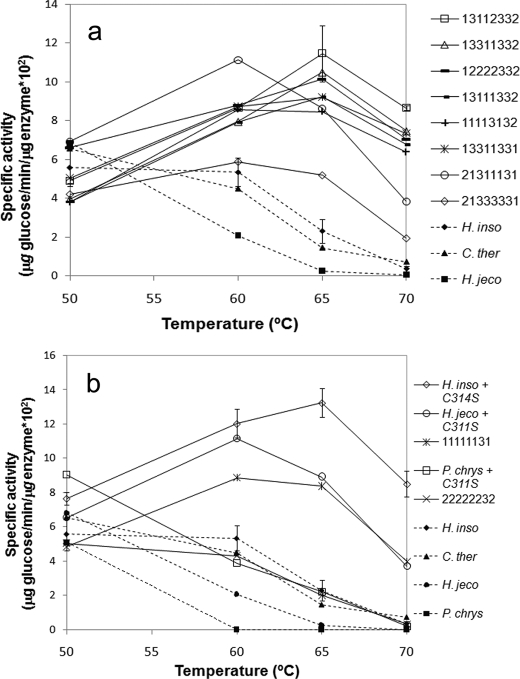
These same eight thermostable chimeras (T50 2–10 °C higher than the most stable parent) were then tested for activity on crystalline cellulose during a 16-h incubation over a range of temperatures. Fig. 2a shows that seven of eight tested thermostable chimeras were maximally active toward Avicel at 60–65 °C, with all eight chimeras retaining activity at 70 °C, the highest temperature tested. In contrast, the three parent CBH IIs show maximum activity at 50 °C and are either completely or almost completely inactive at 70 °C. Additionally, the seven chimeras with increased optimum activity temperatures hydrolyze significantly more Avicel than any of the parent CBH II enzymes. As shown in Fig. 2b, similar behaviors are observed for the H. insolens and H. jecorina parents containing the Cys-Ser mutation. The Cys-Ser mutation also increased the Avicel hydrolysis and maximum operating temperature for the P. chrysosporium CBH II. The P3B7 block substitution in the H. insolens and H. jecorina parents increased both the operating temperature and the hydrolysis of the H. insolens CBH II but did not improve overall cellulose hydrolysis by the H. jecorina enzyme.
FIGURE 2.
CBH II specific activities toward Avicel as a function of temperature. a, CBH II parent and chimera specific activities. b, CBH II parent, C311S mutant, and B7P3 single block substitution chimera specific activities. Reactions were run for 16 h in 50 mm sodium acetate, pH 4.8, with an Avicel concentration of 15 mg/ml. Data presented are averages of two independent replicates with error bars indicating the duplicate activity values for each temperature point. H. inso, H. insolens; C. ther, C. thermophilum; H. jeco, H. jecorina; P. chrys. P. chrysosporium.
We have observed low (<1 mg/liter) secretion of wild type H. jecorina CBH II from the heterologous S. cerevisiae expression host (2). The C311S mutation in the wild type H. jecorina CBH II enzyme markedly increases total secreted CBH II activity (supplemental Table 5). In synthetic (SDCAA) medium, the C311S and B7P3 substitutions increase H. jecorina CBH II total secreted activity by a factor of two, whereas in rich (YPD) medium, the activity increase is 10-fold. For the H. insolens CBH II parent, which is expressed at much higher levels than the other two parent CBH IIs, the C314S mutation increased secreted activity by a factor of ∼1.5, whereas the B7P3 block substitution decreased it. Because the H. insolens and H. jecorina wild type and Cys-Ser mutants all have similar specific activities (Table 1), we conclude that the increase in total secreted cellulase activity is the result of improved secretion of the functional enzyme. A correlation between S. cerevisiae heterologous protein secretion and protein stability has been observed (6), suggesting that the increased secretion of the Cys-Ser mutant CBH IIs might reflect their higher stabilities.
To model the Cys-Ser mutation, we employed the high resolution H. insolens CBH II (Protein Data Bank (PDB) entry 1ocn) crystal structure (7). First, we optimized the hydrogen-bond network with REDUCE (8). Cys-314 was predicted to form a hydrogen bond to the carbonyl of Pro-339. To confirm this prediction, we optimized side chain packing using the modeling platform SHARPEN (9). Ser-314 is predicted to make the similar interactions to Cys-314, resulting in stronger hydrogen bonding and a more favorable geometry (supplemental Fig. 5).
DISCUSSION
Structure-guided SCHEMA recombination of three fungal cellobiohydrolase II (Cel6) enzymes has generated a set of functional CBH II chimeras having a high level of sequence diversity. The relatively low sequence complexity of this synthetic enzyme family (the eight sequence blocks come from one of three parent enzymes for a total of 24 sequence elements) allows important sequence-function relationships to be extracted from functional data (1). We previously showed that SCHEMA blocks contributed additively to the stability of cytochrome P450 chimeras; based on those results, we predicted that accurate models can be built from data on a small number (∼35) of sampled genes (3). To test whether a quantitative sequence-stability model can also be generated for CBH II chimeras, we obtained a data set of 54 sequence-stability measurements and used it to generate the model. The excellent fit of the data to a simple linear model (Fig. 1) suggests that the observable portion of the chimera stability landscape may be explained with additive, context-independent stability contributions from each chimera block.
This result may appear surprising as such modularity runs counter to the commonly emphasized tendency of protein mutations to exhibit coupling effects (10). The observed lack of coupling may arise from the SCHEMA chimera library design algorithm, which used dynamic programming to select recombination sites that minimize the number of disruptions (defined as residue-residue interactions not seen in parental sequences) (11). It is difficult to conclusively determine the role played by the library design, however, because we lack a counterexample library with evident block-block coupling. We therefore rationalize the apparent absence of block-block coupling in the CBHII chimeras by considering two general mechanisms by which coupling might arise.
First, we hypothesize that novel residue contacts not present in the parent structures are a potential cause of non-additive stability effects. Such contacts can only occur across a block-block interface, and a novel unfavorable residue pair (a more likely event than a novel favorable interaction) might lead to unfavorable block-block coupling. The CBH II library has fewer novel residue pairs, and therefore fewer potential disruptions (average number of novel contacting residue pairs 〈E〉 = 14.6), than the cytochrome P450 library (3) (〈E〉 = 29.5 for folded chimeras, 34.8 for unfolded chimeras), even if normalized on a per residue basis (CBH II has 363 amino acids, cytochrome P450 has 467).
The CBH II library has fewer potential disruptions for several reasons. In addition to the higher identity of the CBH II parent sequences, the barrel topology of the CBH II fold limits the number of long range contacts that can be broken by recombination. Between block contacts (heavy atoms within 4.5 Å) comprise only 27% (503/1831) of the total in a contact map derived from H. insolens structure (PDB entry 1ocn) (7). When only counting contacts for which novel residue pairs are possible in chimeras, the interblock total is reduced to 23% (68/294). Furthermore, most of these interactions are between residues on the protein surface, and the possibility of solvent screening further decreases the chances of dramatic disruptive residue-residue interactions (supplemental Fig. 5a). One exception, a buried interaction between positions 176 and 256, is illustrated in supplemental Fig. 5b. At this site, chimeras with B6P2 and either B5P1 or B5P3 pair Met-173–Trp-253 (larger amino acid than parental pairs Met-176–Phe-256 or Leu-173–Trp-253). Nevertheless, upon inspection of the parental crystallographic models, we deem a steric clash at this position unlikely due to movement in the portion of the protein backbone, which positions Trp-253 and the intrinsic flexibility of Met side chains. Notably, one characterized chimera fits this pattern (13333232) and is more stable than the parents (67 °C), in accord with the regression model fit (68 °C).
Another mechanism by which coupling could arise, block structural divergence, does not depend on the presence of novel residue pairs at block interfaces. Instead, as parental sequences diverge, intrinsic block structures may diverge, hindering modular block transplants. In the case of the CBH II library, the high parent pair sequence identifies (82, 66, and 64%) suggest that only minor structure deviations are likely (<1 Å r.m.s.d.) (12). We can explicitly evaluate this possibility by comparing crystallographic structures for H. insolens and H. jecorina CBH II (C. thermophilum CBH II lacks a crystal structure but is 82% identical to H. insolens). Aligning blocks from structures for each parent (PDB entries 1ocn and 1cb2 (12)) generates low α-carbon r.m.s.d. values (0.5, 0.5, 0.6, 0.5, 0.3, 0.7, 0.3, and 0.4 Å r.m.s.d.). H. jecorina blocks superimposed onto H. insolens are illustrated in supplemental Fig. 5c. To check for context-dependent effects, we performed in silico structural recombination, splicing each aligned block onto the opposing host structure. We found that it is possible to construct non-clashing structural models (α-carbons > 3 Å apart) for all single-block substitution chimeras (e.g. 11112111 or 22122222), with the exception of a minor clash (2.65 Å) when using B7P2 (11111121) due to the Asn insertion between blocks six and seven (supplemental Fig. 5d).
Another important factor behind the success of the linear regression model is the fairly context-independent, dominant stabilizing contribution made by block 7 parent 3 (B7P3), whose contribution is 8.5 °C relative to B7P1 (supplemental Table 3). Reverting each of the 10 amino acids that differ in B7P3 and B7P1 to the residue in parent 1 in the background of the most thermostable CBH II chimera, 21311131, identified S313C as having an effect comparable with that of the entire B7P3 block (supplemental Fig. 3). Ser is present at this position in CBH II parent 3 (C. thermophilum) but not parents 1 (H. insolens) and 2 (H. jecorina). Mutating Cys to Ser in the wild type H. insolens and H. jecorina enzymes increased their stabilities by ∼5 and 8 °C, respectively. Making this Cys to Ser mutation in native P. chrysosporium CBH II (not included in the recombination parent set) increased the thermostability of the enzyme by 10 °C.
A number of effects might explain why the Cys-Ser mutation stabilizes a broad range of CBH IIs, including native CBH IIs and chimeras. Cys and Ser are similar (although not isosteric), and these two amino acids dominate sequence alignments at this position when compared with other alternatives (see below). The hydrogen-bonding partners for this residue are backbone elements (the amide of Gly-316 and the carbonyl of Pro-339) and are therefore less likely to be dependent on third party amino acid variations. Furthermore, the immediate neighboring side chains for this pocket (Asn-283, Pro-339, Phe-345) are conserved among all four native CBH II cellulases studied.
We wished also to identify the biophysical basis for the stabilizing effect of the Cys-Ser substitution. Our first hypothesis was that it removed an oxidative damage pathway. Oxidation of unpaired Cys thiol groups has been observed, for example, to lead to irreversible thermal inactivation of T4 lysozyme (13). We tested whether removing the unpaired Cys-311 side chain and replacing it with other side chains of similar size and hydrophobicity would prevent such oxidation without introducing a large structural disruption by substituting Cys-311 to Ala, Leu, and Met as well as Ser in H. jecorina CBH II. Secretion of C311L and C311M mutants was too low to allow T50 determination, whereas the T50 value for the H. jecorina C311A mutant was ∼2 °C lower than for wild type (data not shown), suggesting that oxidation of the unpaired Cys-311 side chain residue is not a dominant factor for the stabilizing effect of C311S.
The high resolution (1.3 Å) H. insolens crystal structure (PDB entry 1ocn) shows that Cys-314 is part of a hydrogen-bonding network (supplemental Fig. 6). The increased hydrogen-bonding capacity of Ser relative to Cys may suggest a role for stronger hydrogen-bonding interactions in the stabilization. The crystal structure also suggests that Ser may be preferred for steric reasons. Specifically, when the Cys side chain is rebuilt with canonical bond angles, a 6° bend is removed, and Cys is pushed closer to the carbonyl of Pro-339, creating an unfavorable steric interaction.
The stabilizing Cys-Ser mutation might have been found by consensus analysis (14), but such an analysis also generates many other equally likely hypotheses. We aligned the 196 protein sequences sharing the greatest identity to the H. jecorina CBH II. Fifty-four of the 250 most identical sequences were excluded from the alignment (supplemental Fig. 7) due to redundancy (i.e. point mutants for structural studies or >95% identical isoforms). There is a bias in favor of Ser-311; 158 sequences have Ser, 20 have Ala, 10 have Cys, 5 have a deletion, and 3 have Gly. However, there are 42 other positions where the most frequent choice occurs with greater than twice the frequency of the H. jecorina amino acid.
The large stabilizing effect of the Cys-Ser mutation raises the possibility that Ser at this position is a global indicator of native cellulase thermostability. However, the T50 of 64.8 °C for H. insolens CBH II, which features Cys at this position, is greater than that of the C. thermophilum CBH II (64.0 °C), indicating that Ser is not the only stability determinant.
Thermostability is not the only property of interest for industrial cellulases. Specific activity, changes to cellulose binding and effects on expression and product inhibition are all important as well. We found that recombination yields CBH II chimeras whose improved thermostability comes without cost to specific activity measured in short time (i.e. 2-h) cellulose hydrolysis assays. Similar observations were made for CBH IIs containing the thermostabilizing Cys-Ser mutation. In 16-h hydrolysis assays, several of the CBH II chimeras and all three tested Cys-Ser mutant CBH IIs hydrolyzed more cellulose than the native CBH IIs. This superior performance is likely the result of having specific activity comparable with that of the parent CBH IIs along with greater thermostability that allows the enzyme to continue to function for a longer time at the elevated temperatures.
In conclusion, we have demonstrated that stabilizing blocks can be recombined to create novel highly stable, active cellulases. The stability regression model predicts that the CBH II SCHEMA library contains 2,026 chimeras that are more stable than the most stable parent enzyme. These chimeras are diverse and distinct from the native cellulases; they differ from the parents by between 8 and 72 mutations (an average of 50) and from each other by an average of 63 mutations. A total of 31 genes from this set were synthesized and expressed in S. cerevisiae; every one of these chimeric CBH IIs was found to be more stable than the most stable parent cellulase, from the thermophilic fungus H. insolens, as measured either by its half-life of inactivation at 63 °C (previous work (Ref. 2)) or by T50 (this work). Reducing the sequence complexity by making chimeras of only eight blocks allowed us to construct the sequence-stability model and identify a single highly stabilizing sequence block. By testing only 10 amino acid substitutions in this block, we were able to identify a single, highly stabilizing substitution. The very large stabilizing effect of the C313S substitution observed across the chimeras and in the native P. chrysosporium, H. insolens, and H. jecorina CBH II enzymes suggests that mutation of any residue at this position to Ser may stabilize any family 6 cellulase into which it is introduced. These findings demonstrate the value of using structure-guided recombination to discover important sequence-function relationships and efficiently generate whole families of highly stable enzymes.
Supplementary Material
This work was supported by grants from the Army-Industry Institute for Collaborative Biotechnologies and the Caltech Innovation Institute.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7 and Tables S1–S5.
- CBH II
- cellobiohydrolase class II
- PASC
- phosphoric acid swollen cellulose
- SDCAA
- synthetic dextrose casamino acids
- YPD
- yeast peptone dextrose
- r.m.s.d.
- root mean square deviation.
REFERENCES
- 1.Otey C. R., Landwehr M., Endelman J. B., Hiraga K., Bloom J. D., Arnold F. H. (2006) PLoS Biol. 4, e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heinzelman P., Snow C. D., Wu I., Nguyen C., Villalobos A., Govindarajan S., Minshull J., Arnold F. H. (2009). Proc. Natl. Acad. Sci. U.S.A. 106, 5610–5615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li Y., Drummond D. A., Sawayama A. M., Snow C. D., Bloom J. D., Arnold F. H. (2007) Nat Biotechnol 25, 1051–1056 [DOI] [PubMed] [Google Scholar]
- 4.Voutilainen S. P., Puranen T., Siika-Aho M., Lappalainen A., Alapuranen M., Kallio J., Hooman S., Viikari L., Vehmaanperä J., Koivula A. (2008) Biotechnol Bioeng. 101, 515–528 [DOI] [PubMed] [Google Scholar]
- 5.Turner P., Mamo G., Karlsson E. N. (2007) Microb. Cell Fact. 6, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shusta E. V., Kieke M. C., Parke E., Kranz D. M., Wittrup K. D. (1999) J. Mol. Biol. 292, 949–956 [DOI] [PubMed] [Google Scholar]
- 7.Varrot A., Macdonald J., Stick R. V., Pell G., Gilbert H. J., Davies G. J. (2003) Chem. Commun. (Camb.) 946–947 [DOI] [PubMed] [Google Scholar]
- 8.Word J. M., Lovell S. C., Richardson J. S., Richardson D. C. (1999) J. Mol. Biol. 285, 1735–1747 [DOI] [PubMed] [Google Scholar]
- 9.Loksha I. V., Maiolo J. R., 3rd, Hong C. W., Ng A., Snow C. D. (2009) J Comput Chem. 30, 999–1005 [DOI] [PubMed] [Google Scholar]
- 10.Fodor A. A., Aldrich R. W. (2004) J. Biol. Chem. 279, 19046–19050 [DOI] [PubMed] [Google Scholar]
- 11.Endelman J. B., Silberg J. J., Wang Z. G., Arnold F. H. (2004) Protein Eng. Des. Sel. 17, 589–594 [DOI] [PubMed] [Google Scholar]
- 12.Koivula A., Reinikainen T., Ruohonen L., Valkeajärvi A., Claeyssens M., Teleman O., Kleywegt G. J., Szardenings M., Rouvinen J., Jones T. A., Teeri T. T. (1996) Protein Eng. 9, 691–699 [DOI] [PubMed] [Google Scholar]
- 13.Perry L. J., Wetzel R. (1987) Protein Eng. 1, 101–105 [DOI] [PubMed] [Google Scholar]
- 14.Lockless S. W., Ranganathan R. (1999) Science 286, 295–299 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.