Abstract
Bloom syndrome caused by inactivation of the Bloom DNA helicase (Blm) is characterized by increases in the level of sister chromatid exchange, homologous recombination (HR) associated with cross-over. It is therefore believed that Blm works as an anti-recombinase. Meanwhile, in Drosophila, DmBlm is required specifically to promote the synthesis-dependent strand anneal (SDSA), a type of HR not associating with cross-over. However, conservation of Blm function in SDSA through higher eukaryotes has been a matter of debate. Here, we demonstrate the function of Blm in SDSA type HR in chicken DT40 B lymphocyte line, where Ig gene conversion diversifies the immunoglobulin V gene through intragenic HR between diverged homologous segments. This reaction is initiated by the activation-induced cytidine deaminase enzyme-mediated uracil formation at the V gene, which in turn converts into abasic site, presumably leading to a single strand gap. Ig gene conversion frequency was drastically reduced in BLM−/− cells. In addition, BLM−/− cells used limited donor segments harboring higher identity compared with other segments in Ig gene conversion event, suggesting that Blm can promote HR between diverged sequences. To further understand the role of Blm in HR between diverged homologous sequences, we measured the frequency of gene targeting induced by an I-SceI-endonuclease-mediated double-strand break. BLM−/− cells showed a severer defect in the gene targeting frequency as the number of heterologous sequences increased at the double-strand break site. Conversely, the overexpression of Blm, even an ATPase-defective mutant, strongly stimulated gene targeting. In summary, Blm promotes HR between diverged sequences through a novel ATPase-independent mechanism.
The RecQ helicases, a subfamily of DNA helicases, carry out the unwinding of duplex DNA in the 3′ to 5′ direction. Homologs of RecQ have been identified in a wide range of organisms, from budding yeast to humans (reviewed in Ref. 1). There are five human RecQ family proteins: Blm, Wrn, RecQ1, RecQ4, and RecQ5. The BLM, WRN, and RECQ4 genes are mutated in Bloom syndrome, Werner syndrome, and Rothmund-Thomson syndrome, respectively (1–3). A hallmark of Bloom syndrome cells is the drastic increase in the level of sister chromatid exchange (SCE),4 which results from homologous recombination (HR) associated with cross-over of the DNA damage caused during DNA replication (4, 5). It is therefore believed that Blm acts as an anti-recombination factor and inhibits aberrant recombination. This idea is supported by the observation that Sgs1, the yeast ortholog of Blm, facilitates the resolution of aberrant joint molecules during meiotic HR (6, 7) and following replication blockage (8).
HR plays a critical role in the maintenance of genome stability by repairing DNA double-strand breaks (DSBs) and releasing replication blockages at damaged template strands (9, 10). The current model for HR-mediated DSB repair is that DSBs are processed to produce a 3′ single-stranded overhang, along which Rad51 is polymerized (11, 12). The resulting Rad51-DNA filament undergoes homology search and strand invasion into intact homologous duplex DNA, leading to the formation of the D-loop structure. DNA synthesis from the invading strand followed by dissociation from the homologous duplex DNA and subsequent re-annealing of the newly synthesized strand with the other end of the DSB completes the repair. This type of HR, referred to as synthesis-dependent strand anneal (SDSA), results in sequence transfer from the intact template sequence (donor) to the damaged DNA (recipient), and accounts for the majority of mitotic HR (11, 13). Extensive strand exchange of the D-loop, on the other hand, leads to the generation of Holliday junction (HJ) intermediates. SDSA does not cause cross-overs, whereas HR involving the Holliday junction often causes cross-overs, such as SCE and meiotic HR. An increase in the level of SCE in Bloom syndrome cells therefore supports the idea that Blm suppresses the formation of HJ as well as recombinogenic DNA lesions. This idea is supported by the biochemical evidence of the Blm-dependent resolution of Holliday junctions (14). On the other hand, in Drosophila, DmBlm is known to facilitate the repair of DSB by promoting SDSA (15, 16). However, the role of Blm in SDSA in the other higher eukaryotic cells has not been defined.
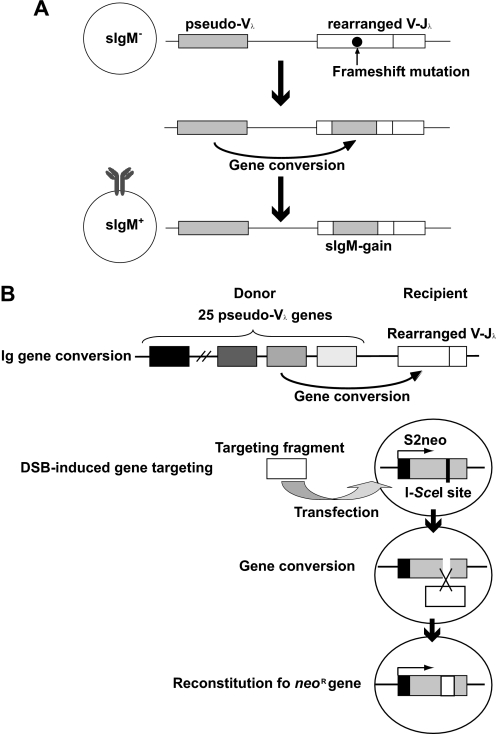
BLM−/− cells established from the chicken DT40 B lymphocyte line exhibit a marked increase in the frequency of both SCE and targeted integration (17–19), as do human Bloom syndrome cells (20, 21). In this study, using the chicken DT40 cells, we investigated the role of Blm in SDSA induced by defined DNA damage. To this end, we evaluated this type of SDSA using two phenotypic assays designed to analyze Ig gene conversion and DSB-induced gene targeting. Ig gene conversion diversifies the Ig variable (V) gene through HR during in vitro passage. This reaction is initiated by activation-induced cytidine deaminase-mediated uracil formation at the functional rearranged V-region (22–24). Uracil is converted to an abasic site, probably leading to a single-strand gap (25). This lesion in the functional rearranged VJλ stimulates the nonreciprocal sequence transfer of a single nucleotide to several hundred nucleotides, from an array of “pseudo-Vλ” regions (donor), located upstream from the functional rearranged VJλ, to the rearranged V region (recipient) (26–28) (see Fig. 1A). Because donor and recipient segments have an ∼10% sequence divergence, sequential Ig gene conversion events are able to substantially diversify Ig V segments. Ig gene conversion is raised only by SDSA without the formation of a Holliday junction. Hence, phenotypic analysis of Ig gene conversion provides a unique opportunity to selectively examine the role of Blm in activation-induced cytidine deaminase-induced SDSA. Moreover, nucleotide sequence analysis of Ig gene conversion products can evaluate the accuracy of HR. Like Ig gene conversion, DSB-induced gene targeting is mediated only by SDSA. The induction of DSBs by a rare-cutting endonuclease, I-SceI, at the endogenous locus, increases the frequency of gene targeting by 3 orders of magnitudes, and the frequency of gene targeting can be evaluated by measuring the reconstitution of a marker gene (29) (see Fig. 1B).
FIGURE 1.
Schematic diagram of assay systems used in this study. A, principle of the Ig gene conversion assay. The predominantly sIgM-negative DT40 clone contains a frameshift in its rearranged V-Jλ segments, which can be repaired by pseudogene-templated conversion events. The rate of Ig gene conversion can be measured in subclones by flow cytometric analysis of sIgM staining. B, phenotypic assays of Ig gene conversion and DSB-induced gene targeting. Pseudo-V genes and the targeting fragment act as donors for the rearranged Vλ segment and S2neo, respectively.
We here show that the loss of Blm drastically reduces the rate of Ig gene conversion without compromising its accuracy or affecting the length of the gene conversion tracts, indicating that Blm plays a role in the promotion of SDSA. This is an unexpected result, because Blm is in fact believed to suppress general HR reactions, particularly recombination between diverged homologous sequences. To understand the function of Blm in SDSA, we analyzed the effect of heterologous sequences near a DSB site on HR-dependent DSB repair. The data demonstrate that Blm can promote SDSA when there is sequence divergence between the damaged recipient DNA and the homologous donor sequence. Thus, Blm has both positive and negative effects on HR, depending upon the type of DNA damage and the step of the HR reaction.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
Gene targeting substrates were amplified with primers listed in supplemental Table S1. Amplified PCR products were cloned into the pCR2.1-TOPO vector (Invitrogen).
Cell Culture, DNA Transfection, and γ-Irradiation
The cells were cultured in RPMI 1640 supplemented with 10−5 m β-mercaptoethanol, 10% fetal calf serum, and 1% chicken serum (Sigma) at 39.5 °C. Methods of DNA transfection for producing stable transfectants and genotoxic treatments were as described previously (30). 137Cs (0.02 gray/s; Gammacell 40, Nordion, Kanata, Canada) was used for γ-irradiation.
Analysis of Ig Gene Conversion
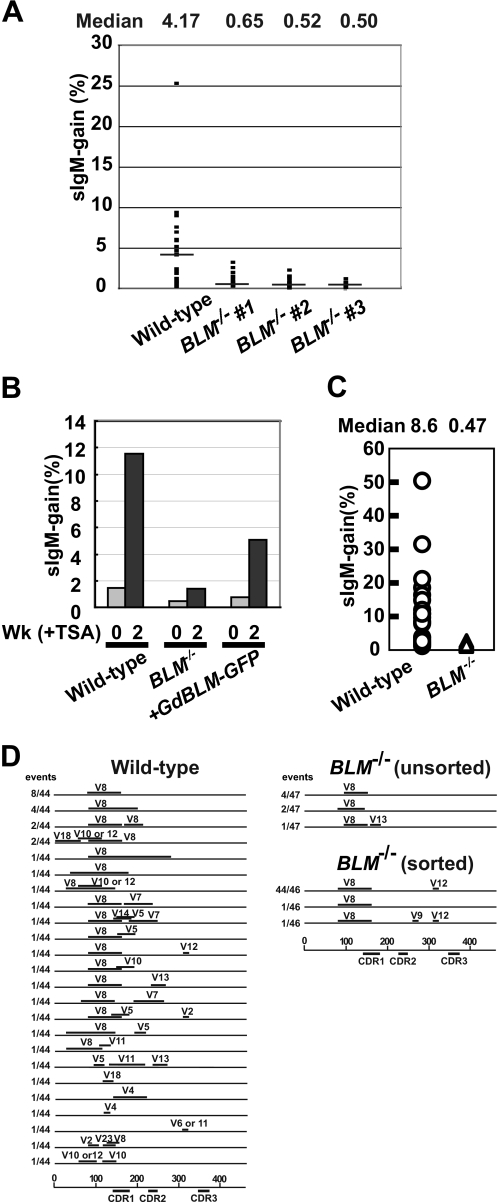
BLM−/− cells were established from CL18, a subclone of DT40 cells that is negative for sIgM (26). We confirmed that BLM−/− cells retained the same frameshift mutation as wild type CL18 cells by sequencing the Ig Vλ region. The rate of Ig gene conversion was assessed by measuring the gain of sIgM expression as described previously (27). Briefly, sIgM gain revertants were monitored by flow cytometric analysis of cells that had been expanded for 3 weeks after subcloning and then stained with fluorescein isothiocyanate-conjugated goat anti-chicken IgM (Bethyl, Montgomery, TX). To enhance Ig gene conversion, tricostatin A (TSA) (Wako Osaka; concentration, 1.25 ng/ml) was added to a mixture of sIgM-negative subclones (see Fig. 2). In each analysis, the abundance of sIgM-positive cells was determined as the percentage of live cells whose fluorescein isothiocyanate-conjugated fluorescence intensity is at least eight times higher than the fluorescein isothiocyanate-conjugated fluorescence peak of sIgM-negative cells.
FIGURE 2.
Reduced Ig gene conversion rate in BLM−/− cells. A, the frequency of sIgM gain revertants was determined in wild type and BLM−/− cells that were clonally expanded for 3 weeks. The median value was calculated using 23 independent subclones for wild type, and 24, 24, and 15 subclones for three independently established BLM−/− clones. The horizontal bars in the panel indicate median percentages. B, cells were cultured in medium containing 1.25 ng/ml TSA, and the frequency of sIgM gain revertants was determined at 14 days. C, the abundance of sIgM gain variants was determined in ∼24 parallel cultures derived from sIgM-negative single cells after clonal expansion for 12 days exposure to TSA. D, gene conversion tract spectra, showing the average tract length in the rearranged Vλ segments from the sIgM-positive revertants. The horizontal lines represents the rearranged Vλ (450 bp); the horizontal bars above the lines represent gene conversion tract.
Analysis of Ig Vλ Nucleotide Sequence
DNA was extracted from expanded subclones at 4 weeks after TSA treatment. PCR-amplified fragments of Vλ segments were cloned into a plasmid and subjected to base sequence analysis. Rearranged Vλ was amplified by PCR with Pyrobest DNA polymerase (Takara Bio) (30 cycles of 94 °C for 30 s, 60 °C for 1 min, and 72 °C for 1 min) with 5′-CAGGAGCTCGCGGGGCCGTCACTGATTGCCG-3′ and 5′-GCGCAAGCTTCCCCAGCCTGCCGCCAAGTCCAAG-3′ primers, as described previously (31). PCR products were cloned into the TOPO pCR2.1 cloning vector (Invitrogen) and sequenced with the M13 forward (−20) or reverse primer using an ABI PRISM 3100 sequencer (Applied Biosystems). Sequence alignment, using GENETYX-MAC (Software Development, Tokyo, Japan), allowed the identification of changes from the parental sequences in each clone. Differentiating between nontemplated nucleotide substitutions and gene conversion was carried out as described previously (31).
I-SceI-induced Gene Targeting
107 cells were suspended in 0.1 ml of Nucleofector Solution T (Amaxa Biosystems) and electroporated using Amaxa program B-23 (Amaxa Biosystems). 2 μg of targeting DNA fragment with expression vectors (Gd-BLM-GFP, EXO1, and human BLM) and nuclease expression vector were transfected. pBluescript II KS+ was used as a negative control. The targeting DNA fragment was amplified by PCR from the Mneo-1 to Mneo-4 substrate plasmid using Pyrobest DNA polymerase (Takara Bio) (30 cycles at 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 2 min), with the 5′-GGATCGGCCATTGAACAAGATGGATTGCAC-3′ and 5′-GGAAACAGCTATGACCATGATTACGCCAAG-3′ primers. Twenty-four hours after electroporation, the number of live cells was counted using fluorescence-activated cell sorter, and the cells were transferred to 96-well cluster trays with or without 2.0 mg of G418 per ml. The cells were grown for 7–10 days, and the HR frequencies were calculated by the following equation: HR frequency (colonies/cells) = number of G418-resistant colonies/(plating efficiency of transfected cells in the absence of G418 × number of live cells determined by fluorescence-activated cell sorter 24 h after electroporation).
Measurement of Chromosome Aberrations
Chromosome aberrations were measured as described previously (32).
RESULTS
Analysis of Ig Gene Conversion in BLM−/− Cells
To investigate the role of Blm in SDSA, we examined the rate of Ig gene conversion. To this end, we measured the gain of surface IgM (sIgM) expression in subclones derived from wild type and BLM−/− cells that have a frameshift mutation in the IgVλ segment, as documented previously (25–27). Because the frameshift mutation is removed by gene conversion from pseudo-V segments to Ig Vλ, the level of sIgM gain may reflect the frequency of gene conversion (Fig. 1A). We cultured multiple subclones derived from limiting dilution culture of wild type and BLM−/− strains. During 3 weeks of clonal expansion, only ∼0.6% of BLM−/− cells showed a gain in sIgM, whereas ∼4% of wild type cells showed a gain in sIgM+ (Fig. 2A). Because the sIgM staining in the BLM−/− cells was close to the background signal of flow cytometric analysis, we cultured the wild type and BLM−/− cell in the presence of a histone deacetylase inhibitor, TSA, which increases the frequency of Ig gene conversion in DT40 cells 50–100-fold (33), to detect possible rare gene conversion events. Over the course of 2 weeks of expansion in the presence of TSA, 10% of the wild type cells acquired sIgM expression, whereas only 0.92% of BLM−/− cells were converted to sIgM-positive (Fig. 2B). Similarly, fluctuation analysis of sIgM gain in subclones indicated that the median of sIgM gain was 8.6% for wild type cells and only 0.47% for BLM−/− cells (Fig. 2C). These results suggest that Blm might augment the rate of Ig gene conversion. Alternatively, Blm might increase the accuracy of gene conversion and thereby contribute to the restoration of functional Ig V genes.
Blm Increases the Rate of Ig Gene Conversion
To evaluate the accuracy of Ig gene conversion, we determined the nucleotide sequences of Ig Vλ segments. We incubated cells for 4 weeks in the presence of TSA, amplified the Ig Vλ segments from the expanded cells by PCR, and then determined the nucleotide sequences of the amplified fragments. For the wild type cells, gene conversion tracts were detected in 37 of 44 Ig Vλ sequences (84%). By contrast, gene conversion events were detectable in only 7 of 47 Ig Vλ sequences in the BLM−/− cells (15%) (Fig. 2D). Aberrant gene conversion events were not observed in the BLM−/− cells, indicating that the accuracy of HR is not compromised by the loss of Blm. These results indicate that Blm may increase the rate of Ig gene conversion in DT40 cells.
To explore the role of Blm in Ig gene conversion, we analyzed the length of the gene conversion tracts and the usage of pseudo-Vλ donors, focusing on the gene conversion events that caused the sIgM gain. We sorted sIgM-positive cells from 23 independent BLM−/− subclones that had been independently exposed to TSA for 4 weeks and determined the nucleotide sequences of 46 segments from each genotype. Wild type cells used a variety of pseudo-Vλ segments as a donor (Fig. 2D). However, BLM−/− cells used only limited species of the pseudo-V segments; of the 92 gene conversion events, 46 used the pseudo-V8 segment and 45 used the pseudo-V12 segment (Fig. 2D). The nucleotide sequences of pseudo-V8 and pseudo-V12 segments have a higher identity with the rearranged Vλ segment, compared with segments that were used only in wild type cells. Thus, Blm may promote gene conversion with diverged donor segments, thereby contributing to the extensive diversification of Ig V genes.
Blm Does Not Play a Major Role in HR between Identical Sequences
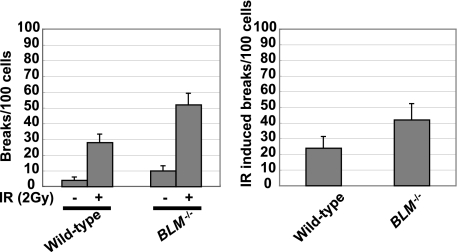
To determine whether the marked reduction in the frequency of Ig gene conversion in BLM−/− cells is simply caused by general defects in HR, we tested whether Blm is required for efficient HR between identical sequences. For this purpose, we measured sister-mediated HR repair by counting the number of chromosomal breaks in cells following ionizing radiation (IR) in the G2 phase, when DSBs are preferentially repaired by HR using the intact sister chromatid rather than by nonhomologous end joining in DT40 cells (30). Because after IR, cells in the G2 phase, but not in the S phase, can enter the M phase within 3 h, we can selectively evaluate HR between sister chromatids (sister HR) by analyzing chromosomal breaks in mitotic cells at 3 h after IR (34). BLM−/− cells showed increased levels of spontaneously arising chromosomal breaks (Fig. 3, left panel), as previously reported (17–19). We measured the number of chromosomal breaks at 3 h after 2 grays of γ-ray irradiation. We subtracted the number of spontaneous chromosomal breaks from this number to obtain the number of IR-induced breaks (Fig. 3, right panel). The number of IR-induced breaks increased only slightly (1.5-fold) in BLM−/− cells, compared with wild type cells (Fig. 3, right panel). This observation is in marked contrast with the dramatic increase (11-fold) in the number of IR-induced breaks in RAD54−/− cells (30). Thus, Blm plays only a minor role, if any, in HR using the intact sister chromatid. Taking the critical role played by Blm in Ig gene conversion into account, this slight reduction of sister HR-dependent DSB repair in BLM−/− cells suggests that Blm is important for HR between diverged sequences but not for HR between identical sequences.
FIGURE 3.
Proficient DSB repair mediated by HR between sister chromatids even in the absence of Blm. Chromosomal breaks without γ-rays (−) or following exposure to 2-gray γ-rays (+) of wild type and BLM−/− cells are shown. Immediately after ionizing radiation, mitotic cells were enriched by colcemid treatment for the last 3 h before cell harvest. The data are represented as macrochromosomal (1–5 and Z) breaks/100 metaphase spreads. S.E. was calculated as (the square root of the number of breaks)/(the number of metaphase spreads analyzed). One hundred metaphase spreads were analyzed in each analysis.
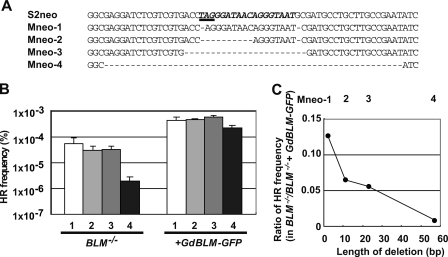
Blm Is Required for Efficient DSB-induced Gene Targeting When Heterologous Sequences Are Present near the DSB Site
We next tested whether Blm promotes HR between diverged homologous sequences. To address this issue, we employed a DSB-induced gene targeting assay (Fig. 1B). We examined the efficiency of DSB-induced gene targeting using a variety of targeting DNA fragments (donor; Mneo-1 to Mneo-4) (Fig. 4A) (35). We engineered BLM−/− cells carrying the S2neo construct at the Ovalbumin locus. Transient transfection of the I-SceI restriction enzyme cuts a cleavage site at S2neo. This cleavage stimulates the targeted integration of co-transfected donor DNA fragments into S2neo and removes the termination codon present in S2neo (underlined sequence in Fig. 4A), thereby leading to the reconstitution of the functional neomycin resistance gene. Of the series of gene targeting donor fragments, Mneo-1 has the highest homology to the recipient S2neo, whereas Mneo-4 has the largest deletion near the S2neo I-SceI site (Mneo-1 to Mneo-4 in Fig. 4A). Therefore, there are 11, 23, and 56 heterologous sequences near the I-SceI site of S2neo, when it recombines with Mneo-2, Mneo-3, and Mneo-4, respectively. The knock-in of these donor fragments into S2neo results in the deletion of amino acid sequences from the coding sequences of the neomycin resistance gene. We verified that the deletion does not affect the activity of the gene (data not shown), as previously reported (35). We transfected individual targeting donor fragments together with the I-SceI expression vector into the BLM−/− clone carrying S2neo. We then measured the frequency of gene targeting by counting the number of neomycin-resistant colonies. Gene targeting events in the BLM−/− cells for the Mneo-1 fragment were about 10-fold less frequent than the control cells complemented by the transient expression of a BLM transgene (Fig. 4B). Remarkably, gene targeting events in BLM−/− cells for the most divergent donor construct, Mneo-4, were about 100-fold less frequent than the control cells. We calculated the ratio of the gene targeting frequency in BLM−/− cells relative to that in the control cells (Fig. 4C) and found that the BLM−/− cells showed a more pronounced reduction of gene targeting efficiency in the presence of larger numbers of heterologous sequences in the recipient S2neo, when compared with the complemented control cells. These results, together with the data for Ig gene conversion (Fig. 2), strongly suggest that Blm facilitates HR between diverged homologous sequences.
FIGURE 4.
Heterologous sequences at a DSB reduce the DSB-induced HR more prominently in BLM−/− cells than in control Blm reconstituted cells. A, base sequence alignment around the I-SceI site in a series of targeting fragments (Mneo-1 to Mneo-4). Donor fragments contain the sequences that are missing in the corresponding sequences of the S2neo recipient (shown by hyphens). Underlined characters show stop codons. Italicized characters indicate I-SceI recognition sequences. B, the gene targeting frequency of the Mneo-1 to Mneo-4 targeting fragments in BLM−/− cells is shown on the y axis. Targeting fragments shown by number (bars 1–4) were transfected into cells carrying S2neo. +GdBLM-GFP indicates BLM−/− cells complemented by the transient expression of a chicken BLM transgene. The error bars represent standard deviations. C, relative HR frequencies on the y axis were calculated by dividing the HR frequency of BLM−/− cells (the percentage of G418R cells among the transfected cells) by that of the complemented cells. The x axis shows the number of missing sequences shown in A.
Blm Facilitates Homologous Pairing in HR When There Are Heterologous Sequences around DSB Site
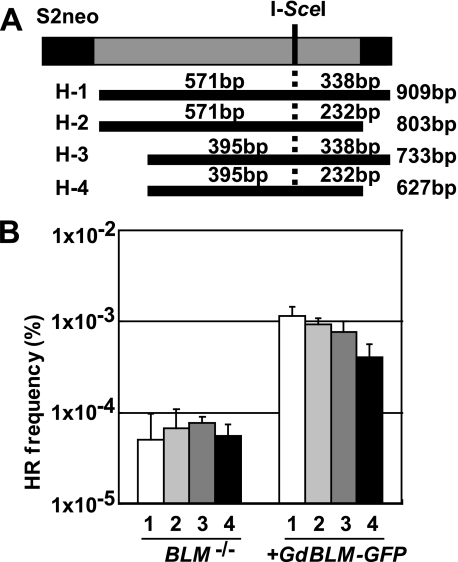
To address how Blm facilitates HR between diverged homologous sequences, we examined the role of Blm in homologous pairing step in HR. To this end, we examined the efficiency of DSB-induced gene targeting using a series of wild type neo fragments carrying variable lengths of homologous arm (Fig. 5A, H-1 to H-4). There are 18-bp heterologous sequences near the I-SceI site of S2neo, when it recombines with all of these targeting fragments. As shown in Fig. 5B, in the control cells complemented by BLM transgene, frequency of the gene targeting increased as the length of homologous arm increases (Fig. 5B). However, gene targeting events in BLM−/− cells were not significantly increased even when a longer homologous arm exists, indicating that limited the length of the homologous arm is used in BLM−/− cells (Fig. 5B). These results suggest that Blm facilitates homologous pairing step in HR, when there are heterologous sequences around the DSB site.
FIGURE 5.
Blm facilitates homologous pairing step in HR. A, donor WTneo fragments carrying various length of homologous arm. The length of left and right arm and total fragments were shown. B, the gene targeting frequency of the H-1 to H-4 targeting fragments in BLM−/− and control Blm reconstituted cells is shown on the y axis. The error bars represent standard deviations.
Relationship of Blm and Exo1 in HR between Diverged Sequences
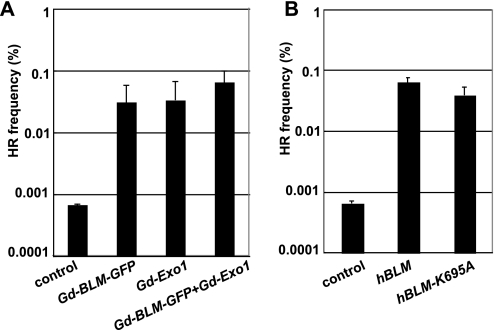
Recently, a biochemical study revealed that the processing of DNA lesions is promoted by the cooperative reaction of Blm and Exo1, a 5′ to 3′ double-strand DNA exonuclease (reviewed in Ref. 36), thereby facilitating later HR events (37). To examine the possible collaboration between Blm and Exo1 in HR between diverged sequences, we overexpressed Blm and Exo1 concurrently in wild type cells harboring the S2neo construct at the Ovalbumin locus and measured gene targeting efficiency using Mneo-3 as a donor sequence. There are 23-bp heterologous sequences near I-SceI site of S2neo, when it recombines with Mneo-3. Surprisingly, the frequency of gene targeting events was drastically increased by the overexpression of either Blm or Exo1 (Fig. 6A). On the other hand, gene targeting efficiency was only increased a few fold by the simultaneous expression of Blm and Exo1, when compared with the overexpression of either Blm or Exo1 alone (Fig. 6A). These results support the idea that overexpression of either Blm or Exo1 can effectively promote HR, presumably at the same step, such as 3′ tail formation at the I-SceI-induced DSB, or subsequent formation of the D-loop structure. According to this scenario, because Exo1 alone can effectively generate an HR intermediate (for example, a D-loop), the additional expression of Blm has a less prominent impact on HR, in comparison with the overexpression of Exo1 alone.
FIGURE 6.
Relationship of Blm and Exo1 and an ATPase-independent promotion of HR between diverged sequences by Blm. A, overexpression of either Blm or Exo1 significantly increases HR between diverged sequences. BLM and/or Exo1 transgenes together with I-SceI expression vector were transfected in wild type cells carrying S2neo with targeting-DNA-fragment Mneo-3. The error bars represent standard deviations. The data for the negative control was from B. B, human BLM (wild type and K695A) transgenes together with I-SceI expression vector were transfected in wild type cells carrying S2neo with targeting-DNA-fragment Mneo-3. Gene targeting frequency and standard deviation are also presented.
Blm Stimulates DSB-induced Gene Targeting through an ATPase Activity-independent Mechanism
To explore the molecular mechanism underlying Blm-dependent enhancement of HR, we tested whether ATPase activity of Blm is required for HR between diverged sequences. To this end, we overexpressed a human Blm K695A mutant (ATPase-dead) in wild type cells harboring the S2neo construct and measured gene targeting efficiency using Mneo-3 as the donor sequence. As expected, wild type human Blm increased the gene targeting efficiency to the same extent as did chicken Blm (Fig. 6B). Surprisingly, ectopic expression of the Blm K695A mutant also raised the gene targeting efficiency as effectively as the wild type Blm (Fig. 6B). This observation is in agreement with the biochemical data indicating that the K695A Blm mutant as well as wild type Blm stimulates Rad51-mediated strand exchange between single-stranded DNA and homologous duplex DNA (52). In summary, Blm may promote HR between diverged sequences through an ATPase activity-independent mechanism.
DISCUSSION
It has been shown that Blm acts as an anti-recombinase and prevents the generation of HJ recombination intermediates (38) as well as aberrant recombination intermediates following replication blockage (9, 10). The fact that both SCE and gene targeting frequencies are markedly augmented in the absence of Blm is consistent with the idea that Blm plays a role in the suppression of HR (20, 21). However, it is unclear whether this marked augmentation is caused by the increased efficiency of HR or, alternatively, the increase in the amount of recombinogenic substrate DNA. By using unique phenotypic assays for HR, we were able to show that Blm plays a critical role in the promotion of HR. We employed two SDSA-mediated HR reactions: Ig gene conversion and DSB-induced gene targeting. It should be noted that neither HR reaction is associated with the formation of the HJ, whereas both are initiated by enzyme-mediated DNA damage, activation-induced cytidine deaminase-induced abasic sites, and I-SceI-induced DSBs. Thus, we are able to selectively evaluate the function of Blm in HR by excluding Blm as a factor in the formation of the HJ or in the release of replication blockage. We here show that Blm is required for efficient Ig gene conversion, particularly when diverged pseudo-V segments are used as a donor (Fig. 2). In contrast, BLM−/− cells can perform HR-mediated repair with a nearly normal level when HR is carried out between identical sequences, i.e. sister chromatids (Fig. 3). These results suggest that Blm is required for efficient recombination between diverged sequences but not necessary for HR between identical sequences. This idea was verified by data showing that the loss of Blm decreased the frequency of DSB-induced gene targeting to a greater extent as the number of heterologous sequences near the DSB site increased (Fig. 4). Moreover, we showed that the limited length of homologous arm is used in BLM−/− cells (Fig. 5), suggesting that Blm facilitates homologous pairing step of HR. We therefore conclude that Blm promotes homologous pairing and thereby participating in HR when there is sequence variation between donor and recipient sequences.
A mismatch between donor and recipient sequences interferes with HR in the following two ways. First, the mismatch interferes with strand invasion into the homologous donor sequences and thereby with the formation of the D-loop structure. Second, the mismatch strongly destabilizes the D-loop. According to our data, in both cases, Blm can counteract the inhibitory effect of the mismatch on HR. In the initial step of HR, broken DNA ends are processed into long 3′ single-stranded DNA overhangs by nuclease(s) as well as by DNA helicase(s) (37, 39–42). Recent studies have revealed the two-step mechanism of DSB end resection in budding yeast: the MRX complex and Sae2-dependent initial minimum resection followed by extensive resection by two nucleases, Exo1 nuclease and Dna2 nuclease, which act cooperatively with Sgs1, the yeast homolog of Blm (43, 44). Hence, a possible scenario is that Blm may facilitate the nuclease-dependent resection of DSBs, and the resulting longer 3′ tails might be able to effectively perform strand invasion in the presence of the heterologous 3′ ends (45). This might explain the role of Blm in the promotion of DSB-dependent gene targeting (Fig. 4), because Blm might facilitate D-loop formation by increasing the length of the 3′ tail (Fig. 6A). The removal of the heterologous 3′ end can be carried out by the Rad1/Rad10 endonuclease and the Srs2 helicase in yeast (46) and by the Fen-1 endonuclease in vertebrate cells (35).
An alternative (but not mutually exclusive) possibility is that Blm stabilizes the D-loop structure and thereby allows extensive DNA synthesis. This model agrees with the data on DSB-induced gene targeting and Ig gene conversion. In both reactions, although the heterologous sequences may strongly destabilize the D-loop structure, Blm may counteract the negative effect of the heterologous sequences on HR. This idea is supported by the accompanying paper by Bugreev et al. (52), in which they show that human Blm enhances D-loop formation by about 4-fold on DNA substrates containing 10% mismatches in vitro. This view is also supported by the several studies in Drosophila. DmBLM mutant (mus309) in Drosophila shows a defect in extensive DNA repair synthesis in the SDSA mechanism, generating short DNA synthesis tracts (15, 16). In addition, a biochemical study using purified protein from Drosophila revealed that DmBlm stimulates strand annealing without ATP hydrolysis, suggesting the possible role of Blm to facilitate SDSA (47). In summary, Blm can increase the efficiency of HR between diverged homologous sequences by promoting both the resection of DSBs and the formation of D-loops.
We showed that Blm promotes HR between diverged homologous sequences through an ATPase-independent mechanism (Fig. 6B). Because the processing of DNA lesions in vitro is promoted by a cooperative reaction between purified human Blm and Exo1, even in the absence of ATP (37), this may account for the ATPase-independent promotion of HR in vivo (Fig. 6B). In addition, the stabilization of the D-loop by Blm may account for the ATPase-independent promotion (Fig. 6B). This idea is supported by the accompanying paper by Bugreev et al. (52), which shows that human Blm alone can enhance Rad51-dependent D-loop formation through an ATPase-independent mechanism in vitro. Moreover, a previous biochemical study showed that DmBlm stimulates strand annealing by an ATPase-independent mechanism (47), raising the possibility that Blm promotes strand annealing and thereby stabilizes formed D-loop.
Our results demonstrate that Blm can significantly promote some HR reactions, including HR-dependent diversification of Ig V genes. This reaction plays a considerably more important role than does V(D)J site-specific recombination in the diversification of B-lymphocyte precursors in birds and domestic animals such as rabbit, sheep, pig, and cattle (reviewed in Refs. 48 and 49). Because HR needs to perform more complex tasks in vertebrates, Blm has adapted to promote HR between diverged homologous sequences in B-cell precursors. In addition to Ig V diversification, Blm-dependent promotion of HR may contribute to cellular tolerance to topoisomerase poisons such as camptothecin and etoposide. These chemotherapeutic agents stabilize the topoisomerase-DNA cleavage complex, thereby inducing DSBs (reviewed in Ref. 50). 3′ tails covalently bound by topoisomerase I are unable to efficiently undergo homology search and D-loop formation, because heterologous sequences at DSBs interfere with HR. The present study suggests that the Blm-dependent promotion of HR may account for the hypersensitivity of Blm-deficient cells to topoisomerase poisons (17, 51).
Supplementary Material
Acknowledgments
We thank M. Nishikawa, R. Ohta, Y. Satoh, S. Tanaka, A. Noguchi, S. Ohsako, A. Shodai, and R. Tsutsui for technical support.
This work was supported in part by funds from the Program for Promotion of Basic Research Activities for Innovative Biosciences (to S. T.), grants from the Uehara Memorial Foundation and the Naito Foundation (to S. T.), and grants from the Kanae Foundation (to K. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- SCE
- sister chromatid exchange
- HR
- homologous recombination
- DSB
- double-strand break
- SDSA
- synthesis-dependent strand anneal
- HJ
- Holliday junction
- TSA
- tricostatin A
- sIgM
- surface IgM
- IR
- ionizing radiation.
REFERENCES
- 1.Cobb J. A., Bjergbaek L. (2006) Nucleic Acids Res. 34, 4106–4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hickson I. D. (2003) Nat. Rev. Cancer 3, 169–178 [DOI] [PubMed] [Google Scholar]
- 3.Kitao S., Shimamoto A., Goto M., Miller R. W., Smithson W. A., Lindor N. M., Furuichi Y. (1999) Nat. Genet. 22, 82–84 [DOI] [PubMed] [Google Scholar]
- 4.Chaganti R. S., Schonberg S., German J. (1974) Proc. Natl. Acad. Sci. U.S.A. 71, 4508–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonoda E., Sasaki M. S., Morrison C., Yamaguchi-Iwai Y., Takata M., Takeda S. (1999) Mol. Cell. Biol. 19, 5166–5169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jessop L., Lichten M. (2008) Mol. Cell 31, 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh S. D., Lao J. P., Taylor A. F., Smith G. R., Hunter N. (2008) Mol. Cell 31, 324–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Branzei D., Sollier J., Liberi G., Zhao X., Maeda D., Seki M., Enomoto T., Ohta K., Foiani M. (2006) Cell 127, 509–522 [DOI] [PubMed] [Google Scholar]
- 9.San Filippo J., Sung P., Klein H. (2008) Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 10.Branzei D., Vanoli F., Foiani M. (2008) Nature 456, 915–920 [DOI] [PubMed] [Google Scholar]
- 11.Pâques F., Haber J. E. (1999) Microbiol. Mol. Biol. Rev. 63, 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Symington L. S. (2002) Microbiol. Mol. Biol. Rev. 66, 630–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson R. D., Jasin M. (2000) EMBO J. 19, 3398–3407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu L., Hickson I. D. (2003) Nature 426, 870–874 [DOI] [PubMed] [Google Scholar]
- 15.Adams M. D., McVey M., Sekelsky J. J. (2003) Science 299, 265–267 [DOI] [PubMed] [Google Scholar]
- 16.McVey M., Larocque J. R., Adams M. D., Sekelsky J. J. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 15694–15699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kohzaki M., Hatanaka A., Sonoda E., Yamazoe M., Kikuchi K., Vu Trung N., Szüts D., Sale J. E., Shinagawa H., Watanabe M., Takeda S. (2007) Mol. Cell. Biol. 27, 2812–2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imamura O., Fujita K., Itoh C., Takeda S., Furuichi Y., Matsumoto T. (2002) Oncogene 21, 954–963 [DOI] [PubMed] [Google Scholar]
- 19.Otsuki M., Seki M., Inoue E., Yoshimura A., Kato G., Yamanouchi S., Kawabe Y., Tada S., Shinohara A., Komura J., Ono T., Takeda S., Ishii Y., Enomoto T. (2007) J. Cell Biol. 179, 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.So S., Nomura Y., Adachi N., Kobayashi Y., Hori T., Kurihara Y., Koyama H. (2006) Genes Cells 11, 363–371 [DOI] [PubMed] [Google Scholar]
- 21.So S., Adachi N., Lieber M. R., Koyama H. (2004) J. Biol. Chem. 279, 55433–55442 [DOI] [PubMed] [Google Scholar]
- 22.Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. (2000) Cell 102, 553–563 [DOI] [PubMed] [Google Scholar]
- 23.Harris R. S., Sale J. E., Petersen-Mahrt S. K., Neuberger M. S. (2002) Curr. Biol. 12, 435–438 [DOI] [PubMed] [Google Scholar]
- 24.Arakawa H., Hauschild J., Buerstedde J. M. (2002) Science 295, 1301–1306 [DOI] [PubMed] [Google Scholar]
- 25.Nakahara M., Sonoda E., Nojima K., Sale J. E., Takenaka K., Kikuchi K., Taniguchi Y., Nakamura K., Sumitomo Y., Bree R. T., Lowndes N. F., Takeda S. (2009) PLoS Genet. 5, e1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buerstedde J. M., Reynaud C. A., Humphries E. H., Olson W., Ewert D. L., Weill J. C. (1990) EMBO J. 9, 921–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawamoto T., Araki K., Sonoda E., Yamashita Y. M., Harada K., Kikuchi K., Masutani C., Hanaoka F., Nozaki K., Hashimoto N., Takeda S. (2005) Mol. Cell 20, 793–799 [DOI] [PubMed] [Google Scholar]
- 28.Saberi A., Nakahara M., Sale J. E., Kikuchi K., Arakawa H., Buerstedde J. M., Yamamoto K., Takeda S., Sonoda E. (2008) Mol. Cell. Biol. 28, 6113–6122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rouet P., Smih F., Jasin M. (1994) Mol. Cell. Biol. 14, 8096–8106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takata M., Sasaki M. S., Sonoda E., Morrison C., Hashimoto M., Utsumi H., Yamaguchi-Iwai Y., Shinohara A., Takeda S. (1998) EMBO J. 17, 5497–5508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sale J. E., Calandrini D. M., Takata M., Takeda S., Neuberger M. S. (2001) Nature 412, 921–926 [DOI] [PubMed] [Google Scholar]
- 32.Fujimori A., Tachiiri S., Sonoda E., Thompson L. H., Dhar P. K., Hiraoka M., Takeda S., Zhang Y., Reth M., Takata M. (2001) EMBO J. 20, 5513–5520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seo H., Masuoka M., Murofushi H., Takeda S., Shibata T., Ohta K. (2005) Nat. Biotechnol. 23, 731–735 [DOI] [PubMed] [Google Scholar]
- 34.Sonoda E., Okada T., Zhao G. Y., Tateishi S., Araki K., Yamaizumi M., Yagi T., Verkaik N. S., van Gent D. C., Takata M., Takeda S. (2003) EMBO J. 22, 3188–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kikuchi K., Taniguchi Y., Hatanaka A., Sonoda E., Hochegger H., Adachi N., Matsuzaki Y., Koyama H., van Gent D. C., Jasin M., Takeda S. (2005) Mol. Cell. Biol. 25, 6948–6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tran P. T., Erdeniz N., Symington L. S., Liskay R. M. (2004) DNA Repair 3, 1549–1559 [DOI] [PubMed] [Google Scholar]
- 37.Nimonkar A. V., Ozsoy A. Z., Genschel J., Modrich P., Kowalczykowski S. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 16906–16911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karow J. K., Constantinou A., Li J. L., West S. C., Hickson I. D. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 6504–6508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gravel S., Chapman J. R., Magill C., Jackson S. P. (2008) Genes Dev. 22, 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sonoda E., Hochegger H., Saberi A., Taniguchi Y., Takeda S. (2006) DNA Repair 5, 1021–1029 [DOI] [PubMed] [Google Scholar]
- 41.Takeda S., Nakamura K., Taniguchi Y., Paull T. T. (2007) Mol. Cell 28, 351–352 [DOI] [PubMed] [Google Scholar]
- 42.Hopkins B. B., Paull T. T. (2008) Cell 135, 250–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G. (2008) Cell 134, 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mimitou E. P., Symington L. S. (2008) Nature 455, 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inbar O., Kupiec M. (1999) Mol. Cell. Biol. 19, 4134–4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pâques F., Haber J. E. (1997) Mol. Cell. Biol. 17, 6765–6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weinert B. T., Rio D. C. (2007) Nucleic Acids Res. 35, 1367–1376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lanning D., Zhu X., Zhai S. K., Knight K. L. (2000) Immunol. Rev. 175, 214–228 [PubMed] [Google Scholar]
- 49.Flajnik M. F. (2002) Nat. Rev. Immunol. 2, 688–698 [DOI] [PubMed] [Google Scholar]
- 50.Pommier Y. (2006) Nat Rev Cancer 6, 789–802 [DOI] [PubMed] [Google Scholar]
- 51.Otsuki M., Seki M., Kawabe Y., Inoue E., Dong Y. P., Abe T., Kato G., Yoshimura A., Tada S., Enomoto T. (2007) Biochem. Biophys. Res. Commun. 355, 477–482 [DOI] [PubMed] [Google Scholar]
- 52.Bugreev D. V., Mazina O. M., Mazin A. V. (2009) J. Biol. Chem. 284, 26349–26359 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.