Abstract
In the presence of ample tryptophan, transcription from the Bacillus subtilis trp operon promoter terminates to give a 140-nucleotide trp leader RNA. Turnover of trp leader RNA has been shown to depend on RNase J1 cleavage at a single-stranded, AU-rich region just upstream of the 3′ transcription terminator. The small size of trp leader RNA and its strong dependence on RNase J1 cleavage for decay make it a suitable substrate for analyzing the requirements for RNase J1 target site specificity. trp leader RNAs with nucleotide changes around the RNase J1 target site were more stable than wild-type trp leader RNA, showing that sequences on either side of the cleavage site contribute to RNase J1 recognition. An analysis of decay intermediates from these mutants suggested limited 3′-to-5′ exonuclease processing from the native 3′ end. trp leader RNAs were designed that contained wild-type or mutant RNase J1 targets elsewhere on the molecule. The presence of an additional RNase J1 cleavage site resulted in faster RNA decay, depending on its location. Addition of a 5′ tail containing 7 A residues caused destabilization of trp leader RNAs. Surprisingly, addition at the 5′ end of a strong stem loop structure that is known to stabilize other RNAs did not result in a longer trp leader RNA half-life, suggesting that the RNase J1 cleavage site may be accessed directly. In the course of these experiments, we found evidence that polynucleotide phosphorylase processivity was inhibited by a GCGGCCGC sequence.
Protective features of the 5′ and 3′ ends of prokaryotic mRNAs explain why these RNA molecules are not degraded immediately by the multiple ribonucleases that are present in a prokaryotic cell. The presence of a nucleoside triphosphate at the 5′ end renders this end a poor substrate for 5′-to-3′ exonucleolytic decay (1–3) or 5′ end-dependent endonucleolytic activity (4–6). The strong stem loop structure found at the 3′ end of many prokaryotic mRNAs, which is the transcription terminator structure, is resistant to 3′-to-5′ exonucleolytic decay (7, 8). A general model for the initiation of mRNA decay in prokaryotes, which is based on numerous studies in Escherichia coli, is as follows: initiation of decay occurs by an endonucleolytic cleavage in the body of the message. Such cleavage generates an upstream fragment with an unprotected 3′ end, which is a good substrate for 3′-to-5′ exonucleases, and a downstream fragment with a monophosphate nucleoside 5′ end, which is a good substrate for additional endonuclease cleavages (9–11). The downstream fragment could also be a good substrate for a 5′-to-3′ exonuclease activity; however, such an activity is not known to exist in E. coli.
The major decay-initiating endonuclease in E. coli is believed to be RNase E, a 5′ end-dependent endonuclease. That is, RNase E endonuclease activity is usually contingent on prior binding to the 5′ terminus, after which the enzyme tracks or loops to its target cleavage site (5, 12). RNase E vastly prefers an RNA substrate with a monophosphate 5′ end (4, 5, 13), and it is thought that conversion of the initial triphosphate 5′ end to a monophosphate 5′ end by a pyrophosphatase activity plays a significant role in RNase E-mediated initiation of decay (14, 15).
The Bacillus subtilis genome has no sequence homologue of RNase E. Instead, the recently discovered RNase J1 (16) is thought to be a major player in initiation of mRNA decay in B. subtilis. RNase J1 is essential, and growth of B. subtilis under conditions of reduced RNase J1 results in a general increase in mRNA half-life (16). A similar enzyme, named RNase J2, is not essential, and a strain with an RNase J2 gene knock-out shows no obvious phenotype. A number of RNAs are now known to be cleaved at specific sites by RNase J1 (1, 16–18), and a recent microarray study showed that the level of many RNAs is affected in a strain deleted for RNase J2 and having reduced expression of RNase J1 (19). Although it was believed initially that RNase J1 was exclusively an endonuclease, more recently it was discovered that RNase J1 has, in addition, a 5′-to-3′ exonuclease activity (2, 3), which requires a 5′-monophosphate end. This discovery is the basis for an alternative to the E. coli model for mRNA decay that would apply to organisms that express RNase J1: mRNA could be degraded directly from the 5′ end, after pyrophosphate removal, or RNase J1 could act as both an endonuclease to cleave the initial transcript and as a 5′ exonuclease on the downstream product of such cleavage (2, 20).
We have used the B. subtilis trp leader RNA to study aspects of RNA decay. In the presence of ample tryptophan, an 11-mer complex of the regulatory protein trp RNA-binding attenuation protein (TRAP)2 is activated and binds to 11 triplet repeats on the nascent trp operon transcript. Binding of TRAP allows formation of a transcription terminator structure such that transcription terminates before RNA polymerase enters into the trp operon protein coding sequences (21, 22). The terminated transcript is 140-nucleotides (nt) long (Fig. 1A), which is a useful size for studying the mechanism of RNA decay. Based on an analysis of trp leader RNA decay in ribonuclease mutant strains, and of a mutant trp leader RNA, we proposed that initiation of trp leader RNA decay in the presence of bound TRAP is dependent on RNase J1 endonuclease cleavage (23). Most of the trp leader RNA is in a form that should be protected from ribonuclease attack: the 5′ end is in a stem loop structure (5′SL), the 3′ end is in a strong transcription terminator structure (3′ TT), and a large part of the internal sequence is bound by the TRAP 11-mer (Fig. 1A, TBS). There is, however, a small, single-stranded, AU-rich sequence from nt 93–107, and we have shown that RNase J1 cleaves in this region in vivo and in vitro (1). Altering nucleotides at this site to give a GC-rich sequence resulted in a 4-fold increase in trp leader RNA half-life (23). RNase J1 cleavage is followed by 3′-to-5′ degradation of the upstream fragment by polynucleotide phosphorylase (PNPase, Ref. 24) and 5′-to-3′ degradation of the downstream fragment by RNase J1 itself (1). There is also evidence for a minor RNase J1 cleavage in the upstream end of the TBS (1).
FIGURE 1.
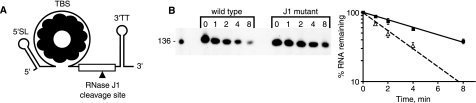
A, schematic diagram of trp leader RNA, showing structural features and location of primary RNase J1 cleavage site. B, Northern blot analysis of trp leader RNA in wild-type and RNase J1 conditional mutant strains grown in the presence of 1 mm IPTG. Time after rifampicin addition (min) is indicated above each lane. Migration of the 136-nt marker (lane M) is indicated at left. Semi-log plot of % RNA remaining versus time is shown at right. Open triangles, wild type; closed squares, RNase J1 mutant.
For the current study, we made a number of trp leader RNA constructs that were designed to probe the recognition requirements for RNase J1 cleavage, including changing the nucleotide sequence at the cleavage site, changing the location of the cleavage site relative to other RNA structural features, and adding an additional cleavage site. To our knowledge, this is the first fine-scale analysis of an RNase J1 cleavage site.
EXPERIMENTAL PROCEDURES
Bacterial Strains
BG626 was the host strain for plasmids carrying the wild type or mutant trp leader constructs. BG626 is trpC2 thr-5 and carries a spectinomycin resistance gene that replaces chromosomal sequences from the end of the aroH coding sequence (last 63 codons missing), through the trp leader sequence, to near the start of the trpE coding sequence (first 40 codons missing). Construction of this strain was described previously (23). For experiments with reduced RNase J1 levels, BG626 was transformed to erythromycin resistance using chromosomal DNA from the RNase J1 conditional mutant (25). In this strain, expression of RNase J1 is under control of the IPTG-inducible pspac promoter. The RNase J1 conditional mutant strain also contained plasmid pMAP65, which carries extra copies of the lacI gene (26). The host for cloning of mutant trp leader constructs was E. coli DH5α (27).
Plasmids
Plasmid pGD5 contains the wild type trp leader sequence and the mtrB gene encoding TRAP (23). The trp leader sequence is on an SphI-EcoRI fragment. A two-step PCR protocol was used to generate the mutant trp leader constructs. Oligonucleotide primers containing complementary sequences including the mutated nucleotides were used to amplify the sequence upstream of the trp leader (with a primer that included the SphI site) and the sequence downstream of the trp leader (with a primer that included the EcoRI site). The two amplicons were annealed to each other and used in a second round of PCR to generate a product that contained the mutated trp leader sequence on a fragment that had SphI and EcoRI recognition sites at the ends. This product was cloned between the SphI and EcoRI sites of the pGD5 vector. The pGD5 derivative bearing the mutant trp leader sequence was used to transform BG626 to tetracycline resistance (10 μg/ml).
PNPase Assay
5′ End-labeled trp leader RNA bearing a monophosphate 5′ end, used for PNPase assays, was prepared as described (1). PNPase reactions were done as described (23) using 1 nm RNA substrate, of which 10% was labeled, and 2.5 nm PNPase.
Northern Blot Analysis
RNA isolation and Northern blot analysis were performed as described (1). For determination of half-life, time points up to two half-lives were used. In all cases, the R2 values were greater than 0.9. Comparison of the half-lives between two trp leader RNAs was used in a two-sample t test to derive p values. A p value < 0.05 was considered significant.
RESULTS AND DISCUSSION
Stabilization of trp Leader RNA in a Strain with a Reduced Level of RNase J1
Previously, a trp leader RNA had been constructed in which the AU-rich, RNase J1 target sequence around nt 101 was changed to an 8-nt GC-rich sequence (23). (As is demonstrated below, cleavage appears to occur mainly after nt 101, not after nt 100, as previously mapped. Therefore, we refer to this RNase J1 cleavage as occurring at nt 101.) The trp leader RNA with the GC-rich sequence is called “NotI RNA,” because the 8-bp sequence encoding this GC-rich segment constitutes a NotI restriction endonuclease site in the DNA encoding the trp leader. Because this change resulted in a 4-fold increase of trp leader RNA half-life to about 10 min, we inferred that RNase J1 cleavage at nt 101 is important for initiating trp leader RNA decay. To show directly that the cellular level of RNase J1 could affect trp leader RNA half-life, we performed Northern blot analysis of trp leader RNA decay in wild-type and RNase J1 conditional mutant strains. In the latter strain, RNase J1 expression is under control of an IPTG-inducible promoter. Addition of IPTG results in a level of RNase J1 that is ∼5-fold lower than in the wild type strain (28). The result in Fig. 1B shows that, indeed, trp leader RNA half-life increases from 2.5 min in the wild type to 5.5 min in the RNase J1 mutant. Thus, merely reducing the level of RNase J1, without eliminating it, results in a 2-fold increase in trp leader RNA half-life. The previous and current results indicate that trp leader RNA half-life is determined primarily by RNase J1 cleavage at nt 101. We focused attention on the sequence requirements for this decay-initiating cleavage.
Effect of Mutations in the RNase J1 Target Site on trp Leader RNA Half-life
trp leader constructs were made that changed fewer nucleotides upstream or downstream of the 101-nt position than were changed in the NotI construct, and these were designated by the restriction endonuclease site that the change generated (EaeI, SacII, and NaeI constructs, Fig. 2A). Mutant trp leader constructs were introduced on a high-copy plasmid, which also contains the mtrB gene encoding TRAP, into a B. subtilis strain that is deleted for the endogenous trp leader region. According to the Zuker mfold structure prediction program (29), the 3′-TT structure was not affected by the changes that were introduced. In fact, wild-type sized trp leader RNA was detected for each of the constructs, indicating that the mutant sequences did not affect conformation of the transcription terminator structure or binding of TRAP, which is required for termination.
FIGURE 2.
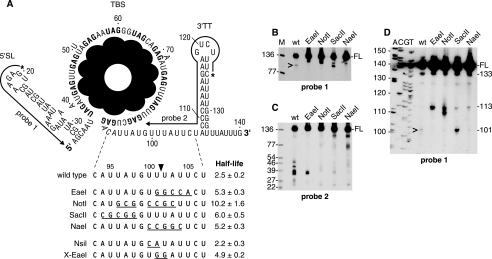
A, trp leader RNA sequence, showing mutations in the RNase J1 target site. Extent of complementarity of 5′ end-labeled probes to sequences near the 5′ and 3′ ends is indicated. Below the trp leader RNA schematic is shown the wild-type and mutant sequences of the RNase J1 target site. Half-lives of each RNA (min) are shown. The major site of RNase J1 cleavage after nt 101 is indicated by the arrowhead. B–D, Northern blot analysis of wt and mutant trp leader RNA, using probe 1 on low-resolution (B) and high-resolution (D) blots, and probe 2 (C). FL, full-length trp leader RNA. Leftmost lane (M) in B and C contained 5′ end-labeled TaqI fragments of plasmid pSE420 DNA (35), and the sizes of these fragments are indicated on the left. Sequencing ladder in D was generated on M13mp18 single-stranded DNA. Sizes of RNA bands detected by probe 1 in D and discussed in the text are indicated on the right. In B and D, the 101-nt upstream product of RNase J1 cleavage is indicated by the caret.
trp leader RNA half-life for each of the constructs was determined by Northern blot analysis after addition of rifampicin. The half-lives shown in Fig. 2A are the average of at least three determinations. The substitution of GC-rich sequence in each of the new constructs resulted in a trp leader RNA half-life increase from about 2.5 min for the wild type to 5–6 min for the mutant.
Analysis of Upstream and Downstream RNase J1 Cleavage Products
The steady-state pattern of fragments from wild-type and mutant trp leader RNAs was probed with upstream and downstream oligonucleotide probes (probes 1 and 2 in Fig. 2A). We have shown previously that, using these probes, we can detect the upstream and downstream products of RNase J1 cleavage from trp leader RNA expressed from a multicopy plasmid (23). An upstream fragment of ∼101 nt is detected by the upstream probe but is present in very low amounts, most likely because it has an unprotected 3′ end, which we have shown is rapidly attacked by PNPase (24). The downstream fragment of ∼39 nt, which contains the 3′-TT, is detected by the 3′-terminal probe but additional bands are also detected (see Fig. 2C, wt lane and below).
Using probe 1, the full-length RNA was detected for all strains (Fig. 2B), and the amount of full-length wild type trp leader RNA was consistently 2–3-fold less than that of the mutant RNAs, which was anticipated in view of its shorter half-life. The expected ∼101-nt band, representing the upstream product of RNase J1 cleavage, was observed in the wild type, but was not present in the EaeI, NotI, and NaeI RNAs. Instead, a band of about 110 nt was observed. For the SacII RNA, a ∼101-nt band was detected, in addition to the ∼110-nt band.
The same RNAs were probed with a downstream probe (probe 2 in Fig. 2A). For the wild type, the previously observed pattern was obtained: a group of fragments running between 25 and 50 nt, with a major band at ∼39 nt (Fig. 2C). These are the downstream products resulting from RNase J1 endonuclease cleavage at nt 101. Bands that are larger than 39 nt likely arise from processed/partially degraded readthrough transcripts, which resolve well for these small fragments but are not apparent at the level of full-length RNA. (In the high-resolution blot shown in Fig. 2D, transcripts that are several nt longer than full-length trp leader RNA are detectable.) We surmise that bands smaller than 39 nt result from RNase J1 5′ exonucleolytic activity on the downstream cleavage product that is hindered by the strong 3′-TT structure, as we have shown occurs in vitro (1). For three of the four mutant RNAs, the NotI, SacII, and NaeI RNAs, the 3′ probe detected only faint bands in the 25-50-nt region, indicating that RNase J1 cleavage at the 101-nt position was occurring at very low levels, if at all. For the EaeI construct, a band of 35–39 nt was detected, which, relative to the amount of full-length RNA, was about 30% of the corresponding bands in the wild type lane. This indicates that there is weak RNase J1 cleavage in the EaeI RNA, not enough to detect the unstable 101-nt upstream product using the 5′ probe but enough to detect the most prominent downstream fragments using the 3′ probe. The amount of 3′ product detected at steady state will depend not only on the level of RNase J1 cleavage but also on the ability of RNase J1 to degrade the downstream product, and this may differ depending on the nucleotide sequence. Thus, we cannot use the amount of 3′ product observed as a measure of the amount of cleavage taking place.
To examine more precisely the upstream fragments detected by the 5′ probe, a Northern blot analysis was performed on RNAs separated on a high-resolution gel (Fig. 2D). In all strains, a band of 133 nt was observed. This likely represents 3′ exonuclease nibbling from the 3′ end of trp leader RNA up to the downstream side of the 3′-TT stem. We have shown previously in vitro that the processivity of PNPase, the major B. subtilis 3′-to-5′ exonuclease, is hindered by the trp leader RNA 3′-TT structure (23). Importantly, the amount of this band, relative to full-length RNA, was approximately the same in all strains, about 3–4%, which indicated that the mutations in the single-stranded region upstream of the 3′-TT did not affect accumulation of this intermediate. Similarly, in all strains a band of 113 nt was observed (not visible in this exposure in the wild type lane but visible on overexposure). Based on results presented below that demonstrate an inhibition of PNPase processivity by a GC-rich sequence, we believe this band results from a slowing of PNPase caused by the three C residues at nt 109–111. In all strains, the quantity of this band, relative to full-length RNA, was 1–2% of full-length RNA. The 101-nt upstream product of RNase J1 cleavage was detected in the strain carrying the wild type at a level that was about 5% of full-length RNA. A similar-sized band was detected in the strain carrying the SacII construct, at a level that was about 8% of full-length RNA. Because the SacII RNA gave very little of the 3′ products that would result from RNase J1 cleavage (Fig. 2C), we suggest that the 101-nt RNA detected in the SacII construct strain is not a result of RNase J1 cleavage but represents a block to PNPase processivity at the GC-rich SacII sequence that abuts the TBS. Similarly, the bands detected in the NotI construct strain running below the 113-nt RNA likely represent a block to PNPase processivity imparted by the GC-rich NotI sequence. It is not clear why additional bands under the 113-nt band would not be detected also for the EaeI and NaeI constructs. More in vitro studies with these mutant RNAs and purified PNPase will be needed to clarify the “rules” of hindrances to PNPase processivity. Note that it is not possible to test our hypotheses about the nature of these bands by probing trp leader RNA in a PNPase mutant strain. In such a strain, the upstream product of RNase J1 cleavage is not degraded, which leads to titration of free TRAP and a lack of terminated trp leader RNA (24).
It is reasonable to conclude from these experiments that RNase J1 recognizes an AU-rich region from nt 94–105, and cleaves within this region to initiate decay. The fact that the NotI mutation had the most dramatic effect on mRNA half-life, almost 2-fold more stable than any of the other mutants, suggested that the change of AU sequence on both sides of the 101-nt position in the NotI RNA severely affected the ability of RNase J1 to initiate decay by cleaving at this site. The other RNAs were mutated on only one or the other side of the 101-nt position, and these gave less dramatic increases in trp leader RNA half-life.
Second Generation Mutations
Two additional mutants were constructed, in which only two nucleotides were changed (Fig. 2A, bottom). When nucleotides 100–101 were changed from UU to CA (NsiI mutant), there was no significant effect on trp leader RNA half-life. However, when nucleotides 101–102 were changed from UU to GG (X-EaeI mutant), the half-life went up to about 5 min. Thus, the presence of U residues upstream and downstream of the cleavage site is not required for RNase J1 recognition, but G residues next to the cleavage site seem to be negatively affect RNase J1 recognition. Future constructions with replacement of single residues in this region may reveal more about RNase J1 specificity.
trp Leader RNAs with an Upstream RNase J1 Target Site
The mapped RNase J1 cleavage at nt 101 is in a single-stranded region that is surrounded by the TBS upstream and the 3′-TT structure downstream. To explore the specificity of RNase J1 target site location relative to RNA structural elements, trp leader RNA constructs were made that had either an additional RNase J1 target site or replacement of the native site with one located elsewhere on the RNA. The readout for possible effects of these changes on RNase J1 activity was a change in trp leader RNA half-life, as determined by Northern blot analysis (at least three independent repeats for each half-life determination). The constructs that were made are shown in Fig. 3 and are referred to in the text below by the numbering in that figure. RNAs A1 and A2 are the previously reported wild-type and NotI mutant RNAs, respectively.
FIGURE 3.
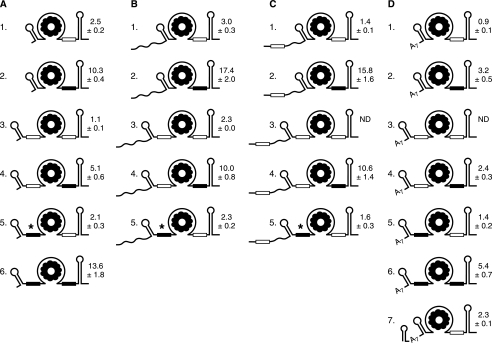
trp leader RNA mutant constructs. Open rectangle denotes RNase J1 target site, nt 96–103. Filled rectangle denotes target site mutated to contain the NotI sequence. The asterisk in RNAs A5, B5, and C5 indicates accumulation of small decay intermediate consisting of 5′ proximal sequence. Half-life of each RNA (average of at least three independent experiments, with S.D.) is shown to the right of the schematic. ND, not done.
First, an additional RNase J1 target sequence was inserted in trp leader RNA upstream of the TBS (RNA A3). For this, the sequence from nt 93–106 was used to replace nt 34 in the segment between the 5′SL and the TBS, resulting in an RNA with two potential RNase J1 target sites. The half-life of RNA A3 was 1.1 min, less than half that of wild-type trp leader RNA (p value < 0.01). When the RNase J1 target upstream of the TBS was on the same RNA with the NotI site at nt 101 (RNA A4) the half-life was 5.1 min, which was half that of the NotI RNA itself (RNA A2). Conversely, when the upstream site was the NotI sequence and the downstream site was wild type (RNA A5), the half-life was indistinguishable from wild type (p value = 0.06). This latter result excludes the possibility that the destabilization observed with RNA A3 was a result of increasing the distance between the 5′SL and the TBS, independent of sequence, rather than the result of inserting an additional RNase J1 cleavage site. When both RNase J1 targets were changed to the NotI sequence (RNA A6) the trp leader RNA had a 13.6-min half-life, which was similar to the half-life for RNA A2 (borderline p value of 0.05).
Taken together, the results with the A series of constructs suggested the following: (i) RNA half-life decreased when an additional RNase J1 target site was present; (ii) the location of an RNase J1 target site did not need to be immediately upstream of the 3′-TT, although in both cases so far the AU-rich sequence was located between a structured RNA (5′SL or 3′TT) and the TBS; (iii) the RNase J1 target site was recognized more efficiently when it was located downstream of the TBS than when it was located upstream.
Accumulation of RNA Decay Intermediates Due to Inhibition of PNPase Processivity
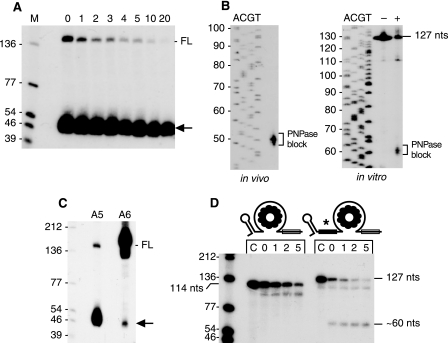
Interestingly, Northern blot analysis of RNA A5 revealed an additional intense set of bands running between the 46- and 54-nt markers (Fig. 4A). This is denoted by the asterisk in Fig. 3A. This set of RNAs had a half-life of 10.8 ± 1.4 min. The precise size of these bands was determined by Northern blot analysis from a high-resolution gel (Fig. 4B, left). A ladder of four bands could be detected, between 48 and 51 nt. Henceforth, this set of bands is referred to as the “∼50-nt RNA.” When the downstream RNase J1 site in RNA A5 was changed to a NotI site (RNA A6), the ∼50-nt RNA was present at a much lower intensity (Fig. 4C). Our earlier studies showed that rapid degradation of TRAP-bound trp leader RNA is dependent primarily on PNPase 3′-to-5′ exonuclease activity (24). It is likely that this decay initiates at the RNase J1 cleavage site, since the 3′-TT structure hinders PNPase decay starting from the native 3′ end (23). To explain the appearance of the intense ∼50-nt RNA in the strain carrying RNA A5, we hypothesized that the NotI sequence (GCGGCCGC) upstream of the TBS blocked PNPase processivity. The end of the NotI sequence is nt 44 in this RNA, and the block to PNPase processing would lead to accumulation of fragments that were somewhat larger than 44 nt. In the case of RNA A6, RNase J1 cleavage at the nt-101 site is abolished, thereby giving little or no RNA fragment with a susceptible 3′ end from which to initiate PNPase degradation. The small amount of the ∼50-nt RNA that was observed for RNA A6 may be due to PNPase degradation that initiates from the 3′ end, which the results in Fig. 2D suggest is occurring to some extent.
FIGURE 4.
A, half-life of ∼50-nt processing intermediate (arrow) in the RNA with the upstream NotI site (RNA A5). B, sizing of the ∼50-nt RNA on a high-resolution gel by Northern blot analysis of in vivo RNA isolated from strain carrying RNA A5 (left) and after PNPase processing in vitro (right). The in vitro samples were incubated without (−) or with (+) PNPase. C, comparison of amount of full-length (FL) and ∼50-nt processing intermediate (arrow) in A5 and A6 RNAs. D, in vitro PNPase activity on trp leader RNAs without and with the upstream NotI sequence. Sizes of full-length substrates (114 nt for wild-type, 127 nt for RNA A5) are indicated, as well as the approximate size of the PNPase digestion product for the RNA with the upstream NotI sequence. The control lane C contained no added PNPase. Above each lane is indicated time (min) after PNPase addition.
To test this explanation for the appearance of the ∼50-nt RNA, we analyzed the ability of purified PNPase to degrade through the NotI sequence in vitro. trp leader RNA versions A1 (wild type) and A5 (upstream NotI sequence) were synthesized in vitro, ending at the 3 Cs in the 3′-TT sequence (i.e. until nt 111 in the wild type trp leader RNA sequence; see Fig. 2A). To enhance the in vitro transcription reaction, three Gs were included at the 5′ end, giving an RNA size of 114 nt for the wild type and 127 nt for the NotI RNA. The 3′ end of these in vitro transcripts is predicted to be unstructured, which is a requirement for efficient 3′ exonuclease activity by PNPase (23). The trp leader RNAs were labeled at the 5′ end and were pre-incubated with TRAP before addition of purified PNPase. As seen in Fig. 4D, PNPase was able to digest wild type trp leader RNA with time. An accumulation of a processing intermediate just below the 114-nt full-length band likely represents a pause at the downstream edge of the TBS. In the case of trp leader RNA with the upstream NotI sequence (RNA A5), additional bands of about 60 nt were observed. The sizes of these bands were determined from a high-resolution gel (Fig. 4B, right), and were found to be a ladder of three bands between 59–61 nt. Taking into account the three extra Gs at the 5′ end, the location of the block to PNPase would be at nt 56–58, somewhat downstream from what was observed in vivo (Fig. 4B, left). This difference may be due to the presence of other 3′ exonucleases in vivo, which could nibble at the 3′ end after PNPase release. We conclude that PNPase has difficulty degrading past this inserted sequence. This observation was useful in determining decay pathways in other RNAs (see below).
trp Leader RNAs with a 27-nt Sequence Added at the 5′ End
Based on many studies that show a 5′ end dependence of RNA decay in B. subtilis (20), one might have expected that the presence of the stem loop structure at the 5′ end of trp leader RNA would confer a relatively long half-life. The 2.5-min half-life of trp leader RNA is, in fact, rather short; in a survey of nearly 1500 B. subtilis mRNAs, only 10% were found to have half-lives of less than 3 min (30). To assess the influence of 5′ determinants on trp leader RNA stability, a 27-nt sequence was added to the 5′ end (Table 1 and Fig. 3B). The 27-nt sequence was predicted to exist in a single-stranded conformation, based on the Zuker folding prediction software. RNAs B1-B4 had the same trp leader RNA sequence as RNAs A1–A4 except for the added 27-nt sequence. Somewhat unexpectedly, in all these cases the added 5′ sequence had a stabilizing effect. For the wild type trp leader RNA, the 5′ addition (RNA B1) had a small but statistically significant effect on stability, increasing the half-life from 2.5 to 3.0 min (p value = 0.04). In the context of the stabilizing NotI sequence at nt 101 (RNA B2), the added 5′ sequence gave a highly stable RNA, with a half-life of 17.4 min. The unstable RNA with two RNase J1 target sites (RNA A3) was stabilized 2-fold by the addition of the 27-nt sequence at the 5′ end (RNA B3). Similarly, the intermediate stability of RNA A4, with the upstream RNase J1 target and the downstream NotI site, increased 2-fold by the addition of the 27-nt sequence (RNA B4).
TABLE 1.
Sequences added to the 5′ end of trp leader RNA
Nucleotides 93–106, which are included in the 5′ addition in the C series, are italicized. Underlined nucleotides in the D7 RNA are complementary sequences that are predicted to form a stable stem structure.
| RNA | Sequence |
|---|---|
| B1-B5 | GGAUCCAAAAACAUGCAAGUCGAAACG |
| C1-C5 | GGUACCUUUCAUUAUGUUUAUUCAACG |
| D7 | GAUCAUGAUAAUAGCUAUUAUCAUGAUAAAAAAA |
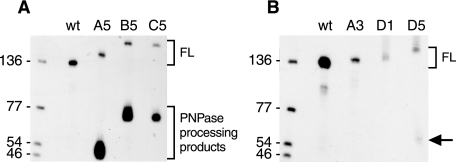
One more construct with this 5′ end sequence was made to test our explanation for the accumulation of the ∼50-nt RNA observed with RNA A5 (Fig. 3A). If the ∼50-nt RNA was the result of a block to PNPase processivity, then addition of a 27-nt sequence at the 5′ end of the A5 RNA should give a similarly intense decay intermediate that was ∼77-nt long. This was indeed observed, as shown in Fig. 5A (RNA B5). Unlike RNAs B1-B4, however, the addition of the 27-nt sequence in RNA B5 did not result in any significant increase in stability, compared with RNA A5.
FIGURE 5.
Steady state pattern of selected trp leader RNAs. Full-length (FL) and processed products detected by probe 1 in strains carrying the construct indicated above each lane.
We had expected that addition of the 27-nt single-stranded tail to the 5′ end might cause instability of the trp leader RNA, either because the accessible 5′ end could be dephosphorylated and serve as a substrate for 5′ exonuclease activity, or because the 5′ end could provide a good binding site for RNase J1, whose endonuclease activity may be 5′ end-dependent. It was possible that the primary sequence at the very 5′ end (GG) was not a good substrate for either of these activities, or that, by an unknown mechanism, addition of this 5′ tail hindered RNase J1 access to, or recognition of, the primary target site at nt 101.
As a variation on the experiments with the B series constructs, additional constructs were made (Fig. 3C) bearing a 5′ single-stranded tail that contained the sequence constituting the downstream RNase J1 target site (nt 93–106). We reasoned that a 5′ tail with an RNase J1 recognition site might function better to destabilize. Furthermore, although the results with RNAs A3 and A4 had suggested that insertion of an additional RNase J1 target site could decrease trp leader RNA stability, in those cases, the additional site was surrounded by the 5′SL on one side and the TBS on the other. The C series constructs were useful in addressing whether the presence of an RNase J1 site in a region not bounded by structured RNA could function in modulating RNA half-life.
The 93–106 nt sequence was used to replace 14 nt of the 27-nt 5′ addition that had been studied in the B series. Because this replacement was predicted to give some secondary structure, the sequence at the 5′ end of the 27-nt addition was altered such that there was no more predicted secondary structure (Table 1). RNA C1, containing the additional RNase J1 target sequence in the upstream segment, was indeed significantly less stable than the wild type (p value < 0.01). Similarly, RNA C5 was significantly less stable than RNA A5 (p value = 0.04). However, RNAs C2 and C4, which had the same 5′ sequence but in the context of the NotI mutation at nt 101, gave half-lives that were much more stable than the corresponding RNAs A2 and A4, and that had similar stability to RNAs B2 and B4 containing the 27-nt 5′ sequence without the RNase J1 target site (p value for B2 versus C2 = 0.17). Thus, the 5′ addition in the C series was stabilizing in two cases and destabilizing in two other cases. At present, we have no reasonable explanation for these observations.
RNA C5 had the added upstream NotI sequence that was capable of blocking PNPase, in the context of the 5′ tail containing the RNase J1 cleavage site. This RNA was used to determine whether the C series RNAs were being degraded exonucleolytically. As we had found with RNA B5, RNA C5 gave an accumulation of a 5′-terminal ∼77-nt RNA (Fig. 5A). However, the accumulation of this RNA in the case of RNA C5 was about 2-fold less (relative to full-length RNA) than for RNAs A5 and B5, suggesting some 5′ exonuclease degradation. These results suggested that the upstream RNase J1 recognition site, although not surrounded by structured RNA on both sides, may be recognized weakly for endonuclease cleavage, allowing 5′ exonucleolytic decay from the newly generated monophosphate 5′ end. Addition of this 5′ sequence in the case of RNAs C1 and C5 accelerated decay of relatively unstable trp leader RNAs that undergo efficient RNase J1 cleavage at nt 101. It is not clear, however, why this alternate pathway to access trp leader RNA did not result in a detectable decrease in half-life of the RNAs that have the NotI sequence at nt 101 (RNAs C2 and C4).
trp Leader RNAs with an A7 Tail Added at the 5′ End
We next turned to a less complex addition at the 5′ end, an A7 sequence (Fig. 3D). We observed previously that placement of a 7-nt tail upstream of a stabilizing 5′ stem loop structure could render the RNA unstable (31). Addition of the A7 tail to the wild-type trp leader RNA (RNA D1) resulted in an extremely unstable trp leader RNA: Northern blot analysis of RNAs extracted at increasing times after rifampicin addition gave a half-life of less than 1 min, and a comparison of the steady-state pattern of wild type trp leader RNA with unstable RNAs A3 and D1 showed a sharp decrease in concentration (Fig. 5B). When the A7 tail was added to the stable NotI mutant (RNA A2) to give RNA D2, the half-life decreased from 10.3 min to 3.2 min, i.e. an even greater destabilizing effect than the effect of inserting an upstream RNase J1 target site (RNA A4). Similarly, adding the A7 tail to RNA A4 to give RNA D4 resulted in a 2-fold reduction in half-life. These results demonstrate the destabilizing effect of a 5′ A7 sequence, and further experiments will be needed to understand the difference between the results with the B series and the D series (e.g. would a G2A5 5′ tail have the same effect as an A7 tail?).
Another RNA of interest was the D5 RNA, in which the A7 sequence was added to the 5′ end of RNA A5. RNA A5 contained the wild-type downstream RNase J1 site and the upstream NotI sequence, and gave a wild type half-life, as well as intense accumulation of the ∼50-nt RNA (Fig. 4B). In the case of RNA D5, the half-life of the full-length RNA was reduced to 1.4 min (p value relative to wild type < 0.01), and while the expected ∼57-nt 5′ fragment was observed (arrow in Fig. 5B) it did not accumulate to the levels seen with RNAs A5 and B5 (compare Fig. 5A). We hypothesize that addition of the A7 tail makes trp leader RNA a substrate for 5′ exonuclease activity, resulting in little accumulation of the RNA fragment that has a block to PNPase processivity at its 3′ end. Similarly, addition of the A7 tail to the highly stable RNA A6, which had two NotI sites and a 13.6-min half-life, resulted in a reduction in half-life to 5.4 min (RNA D6).
trp Leader RNA with a 5′ Stabilizer
Experiments in our laboratory and others (31–33), as well as a survey of stable B. subtilis mRNAs (20), have indicated that a 5′-terminal stem loop structure should have a strong stabilizing effect. We predicted that addition of such a structure would stabilize greatly the extremely unstable D1 RNA. Accordingly, a final construct in the D series was made (RNA D7) that contained the 5′-terminal stem loop structure of mutant mdr (or bmr3) RNA (ΔGo = −10.0 kcal mol−1), which was shown to increase mdr mRNA stability at least 4-fold (33), and which we have found confers extreme stability to other RNAs.3 Surprisingly, while this addition did stabilize RNA D1, it only brought it back to a 2.3-min half-life, which was not significantly different from wild type (p value = 0.11). This latter result suggests the following: when the A7 tail is accessible at the 5′ end, it can serve, perhaps after dephosphorylation of the 5′-triphosphate end, as a binding site for RNase J1 and allow processive 5′-to-3′ exonuclease activity. However, when the A7 tail is occluded by a strong secondary structure, decay of trp leader RNA reverts to the pathway for decay of the wild type, which is not 5′ end-dependent and relies on direct access of RNase J1 to its target at the nt 101 site.
The panel of trp leader RNA mutants constructed in this study demonstrates the power of using this small RNA as a model substrate to probe the function of RNase J1. It should be emphasized that the prompt degradation of trp leader RNA to release TRAP, which then binds nascent transcripts being synthesized from the constitutive trp promoter, is an important feature of the trp regulatory system. Without rapid trp leader RNA degradation, the limiting number of TRAP molecules become trapped, allowing new trp transcription to continue into the trp operon coding sequences, even in the presence of ample tryptophan (24). The presence of the 5′SL is also an important feature of trp transcription regulation, as it has been shown that efficient TRAP binding depends, in part, on this structure (34). In general, however, a stem loop structure located at the 5′ terminus is known to stabilize RNA (20). Thus, the trp leader system requires rapid RNA turnover, but appears to contain a 5′ RNA stabilizer. We suggest that this dilemma may be solved by efficient, 5′ end-independent binding of RNase J1 to its 3′ proximal target site, followed by cleavage at nt 101 and PNPase degradation from the newly generated 3′ end.
Our study allows some tentative conclusions regarding the specificity of RNase J1 endonuclease activity, including: (i) a 12-nt AU-rich sequence is recognized for RNase J1 cleavage; substitution with G or C nucleotides upstream or downstream of the cleavage site severely affects RNase J1 endonucleolytic activity. (ii) an RNase J1 target site is efficiently utilized when located between structured RNA regions. (iii) at least for this small molecule, an additional RNase J1 cleavage site translates into more rapid decay (Fig. 3A, series). In addition, there is a hint that RNase J1 can act both exonucleolytically from the native 5′ end and endonucleolytically on the same RNA substrate (Fig. 3D, series), and RNase J1 endonuclease activity may not be 5′ end-dependent (RNA D7). One aim of future studies will be to complement our in vivo findings with biochemical assays of RNase J1 target site preference in vitro. In addition, further experiments are planned to understand how different sequences/structures located at the 5′ end affect RNase J1 access to its internal target site.
Acknowledgment
We thank Paul Babitzke for the purified TRAP protein and for helpful comments on the manuscript.
This work was supported, in whole or in part, by United States Public Health Service Grant GM48804 (to D. H. B.) from the National Institutes of Health.
S. Yao and D. H. Bechhofer, unpublished experiments.
- TRAP
- trp RNA-binding attenuation protein
- nt
- nucleotide
- PNPase
- polynucleotide phosphorylase
- 5′SL
- 5′ stem loop structure
- TBS
- TRAP binding site
- 3′-TT
- 3′-transcription terminator structure
- IPTG
- isopropyl-1-thio-β-d-galactopyranoside
- wt
- wild type.
REFERENCES
- 1.Deikus G., Condon C., Bechhofer D. H. (2008) J. Biol. Chem. 283, 17158–17167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li de la Sierra-Gallay I., Zig L., Jamalli A., Putzer H. (2008) Nat. Struct. Mol. Biol. 15, 206–212 [DOI] [PubMed] [Google Scholar]
- 3.Mathy N., Bénard L., Pellegrini O., Daou R., Wen T., Condon C. (2007) Cell 129, 681–692 [DOI] [PubMed] [Google Scholar]
- 4.Jiang X., Belasco J. G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 9211–9216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mackie G. A. (1998) Nature 395, 720–723 [DOI] [PubMed] [Google Scholar]
- 6.Tock M. R., Walsh A. P., Carroll G., McDowall K. J. (2000) J. Biol. Chem. 275, 8726–8732 [DOI] [PubMed] [Google Scholar]
- 7.McLaren R. S., Newbury S. F., Dance G. S., Causton H. C., Higgins C. F. (1991) J. Mol. Biol. 221, 81–95 [PubMed] [Google Scholar]
- 8.Spickler C., Mackie G. A. (2000) J. Bacteriol. 182, 2422–2427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpousis A. J. (2007) Annu Rev. Microbiol. 61, 71–87 [DOI] [PubMed] [Google Scholar]
- 10.Coburn G. A., Mackie G. A. (1999) Prog Nucleic Acid Res. Mol. Biol. 62, 55–108 [DOI] [PubMed] [Google Scholar]
- 11.Kushner S. R. (2002) J. Bacteriol. 184, 4658–4665; discussion 4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackie G. A. (2000) J. Biol. Chem. 275, 25069–25072 [DOI] [PubMed] [Google Scholar]
- 13.Jiang X., Diwa A., Belasco J. G. (2000) J. Bacteriol. 182, 2468–2475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Celesnik H., Deana A., Belasco J. G. (2007) Mol. Cell 27, 79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deana A., Celesnik H., Belasco J. G. (2008) Nature 451, 355–358 [DOI] [PubMed] [Google Scholar]
- 16.Even S., Pellegrini O., Zig L., Labas V., Vinh J., Bréchemmier-Baey D., Putzer H. (2005) Nucleic Acids Res. 33, 2141–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choonee N., Even S., Zig L., Putzer H. (2007) Nucleic Acids Res. 35, 1578–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yao S., Blaustein J. B., Bechhofer D. H. (2007) Nucleic Acids Res. 35, 4464–4473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mäder U., Zig L., Kretschmer J., Homuth G., Putzer H. (2008) Mol. Microbiol. 70, 183–196 [DOI] [PubMed] [Google Scholar]
- 20.Bechhofer D. H. (2009) Prog. Mol. Biol. Transl. Sci. 85, 231–273 [DOI] [PubMed] [Google Scholar]
- 21.Babitzke P., Gollnick P. (2001) J. Bacteriol. 183, 5795–5802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Henkin T. M., Yanofsky C. (2002) Bioessays. 24, 700–707 [DOI] [PubMed] [Google Scholar]
- 23.Deikus G., Bechhofer D. H. (2007) J. Biol. Chem. 282, 20238–20244 [DOI] [PubMed] [Google Scholar]
- 24.Deikus G., Babitzke P., Bechhofer D. H. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2747–2751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Britton R. A., Wen T., Schaefer L., Pellegrini O., Uicker W. C., Mathy N., Tobin C., Daou R., Szyk J., Condon C. (2007) Mol. Microbiol. 63, 127–138 [DOI] [PubMed] [Google Scholar]
- 26.Petit M. A., Dervyn E., Rose M., Entian K. D., McGovern S., Ehrlich S. D., Bruand C. (1998) Mol. Microbiol. 29, 261–273 [DOI] [PubMed] [Google Scholar]
- 27.Grant S. G., Jessee J., Bloom F. R., Hanahan D. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 4645–4649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daou-Chabo R., Mathy N., Benard L., Condon C. (2009) Mol. Microbiol. 71, 1538–1550 [DOI] [PubMed] [Google Scholar]
- 29.Zuker M. (2003) Nucleic Acids Res. 31, 3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hambraeus G., von Wachenfeldt C., Hederstedt L. (2003) Mol. Genet. Genomics 269, 706–714 [DOI] [PubMed] [Google Scholar]
- 31.Sharp J. S., Bechhofer D. H. (2005) Mol. Microbiol. 57, 484–495 [DOI] [PubMed] [Google Scholar]
- 32.Hambraeus G., Karhumaa K., Rutberg B. (2002) Microbiology 148, 1795–1803 [DOI] [PubMed] [Google Scholar]
- 33.Ohki R., Tateno K. (2004) J. Bacteriol. 186, 7450–7455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McGraw A. P., Bevilacqua P. C., Babitzke P. (2007) RNA 13, 2020–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brosius J. (1992) Methods Enzymol. 216, 469–483 [DOI] [PubMed] [Google Scholar]