Abstract
The Ecdysoneless (Ecd) protein is required for cell-autonomous roles in development and oogenesis in Drosophila, but the function of its evolutionarily conserved mammalian orthologs is not clear. To study the cellular function of Ecd in mammalian cells, we generated Ecdlox/lox mouse embryonic fibroblast cells from Ecd floxed mouse embryos. Cre-mediated deletion of Ecd in Ecdlox/lox mouse embryonic fibroblasts led to a proliferative block due to a delay in G1-S cell cycle progression; this defect was reversed by the introduction of human Ecd. Loss of Ecd led to marked down-regulation of E2F target gene expression. Furthermore, Ecd directly bound to Rb at the pocket domain and competed with E2F for binding to hypophosphorylated Rb. Our results demonstrate that mammalian Ecd plays a role in cell cycle progression via the Rb-E2F pathway.
Precisely regulated cell proliferation is essential for embryonic development as well as for homeostasis in adult organs and tissues, whereas uncontrolled cell proliferation is a hallmark of cancer. Thus, understanding how the cell cycle machinery is controlled is an important area of research.
Studies with viral oncogenes that induce dominant cellular transformation have led to the identification and elucidation of a number of biochemical pathways that are perturbed in human cancer (1). One group of tumor viruses directly implicated in the pathogenesis of human cancer is the “high-risk” subgroup of human papillomaviruses (HPVs).7 In vitro studies have defined two HPV oncogenes, E6 and E7, that are nearly always expressed in HPV-associated carcinomas and cell lines derived from them (2). The ability of the HPV E6 and/or E7 oncogene to induce the immortalization of human epithelial cells in vitro has provided an invaluable tool to elucidate the mechanisms by which these oncogenes function (1–3).
Recently, we identified the human ortholog of Drosophila Ecdysoneless (hEcd) as an E6-binding partner, and our initial studies indicated that hEcd interacts with and stabilizes p53. Furthermore, its overexpression in mammalian cells enhances p53 target gene transcription, whereas its transient knockdown has the opposite effect (4). However, the physiological role of hEcd remains unknown. Notably, unlike stable knockdown of p53 (which extends the life span of normally senescing cells), stable hEcd knockdown leads to a proliferative block, suggesting a p53-independent role for hEcd in cell survival/proliferation.
The Ecd gene was defined nearly 30 years ago based on Drosophila ecdysoneless (ecd) mutations; temperature-sensitive ecd mutant embryos arrest at the second larval instar stage at the restrictive temperature, apparently due to reduced levels of ecdysone (5). The protein product of this locus was molecularly identified only recently, and ecd mutants were found to exhibit a general defect in cell survival in addition to the reproductive and developmental defects; the nature of the survival defect or its biochemical basis remains unknown (6). Two studies in yeast suggest that Ecd (called human SGT1 in these studies) might be involved in the transcription of glycolytic genes. Sato et al. (7) identified hEcd using a cross-species complementation where hEcd was shown to rescue the growth defect of the gcr2 mutation, in which transcription of glycolytic genes is dysregulated. The authors concluded that human Ecd/SGT1 could have a role in gene transcription; however, human Ecd/SGT1 has no sequence similarity to the GCR2 gene, whose deficiency it complemented, and in fact, Saccharomyces cerevisiae does not have a hEcd ortholog. In a follow-up study, the same investigators identified an Ecd ortholog in Schizosaccharomyces pombe and found it to be essential for yeast survival and growth; the mechanisms of its function remain unknown, however (8).
Given the limited understanding of the evolutionarily conserved Ecd protein family, we generated conditional Ecd knock-out mice and examined the consequences of Ecd deletion. We observed that homozygous Ecd deletion in mice leads to early embryonic lethality.8 To examine the role of Ecd at the cellular level, we generated Ecd-null mouse embryonic fibroblasts (MEFs) and then deleted Ecd by Cre adenovirus infection of Ecdlox/lox MEFs. Ecd-deleted cells showed a delay in G1-S cell cycle progression with a delay in Rb (retinoblastoma) phosphorylation and reduced expression of E2F target genes.
The Rb protein family, consisting of three related proteins, Rb/p105, p107, and Rb2/p130, has emerged as a key controller of cell cycle progression, and these proteins play critical roles in development and differentiation of various tissues (9–16). Precisely how the Rb family proteins regulate cell proliferation is still incompletely understood. A large body of evidence has established a basic paradigm of how these proteins contribute to the control of cell cycle progression. Unphosphorylated Rb proteins interact with E2F transcription factors and prevent the transcription of genes regulated by E2F proteins (17, 18). During cell cycle progression, cyclin-CDK complexes mediate hyperphosphorylation of Rb, an event that leads to loss of its interaction with E2F; this allows E2F target gene transcription and cell cycle progression.
In this work, we provide evidence that Ecd directly interacts with Rb at the pocket domain, competes with E2F for association with hypophosphorylated Rb, and regulates E2F target gene expression and cell cycle progression. Thus, this study demonstrates that mammalian Ecd plays a role in cell cycle progression via the Rb-E2F pathway.
EXPERIMENTAL PROCEDURES
Establishment of MEFs
Embryonic day 13.5 embryos were dissected from Ecd+/lox intercrossed females, and MEFs were isolated and immortalized following the 3T3 protocol (19). MEFs were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. Ecdlox/lox MEFs expressing hEcd and HPV16 E7 were generated by infecting retrovirus encoding full-length human Ecd and HPV16 E7 genes, respectively. E7 expression was confirmed by reverse transcription-PCR using primer set 5′-GATCTCTACTGTTATGAGCA-3′ and 5′-TAACAGGTCTTCCAAAGTAC-3′.
Plasmids
N-terminally FLAG-tagged full-length Ecd (amino acids 1–644) plasmid has been described previously (4). N-terminally FLAG-tagged Ecd fragments were generated by PCR amplification and subcloned into the EcoRV and NotI sites of pcDNA3.1 (Invitrogen). For expression of GST-fused Ecd, amino acids 1–644, 150–644, and 439–644 of Ecd cDNA were PCR-amplified and subcloned into the SalI and NotI sites of pGEX-6p-1 (GE Healthcare). pGEX6p-1-Rb (amino acids 379–928, 379–792, 768–928, and 792–928) and pGEX6p-1-p130 (amino acids 417–1139) were generated by PCR cloning. For expression of C-terminally His6-tagged Ecd, full-length Ecd coding sequence was cloned into the SalI and NotI sites of the pFastBac1 vector (Invitrogen). Protein was expressed in Sf21 cells and subjected to histidine affinity purification.
Growth Curve, Colony Formation Assay, and Cell Cycle Analysis
Adenoviruses encoding enhanced green fluorescent protein-Cre or enhanced green fluorescent protein were purchased from the University of Iowa Gene Transfer Vector Core. Adenovirus-infected cells were plated (1 × 104 cells/well of 6-well plates) and counted at the indicated time points. For colony formation assay, the cells were trypsinized 2 days after infection and plated at 5000 cells/100-mm dish in triplicate or at 1000 cells/well of a 6-well plate. After 10 days, the colonies were stained with crystal violet (0.5% crystal violet in 25% methanol). The stain retained in the colonies was solubilized in 10% acetic acid, and the absorbance was measured at 590 nm. For BrdUrd staining, adenovirus-infected cells on coverslips were starved in Dulbecco's modified Eagle's medium containing 0.2% fetal calf serum for 72 h and stimulated with complete Dulbecco's modified Eagle's medium containing 10% fetal calf serum. Cells were labeled with 10 μm BrdUrd for 1 h and analyzed using mouse anti-BrdUrd antibody (555627, Pharmingen). Cells on coverslips were fixed in 1% paraformaldehyde for 20 min and then subjected to permeabilization (in PBS containing 0.5% Triton X-100 for 15 min at room temperature) and blocking (in PBS containing 1% bovine serum albumin for 30 min at room temperature). After blocking, cells were incubated for 1 h with mouse anti-BrdUrd antibody (1:1000 dilution in PBS containing 0.1% Tween 20, 0.15 m NaCl, 4.2 mm MgCl2, and 10 K units/ml DNase I) and then with Alexa Fluor 564-labeled anti-mouse IgG (1:1000 dilution; Molecular Probes) in PBS containing 0.1% Tween 20. After washing in PBS containing 0.1% Tween 20, cells on coverslips were mounted using VECTASHIELD solution containing 4′,6-diamidino-2-phenylindole (Vector Laboratories) and visualized under a fluorescence microscope. TUNEL assay was performed using an in situ cell death detection kit (POD, Roche Applied Science).
Immunoblotting and Immunoprecipitation
Cell extracts were prepared in lysis buffer (80 mm Tris-HCl (pH 6.8), 2% SDS, and 5% glycerol) and boiled at 95 °C for 5 min, and protein concentration was measured using the BCA protein assay reagent (Pierce). Immunoblotting was performed with primary antibodies against Ecd (4A8, generated by the Monoclonal Antibody Facility at the Lurie Cancer Center, Northwestern University, Chicago); Rb (554136, Pharmingen); p107 (sc-318), p130 (sc-317), cyclin A (sc-596), cyclin B1 (sc-752), cyclin E (sc-481), cyclin D1 (sc-20044), CDK2 (sc-6248), CDK4 (sc-260), and CDK6 (sc-53638) (Santa Cruz Biotechnology, Santa Cruz, CA); and α-tubulin (T6199, Sigma). For immunoprecipitation, cell extracts were prepared in lysis buffer (20 mm Tris-HCl (pH 7.5), 200 mm NaCl, 0.5% Nonidet P-40, and protease inhibitor mixture (Roche Applied Science)) and immunoprecipitated with 2 μg of antibodies against Rb (sc-50 and sc-7905), E2F1 (sc-193), E2F2 (sc-633), and E2F3 (sc-878) (Santa Cruz Biotechnology) for 2 h to overnight at 4 °C. The immunocomplexes were pulled down with protein A/G-agarose (sc-2003, Santa Cruz Biotechnology) for an additional 2 h.
In Vitro Kinase Assay
In vitro kinase assay was performed using purified GST-Rb (amino acids 379–928) as a substrate. Adenovirus-infected MEFs were starved for 3 days and stimulated with serum. Cells were harvested in lysis buffer (20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.5% Nonidet P-40, 0.1 mm Na4VO3, 1 mm NaF, and protease inhibitor mixture), and CDK complex was recovered by immunoprecipitation with 2 μg of anti-CDK4 (sc-56277) or CDK2 (sc-6248) antibodies (Santa Cruz Biotechnology). To examine the effects of added Ecd, 2 μg of purified Ecd protein was added to 500 μg of Ecd-null MEF extracts before immunoprecipitation for 2 h at 4 °C. CDK2 complex was captured with protein A/G-agarose for 1 h and washed with lysis buffer followed by one wash with kinase buffer (50 mm Tris-HCl (pH 7.5), 7.5 mm MgCl2, 1 mm dithiothreitol, 0.1 mm Na4VO3, and 1 mm NaF). CDK2 complex and GST-Rb (500 ng) were incubated in kinase buffer containing 10 mm β-glycerophosphate, 33 μm ATP, and 10 μCi of [γ-32P]ATP (10 mCi/ml, 6000 Ci/mmol) at room temperature for 20 min. The products were subjected to SDS-PAGE, transferred to polyvinylidene difluoride membranes, and autoradiographed.
In Vitro Binding Assay
Various GST fusion proteins were purified using glutathione-Sepharose 4B beads (GE Healthcare). The GST part was removed by cleavage with PreScission protease (GE Healthcare). GST pulldown assays were performed in lysis buffer (20 mm Tris-HCl (pH 7.5), 150 mm NaCl, 0.5% Nonidet P-40, and protease inhibitor mixture) for 2 h at 4 °C. FLAG-tagged Ecd or E2F1 proteins for GST pulldown assays were transiently expressed in 293T or U2OS cells. His pulldown assay was performed in lysis buffer containing 5 mm imidazole with nickel beads (Invitrogen).
Real-time PCR
RNA was isolated from MEFs infected with control virus or Cre adenovirus using TRIzol reagent (Invitrogen), and 2 μg of total RNA was used for reverse transcriptase reaction using SuperScriptTM II reverse transcriptase (Invitrogen). PCR amplification was performed with specific primer sets (supplemental Table 1).
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assays were performed using a ChIP-ITTM Express assay kit (Active Motif, Carlsbad, CA). Two micrograms of antibodies was used for ChIP, and PCR amplifications were performed using 35–40 cycles. The following antibodies were used: control IgG (sc-2027), Rb (sc-50), and p130 (sc-317) (Santa Cruz Biotechnology). The PCR primers for B-myb and cdc2 were described previously (20). PCR was performed with different PCR cycle numbers to confirm PCR results from the linear range.
RESULTS
Ecd Deletion Leads to a Proliferation Defect in MEFs
To assess the impact of Ecd deletion at the whole organism level, we generated an Ecd floxed allele by introducing loxP sites flanking exons 4–7 to make Ecd knock-out mice. When heterozygous Ecd+/− mice were intercrossed, no Ecd-null pups were observed among >200 live born pups screened, suggesting that homozygous Ecd knock-out mice are lethal.8
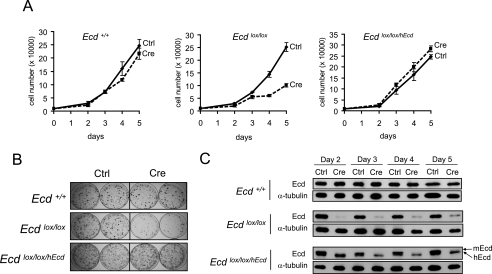
To study the role of Ecd at the cellular level, we established spontaneously immortal MEF lines from Ecd+/+ and Ecdlox/lox mice and generated Ecd-null MEFs from the latter by Cre adenovirus infection. In several experiments, we noted that adenoviral Cre expression in Ecdlox/lox but not Ecd+/+ MEFs led to a proliferative block, and any residual proliferating cells were found to express Ecd protein, reflecting cells that were not adenovirally infected (data not shown). To assure that the proliferative block was indeed a result of Ecd deletion, we engineered an Ecdlox/lox/hEcd MEF line by retrovirally introducing the human Ecd coding sequence into Ecdlox/lox MEFs. Compared with wild-type MEFs, Cre-mediated deletion of Ecd in Ecdlox/lox MEFs induced a clear proliferative block (Fig. 1A). In contrast, proliferation of Ecdlox/lox/hEcd MEFs was minimally affected (Fig. 1A) even though the endogenous (murine) Ecd was fully deleted (Fig. 1C, upper band). Colony formation assays further confirmed the proliferative defect upon Ecd deletion and rescue with hEcd (Fig. 1B). These results demonstrate an essential role for Ecd in cell proliferation.
FIGURE 1.
Ablation of Ecd causes a proliferation defect in MEFs. A, shown are the growth curves of Ecd+/+, Ecdlox/lox, and Ecdlox/lox/hEcd MEFs after control (Ctrl) virus or Cre adenovirus infection. Each time point shows the average cell number of triplicates. A representative experiment of three is shown. B, for colony formation assay, cells infected with control virus or Cre adenovirus were grown for 10 days and stained with crystal violet. C, shown are Ecd expression levels at different time points after Cre adenovirus infection. Note that reconstituted control cells expressed both mouse (m; upper band) and human (lower band) Ecd, whereas only hEcd was seen in Cre adenovirus-infected cells.
Ecd Deletion Leads to a Delay in G1-S Progression
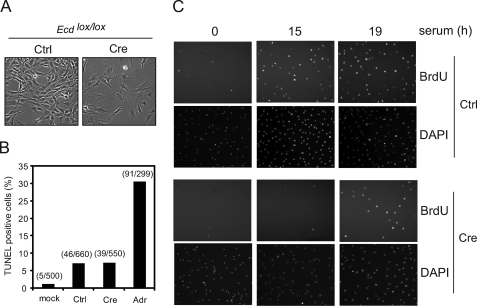
We reasoned that the defect in proliferation in Ecd-null MEFs could be due to either increased cell death or impairment of cell cycle progression. We excluded the first possibility because we did not observe the characteristic morphological features of apoptosis (Fig. 2A) or any evidence of apoptosis by TUNEL staining (Fig. 2B) in Ecd-null MEFs. Notably, Ecd-null MEFs exhibited an enlarged and flattened morphology that is typical of G1-arrested cells (Fig. 2A). We therefore compared the G1-S cell cycle progression of Ecd-null versus control MEFs by BrdUrd staining. Ecdlox/lox MEFs were infected with control virus or Cre adenovirus, synchronized by serum deprivation, and released into the cell cycle by adding serum-containing medium. Compared with serum-stimulated Ecdlox/lox MEFs infected with control virus, Cre adenovirus-infected cells exhibited a delay in G1-S progression (Fig. 2C). These results indicate that the cell proliferation defect found in Ecd-null MEFs is due to a delay in G1-S progression.
FIGURE 2.
Ecd disruption leads to G1-S delay in MEFs. A, phase-contrast microscopic images of Ecdlox/lox MEFs 4 days after control (Ctrl) virus or Cre adenovirus infection. B, TUNEL assay of Ecdlox/lox MEFs 5 days after Cre adenovirus infection. At least 500 cells were counted, and the percentage of apoptotic cells is shown in the graph. MEFs treated with adriamycin (Adr; 33 μm, 24 h) served as positive controls. C, BrdUrd (BrdU) staining of control virus- or Cre adenovirus-infected Ecdlox/lox MEFs after serum stimulation. DAPI, 4′,6-diamidino-2-phenylindole.
Ecd Deletion Leads to Impairment of Cell Cycle-associated Rb Phosphorylation
Given our observation that lack of Ecd imposed a block in G1-S transition, we examined the status of key proteins known to control this transition. It is well established that a critical event during G1-S transition is the phosphorylation of Rb by CDK4/6 and CDK2, which leads to the release of E2F family members from the Rb-E2F complex and allows E2F proteins to facilitate the expression of E2F-responsive genes (21, 22). Therefore, we compared the phosphorylation status of Rb family proteins in control and Ecd-null MEFs. Analysis of unsynchronized cells revealed a relatively moderate increase in the levels of hypophosphorylated compared with hyperphosphorylated Rb in Ecd-deleted versus control MEFs (Fig. 3A). When synchronized MEFs were serum-stimulated, an expected time-dependent increase in the proportion of hyperphosphorylated Rb family proteins was seen in control cells, whereas the emergence of hyperphosphorylated forms was delayed in Ecd-null MEFs (Fig. 3B). In addition, the level of p107 was reduced in Ecd-null cells; as discussed below, this is likely because p107 is a transcriptional target of E2F (18).
FIGURE 3.
Reduced Rb phosphorylation and CDK2 expression in Ecd-null MEFs. A and B, phosphorylation of Rb in asynchronous and synchronized/serum-stimulated MEFs. Open and closed arrowheads indicate hypo- and hyperphosphorylated Rb proteins, respectively. C, expression of CDKs and CDK inhibitors in Ecd-null MEFs. D and E, in vitro kinase assay (KA) of extracts of serum-restimulated MEFs using anti-CDK4 or anti-CDK2 immunoprecipitates (IP). Anti-CDK4 or anti-CDK2 immunoprecipitates using 500 μg of Ecdlox/lox MEF extracts infected with control (Ctrl) virus or Cre adenovirus were subjected to in vitro kinase assay. In one lane of E, purified Ecd protein was added to Ecd-null MEF extracts before immunoprecipitation. WB, Western blot.
Next, we compared the control and Ecd-null MEFs for the levels of CDK2, CDK4, and CDK6 kinases known to mediate Rb phosphorylation during G1-S progression. Although CDK4 and CDK6 levels were not altered by Ecd deletion, CDK2 expression was reduced in Ecd-null MEFs (Fig. 3C). In vitro kinase assay showed that whereas CDK4 kinase activity was comparable between control and Ecd-null MEFs (Fig. 3D), the total levels of CDK2 kinase activity in Ecd-deleted MEFs were substantially reduced in Ecd-null MEFs compared with control MEFs (Fig. 3E). The addition of purified Ecd to Ecd-null MEF extracts prior to CDK2 immunoprecipitation did not affect the level of CDK2 kinase activity (Fig. 3E). This result suggests that Ecd does not influence the activity of CDK2 directly by altering its association with cyclins or CDK inhibitor proteins. Furthermore, the expression levels of CDK inhibitors p16, p21, and p27 were comparable between control and Ecd-null MEFs (Fig. 3C). Taken together, these results support the notion that depletion of Ecd protein reduces the expression and activity of CDK2. As CDK2 is a known E2F target and the CDK2 promoter has an E2F-binding element (23–25), this finding led us to analyze further whether other E2F target genes are down-regulated in Ecd-null cells.
Expression of E2F Target Genes Is Reduced in Ecd-null MEFs due to Inefficient Rb Protein Dissociation from Cell Cycle Gene Promoters
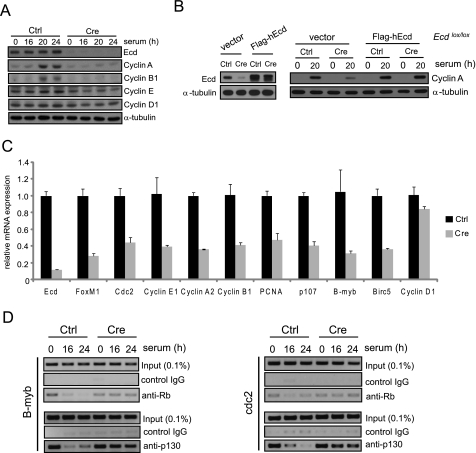
In view of the delay in cell cycle progression and Rb hyperphosphorylation upon Ecd deletion and the decreased CDK2 levels and kinase activity in Ecd-deleted cells, we examined the expression of several cell cycle regulatory proteins, including those known to be induced through E2F-dependent transcription. The levels of cyclins E, A, and B1 were substantially lower in Ecd-deleted MEFs (Fig. 4A). Reconstitution of Ecd expression in Ecdlox/lox MEFs infected with Cre adenovirus restored cyclin A expression (Fig. 4B). Real-time PCR analysis showed a parallel reduction in the mRNA levels of these cyclins (Fig. 4C). However, the level of cyclin D1, a regulatory component for CDK4, was comparable in control versus Ecd-null MEFs (Fig. 4A). Because cyclins E, A, and B1 are known E2F target genes, these results (together with the reduction in CDK2 levels and Rb phosphorylation) suggested a role for Ecd in regulating the Rb-E2F pathway.
FIGURE 4.
Reduced expression of cell cycle-related genes in Ecd-null MEFs. A, shown is the immunoblotting for cell cycle regulatory proteins detected in the indicated serum-stimulated Ecdlox/lox MEFs infected with control (Ctrl) virus or Cre adenovirus. B, reconstitution with FLAG-hEcd restored cyclin A expression in Ecdlox/lox MEFs infected with Cre adenovirus. The level of Ecd expression in control vector- or FLAG-hEcd-expressing Ecdlox/lox MEFs after Cre adenovirus infection is shown (left). C, Ecdlox/lox MEFs were infected with control virus or Cre adenovirus, and 3 days after infection, cells were collected for real-time PCR of the indicated genes. Error bars represent the mean ± S.D. from three independent PCRs. D, wild-type and Ecd-null MEFs were starved for 3 days and serum-stimulated. At different time points, samples were collected and used for ChIP assay with antibodies against Rb and p130, followed by PCR amplification of the indicated E2F target promoters. PCNA, proliferating cell nuclear antigen.
To address this notion further, we examined the expression of other known E2F target genes: proliferating cell nuclear antigen, B-myb, FoxM1, p107, birc5, and cdc2. Notably, these E2F target genes were also down-regulated in Ecd-null MEFs, whereas cyclin D1 and glyceraldehyde-3-phosphate dehydrogenase were not affected, suggesting the specific effect of Ecd deletion on E2F-specific targets (Fig. 4C). These results clearly indicate that Ecd regulates E2F target gene expression at the transcriptional level.
Given that Ecd deletion led to a down-regulation of E2F target genes and reduced the cell cycle-associated phosphorylation of Rb proteins, we reasoned that lack of Ecd may promote continued association of Rb with E2F proteins. To address this possibility in a physiological context, we examined the in vivo occupancy of Rb proteins on known E2F target promoters using the ChIP assay.
We carried out ChIP assays in synchronized cells after serum stimulation. As expected, the occupancy of Rb and p130 on B-myb and cdc2 promoters steadily declined as control MEFs transitioned out of G0/G1 phase (Fig. 4D). In contrast, Rb and p130 continued to remain associated with E2F target promoters at 16 and 24 h after serum addition in Ecd-null MEFs (Fig. 4D). These results show that loss of Ecd protein results in the reduction of the dissociation of Rb proteins from E2F proteins and suggest a role for Ecd in cell cycle progression.
Ecd Interacts with Rb Family Proteins
Although a number of potential mechanisms could be hypothesized to account for the role of Ecd in Rb-E2F dissociation implied by experiments presented above, a simple model would involve a potential interaction of Ecd with Rb proteins at or near the site where E2F proteins bind. To explore this model, we first tested the potential interaction of purified recombinant Ecd and Rb using a GST pulldown assay. Purified Ecd was pulled down by a GST-Rb fusion protein that incorporated the A/B and C pocket domains (amino acids 379–928) (Fig. 5A). Consistent with this in vitro interaction, immunoprecipitation analysis showed that endogenous Ecd and Rb proteins formed a complex in MEFs (Fig. 5B). Reciprocal experiments using anti-Ecd antibodies to immunoprecipitate the complexes did not co-immunoprecipitate Rb. Further analysis revealed that the currently available anti-Ecd antibodies inhibit the Rb-Ecd interaction (data not shown).
FIGURE 5.
Ecd interacts with Rb and inhibits Rb-E2F interaction. A, shown are the results from in vitro binding assay with purified GST-Rb and Ecd. B, shown is the in vivo binding of Rb and Ecd in MEF extracts. Anti-Rb immunoprecipitates from extracts of MEFs were immunoblotted with an anti-Ecd antibody. C, shown are the results from in vitro binding assay with purified Rb family proteins and full-length Ecd. D, GST or GST-Rb (amino acids 379–928) was incubated with lysates of 293T cells expressing the indicated truncated forms of FLAG-Ecd and subjected to anti-FLAG immunoblotting. E, Ecd preferentially bound to the hypophosphorylated form of Rb. In vitro binding assay was performed with purified GST-Ecd (amino acids 150–644), GST-E2F1, and MEF cell extracts (a mixture of serum-starved and asynchronous cells). Open and closed arrowheads indicate hypo- and hyperphosphorylated Rb forms, respectively. F, shown are the results from in vitro binding assay with purified GST-Rb (amino acids 379–928, 379–792, and 768–928) and Ecd. U2OS cell extracts expressing FLAG-hEcd or E2F1 were incubated with purified GST-Rb proteins. G, purified GST-E2F1 (200 ng) and Rb (amino acids 379–928; 50 ng) were incubated in the absence or presence of Ecd (0.2, 0.5, 1, or 2 μg). H, 1 μg of full-length (amino acids 1–644) or N-terminally truncated (amino acids 439–644) Ecd protein was used to compete with GST-E2F1 for binding to Rb. The purity of Ecd proteins is shown in right panel. I, purified Rb (50 ng) and Ecd-His (100 ng) were pulled down with nickel beads in the absence or presence of GST or GST-E2F1 (0.2 or 1 μg). J, endogenous Rb-E2F complex could be dissociated by Ecd protein. Purified Ecd protein (0.2 or 2 μg) was added to 100-μg aliquots of T98G cell extracts prior to immunoprecipitation using a mixture of anti-E2F1–3 antibodies. E2F-associated Rb was detected by immunoblotting. Arrowheads in Ponceau S stain panels indicate the purified GST/GST fusion proteins.
As a next step, we examined the binding of Ecd with other Rb family proteins, p107 and p130, in comparison with Rb. Notably, the in vitro studies indicated that Ecd interacted with all Rb family proteins, although its interaction with Rb and p130 appeared more robust (Fig. 5C). Analysis of a series of truncation mutants of Ecd showed that only full-length Ecd (amino acids 1–644) and the fragment encompassing amino acids 150–644 interacted with Rb (Fig. 5D). Furthermore, GST pulldown analysis using MEF cell lysates showed that Ecd interacted with the hypophosphorylated form of Rb, similar to E2F1 (Fig. 5E). GST pulldown assay with various fragments of Rb showed that Ecd interacted with the same region of Rb as did E2F1 (Fig. 5F). These results demonstrate that Ecd can directly bind to Rb family proteins and that the region of Rb where Ecd binds likely overlaps with the E2F interaction region.
Ecd Inhibits Rb-E2F1 Binding
The existence of an Rb-Ecd complex in cells and the in vitro demonstration that Ecd interacts with the pocket domains of Rb raised the possibility that Ecd may play a role in the dissociation of Rb from E2F. To test this idea, we first examined whether Ecd can compete with E2F for binding to Rb. We assessed the effect of purified Ecd on Rb-E2F1 interaction using a pulldown assay with GST-E2F1 and purified Rb and Ecd proteins. Notably, the addition of increasing amounts of Ecd in Rb-E2F binding reactions led to a diminution of Rb-E2F1 binding (Fig. 5G). In contrast to full-length Ecd, a C-terminal fragment (amino acids 439–644) of Ecd that showed little binding to Rb had no effect on Rb-E2F1 binding (Fig. 5H). In a converse experiment to assess whether both Ecd and E2F1 bind to a shared region on Rb, we examined the effect of purified GST-E2F1 on in vitro Rb-Ecd interaction using a nickel bead pulldown of His6-tagged Ecd. The presence of increasing amounts of GST-E2F1 but not of GST reduced the Rb-Ecd binding (Fig. 5I). As expected, HPV16 E7 competed with Ecd for binding to Rb, which further supports the observation that Ecd binds to Rb through its pocket domain (data not shown). Finally, we assessed the effects of adding purified Ecd on Rb-E2F complexes isolated from cells. The addition of increasing amounts of Ecd to cell lysates led to a disruption of endogenous Rb-E2F complexes when analyzed by Rb co-immunoprecipitation with a mixture of anti-E2F1–3 antibody (Fig. 5J). Taken together, these results clearly demonstrate that Ecd competes with E2F for binding to hypophosphorylated Rb and can facilitate disruption of Rb-E2F complexes.
HPV16 E7 Overcomes Ecd Deletion-induced Proliferative Block
According to the model deduced from analyses presented above that Ecd promotes Rb-E2F dissociation during cell cycle progression, we predicted that the proliferative block imposed by Ecd deletion would be reversed if the stabilized Rb-E2F complex could be forced to dissociate. Viral oncoproteins such as HPV16 E7 bind to pocket domains of Rb family proteins and induce their dissociation from E2F proteins (26). Indeed, expression of HPV16 E7 has been shown to rescue the impaired proliferation due to defects in the Rb-E2F pathway in CDK2 and CDK4 double knock-out MEFs (27). Therefore, we stably expressed HPV16 E7 in Ecdlox/lox MEFs using retroviral infection (Fig. 6A) and subsequently infected the cells with Cre adenovirus. Cre-mediated deletion of Ecd was confirmed by immunoblotting (Fig. 6B). Notably, E7-expressing MEFs did not undergo a proliferative block despite Cre-mediated Ecd deletion (Fig. 6C). These analyses functionally validate the model that Ecd plays a role in cell cycle progression by promoting the dissociation of Rb-E2F complexes.
FIGURE 6.
HPV16 E7 overcomes Ecd deletion-induced proliferative block. A, reverse transcription (RT)-PCR of E7 expression in Ecdlox/lox MEFs infected with HPV16 E7. Vec, vector; GAPDH, glyceraldehyde-3-phosphate dehydrogenase. B, expression level of Ecd in Ecdlox/lox MEFs expressing FLAG-hEcd and HPV16 E7 after Cre adenovirus infection. Exogenously expressed FLAG-hEcd migrated slightly slower than endogenous mouse Ecd. C, colony formation assay of FLAG-hEcd- or E7-expressing Ecdlox/lox MEFs after control (Ctrl) virus or Cre adenovirus infection. Cells were stained with crystal violet, and the solubilized dye absorbance was measured at 590 nm. The graph shows the relative rescue efficiency compared with vector cells. Error bars represent the mean ± S.D. from three independent plates.
DISCUSSION
Dissociation of Rb-E2F complexes is widely viewed as a critical event in G1-S cell cycle transition, and the basic components and regulators of this pathway are commonly altered in a variety of human cancers (28–32). This pathway is also a prominent target of viral oncogenes implicated in human cancer (33–35). Further mechanistic insights into the control of Rb-E2F dissociation during cell cycle progression are therefore of substantial interest in cell, developmental, and cancer biology. Here, we have identified the little understood Ecd protein, the product of the mammalian ortholog of the Drosophila ecdysoneless gene, as a novel cell cycle regulator and have demonstrated that Ecd functions by interacting with Rb and facilitating Rb-E2F dissociation during cell cycle progression.
Our conclusion that Ecd plays a role in cell cycle progression is based on clear evidence from cell cycle analyses using MEFs in which floxed Ecd was conditionally deleted using Cre recombinase. The proliferative arrest imposed by conditional deletion of Ecd was reversed by ectopically introduced hEcd, thereby clearly establishing that loss of Ecd itself rather than any alteration of neighboring gene products was responsible for the observed cell cycle phenotype. In addition, Ecd deletion led to delay of G1-S cell cycle progression. This represents the first formal demonstration of a role for Ecd in cell cycle progression.
Although a number of mechanisms could be envisioned for a role of Ecd in cell cycle progression, analyses of biochemical events known to accompany G1-S cell cycle progression strongly implicated its role in the Rb-E2F pathway. These included the substantial delay and overall reduction in the conversion of Rb from its hypophosphorylated to hyperphosphorylated state normally seen during G1-S transition. In addition, we observed a reduction in the total levels and activity of CDK2, whereas CDK4 levels and activity were unaltered. The latter observation prompted us to investigate if Ecd is required for cell cycle-associated E2F target gene expression. Biochemical analyses of E2F target gene products in Ecd-deleted versus parental Ecdlox/lox MEFs firmly established that Ecd is required for cell cycle-associated E2F target gene expression and provide a logical basis for impaired cell cycle transit in cells deficient in Ecd.
A possible model of how Ecd could facilitate the E2F-dependent transcription, one for which our studies provide supportive evidence, is that Ecd facilitates Rb-E2F dissociation and thereby helps relieve E2F transcription factors from repression by Rb family proteins. Several key findings reported here support this model. First, Ecd deletion resulted in a drastic delay and overall reduction in Rb phosphorylation, a modification that has been established as a key link in Rb dissociation from E2F as cells progress through G1-S transition. Second, we showed, using ChIP assay, that more Rb, as well as p130, was associated with E2F target promoters in Ecd-null MEFs. Third, CDK4 activity in Ecd-null cells was not altered, which suggests that Ecd does not function upstream of Rb phosphorylation. The decrease in Rb hyperphosphorylation in Ecd-null cells is likely to reflect the reduced levels of E2F targets such as cdc2, CDK2, and cyclins E, A, and B, which are transcriptionally down-regulated in Ecd-null cells. Fourth, we showed that Ecd directly interacts with Rb as well as with other Rb family members. Fifth, we established that binding to Ecd prevents Rb from interacting with E2F1 and thereby facilitates the disruption of in vitro assembled as well as endogenous cellular Rb-E2F complexes. Finally, HPV16 E7, an Rb pocket-binding oncoprotein known to induce the dissociation of Rb-E2F complexes, reversed the cell cycle block imposed by Ecd deficiency, strongly arguing that the functional effects of Ecd deficiency on the cell cycle are due to impaired Rb-E2F dissociation. Collectively, the biochemical evidence, together with functional effects of Ecd deficiency on E2F-mediated gene expression and cell cycle progression, strongly supports the model that Ecd plays a physiological role to facilitate the dissociation of Rb family proteins from E2F transcription factors, thereby helping to switch E2F proteins to a transcriptional activation mode for cell cycle progression.
When we examined in vivo promoter occupancy by Rb proteins, we found that more Rb proteins were associated with the E2F target promoters in the Ecd-null MEFs. This demonstrates that the suppressed target gene expression in Ecd-deleted MEFs was caused by the association of the repressor Rb complex in these target gene promoters. Because Rb proteins associate with target promoters only through interaction with E2F proteins, we next studied how Ecd affects Rb-E2F interaction to understand how Ecd functions in cell cycle regulation. Rb family proteins possess a “pocket” domain through which it interacts with numerous cellular proteins and viral oncoproteins (36). Many proteins including E1A, E7, and TAg oncogenic proteins have an LxCxE sequence motif to interact with the Rb pocket domain (33). Competitive Rb interaction with cellular proteins is one of the mechanisms by which viral oncogenes dysregulate the normal cell cycle. For example, E1A displaces E2F1 on Rb by competing for binding to the pocket region, which disturbs the normal cell cycle. Here, we have shown that Ecd binds to Rb directly. The requirement of an A/B pocket for Rb-Ecd interaction gave us the hint that Ecd may compete with E2F1 to bind to Rb because E2F1 also interacts with Rb through the same pocket region. In vitro and in vivo binding experiments have demonstrated that Ecd can displace E2F1 by competing with it for Rb binding. Given that Ecd inhibits Rb interaction with E2F1, Ecd may have a role in regulating E2F transcription by modulating Rb-E2F1 interaction.
In conclusion, we have demonstrated that the previously little understood but evolutionarily conserved Ecd protein plays an important role in cell cycle progression. The cell cycle role of Ecd appears to involve its binding to hypophosphorylated Rb, thereby facilitating Rb-E2F dissociation and cell cycle progression. Thus, our studies identify Ecd as a novel component for physiological regulation of the mammalian cell cycle.
Supplementary Material
Acknowledgments
We thank Dr. Pradip Raychaudhri (University of Illinois at Chicago) for GST-Rb and GST-E2F1 constructs, Dr. Amy Yee (Tufts University Medical School, Boston) for GST-p107 and CMV-p130 constructs, Dr. Karl Munger (Harvard University, Boston) for the GST-HPV16E7 construct, and Dr. Izolda Papova (Northwestern University) for help in generating anti-Ecd monoclonal antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 CA96844-06 and CA94143 (to V. B.) and Grants CA87986, CA105489, CA99900, CA99163, and CA116552 (to H. B.). This work was also supported by Department of Defense Grant W81XWH-07-1-0351 (to V. B.). The work presented here was initiated while the investigators (J. H. K., C. B. G., M. N., Y. Z., H. B., and V. B.) were at the Department of Medicine, Evanston Northwestern Healthcare Research Institute, Northwestern University.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
C. B. Gurumurthy, M. Naramura, J. H. Kim, H. Band, and V. Band, manuscript in preparation.
- HPV
- human papillomavirus
- hEcd
- human Ecdysoneless
- MEF
- mouse embryonic fibroblast
- CDK
- cyclin-dependent kinase
- GST
- glutathione S-transferase
- BrdUrd
- bromodeoxyuridine
- PBS
- phosphate-buffered saline
- ChIP
- chromatin immunoprecipitation.
REFERENCES
- 1.zur Hausen H. (2002) Nat. Rev. Cancer 2, 342–350 [DOI] [PubMed] [Google Scholar]
- 2.Wise-Draper T. M., Wells S. I. (2008) Front. Biosci. 13, 1003–1017 [DOI] [PubMed] [Google Scholar]
- 3.Tungteakkhun S. S., Duerksen-Hughes P. J. (2008) Arch. Virol. 153, 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang Y., Chen J., Gurumurthy C. B., Kim J., Bhat I., Gao Q., Dimri G., Lee S. W., Band H., Band V. (2006) Cancer Res. 66, 7167–7175 [DOI] [PubMed] [Google Scholar]
- 5.Garen A., Kauvar L., Lepesant J. A. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 5099–5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaziova I., Bonnette P. C., Henrich V. C., Jindra M. (2004) Development 131, 2715–2725 [DOI] [PubMed] [Google Scholar]
- 7.Sato T., Jigami Y., Suzuki T., Uemura H. (1999) Mol. Gen. Genet. 260, 535–540 [DOI] [PubMed] [Google Scholar]
- 8.Kainou T., Shinzato T., Sasaki K., Mitsui Y., Giga-Hama Y., Kumagai H., Uemura H. (2006) Yeast 23, 35–53 [DOI] [PubMed] [Google Scholar]
- 9.Chellappan S. P., Hiebert S., Mudryj M., Horowitz J. M., Nevins J. R. (1991) Cell 65, 1053–1061 [DOI] [PubMed] [Google Scholar]
- 10.Classon M., Harlow E. (2002) Nat. Rev. Cancer 2, 910–917 [DOI] [PubMed] [Google Scholar]
- 11.Classon M., Kennedy B. K., Mulloy R., Harlow E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10826–10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Classon M., Salama S., Gorka C., Mulloy R., Braun P., Harlow E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10820–10825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balciunaite E., Spektor A., Lents N. H., Cam H., te Riele H., Scime A., Rudnicki M. A., Young R., Dynlacht B. D. (2005) Mol. Cell. Biol. 25, 8166–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wikenheiser-Brokamp K. A. (2004) Development 131, 4299–4310 [DOI] [PubMed] [Google Scholar]
- 15.Nguyen D. X., McCance D. J. (2005) J. Cell. Biochem. 94, 870–879 [DOI] [PubMed] [Google Scholar]
- 16.Lipinski M. M., Jacks T. (1999) Oncogene 18, 7873–7882 [DOI] [PubMed] [Google Scholar]
- 17.Giacinti C., Giordano A. (2006) Oncogene 25, 5220–5227 [DOI] [PubMed] [Google Scholar]
- 18.Hurford R. K., Jr., Cobrinik D., Lee M. H., Dyson N. (1997) Genes Dev. 11, 1447–1463 [DOI] [PubMed] [Google Scholar]
- 19.Todaro G. J., Wolman S. R., Green H. (1963) J. Cell. Physiol. 62, 257–265 [DOI] [PubMed] [Google Scholar]
- 20.Rayman J. B., Takahashi Y., Indjeian V. B., Dannenberg J. H., Catchpole S., Watson R. J., te Riele H., Dynlacht B. D. (2002) Genes Dev. 16, 933–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weinberg R. A. (1995) Cell 81, 323–330 [DOI] [PubMed] [Google Scholar]
- 22.Dyson N. (1998) Genes Dev. 12, 2245–2262 [DOI] [PubMed] [Google Scholar]
- 23.Shiffman D., Brooks E. E., Brooks A. R., Chan C. S., Milner P. G. (1996) J. Biol. Chem. 271, 12199–12204 [DOI] [PubMed] [Google Scholar]
- 24.Tikoo R., Zanazzi G., Shiffman D., Salzer J., Chao M. V. (2000) J. Neurosci. 20, 4627–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeGregori J., Leone G., Miron A., Jakoi L., Nevins J. R. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 7245–7250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chellappan S., Kraus V. B., Kroger B., Munger K., Howley P. M., Phelps W. C., Nevins J. R. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4549–4553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berthet C., Klarmann K. D., Hilton M. B., Suh H. C., Keller J. R., Kiyokawa H., Kaldis P. (2006) Dev. Cell 10, 563–573 [DOI] [PubMed] [Google Scholar]
- 28.Nevins J. R. (2001) Hum. Mol. Genet. 10, 699–703 [DOI] [PubMed] [Google Scholar]
- 29.Riley D. J., Lee E. Y., Lee W. H. (1994) Annu. Rev. Cell Biol. 10, 1–29 [DOI] [PubMed] [Google Scholar]
- 30.Liggett W. H., Jr., Sidransky D. (1998) J. Clin. Oncol. 16, 1197–1206 [DOI] [PubMed] [Google Scholar]
- 31.Kaye F. J. (2002) Oncogene 21, 6908–6914 [DOI] [PubMed] [Google Scholar]
- 32.Tsihlias J., Kapusta L., Slingerland J. (1999) Annu. Rev. Med. 50, 401–423 [DOI] [PubMed] [Google Scholar]
- 33.Felsani A., Mileo A. M., Paggi M. G. (2006) Oncogene 25, 5277–5285 [DOI] [PubMed] [Google Scholar]
- 34.Ahuja D., Sáenz-Robles M. T., Pipas J. M. (2005) Oncogene 24, 7729–7745 [DOI] [PubMed] [Google Scholar]
- 35.zur Hausen H., de Villiers E. M. (1994) Annu. Rev. Microbiol. 48, 427–447 [DOI] [PubMed] [Google Scholar]
- 36.Kouzarides T. (1995) Semin. Cancer Biol. 6, 91–98 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.